The Lung Microbiota and Lung Cancer: A Growing Relationship
Abstract
Simple Summary
Abstract
1. Introduction
2. Lung Microbiota and Non-Cancerous Diseases
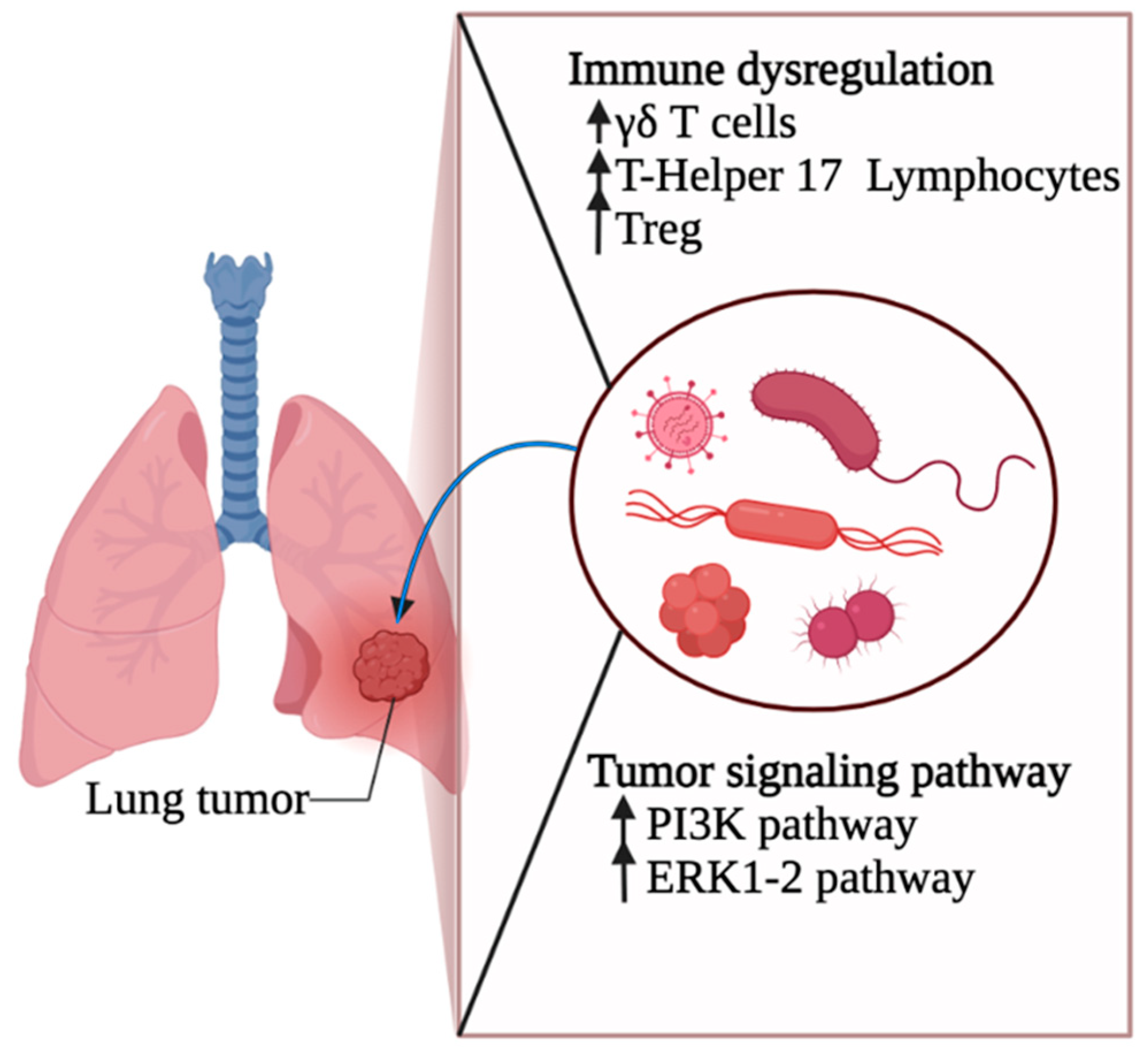
3. Lung Microbiota and Lung Cancer Pathogenesis
3.1. Lung Cancer Initiation
3.2. Lung Cancer Progression
4. Microbiota as a Therapeutic Target in Lung Cancer
4.1. Microbiota and Immunotherapy
4.2. Microbiota and Chemotherapy
5. Future Directions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| BAL | Broncho-alveolar lavage |
| BAX | Bcl-2-associated X protein |
| CDA | Cytidine deaminase |
| CDK1NB | Cyclin-dependent kinase inhibitor 1B |
| COPD | Chronic obstructive lung disease |
| CTLA-4 | Cytotoxic T-lymphocyte-associated protein 4 |
| EC | endothelial cells |
| ERK | Extracellular signal-regulated kinases |
| GEMM | genetically engineered mouse models |
| GZMB | Granzyme B |
| Her2 | human epidermal growth factor receptor 2 |
| FEV | Forced expiratory volume |
| FMT | Fecal microbiota transplant |
| ICI | Immune checkpoint inhibitor |
| IFN-γ | Interferon gamma |
| IL | Interleukin |
| IPF | Idiopathic pulmonary fibrosis |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MAPK | Mitogen-activated protein kinase |
| NF-κB-1 | Nuclear factor kappa B subunit 1 |
| NSCLC | Non-small-cell lung cancer |
| RCC | Renal cell carcinoma |
| PARP1 | Poly [ADP-ribose] polymerase 1 |
| PD-1 | Programmed cell death protein 1 |
| PD-L1 | Programmed death-ligand 1 |
| PFR1 | Perforin 1 |
| PI3K | Phosphoinositide 3-kinase |
| PRRs | Pattern recognition receptors |
| PTPA | protein-tyrosine-phosphatase A |
| TNF | Tumor necrosis factor |
| STAT3 | Signal transducer and activator of transcription 3 |
| TLR4 | Toll-like receptor 4 |
| TNFa | Tumor necrosis factor alfa |
| VEGF | Vascular endothelial growth factor |
References
- Ocáriz-Díez, M.; Cruellas, M.; Gascón, M.; Lastra, R.; Martínez-Lostao, L.; Ramírez-Labrada, A.; Paño, J.R.; Sesma, A.; Torres, I.; Yubero, A. Microbiota and Lung Cancer. Opportunities and Challenges for Improving Immunotherapy Efficacy. Front. Oncol. 2020, 10, 1945. [Google Scholar] [CrossRef] [PubMed]
- Ansaldo, E.; Farley, T.K.; Belkaid, Y. Control of immunity by the microbiota. Annu. Rev. Immunol. 2021, 39, 449–479. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.-N.; Ma, Q.; Ge, Y.; Yi, C.-X.; Wei, L.-Q.; Tan, J.-C.; Chu, Q.; Li, J.-Q.; Zhang, P.; Wang, H. Microbiome dysbiosis in lung cancer: From composition to therapy. NPJ Precis. Oncol. 2020, 4, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Dickson, R.P.; Erb-Downward, J.R.; Martinez, F.J.; Huffnagle, G.B. The microbiome and the respiratory tract. Annu. Rev. Physiol. 2016, 78, 481–504. [Google Scholar] [CrossRef]
- Sender, R.; Fuchs, S.; Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef]
- Fabbrizzi, A.; Amedei, A.; Lavorini, F.; Renda, T.; Fontana, G. The lung microbiome: Clinical and therapeutic implications. Intern. Emerg. Med. 2019, 14, 1241–1250. [Google Scholar] [CrossRef]
- McLean, A.E.; Kao, S.C.; Barnes, D.J.; Wong, K.K.; Scolyer, R.A.; Cooper, W.A.; Kohonen-Corish, M.R. The Emerging Role of the Lung Microbiome and its importance in Non-Small Cell Lung Cancer Diagnosis and Treatment. Lung Cancer 2022, 165, 124–132. [Google Scholar] [CrossRef]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef]
- Segal, L.N.; Clemente, J.C.; Tsay, J.-C.J.; Koralov, S.B.; Keller, B.C.; Wu, B.G.; Li, Y.; Shen, N.; Ghedin, E.; Morris, A. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat. Microbiol. 2016, 1, 1–11. [Google Scholar] [CrossRef]
- Sommariva, M.; Le Noci, V.; Bianchi, F.; Camelliti, S.; Balsari, A.; Tagliabue, E.; Sfondrini, L. The lung microbiota: Role in maintaining pulmonary immune homeostasis and its implications in cancer development and therapy. Cell. Mol. Life Sci. 2020, 77, 2739–2749. [Google Scholar] [CrossRef]
- Bingula, R.; Filaire, M.; Radosevic-Robin, N.; Bey, M.; Berthon, J.-Y.; Bernalier-Donadille, A.; Vasson, M.-P.; Filaire, E. Desired turbulence? Gut-lung axis, immunity, and lung cancer. J. Oncol. 2017, 2017, 5035371. [Google Scholar] [CrossRef] [PubMed]
- O’Dwyer, D.N.; Dickson, R.P.; Moore, B.B. The lung microbiome, immunity, and the pathogenesis of chronic lung disease. J. Immunol. 2016, 196, 4839–4847. [Google Scholar] [CrossRef]
- Dicker, A.J.; Huang, J.T.; Lonergan, M.; Keir, H.R.; Fong, C.J.; Tan, B.; Cassidy, A.J.; Finch, S.; Mullerova, H.; Miller, B.E. The sputum microbiome, airway inflammation, and mortality in chronic obstructive pulmonary disease. J. Allergy Clin. Immunol. 2021, 147, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Dai, Z.; Wang, Z.; Deng, Z.; Zhang, J.; Pu, J.; Cao, W.; Pan, T.; Zhou, Y.; Yang, Z. Gut microbiota dysbiosis contributes to the development of chronic obstructive pulmonary disease. Respir. Res. 2021, 22, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Hufnagl, K.; Pali-Schöll, I.; Roth-Walter, F.; Jensen-Jarolim, E. Dysbiosis of the gut and lung microbiome has a role in asthma. Semin. Immunopathol. 2020, 42, 75–93. [Google Scholar] [CrossRef] [PubMed]
- Frey, D.L.; Boutin, S.; Dittrich, S.A.; Graeber, S.Y.; Stahl, M.; Wege, S.; Herth, F.J.; Sommerburg, O.; Schultz, C.; Mall, M.A. Relationship between airway dysbiosis, inflammation and lung function in adults with cystic fibrosis. J. Cyst. Fibros. 2021, 20, 754–760. [Google Scholar] [CrossRef]
- O’Dwyer, D.N.; Ashley, S.L.; Gurczynski, S.J.; Xia, M.; Wilke, C.; Falkowski, N.R.; Norman, K.C.; Arnold, K.B.; Huffnagle, G.B.; Salisbury, M.L. Lung microbiota contribute to pulmonary inflammation and disease progression in pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2019, 199, 1127–1138. [Google Scholar] [CrossRef]
- Jin, C.; Lagoudas, G.K.; Zhao, C.; Bullman, S.; Bhutkar, A.; Hu, B.; Ameh, S.; Sandel, D.; Liang, X.S.; Mazzilli, S. Commensal microbiota promote lung cancer development via γδ T cells. Cell 2019, 176, 998–1013.e16. [Google Scholar] [CrossRef]
- Chang, S.H.; Mirabolfathinejad, S.G.; Katta, H.; Cumpian, A.M.; Gong, L.; Caetano, M.S.; Moghaddam, S.J.; Dong, C. T helper 17 cells play a critical pathogenic role in lung cancer. Proc. Natl. Acad. Sci. USA 2014, 111, 5664–5669. [Google Scholar] [CrossRef]
- Goto, T. Airway microbiota as a modulator of lung cancer. Int. J. Mol. Sci. 2020, 21, 3044. [Google Scholar] [CrossRef]
- Maddi, A.; Sabharwal, A.; Violante, T.; Manuballa, S.; Genco, R.; Patnaik, S.; Yendamuri, S. The microbiome and lung cancer. J. Thorac. Dis. 2019, 11, 280. [Google Scholar] [CrossRef] [PubMed]
- Apopa, P.L.; Alley, L.; Penney, R.B.; Arnaoutakis, K.; Steliga, M.A.; Jeffus, S.; Bircan, E.; Gopalan, B.; Jin, J.; Patumcharoenpol, P. PARP1 is up-regulated in non-small cell lung cancer tissues in the presence of the cyanobacterial toxin microcystin. Front. Microbiol. 2018, 9, 1757. [Google Scholar] [CrossRef] [PubMed]
- Tsay, J.-C.J.; Wu, B.G.; Badri, M.H.; Clemente, J.C.; Shen, N.; Meyn, P.; Li, Y.; Yie, T.-A.; Lhakhang, T.; Olsen, E. Airway microbiota is associated with upregulation of the PI3K pathway in lung cancer. Am. J. Respir. Crit. Care Med. 2018, 198, 1188–1198. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Chen, E.S.; Zhao, C.; Jin, C. Host-microbiome interaction in lung cancer. Front. Immunol. 2021, 12, 1956. [Google Scholar] [CrossRef]
- Brenner, D.R.; McLaughlin, J.R.; Hung, R.J. Previous lung diseases and lung cancer risk: A systematic review and meta-analysis. PLoS ONE 2011, 6, e17479. [Google Scholar] [CrossRef]
- Seiler, C.L.; Song, J.M.; Fernandez, J.; Abrahante, J.E.; Kono, T.J.; Chen, Y.; Ren, Y.; Kassie, F.; Tretyakova, N.Y. Epigenetic changes in alveolar type II lung cells of A/J mice following intranasal treatment with lipopolysaccharide. Chem. Res. Toxicol. 2019, 32, 831–839. [Google Scholar] [CrossRef]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Alegre, M.-L.; Luke, J.J.; Gajewski, T.F. The commensal microbiome is associated with anti–PD-1 efficacy in metastatic melanoma patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef]
- Budden, K.F.; Shukla, S.D.; Rehman, S.F.; Bowerman, K.L.; Keely, S.; Hugenholtz, P.; Armstrong-James, D.P.; Adcock, I.M.; Chotirmall, S.H.; Chung, K.F. Functional effects of the microbiota in chronic respiratory disease. Lancet Respir. Med. 2019, 7, 907–920. [Google Scholar] [CrossRef]
- Dickson, R.P.; Erb-Downward, J.R.; Freeman, C.M.; McCloskey, L.; Falkowski, N.R.; Huffnagle, G.B.; Curtis, J.L. Bacterial topography of the healthy human lower respiratory tract. MBio 2017, 8, e02287-16. [Google Scholar] [CrossRef]
- Sabbula, B.R.; Anjum, F. Squamous Cell Lung Cancer. In StatPearls [Internet]; StatPearls Publishing: Tampa, FL, USA, 2021. [Google Scholar]
- Mantovani, A.; Allavena, P.; Sica, A.; Balkwill, F. Cancer-related inflammation. Nature 2008, 454, 436–444. [Google Scholar] [CrossRef]
- Torre, L.A.; Siegel, R.L.; Jemal, A. Lung cancer statistics. In Lung Cancer and Personalized Medicine; Springer: Cham, Switzerland, 2016; pp. 1–19. [Google Scholar]
- Herbst, R.S.; Baas, P.; Kim, D.-W.; Felip, E.; Pérez-Gracia, J.L.; Han, J.-Y.; Molina, J.; Kim, J.-H.; Arvis, C.D.; Ahn, M.-J. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): A randomised controlled trial. Lancet 2016, 387, 1540–1550. [Google Scholar] [CrossRef]
- Moffatt, M.F.; Cookson, W.O. The lung microbiome in health and disease. Clin. Med. 2017, 17, 525. [Google Scholar] [CrossRef] [PubMed]
- Gur, C.; Ibrahim, Y.; Isaacson, B.; Yamin, R.; Abed, J.; Gamliel, M.; Enk, J.; Bar-On, Y.; Stanietsky-Kaynan, N.; Coppenhagen-Glazer, S. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 2015, 42, 344–355. [Google Scholar] [CrossRef] [PubMed]
- Chu, S.; Cheng, Z.; Yin, Z.; Xu, J.; Wu, F.; Jin, Y.; Yang, G. Airway Fusobacterium is Associated with Poor Response to Immunotherapy in Lung Cancer. Onco Targets Ther. 2022, 15, 201. [Google Scholar] [CrossRef] [PubMed]
- Qian, K.; Deng, Y.; Krimsky, W.S.; Feng, Y.-G.; Peng, J.; Tai, Y.-H.; Peng, H.; Jiang, L.-H. Airway Microbiota in Patients With Synchronous Multiple Primary Lung Cancer: The Bacterial Topography of the Respiratory Tract. Front. Oncol. 2022, 12, 811279. [Google Scholar] [CrossRef] [PubMed]
- Surette, M.G. The cystic fibrosis lung microbiome. Ann. Am. Thorac. Soc. 2014, 11, S61–S65. [Google Scholar] [CrossRef]
- Segal, L.; Alekseyenko, A.; Clemente, J.; Kulkarni, R.; Wu, B.; Chen, H.; Berger, K.; Goldring, R.; Rom, W.; Blaser, M. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome 2013, 1, 19. [Google Scholar] [CrossRef]
- Atanasova, K.R.; Yilmaz, Ö. Looking in the Porphyromonas gingivalis cabinet of curiosities: The microbium, the host and cancer association. Mol. Oral Microbiol. 2014, 29, 55–66. [Google Scholar] [CrossRef]
- Liu, Y.; Yuan, X.; Chen, K.; Zhou, F.; Yang, H.; Yang, H.; Qi, Y.; Kong, J.; Sun, W.; Gao, S. Clinical significance and prognostic value of Porphyromonas gingivalis infection in lung cancer. Transl. Oncol. 2021, 14, 100972. [Google Scholar] [CrossRef]
- Martins, D.; Mendes, F.; Schmitt, F. Microbiome: A supportive or a leading actor in lung cancer? Pathobiology 2021, 88, 198–207. [Google Scholar] [CrossRef]
- Jungnickel, C.; Schnabel, P.A.; Bohle, R.; Wiewrodt, R.; Herr, C.; Bals, R.; Beisswenger, C. Nontypeable Haemophilus influenzae–Promoted Proliferation of Kras-Induced Early Adenomatous Lesions Is Completely Dependent on Toll-Like Receptor Signaling. Am. J. Pathol. 2017, 187, 973–979. [Google Scholar] [CrossRef] [PubMed]
- Moon, D.C.; Choi, C.H.; Lee, S.M.; Lee, J.H.; Kim, S.I.; Kim, D.S.; Lee, J.C. Nuclear translocation of Acinetobacter baumannii transposase induces DNA methylation of CpG regions in the promoters of E-cadherin gene. PLoS ONE 2012, 7, e38974. [Google Scholar] [CrossRef] [PubMed]
- Hattar, K.; Reinert, C.P.; Sibelius, U.; Gökyildirim, M.Y.; Subtil, F.S.; Wilhelm, J.; Eul, B.; Dahlem, G.; Grimminger, F.; Seeger, W. Lipoteichoic acids from Staphylococcus aureus stimulate proliferation of human non-small-cell lung cancer cells in vitro. Cancer Immunol. Immunother. 2017, 66, 799–809. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Su, H.; She, Y.; Dai, C.; Xie, D.; Narrandes, S.; Huang, S.; Chen, C.; Xu, W. Whole genome sequencing revealed microbiome in lung adenocarcinomas presented as ground-glass nodules. Transl. Lung Cancer Res. 2019, 8, 235. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.H.; He, J.; Su, X.F.; Wen, Y.N.; Zhang, S.J.; Liu, L.Y.; Zhao, H.; Ye, C.P.; Wu, J.H.; Cai, S. The airway microbiota of non-small cell lung cancer patients and its relationship to tumor stage and EGFR gene mutation. Thorac. Cancer 2022, 13, 858–869. [Google Scholar] [CrossRef]
- Higuchi, R.; Goto, T.; Hirotsu, Y.; Otake, S.; Oyama, T.; Amemiya, K.; Mochizuki, H.; Omata, M. Streptococcus australis and Ralstonia pickettii as Major Microbiota in Mesotheliomas. J. Pers. Med. 2021, 11, 297. [Google Scholar] [CrossRef]
- Druzhinin, V.; Matskova, L.; Demenkov, P.; Baranova, E.; Volobaev, V.; Minina, V.; Larionov, A.; Titov, V.; Fucic, A. Genetic damage in lymphocytes of lung cancer patients is correlated to the composition of the respiratory tract microbiome. Mutagenesis 2021, 36, 143–153. [Google Scholar] [CrossRef]
- Li, K.-j.; Chen, Z.-l.; Huang, Y.; Zhang, R.; Luan, X.-q.; Lei, T.-t.; Chen, L. Dysbiosis of lower respiratory tract microbiome are associated with inflammation and microbial function variety. Respir. Res. 2019, 20, 1–16. [Google Scholar] [CrossRef]
- Greathouse, K.L.; White, J.R.; Vargas, A.J.; Bliskovsky, V.V.; Beck, J.A.; von Muhlinen, N.; Polley, E.C.; Bowman, E.D.; Khan, M.A.; Robles, A.I. Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. 2018, 19, 1–16. [Google Scholar] [CrossRef]
- Ran, Z.; Liu, J.; Wang, F.; Xin, C.; Xiong, B.; Song, Z. Pulmonary Micro-Ecological Changes and Potential Microbial Markers in Lung Cancer Patients. Front. Oncol. 2021, 10, 3085. [Google Scholar] [CrossRef]
- Leng, Q.; Holden, V.K.; Deepak, J.; Todd, N.W.; Jiang, F. Microbiota biomarkers for lung cancer. Diagnostics 2021, 11, 407. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Wang, Z.; Wang, J.; Ding, C.; Sun, C.; Liu, P.; Xu, X.; Liu, Y.; Chen, B.; Gu, B. Characterization of the lung microbiome and exploration of potential bacterial biomarkers for lung cancer. Transl. Lung Cancer Res. 2020, 9, 693. [Google Scholar] [CrossRef] [PubMed]
- Perrone, F.; Belluomini, L.; Mazzotta, M.; Bianconi, M.; Di Noia, V.; Meacci, F.; Montrone, M.; Pignataro, D.; Prelaj, A.; Rinaldi, S. Exploring the role of respiratory microbiome in lung cancer: A systematic review. Crit. Rev. Oncol. Hematol. 2021, 164, 103404. [Google Scholar] [CrossRef] [PubMed]
- Picardo, S.L.; Coburn, B.; Hansen, A.R. The microbiome and cancer for clinicians. Crit. Rev. Oncol. Hematol. 2019, 141, 1–12. [Google Scholar] [CrossRef]
- de Groot, P.M.; Wu, C.C.; Carter, B.W.; Munden, R.F. The epidemiology of lung cancer. Transl. Lung Cancer Res. 2018, 7, 220. [Google Scholar] [CrossRef]
- Lin, W.-W.; Karin, M. A cytokine-mediated link between innate immunity, inflammation, and cancer. J. Clin. Investig. 2007, 117, 1175–1183. [Google Scholar] [CrossRef]
- Koshiol, J.; Flores, R.; Lam, T.K.; Taylor, P.R.; Weinstein, S.J.; Virtamo, J.; Albanes, D.; Perez-Perez, G.; Caporaso, N.E.; Blaser, M.J. Helicobacter pylori seropositivity and risk of lung cancer. PLoS ONE 2012, 7, e32106. [Google Scholar] [CrossRef]
- Mao, Q.; Jiang, F.; Yin, R.; Wang, J.; Xia, W.; Dong, G.; Ma, W.; Yang, Y.; Xu, L.; Hu, J. Interplay between the lung microbiome and lung cancer. Cancer Lett. 2018, 415, 40–48. [Google Scholar] [CrossRef]
- Wu, S.; Rhee, K.-J.; Albesiano, E.; Rabizadeh, S.; Wu, X.; Yen, H.-R.; Huso, D.L.; Brancati, F.L.; Wick, E.; McAllister, F. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 2009, 15, 1016–1022. [Google Scholar] [CrossRef]
- Carbonero, F.; Benefiel, A.C.; Alizadeh-Ghamsari, A.H.; Gaskins, H.R. Microbial pathways in colonic sulfur metabolism and links with health and disease. Front. Physiol. 2012, 3, 448. [Google Scholar] [CrossRef]
- Chow, S.C.; Gowing, S.D.; Cools-Lartigue, J.J.; Chen, C.B.; Berube, J.; Yoon, H.W.; Chan, C.H.; Rousseau, M.C.; Bourdeau, F.; Giannias, B. Gram negative bacteria increase non-small cell lung cancer metastasis via toll-like receptor 4 activation and mitogen-activated protein kinase phosphorylation. Int. J. Cancer 2015, 136, 1341–1350. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Gail, M.H.; Consonni, D.; Carugno, M.; Humphrys, M.; Pesatori, A.C.; Caporaso, N.E.; Goedert, J.J.; Ravel, J.; Landi, M.T. Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol. 2016, 17, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Li, J.; Guan, Y.; Lou, Y.; Chen, H.; Xu, M.; Deng, D.; Chen, J.; Ni, B.; Zhao, L. Dysbiosis of the gut microbiome is associated with tumor biomarkers in lung cancer. Int. J. Biol. Sci. 2019, 15, 2381. [Google Scholar] [CrossRef]
- Gustafson, A.M.; Soldi, R.; Anderlind, C.; Scholand, M.B.; Qian, J.; Zhang, X.; Cooper, K.; Walker, D.; McWilliams, A.; Liu, G. Airway PI3K pathway activation is an early and reversible event in lung cancer development. Sci. Transl. Med. 2010, 2, 26ra25. [Google Scholar] [CrossRef] [PubMed]
- Le Noci, V.; Guglielmetti, S.; Arioli, S.; Camisaschi, C.; Bianchi, F.; Sommariva, M.; Storti, C.; Triulzi, T.; Castelli, C.; Balsari, A. Modulation of pulmonary microbiota by antibiotic or probiotic aerosol therapy: A strategy to promote immunosurveillance against lung metastases. Cell Rep. 2018, 24, 3528–3538. [Google Scholar] [CrossRef]
- Wang, J.; Ge, P.; Qiang, L.; Tian, F.; Zhao, D.; Chai, Q.; Zhu, M.; Zhou, R.; Meng, G.; Iwakura, Y. The mycobacterial phosphatase PtpA regulates the expression of host genes and promotes cell proliferation. Nat. Commun. 2017, 8, 1–16. [Google Scholar] [CrossRef]
- Gowing, S.D.; Chow, S.C.; Cools-Lartigue, J.J.; Chen, C.B.; Najmeh, S.; Jiang, H.Y.; Bourdeau, F.; Beauchamp, A.; Mancini, U.; Angers, I. Gram-positive pneumonia augments non-small cell lung cancer metastasis via host toll-like receptor 2 activation. Int. J. Cancer 2017, 141, 561–571. [Google Scholar] [CrossRef]
- Liu, X.; Cheng, Y.; Zang, D.; Zhang, M.; Li, X.; Liu, D.; Gao, B.; Zhou, H.; Sun, J.; Han, X. The role of gut microbiota in lung cancer: From carcinogenesis to immunotherapy. Front. Oncol. 2021, 11, 3118. [Google Scholar] [CrossRef]
- Cui, M.; Xiao, H.; Li, Y.; Zhou, L.; Zhao, S.; Luo, D.; Zheng, Q.; Dong, J.; Zhao, Y.; Zhang, X. Faecal microbiota transplantation protects against radiation-induced toxicity. EMBO Mol. Med. 2017, 9, 448–461. [Google Scholar] [CrossRef]
- Nakashima, S.; Kakugawa, T.; Yura, H.; Tomonaga, M.; Harada, T.; Hara, A.; Hara, S.; Nakano, M.; Yamasaki, E.; Sakamoto, N. Identification of Helicobacter pylori VacA in human lung and its effects on lung cells. Biochem. Biophys. Res. Commun. 2015, 460, 721–726. [Google Scholar] [CrossRef]
- Zhang, E.; Feng, X.; Liu, F.; Zhang, P.; Liang, J.; Tang, X. Roles of PI3K/Akt and c-Jun signaling pathways in human papillomavirus type 16 oncoprotein-induced HIF-1α, VEGF, and IL-8 expression and in vitro angiogenesis in non-small cell lung cancer cells. PLoS ONE 2014, 9, e103440. [Google Scholar] [CrossRef] [PubMed]
- Hamada, S.; Ooshima, T.; Torii, M.; Imanishi, H.; Masuda, N.; Sobue, S.; Kotani, S. Dental Caries Induction in Experimental Animls by Clinical Strains of Streptococcus mutans Isolated from Japanese Children. Microbiol. Immunol. 1978, 22, 301–314. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.S.; Baldwin, D.R. Recent advances in the management of lung cancer. Clin. Med. Lond 2018, 18, s41–s46. [Google Scholar] [CrossRef] [PubMed]
- Walters, S.; Maringe, C.; Coleman, M.P.; Peake, M.D.; Butler, J.; Young, N.; Bergstrom, S.; Hanna, L.; Jakobsen, E.; Kolbeck, K.; et al. Lung cancer survival and stage at diagnosis in Australia, Canada, Denmark, Norway, Sweden and the UK: A population-based study, 2004–2007. Thorax 2013, 68, 551–564. [Google Scholar] [CrossRef]
- Reck, M.; Rodriguez-Abreu, D.; Robinson, A.G.; Hui, R.; Csoszi, T.; Fulop, A.; Gottfried, M.; Peled, N.; Tafreshi, A.; Cuffe, S.; et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2016, 375, 1823–1833. [Google Scholar] [CrossRef] [PubMed]
- Brahmer, J.; Reckamp, K.L.; Baas, P.; Crino, L.; Eberhardt, W.E.; Poddubskaya, E.; Antonia, S.; Pluzanski, A.; Vokes, E.E.; Holgado, E.; et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 373, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Shen, K.; Cui, J.; Wei, Y.; Chen, X.; Liu, G.; Gao, X.; Li, W.; Lu, H.; Zhan, P.; Lv, T.; et al. Effectiveness and safety of PD-1/PD-L1 or CTLA4 inhibitors combined with chemotherapy as a first-line treatment for lung cancer: A meta-analysis. J. Thorac. Dis. 2018, 10, 6636–6652. [Google Scholar] [CrossRef]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Helmink, B.A.; Spencer, C.N.; Reuben, A.; Wargo, J.A. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell 2018, 33, 570–580. [Google Scholar] [CrossRef]
- Zitvogel, L.; Ma, Y.; Raoult, D.; Kroemer, G.; Gajewski, T.F. The microbiome in cancer immunotherapy: Diagnostic tools and therapeutic strategies. Science 2018, 359, 1366–1370. [Google Scholar] [CrossRef]
- Ghanem, S.; Kim, C.; Dutta, D.; Salifu, M.; Lim, S. Antimicrobial therapy during cancer treatment: Beyond antibacterial effects. J. Intern. Med. 2021, 290, 40–56. [Google Scholar] [CrossRef] [PubMed]
- Vetizou, M.; Pitt, J.M.; Daillere, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P.; et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef] [PubMed]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.L.; et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillere, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef]
- Ahmed, J.; Kumar, A.; Parikh, K.; Anwar, A.; Knoll, B.M.; Puccio, C.; Chun, H.; Fanucchi, M.; Lim, S.H. Use of broad-spectrum antibiotics impacts outcome in patients treated with immune checkpoint inhibitors. Oncoimmunology 2018, 7, e1507670. [Google Scholar] [CrossRef] [PubMed]
- Derosa, L.; Hellmann, M.D.; Spaziano, M.; Halpenny, D.; Fidelle, M.; Rizvi, H.; Long, N.; Plodkowski, A.J.; Arbour, K.C.; Chaft, J.E.; et al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Ann. Oncol. 2018, 29, 1437–1444. [Google Scholar] [CrossRef]
- Jin, Y.; Dong, H.; Xia, L.; Yang, Y.; Zhu, Y.; Shen, Y.; Zheng, H.; Yao, C.; Wang, Y.; Lu, S. The Diversity of Gut Microbiome is Associated With Favorable Responses to Anti-Programmed Death 1 Immunotherapy in Chinese Patients With NSCLC. J. Thorac. Oncol. 2019, 14, 1378–1389. [Google Scholar] [CrossRef]
- Vande Voorde, J.; Balzarini, J.; Liekens, S. Mycoplasmas and cancer: Focus on nucleoside metabolism. EXCLI J. 2014, 13, 300–322. [Google Scholar]
- Jiang, S.; Zhang, S.; Langenfeld, J.; Lo, S.C.; Rogers, M.B. Mycoplasma infection transforms normal lung cells and induces bone morphogenetic protein 2 expression by post-transcriptional mechanisms. J. Cell Biochem. 2008, 104, 580–594. [Google Scholar] [CrossRef]
- Apostolou, P.; Tsantsaridou, A.; Papasotiriou, I.; Toloudi, M.; Chatziioannou, M.; Giamouzis, G. Bacterial and fungal microflora in surgically removed lung cancer samples. J. Cardiothorac. Surg. 2011, 6, 137. [Google Scholar] [CrossRef]
- Ciccolini, J.; Serdjebi, C.; Peters, G.J.; Giovannetti, E. Pharmacokinetics and pharmacogenetics of Gemcitabine as a mainstay in adult and pediatric oncology: An EORTC-PAMM perspective. Cancer Chemother. Pharmacol. 2016, 78, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Geller, L.T.; Barzily-Rokni, M.; Danino, T.; Jonas, O.H.; Shental, N.; Nejman, D.; Gavert, N.; Zwang, Y.; Cooper, Z.A.; Shee, K.; et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 2017, 357, 1156–1160. [Google Scholar] [CrossRef] [PubMed]
- Gui, Q.F.; Lu, H.F.; Zhang, C.X.; Xu, Z.R.; Yang, Y.H. Well-balanced commensal microbiota contributes to anti-cancer response in a lung cancer mouse model. Genet. Mol. Res. 2015, 14, 5642–5651. [Google Scholar] [CrossRef] [PubMed]
- Daillere, R.; Vetizou, M.; Waldschmitt, N.; Yamazaki, T.; Isnard, C.; Poirier-Colame, V.; Duong, C.P.M.; Flament, C.; Lepage, P.; Roberti, M.P.; et al. Enterococcus hirae and Barnesiella intestinihominis Facilitate Cyclophosphamide-Induced Therapeutic Immunomodulatory Effects. Immunity 2016, 45, 931–943. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Lim, S.H. Bidirectional interaction between intestinal microbiome and cancer: Opportunities for therapeutic interventions. Biomark. Res. 2020, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Couturier-Maillard, A.; Secher, T.; Rehman, A.; Normand, S.; De Arcangelis, A.; Haesler, R.; Huot, L.; Grandjean, T.; Bressenot, A.; Delanoye-Crespin, A. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J. Clin. Investig. 2013, 123, 700–711. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.; Karpinets, T.; Prieto, P.; Vicente, D.; Hoffman, K.; Wei, S.C. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef]
- Kakihana, K.; Fujioka, Y.; Suda, W.; Najima, Y.; Kuwata, G.; Sasajima, S.; Mimura, I.; Morita, H.; Sugiyama, D.; Nishikawa, H. Fecal microbiota transplantation for patients with steroid-resistant acute graft-versus-host disease of the gut. Blood J. Am. Soc. Hematol. 2016, 128, 2083–2088. [Google Scholar] [CrossRef]
- Qi, X.; Li, X.; Zhao, Y.; Wu, X.; Chen, F.; Ma, X.; Zhang, F.; Wu, D. Treating steroid refractory intestinal acute graft-vs.-host disease with fecal microbiota transplantation: A pilot study. Front. Immunol. 2018, 9, 2195. [Google Scholar] [CrossRef]
- Plovier, H.; Everard, A.; Druart, C.; Depommier, C.; Van Hul, M.; Geurts, L.; Chilloux, J.; Ottman, N.; Duparc, T.; Lichtenstein, L. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 2017, 23, 107–113. [Google Scholar] [CrossRef]
- Depommier, C.; Everard, A.; Druart, C.; Plovier, H.; Van Hul, M.; Vieira-Silva, S.; Falony, G.; Raes, J.; Maiter, D.; Delzenne, N.M. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: A proof-of-concept exploratory study. Nat. Med. 2019, 25, 1096–1103. [Google Scholar] [CrossRef] [PubMed]

| Bacterial Classification | Sampling Method | Association with Lung Cancer | Potential Mechanism of Association with Lung Cancer |
|---|---|---|---|
| Pseudomonas [24,25,26,27,28,29] | Lung explants, BAL, Brush specimen, lung tissues | Marked presence in lung adenocarcinoma | Associated with an increase in macrophage population and IFN-γ in BAL sample. Amplification of neutrophil elastase activity. Enhanced/diminished population associated with smoking |
| Streptococcus [24,25,27,30,31,32,33,34] | Lung explants, BAL, Brush specimen, lung tissues | Marked presence in lung cancer, specifically adenocarcinoma, squamous cell carcinoma, and increased risk of hepatic metastasis from NSCLC Increased levels in the gut of small cell lung cancer patients | Enhanced ERK and PI3K pathway. Increased presence of Th17 cells and neutrophils. Enhanced/ diminished population in smokers |
| Prevotella [24,25,30,31,32,33,34] | Lung explants, BAL, Brush specimen | Low presence in the gut of NSCLC patients. Marked presence in the gut of squamous cell carcinoma patients Marked presence in lung cancer and adenocarcinoma | Correlated to an inflammatory phenotype, including an enhanced Th17 lymphocyte and neutrophil response Enhancement of ERK and PI3K pathway |
| Fusobacterium [25,31,32,33,35,36,37] | Lung explants, BAL, Brush specimen | Poor response in lung cancer to anti-PD-1 therapy if Fusobacterium is present in the airway. One of the most abundant specific bacterial community members detected in synchronous multiple primary lung cancer sMPLC lesions. | Fap2 protein of Fusobacterium inhibits natural killer cell killing by interacting with an inhibitory receptor present on all human NK cells and on various T cells (TIGIT receptor) |
| Veillonella [24,25,27,31,38,39] | Lung explants, BAL, Brush specimen, lung tissues, saliva | Observed in both small cell lung cancer and adenocarcinoma | Correlated to an inflammatory phenotype, including an enhanced Th17 lymphocyte and neutrophil response. Most abundant agent driving dysbiosis and amplification of IL17, PI3K, MAPK, and ERK pathways in the airway transcriptome |
| Prophyromonas [25,26,31,40,41] | Lung explants, BAL, Brush specimen | Higher P. gingivalis staining in carcinoma tissues of patients with small cell lung cancer, lung adenocarcinoma, and lung squamous cell carcinoma (35.00%, 26.89%, and 39.00%, respectively) compared to the adjacent lung tissues | Activation of cancer-associated transcription factors by modulating ATP-induced cytosolic, mitochondrial ROS, and antioxidant glutathione response through the inhibition of ATP/ P2X7-induced cell death by P. gingivalis. |
| Neisseria [25,42] | Lung explants, BAL, Brush specimen, saliva | Reduced presence in lung cancer | Suppress cell growth |
| Haemophilus [25,26,43] | Lung explants, BAL | Stimulates proliferation of early adenomatous lesions leading to alveolar adenomatous hyperplasia and adenocarcinoma | Upregulation of IL-17C and neutrophil infiltration. Can also promote metastatic progression in combination with cigarette smoke (8) |
| Sphingomonas [26,27] | BAL, Brush specimen, lung tissues | Marked presence in adenocarcinoma | Associated with an increase in macrophage population and prominent IFN-γ population |
| Acinetobacter [25,26,34,44] | Lung explants, BAL | Marked presence in lung cancer and adenocarcinoma | DNA methylation of CpG regions in the promoters of E-cadherin gene induced by A. baumannii transposase (Tnp) and down-regulation of this gene |
| Staphylococcus [25,28,45] | Lung explants, BAL, Brush specimen, lung tissues | Marked presence in the gut of NSCLC patients responsive to Nivolumab | Lipoteichoic acid induced cellular proliferation and liberation of interleukin (IL)-8. |
| Corynebacterium [25,26,31,46] | Lung explants, BAL, Brush specimen | No significant difference in microbiota composition between ground glass pulmonary nodules and normal tissues except in adenocarcinoma (AD) | - |
| Lactobacillus [24,26,27,31] | BAL, Brush specimen, lung tissues | Low levels in the gut of NSCLC patients (8). High abundance in the gut of squamous cell carcinoma patients | Gut microbiota’s role in regulating the lung’s immune response. |
| Actinobacillus [24,26,31,47] | Lung explants, BAL, saliva | Increase in the gut of lung cancer patients and commonly found in the lungs and sputum of lung cancer patients | Actinobacillus’ presence in the airway leads to chronic lung inflammation promoting the initiation and early development of lung cancer. |
| Propionibacterium [26,47] | BAL | Marked presence in the gut of NSCLC patients responsive to Nivolumab Marked presence in squamous cell carcinoma | Gut microbiota’s role in regulating the lung’s immune response. |
| Ralstonia [29,48] | Lung tissues | Marked presence in adenocarcinoma | Plays a role in impairing the tumor microenvironment’s immunity |
| Megasphaera [39,49] | BAL | Marked presence in lung cancer patients | Promotes somatic cell genome instability via high levels of chromosomal aberrations (CAs) and micronuclei (MN) frequency seen in peripheral blood lymphocytes of patients with lung cancer |
| Acidovorax [50,51] | Lung tissues | A marked presence in squamous cell carcinoma patients compared with adenocarcinoma. Specific taxa are more common in smokers with TP53 mutation | Degrades tobacco smoke compounds and thus promoting survival of transformed cells and subsequent tumor development. Malignant transformation of the lung epithelium via DNA damage and mutations in TP53, mediated by microbial toxins or reactive oxygen/nitrogen. |
| Capnocytophaga [52,53,54] | Saliva | A marked increase in lungs of lung cancer patients compared to control | Stimulation of chronic inflammation, thus promoting the development of lung cancer, especially lung squamous cell carcinoma |
| Cyanobacteria [52,55] | Lung tissue | A marked increase in lungs of adenocarcinoma patients | Cyanobacteria toxin microcystin is associated with reduced CD36 and increased levels of PARP1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bou Zerdan, M.; Kassab, J.; Meouchy, P.; Haroun, E.; Nehme, R.; Bou Zerdan, M.; Fahed, G.; Petrosino, M.; Dutta, D.; Graziano, S. The Lung Microbiota and Lung Cancer: A Growing Relationship. Cancers 2022, 14, 4813. https://doi.org/10.3390/cancers14194813
Bou Zerdan M, Kassab J, Meouchy P, Haroun E, Nehme R, Bou Zerdan M, Fahed G, Petrosino M, Dutta D, Graziano S. The Lung Microbiota and Lung Cancer: A Growing Relationship. Cancers. 2022; 14(19):4813. https://doi.org/10.3390/cancers14194813
Chicago/Turabian StyleBou Zerdan, Maroun, Joseph Kassab, Paul Meouchy, Elio Haroun, Rami Nehme, Morgan Bou Zerdan, Gracia Fahed, Michael Petrosino, Dibyendu Dutta, and Stephen Graziano. 2022. "The Lung Microbiota and Lung Cancer: A Growing Relationship" Cancers 14, no. 19: 4813. https://doi.org/10.3390/cancers14194813
APA StyleBou Zerdan, M., Kassab, J., Meouchy, P., Haroun, E., Nehme, R., Bou Zerdan, M., Fahed, G., Petrosino, M., Dutta, D., & Graziano, S. (2022). The Lung Microbiota and Lung Cancer: A Growing Relationship. Cancers, 14(19), 4813. https://doi.org/10.3390/cancers14194813





