Oncogenic Dysregulation of Circulating Noncoding RNAs: Novel Challenges and Opportunities in Sarcoma Diagnosis and Treatment
Abstract
Simple Summary
Abstract
1. Introduction
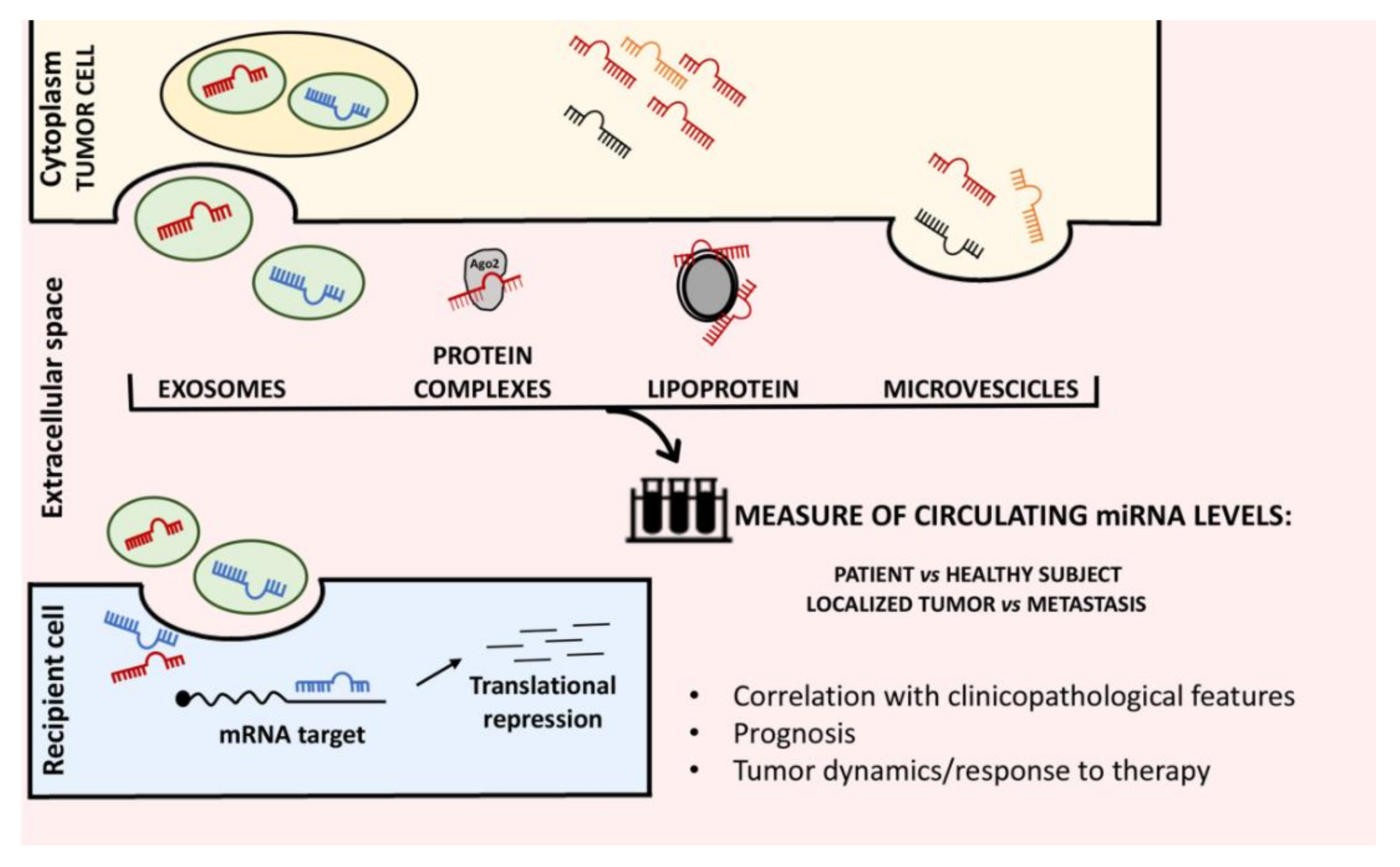
2. Circulating miRNAs Level May Function as Indicator of the Disease Status in Sarcoma Patients
2.1. Increased Circulating miRNAs Expression Associates with Worse Prognosis in Sarcoma Patients
2.2. Down-Regulation of Specific Circulating miRNAs Correlates with Sarcoma Pathogenesis
3. Circulating lncRNAs as Novel Prognostic Factors for Sarcoma Malignancies
| LncRNA Name | Expression Pattern | Sarcoma Type/ No. of Patients | Similar Expression in Tumor Tissue | Disease Monitoring | Circulating Expression also in Other Cancers |
|---|---|---|---|---|---|
| TUG1 | Upregulated (plasma) | OS [78,79]/ 40,76 | yes | Decreased expression in post-operative patients Increased level in case of relapse | LAD [98] Breast cancer [99] MM [100] |
| HNF1A-AS1 | Upregulated (serum) | OS [80]/72 | yes | Decreased expression in patients with post-operative chemotherapy Increased level in case of relapse | ESCC [101] |
| FAL1 | Upregulated (serum) | OS [81]/39 | yes | Decreased expression in post-operative patients | GC [102] HCC [103] |
| EPEL | Upregulated (serum) | OS [82]/39 | yes | Not analyzed | No data available |
| MALAT1 | Upregulated (serum) | OS [83]/46 | yes | Not analyzed | Breast cancer [104] MM [105] gastric adenocarcinoma [106] NSLC [107] glioblastoma multiforme [108] EOC [109] |
| FGD5-AS1 | Upregulated (serum) | OS [85]/97 | yes | Not analyzed | Thyroid Cancer [110] |
| ATB | Upregulated (serum) | OS [89]/60 | yes | Increased expression in case of relapse | CRC [111] HCC [112] Breast cancer [113] |
| LINC01278 | Upregulated (serum) | OS [94]/66 | yes | Not analyzed | No data available |
| RAMP2-AS1 | Upregulated (serum) | Chondrosarcoma [95]/45 | Not reported | Not analyzed | No data available |
| HAND2-AS1 | Downregulated (serum) | OS [96]/48 | yes | not analyzed | No data available |
| NEF | Downregulated (plasma) | OS [97]/49 | Not reported | Not analyzed | NSCLC [114] IHCC [115] GC [116] Glioma [117] |
| LINC01354 | Upregulated (serum) | OS [93]/30 | yes | Not analyzed | No data available |
| UCA1 | Upregulated (serum) | OS [90,91]/85 | yes | Not analyzed | Bladder cancer [118] HCC [119] PC [120] CRC [121,122] |
| LINK-A | Upregulated (plasma of metastatic patients) | OS [92]/62 | Not reported | Not analyzed | OC [123] |
4. Circulating circRNAs in Sarcoma Diagnosis and Prognosis
4.1. Free Circulating circRNAs in Liquid Biopsies
4.2. Exosomal circRNAs
5. Methods and Technological Challenges in the Early Detection of Circulating RNAs
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Wunder, J.S.; Nielsen, T.O.; Maki, R.G.; O’Sullivan, B.; Alman, B.A. Opportunities for improving the therapeutic ratio for patients with sarcoma. Lancet Oncol. 2007, 8, 513–524. [Google Scholar] [CrossRef]
- Grünewald, T.G.; Alonso, M.; Avnet, S.; Banito, A.; Burdach, S.; Cidre-Aranaz, F.; Di Pompo, G.; Distel, M.; Dorado-Garcia, H.; Garcia-Castro, J.; et al. Sarcoma treatment in the era of molecular medicine. EMBO Mol. Med. 2020, 12, e11131. [Google Scholar] [CrossRef] [PubMed]
- Zöllner, S.K.; Amatruda, J.F.; Bauer, S.; Collaud, S.; de Álava, E.; DuBois, S.G.; Hardes, J.; Hartmann, W.; Kovar, H.; Metzler, M.; et al. Ewing Sarcoma-Diagnosis, Treatment, Clinical Challenges and Future Perspectives. J. Clin. Med. 2021, 10, 1685. [Google Scholar] [CrossRef] [PubMed]
- Dancsok, A.R.; Asleh-Aburaya, K.; Nielsen, T.O. Advances in sarcoma diagnostics and treatment. Oncotarget 2017, 8, 7068–7093. [Google Scholar] [CrossRef] [PubMed]
- Ignatiadis, M.; Sledge, G.W.; Jeffrey, S.S. Liquid biopsy enters the clinic—Implementation issues and future challenges. Nat. Rev. Clin. Oncol. 2021, 18, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Leary, R.J.; Sausen, M.; Kinde, I.; Papadopoulos, N.; Carpten, J.D.; Craig, D.; O’Shaughnessy, J.; Kinzler, K.W.; Parmigiani, G.; Vogelstein, B.; et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci. Transl. Med. 2012, 4, 162ra154. [Google Scholar] [CrossRef]
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238. [Google Scholar] [CrossRef]
- Namløs, H.M.; Boye, K.; Meza-Zepeda, L.A. Cell-free DNA in blood as a noninvasive insight into the sarcoma genome. Mol. Asp. Med. 2020, 72, 100827. [Google Scholar] [CrossRef]
- Klega, K.; Imamovic-Tuco, A.; Ha, G.; Clapp, A.N.; Meyer, S.; Ward, A.; Clinton, C.; Nag, A.; Van Allen, E.; Mullen, E.; et al. Detection of Somatic Structural Variants Enables Quantification and Characterization of Circulating Tumor DNA in Children With Solid Tumors. JCO Precis. Oncol. 2018, 2018, PO.17.00285. [Google Scholar] [CrossRef]
- Shukla, N.N.; Patel, J.A.; Magnan, H.; Zehir, A.; You, D.; Tang, J.; Meng, F.; Samoila, A.; Slotkin, E.K.; Ambati, S.R.; et al. Plasma DNA-based molecular diagnosis, prognostication, and monitoring of patients with. JCO Precis. Oncol. 2017, 2017. [Google Scholar] [CrossRef]
- Anfossi, S.; Babayan, A.; Pantel, K.; Calin, G.A. Clinical utility of circulating non-coding RNAs—An update. Nat. Rev. Clin. Oncol. 2018, 15, 541–563. [Google Scholar] [CrossRef] [PubMed]
- Valihrach, L.; Androvic, P.; Kubista, M. Circulating miRNA analysis for cancer diagnostics and therapy. Mol. Asp. Med. 2020, 72, 100825. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Wang, H.; Yao, X.; Zhang, D.; Xie, Y.; Cui, R.; Zhang, X. Circulating MicroRNAs in Cancer: Potential and Challenge. Front. Genet. 2019, 10, 626. [Google Scholar] [CrossRef] [PubMed]
- Cortez, M.A.; Bueso-Ramos, C.; Ferdin, J.; Lopez-Berestein, G.; Sood, A.K.; Calin, G.A. MicroRNAs in body fluids—The mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 2011, 8, 467–477. [Google Scholar] [CrossRef]
- Wang, H.; Peng, R.; Wang, J.; Qin, Z.; Xue, L. Circulating microRNAs as potential cancer biomarkers: The advantage and disadvantage. Clin. Epigenetics 2018, 10, 59. [Google Scholar] [CrossRef]
- Sohel, M.M.H. Circulating microRNAs as biomarkers in cancer diagnosis. Life Sci. 2020, 248, 117473. [Google Scholar] [CrossRef]
- Mashouri, L.; Yousefi, H.; Aref, A.R.; Ahadi, A.M.; Molaei, F.; Alahari, S.K. Exosomes: Composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol. Cancer 2019, 18, 75. [Google Scholar] [CrossRef]
- De Toro, J.; Herschlik, L.; Waldner, C.; Mongini, C. Emerging roles of exosomes in normal and pathological conditions: New insights for diagnosis and therapeutic applications. Front. Immunol. 2015, 6, 203. [Google Scholar] [CrossRef]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Baranwal, S.; Alahari, S.K. miRNA control of tumor cell invasion and metastasis. Int. J. Cancer 2010, 126, 1283–1290. [Google Scholar] [CrossRef] [PubMed]
- Frixa, T.; Donzelli, S.; Blandino, G. Oncogenic MicroRNAs: Key Players in Malignant Transformation. Cancers 2015, 7, 2466–2485. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Chen, L.; Chen, X.; Sun, W.; Zhou, X. Identification of serum microRNA-21 as a biomarker for chemosensitivity and prognosis in human osteosarcoma. J. Int. Med. Res. 2012, 40, 2090–2097. [Google Scholar] [CrossRef] [PubMed]
- Fulci, V.; Chiaretti, S.; Goldoni, M.; Azzalin, G.; Carucci, N.; Tavolaro, S.; Castellano, L.; Magrelli, A.; Citarella, F.; Messina, M.; et al. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood 2007, 109, 4944–4951. [Google Scholar] [CrossRef]
- Feng, Y.H.; Wu, C.L.; Tsao, C.J.; Chang, J.G.; Lu, P.J.; Yeh, K.T.; Uen, Y.H.; Lee, J.C.; Shiau, A.L. Deregulated expression of sprouty2 and microRNA-21 in human colon cancer: Correlation with the clinical stage of the disease. Cancer Biol. Ther. 2011, 11, 111–121. [Google Scholar] [CrossRef]
- Hua, Y.; Jin, Z.; Zhou, F.; Zhang, Y.Q.; Zhuang, Y. The expression significance of serum MiR-21 in patients with osteosarcoma and its relationship with chemosensitivity. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 2989–2994. [Google Scholar]
- Fujiwara, T.; Uotani, K.; Yoshida, A.; Morita, T.; Nezu, Y.; Kobayashi, E.; Uehara, T.; Omori, T.; Sugiu, K.; Komatsubara, T.; et al. Clinical significance of circulating miR-25-3p as a novel diagnostic and prognostic biomarker in osteosarcoma. Oncotarget 2017, 8, 33375–33392. [Google Scholar] [CrossRef]
- Zhou, G.; Lu, M.; Chen, J.; Li, C.; Zhang, J.; Shi, X.; Wu, S. Identification of miR-199a-5p in serum as noninvasive biomarkers for detecting and monitoring osteosarcoma. Tumor Biol. 2015, 36, 8845–8852. [Google Scholar] [CrossRef]
- Zhang, C.; Yao, C.; Li, H.; Wang, G.; He, X. Combined elevation of microRNA-196a and microRNA-196b in sera predicts unfavorable prognosis in patients with osteosarcomas. Int. J. Mol. Sci. 2014, 15, 6544–6555. [Google Scholar] [CrossRef]
- Li, S.; Gao, Y.; Wang, Y.; Wang, K.; Dai, Z.P.; Xu, D.; Liu, W.; Li, Z.L.; Zhang, Z.D.; Yang, S.H.; et al. Serum microRNA-17 functions as a prognostic biomarker in osteosarcoma. Oncol. Lett. 2016, 12, 4905–4910. [Google Scholar] [CrossRef]
- Wang, T.; Ji, F.; Dai, Z.; Xie, Y.; Yuan, D. Increased expression of microRNA-191 as a potential serum biomarker for diagnosis and prognosis in human osteosarcoma. Cancer Biomark. 2015, 15, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhang, Y.; Zhang, X.; Zhang, M.; Liu, H.; Zhang, S.; Qi, B.; Sun, X. Serum microRNA-221 functions as a potential diagnostic and prognostic marker for patients with osteosarcoma. Biomed. Pharm. 2015, 75, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.D.; Xin, Q.; Tao, C.S.; Sun, P.F.; Xu, P.; Wu, B.; Qu, L.; Li, S.Z. Serum miR-300 as a diagnostic and prognostic biomarker in osteosarcoma. Oncol. Lett. 2016, 12, 3912–3918. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhou, S.; Wang, B.; Hu, J.; Zhou, Y.; Jiang, M.; Wu, M.; Qin, L.; Yang, X. miR-421 is a diagnostic and prognostic marker in patients with osteosarcoma. Tumor Biol. 2016, 37, 9001–9007. [Google Scholar] [CrossRef] [PubMed]
- Fei, D.; Li, Y.; Zhao, D.; Zhao, K.; Dai, L.; Gao, Z. Serum miR-9 as a prognostic biomarker in patients with osteosarcoma. J. Int. Med. Res. 2014, 42, 932–937. [Google Scholar] [CrossRef]
- Tang, J.; Zhao, H.; Cai, H.; Wu, H. Diagnostic and prognostic potentials of microRNA-27a in osteosarcoma. Biomed. Pharm. 2015, 71, 222–226. [Google Scholar] [CrossRef]
- Xie, X.Y.; Chen, X.M.; Shi, L.; Liu, J.W. Increased expression of microRNA-26a-5p predicted a poor survival outcome in osteosarcoma patients: An observational study. Medicine 2021, 100, e24765. [Google Scholar] [CrossRef]
- Hong, Q.; Fang, J.; Pang, Y.; Zheng, J. Prognostic value of the microRNA-29 family in patients with primary osteosarcomas. Med. Oncol. 2014, 31, 37. [Google Scholar] [CrossRef]
- Zhang, Z.Q.; Meng, H.; Wang, N.; Liang, L.N.; Liu, L.N.; Lu, S.M.; Luan, Y. Serum microRNA 143 and microRNA 215 as potential biomarkers for the diagnosis of chronic hepatitis and hepatocellular carcinoma. Diagn. Pathol. 2014, 9, 135. [Google Scholar] [CrossRef]
- Monterde-Cruz, L.; Ramírez-Salazar, E.G.; Rico-Martínez, G.; Linares-González, L.M.; Guzmán-González, R.; Delgado-Cedillo, E.; Estrada-Villaseñor, E.; Valdés-Flores, M.; Velázquez-Cruz, R.; Hidalgo-Bravo, A. Circulating miR-215-5p and miR-642a-5p as potential biomarker for diagnosis of osteosarcoma in Mexican population. Hum. Cell 2018, 31, 292–299. [Google Scholar] [CrossRef]
- Li, Q.; Song, S.; Ni, G.; Li, Y.; Wang, X. Serum miR-542-3p as a prognostic biomarker in osteosarcoma. Cancer Biomark. 2018, 21, 521–526. [Google Scholar] [CrossRef] [PubMed]
- Uotani, K.; Fujiwara, T.; Yoshida, A.; Iwata, S.; Morita, T.; Kiyono, M.; Yokoo, S.; Kunisada, T.; Takeda, K.; Hasei, J.; et al. Circulating MicroRNA-92b-3p as a Novel Biomarker for Monitoring of Synovial Sarcoma. Sci. Rep. 2017, 7, 14634. [Google Scholar] [CrossRef] [PubMed]
- Piano, M.A.; Gianesello, L.; Grassi, A.; Del Bianco, P.; Mattiolo, A.; Cattelan, A.M.; Sasset, L.; Zanovello, P.; Calabrò, M.L. Circulating miRNA-375 as a potential novel biomarker for active Kaposi’s sarcoma in AIDS patients. J. Cell. Mol. Med. 2019, 23, 1486–1494. [Google Scholar] [CrossRef] [PubMed]
- Miyachi, M.; Tsuchiya, K.; Yoshida, H.; Yagyu, S.; Kikuchi, K.; Misawa, A.; Iehara, T.; Hosoi, H. Circulating muscle-specific microRNA, miR-206, as a potential diagnostic marker for rhabdomyosarcoma. Biochem. Biophys. Res. Commun. 2010, 400, 89–93. [Google Scholar] [CrossRef]
- Kohama, I.; Asano, N.; Matsuzaki, J.; Yamamoto, Y.; Yamamoto, T.; Takahashi, R.U.; Kobayashi, E.; Takizawa, S.; Sakamoto, H.; Kato, K.; et al. Comprehensive serum and tissue microRNA profiling in dedifferentiated liposarcoma. Oncol. Lett. 2021, 22, 623. [Google Scholar] [CrossRef]
- Fricke, A.; Cimniak, A.F.V.; Ullrich, P.V.; Becherer, C.; Bickert, C.; Pfeifer, D.; Heinz, J.; Stark, G.B.; Bannasch, H.; Braig, D.; et al. Whole blood miRNA expression analysis reveals miR-3613-3p as a potential biomarker for dedifferentiated liposarcoma. Cancer Biomark. 2018, 22, 199–207. [Google Scholar] [CrossRef]
- Sciandra, M.; De Feo, A.; Parra, A.; Landuzzi, L.; Lollini, P.L.; Manara, M.C.; Mattia, G.; Pontecorvi, G.; Baricordi, C.; Guerzoni, C.; et al. Circulating miR34a levels as a potential biomarker in the follow-up of Ewing sarcoma. J. Cell Commun. Signal. 2020, 14, 335–347. [Google Scholar] [CrossRef]
- Morita, T.; Fujiwara, T.; Yoshida, A.; Uotani, K.; Kiyono, M.; Yokoo, S.; Hasei, J.; Kunisada, T.; Ozaki, T. Clinical relevance and functional significance of cell-free microRNA-1260b expression profiles in infiltrative myxofibrosarcoma. Sci. Rep. 2020, 10, 9414. [Google Scholar] [CrossRef]
- Gong, L.; Bao, Q.; Hu, C.; Wang, J.; Zhou, Q.; Wei, L.; Tong, L.; Zhang, W.; Shen, Y. Exosomal miR-675 from metastatic osteosarcoma promotes cell migration and invasion by targeting CALN1. Biochem. Biophys. Res. Commun. 2018, 500, 170–176. [Google Scholar] [CrossRef]
- Wu, C.; Li, Z.; Feng, G.; Wang, L.; Xie, J.; Jin, Y.; Liu, S. Tumor suppressing role of serum-derived exosomal microRNA-15a in osteosarcoma cells through the GATA binding protein 2/murine double minute 2 axis and the p53 signaling pathway. Bioengineered 2021, 12, 8378–8395. [Google Scholar] [CrossRef]
- Ghamloush, F.; Ghayad, S.E.; Rammal, G.; Fahs, A.; Ayoub, A.J.; Merabi, Z.; Harajly, M.; Zalzali, H.; Saab, R. The PAX3-FOXO1 oncogene alters exosome miRNA content and leads to paracrine effects mediated by exosomal miR-486. Sci. Rep. 2019, 9, 14242. [Google Scholar] [CrossRef] [PubMed]
- Casadei, L.; Calore, F.; Creighton, C.J.; Guescini, M.; Batte, K.; Iwenofu, O.H.; Zewdu, A.; Braggio, D.A.; Bill, K.L.; Fadda, P.; et al. Exosome-Derived miR-25-3p and miR-92a-3p Stimulate Liposarcoma Progression. Cancer Res. 2017, 77, 3846–3856. [Google Scholar] [CrossRef] [PubMed]
- Tian, Q.; Jia, J.; Ling, S.; Liu, Y.; Yang, S.; Shao, Z. A causal role for circulating miR-34b in osteosarcoma. Eur. J. Surg. Oncol. 2014, 40, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Cong, C.; Wang, W.; Tian, J.; Gao, T.; Zheng, W.; Zhou, C. Identification of serum miR-124 as a biomarker for diagnosis and prognosis in osteosarcoma. Cancer Biomark. 2018, 21, 449–454. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Ma, X.; Yue, J.; Chen, T.; Wang, X.Y.; Wang, Z.W.; Pan, J.; Lin, Y. The diagnostic effect of serum miR-139-5p as an indicator in osteosarcoma. Cancer Biomark. 2018, 23, 561–567. [Google Scholar] [CrossRef]
- Yao, Z.S.; Li, C.; Liang, D.; Jiang, X.B.; Tang, J.J.; Ye, L.Q.; Yuan, K.; Ren, H.; Yang, Z.D.; Jin, D.X.; et al. Diagnostic and prognostic implications of serum miR-101 in osteosarcoma. Cancer Biomark. 2018, 22, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhao, X.; Zhang, Y.J.; Fang, G.W.; Xue, Y. MicroRNA-375 as a potential serum biomarker for the diagnosis, prognosis, and chemosensitivity prediction of osteosarcoma. J. Int. Med. Res. 2018, 46, 975–983. [Google Scholar] [CrossRef]
- Cai, H.; Zhao, H.; Tang, J.; Wu, H. Serum miR-195 is a diagnostic and prognostic marker for osteosarcoma. J. Surg. Res. 2015, 194, 505–510. [Google Scholar] [CrossRef]
- Allen-Rhoades, W.; Kurenbekova, L.; Satterfield, L.; Parikh, N.; Fuja, D.; Shuck, R.L.; Rainusso, N.; Trucco, M.; Barkauskas, D.A.; Jo, E.; et al. Cross-species identification of a plasma microRNA signature for detection, therapeutic monitoring, and prognosis in osteosarcoma. Cancer Med. 2015, 4, 977–988. [Google Scholar] [CrossRef]
- Pang, P.C.; Shi, X.Y.; Huang, W.L.; Sun, K. miR-497 as a potential serum biomarker for the diagnosis and prognosis of osteosarcoma. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3765–3769. [Google Scholar]
- Niu, J.; Sun, Y.; Guo, Q.; Niu, D.; Liu, B. Serum miR-95-3p is a diagnostic and prognostic marker for osteosarcoma. SpringerPlus 2016, 5, 1947. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yang, Y.; Xu, J.; Luo, Y.; Xin, Y.; Wang, Y. Downregulation of microRNA-95-3p suppresses cell growth of osteosarcoma via CDKN1A/p21 expression. Oncol. Rep. 2018, 39, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Wu, Q.; Wang, Z.; Che, Y.; Zheng, S.; Chen, Y.; Zhong, X.; Shi, F. miR-223: An Immune Regulator in Infectious Disorders. Front. Immunol. 2021, 12, 781815. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Liu, Y.; Liao, W.; Liu, R.; Shi, P.; Wang, L. miRNA-223 is a potential diagnostic and prognostic marker for osteosarcoma. J. Bone Oncol. 2016, 5, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.N.; Luo, S.; Liu, C.; Piao, Z.; Gou, W.; Wang, Y.; Guan, W.; Li, Q.; Zou, H.; Yang, Z.Z.; et al. miR-491 Inhibits Osteosarcoma Lung Metastasis and Chemoresistance by Targeting αB-crystallin. Mol. Ther. 2017, 25, 2140–2149. [Google Scholar] [CrossRef]
- Shi, L.; Xie, C.; Zhu, J.; Chen, X. Downregulation of serum miR-194 predicts poor prognosis in osteosarcoma patients. Ann. Diagn. Pathol. 2020, 46, 151488. [Google Scholar] [CrossRef]
- Novello, C.; Pazzaglia, L.; Cingolani, C.; Conti, A.; Quattrini, I.; Manara, M.C.; Tognon, M.; Picci, P.; Benassi, M.S. miRNA expression profile in human osteosarcoma: Role of miR-1 and miR-133b in proliferation and cell cycle control. Int. J. Oncol. 2013, 42, 667–675. [Google Scholar] [CrossRef]
- Zhang, C.; Yao, C.; Li, H.; Wang, G.; He, X. Serum levels of microRNA-133b and microRNA-206 expression predict prognosis in patients with osteosarcoma. Int. J. Clin. Exp. Pathol. 2014, 7, 4194–4203. [Google Scholar]
- Luo, Z.; Liu, M.; Zhang, H.; Xia, Y. Association of circulating miR-125b and survival in patients with osteosarcoma-A single center experience. J. Bone Oncol. 2016, 5, 167–172. [Google Scholar] [CrossRef]
- Nie, C.L.; Ren, W.H.; Ma, Y.; Xi, J.S.; Han, B. Circulating miR-125b as a biomarker of Ewing’s sarcoma in Chinese children. Genet. Mol. Res. 2015, 14, 19049–19056. [Google Scholar] [CrossRef]
- Tombolan, L.; Millino, C.; Pacchioni, B.; Cattelan, M.; Zin, A.; Bonvini, P.; Bisogno, G. Circulating miR-26a as Potential Prognostic Biomarkers in Pediatric Rhabdomyosarcoma. Front. Genet. 2020, 11, 606274. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.; Wang, X.; Wang, C.; Li, L. Elevated levels of serum MiR-152 and miR-24 in uterine sarcoma: Potential for inducing autophagy via SIRT1 and deacetylated LC3. Br. J. Biomed. Sci. 2018, 75, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Batista, P.J.; Chang, H.Y. Long noncoding RNAs: Cellular address codes in development and disease. Cell 2013, 152, 1298–1307. [Google Scholar] [CrossRef]
- Song, Z.; Lin, J.; Li, Z.; Huang, C. The nuclear functions of long noncoding RNAs come into focus. Non-Coding RNA Res. 2021, 6, 70–79. [Google Scholar] [CrossRef]
- Uszczynska-Ratajczak, B.; Lagarde, J.; Frankish, A.; Guigó, R.; Johnson, R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 2018, 19, 535–548. [Google Scholar] [CrossRef]
- Qian, Y.; Shi, L.; Luo, Z. Long Non-coding RNAs in Cancer: Implications for Diagnosis, Prognosis, and Therapy. Front. Med. 2020, 7, 612393. [Google Scholar] [CrossRef] [PubMed]
- Moonmuang, S.; Chaiyawat, P.; Jantrapirom, S.; Pruksakorn, D.; Lo Piccolo, L. Circulating Long Non-Coding RNAs as Novel Potential Biomarkers for Osteogenic Sarcoma. Cancers 2021, 13, 4214. [Google Scholar] [CrossRef]
- Sheng, K.; Li, Y. LncRNA TUG1 promotes the development of osteosarcoma through RUNX2. Exp. Ther. Med. 2019, 18, 3002–3008. [Google Scholar] [CrossRef]
- Ma, B.; Li, M.; Zhang, L.; Huang, M.; Lei, J.B.; Fu, G.H.; Liu, C.X.; Lai, Q.W.; Chen, Q.Q.; Wang, Y.L. Upregulation of long non-coding RNA TUG1 correlates with poor prognosis and disease status in osteosarcoma. Tumour. Biol. 2016, 37, 4445–4455. [Google Scholar] [CrossRef]
- Cai, L.; Lv, J.; Zhang, Y.; Li, J.; Wang, Y.; Yang, H. The lncRNA HNF1A-AS1 is a negative prognostic factor and promotes tumorigenesis in osteosarcoma. J. Cell Mol. Med. 2017, 21, 2654–2662. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Z.; Zhang, S.; Li, Z.; Li, D.; Yang, S.; Zhang, H.; Zeng, X.; Liu, J. LncRNA FAL1 is a negative prognostic biomarker and exhibits pro-oncogenic function in osteosarcoma. J. Cell. Biochem. 2018, 119, 8481–8489. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Liu, Z.; Lu, S.; Hu, B. EPEL promotes the migration and invasion of osteosarcoma cells by upregulating ROCK1. Oncol. Lett. 2019, 17, 3133–3140. [Google Scholar] [CrossRef] [PubMed]
- Huo, Y.; Li, Q.; Wang, X.; Jiao, X.; Zheng, J.; Li, Z.; Pan, X. MALAT1 predicts poor survival in osteosarcoma patients and promotes cell metastasis through associating with EZH2. Oncotarget 2017, 8, 46993–47006. [Google Scholar] [CrossRef] [PubMed]
- Malakoti, F.; Targhazeh, N.; Karimzadeh, H.; Mohammadi, E.; Asadi, M.; Asemi, Z.; Alemi, F. Multiple function of lncRNA MALAT1 in cancer occurrence and progression. Chem. Biol. Drug Des. 2022, 100, 1–25. [Google Scholar] [CrossRef]
- Song, Q.H.; Guo, M.J.; Zheng, J.S.; Zheng, X.H.; Ye, Z.H.; Wei, P. Study on Targeting Relationship Between miR-320b and FGD5-AS1 and Its Effect on Biological Function of Osteosarcoma Cells. Cancer Manag. Res. 2020, 12, 13589–13598. [Google Scholar] [CrossRef]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef]
- Bayraktar, R.; Van Roosbroeck, K.; Calin, G.A. Cell-to-cell communication: MicroRNAs as hormones. Mol. Oncol. 2017, 11, 1673–1686. [Google Scholar] [CrossRef]
- Han, F.; Wang, C.; Wang, Y.; Zhang, L. Long noncoding RNA ATB promotes osteosarcoma cell proliferation, migration and invasion by suppressing miR-200s. Am. J. Cancer Res. 2017, 7, 770–783. [Google Scholar]
- Li, W.; Xie, P.; Ruan, W.H. Overexpression of lncRNA UCA1 promotes osteosarcoma progression and correlates with poor prognosis. J. Bone Oncol. 2016, 5, 80–85. [Google Scholar] [CrossRef]
- Wen, J.J.; Ma, Y.D.; Yang, G.S.; Wang, G.M. Analysis of circulating long non-coding RNA UCA1 as potential biomarkers for diagnosis and prognosis of osteosarcoma. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 498–503. [Google Scholar] [PubMed]
- Zhao, B.; Liu, K.; Cai, L. LINK-A lncRNA functions in the metastasis of osteosarcoma by upregulating HIF1α. Oncol. Lett. 2019, 17, 5005–5011. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Luo, Y. LINC01354 Promotes Osteosarcoma Cell Invasion by Up-regulating Integrin β1. Arch. Med. Res. 2020, 51, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.F.; Zhou, B.S.; An, X.C.; An, F.M.; Li, S.H. LINC01278 is Highly Expressed in Osteosarcoma and Participates in the Development of Tumors by Mediating the miR-134-5p/KRAS Axis. Onco Targets Ther. 2021, 14, 683–695. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Zhang, Z.; Cheng, F.; Shao, Z. Exosomal lncRNA RAMP2-AS1 Derived from Chondrosarcoma Cells Promotes Angiogenesis through miR-2355-5p/VEGFR2 Axis. Onco Targets Ther. 2020, 13, 3291–3301. [Google Scholar] [CrossRef]
- Chen, S.; Xu, X.; Lu, S.; Hu, B. Long non-coding RNA HAND2-AS1 targets glucose metabolism and inhibits cancer cell proliferation in osteosarcoma. Oncol. Lett. 2019, 18, 1323–1329. [Google Scholar] [CrossRef]
- Yang, Q.; Yu, H.; Yin, Q.; Hu, X.; Zhang, C. lncRNA-NEF is downregulated in osteosarcoma and inhibits cancer cell migration and invasion by downregulating miRNA-21. Oncol. Lett. 2019, 17, 5403–5408. [Google Scholar] [CrossRef]
- Liu, H.; Zhou, G.; Fu, X.; Cui, H.; Pu, G.; Xiao, Y.; Sun, W.; Dong, X.; Zhang, L.; Cao, S.; et al. Long noncoding RNA TUG1 is a diagnostic factor in lung adenocarcinoma and suppresses apoptosis via epigenetic silencing of BAX. Oncotarget 2017, 8, 101899–101910. [Google Scholar] [CrossRef]
- Alkhathami, A.G.; Hadi, A.; Alfaifi, M.; Alshahrani, M.Y.; Verma, A.K.; Beg, M.M.A. Serum-Based lncRNA ANRIL, TUG1, UCA1, and HIT Expressions in Breast Cancer Patients. Dis. Markers 2022, 2022, 9997212. [Google Scholar] [CrossRef]
- Yin, Q.; Shen, X.; Cui, X.; Ju, S. Elevated serum lncRNA TUG1 levels are a potential diagnostic biomarker of multiple myeloma. Exp. Hematol. 2019, 79, 47–55.e42. [Google Scholar] [CrossRef]
- Tong, Y.S.; Wang, X.W.; Zhou, X.L.; Liu, Z.H.; Yang, T.X.; Shi, W.H.; Xie, H.W.; Lv, J.; Wu, Q.Q.; Cao, X.F. Identification of the long non-coding RNA POU3F3 in plasma as a novel biomarker for diagnosis of esophageal squamous cell carcinoma. Mol. Cancer 2015, 14, 3. [Google Scholar] [CrossRef]
- Zhu, C.H.; Xiao, D.H.; Dai, L.G.; Xu, H.G.; Jiang, Y.H.; Zheng, Z.J. Highly expressed lncRNA FAL1 promotes the progression of gastric cancer by inhibiting PTEN. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8257–8264. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Mao, R.; Liu, C.; Zhang, W.; Tang, Y.; Guo, Z. LncRNA FAL1 promotes cell proliferation and migration by acting as a CeRNA of miR-1236 in hepatocellular carcinoma cells. Life Sci. 2018, 197, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Fan, R.; Chen, L.; Qian, H. Clinical Significance of Long Non-coding RNA MALAT1 Expression in Tissue and Serum of Breast Cancer. Ann. Clin. Lab. Sci. 2016, 46, 418–424. [Google Scholar] [PubMed]
- Liu, N.; Feng, S.; Li, H.; Chen, X.; Bai, S.; Liu, Y. Long non-coding RNA MALAT1 facilitates the tumorigenesis, invasion and glycolysis of multiple myeloma via miR-1271-5p/SOX13 axis. J. Cancer Res. Clin. Oncol. 2020, 146, 367–379. [Google Scholar] [CrossRef]
- Lu, Z.; Luo, T.; Pang, T.; Du, Z.; Yin, X.; Cui, H.; Fang, G.; Xue, X. MALAT1 promotes gastric adenocarcinoma through the MALAT1/miR-181a-5p/AKT3 axis. Open Biol. 2019, 9, 190095. [Google Scholar] [CrossRef]
- Zhang, R.; Xia, Y.; Wang, Z.; Zheng, J.; Chen, Y.; Li, X.; Wang, Y.; Ming, H. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2017, 490, 406–414. [Google Scholar] [CrossRef]
- Chen, W.; Xu, X.K.; Li, J.L.; Kong, K.K.; Li, H.; Chen, C.; He, J.; Wang, F.; Li, P.; Ge, X.S.; et al. MALAT1 is a prognostic factor in glioblastoma multiforme and induces chemoresistance to temozolomide through suppressing miR-203 and promoting thymidylate synthase expression. Oncotarget 2017, 8, 22783–22799. [Google Scholar] [CrossRef]
- Qiu, J.J.; Lin, X.J.; Tang, X.Y.; Zheng, T.T.; Lin, Y.Y.; Hua, K.Q. Exosomal Metastasis-Associated Lung Adenocarcinoma Transcript 1 Promotes Angiogenesis and Predicts Poor Prognosis in Epithelial Ovarian Cancer. Int. J. Biol. Sci. 2018, 14, 1960–1973. [Google Scholar] [CrossRef]
- Liu, B.; Chen, J.; Shang, F.; Lian, M.; Shen, X.; Fang, J. Tumor-Derived Exosome FGD5-AS1 Promotes Angiogenesis, Vascular Permeability, and Metastasis in Thyroid Cancer by Targeting the miR-6838-5p/VAV2 Axis. J. Oncol. 2022, 2022, 4702855. [Google Scholar] [CrossRef]
- Abedini, P.; Fattahi, A.; Agah, S.; Talebi, A.; Beygi, A.H.; Amini, S.M.; Mirzaei, A.; Akbari, A. Expression analysis of circulating plasma long noncoding RNAs in colorectal cancer: The relevance of lncRNAs ATB and CCAT1 as potential clinical hallmarks. J. Cell Physiol. 2019, 234, 22028–22033. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.R.; Kim, G.; Tak, W.Y.; Jang, S.Y.; Kweon, Y.O.; Park, J.G.; Lee, H.W.; Han, Y.S.; Chun, J.M.; Park, S.Y.; et al. Circulating exosomal noncoding RNAs as prognostic biomarkers in human hepatocellular carcinoma. Int. J. Cancer 2019, 144, 1444–1452. [Google Scholar] [CrossRef] [PubMed]
- El-Ashmawy, N.E.; Hussien, F.Z.; El-Feky, O.A.; Hamouda, S.M.; Al-Ashmawy, G.M. Serum LncRNA-ATB and FAM83H-AS1 as diagnostic/prognostic non-invasive biomarkers for breast cancer. Life Sci. 2020, 259, 118193. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Xu, W.; Zhang, Y.; Gong, F. Long non-coding RNA-NEF targets glucose transportation to inhibit the proliferation of non-small-cell lung cancer cells. Oncol. Lett. 2019, 17, 2795–2801. [Google Scholar] [CrossRef]
- Liang, Z.; Zhu, B.; Meng, D.; Shen, X.; Li, X.; Wang, Z.; Li, L. Down-regulation of lncRNA-NEF indicates poor prognosis in intrahepatic cholangiocarcinoma. Biosci. Rep. 2019, 39, BSR20181573. [Google Scholar] [CrossRef]
- Wang, X.; Jiang, X.; Zhou, L.; Wang, Z.; Huang, H.; Wang, M. LncRNA-NEF is involved the regulation of gastric carcinoma cell proliferation by targeting RUNX1. Mol. Med. Rep. 2019, 19, 2051–2056. [Google Scholar] [CrossRef]
- Huang, Q.; Chen, H.; Zuo, B.; Cheng, C.; Yu, W.; Yang, Y. lncRNA NEF inhibits glioma by downregulating TGF-β1. Exp. Ther. Med. 2019, 18, 692–698. [Google Scholar] [CrossRef]
- Pan, J.; Xie, X.; Li, H.; Li, Z.; Ren, C.; Ming, L. Detection of serum long non-coding RNA UCA1 and circular RNAs for the diagnosis of bladder cancer and prediction of recurrence. Int. J. Clin. Exp. Pathol. 2019, 12, 2951–2958. [Google Scholar]
- Zheng, Z.K.; Pang, C.; Yang, Y.; Duan, Q.; Zhang, J.; Liu, W.C. Serum long noncoding RNA urothelial carcinoma-associated 1: A novel biomarker for diagnosis and prognosis of hepatocellular carcinoma. J. Int. Med. Res. 2018, 46, 348–356. [Google Scholar] [CrossRef]
- Yu, Y.; Gao, F.; He, Q.; Li, G.; Ding, G. lncRNA UCA1 Functions as a ceRNA to Promote Prostate Cancer Progression via Sponging miR143. Mol. Ther. Nucleic Acids 2020, 19, 751–758. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, Z.; Pan, D.; Xin, Z.; Bu, F.; Zhang, Y.; Tian, Q.; Feng, X. Circulating lncRNA UCA1 and lncRNA PGM5-AS1 act as potential diagnostic biomarkers for early-stage colorectal cancer. Biosci. Rep. 2021, 41, BSR20211115. [Google Scholar] [CrossRef] [PubMed]
- Luan, Y.; Li, X.; Zhao, R.; Li, Y.; Liu, L.; Hao, Y.; Oleg Vladimir, B.; Jia, L. Circulating lncRNA UCA1 Promotes Malignancy of Colorectal Cancer via the miR-143/MYO6 Axis. Mol. Ther. Nucleic Acids 2020, 19, 790–803. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yao, B.; Tang, S.; Chen, Y. LINK-A Long Non-Coding RNA (lncRNA) Participates in Metastasis of Ovarian Carcinoma and Upregulates Hypoxia-Inducible Factor 1 (HIF1α). Med. Sci. Monit. 2019, 25, 2221–2227. [Google Scholar] [CrossRef] [PubMed]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef]
- Starke, S.; Jost, I.; Rossbach, O.; Schneider, T.; Schreiner, S.; Hung, L.H.; Bindereif, A. Exon circularization requires canonical splice signals. Cell Rep. 2015, 10, 103–111. [Google Scholar] [CrossRef]
- Liang, D.; Tatomer, D.C.; Luo, Z.; Wu, H.; Yang, L.; Chen, L.L.; Cherry, S.; Wilusz, J.E. The Output of Protein-Coding Genes Shifts to Circular RNAs When the Pre-mRNA Processing Machinery Is Limiting. Mol. Cell 2017, 68, 940–954.e943. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef]
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.; Schreiber, A.W.; Gregory, P.A.; Goodall, G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015, 160, 1125–1134. [Google Scholar] [CrossRef]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef]
- Salzman, J.; Chen, R.E.; Olsen, M.N.; Wang, P.L.; Brown, P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013, 9, e1003777. [Google Scholar] [CrossRef]
- Enuka, Y.; Lauriola, M.; Feldman, M.E.; Sas-Chen, A.; Ulitsky, I.; Yarden, Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016, 44, 1370–1383. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Bao, C.; Guo, W.; Li, S.; Chen, J.; Chen, B.; Luo, Y.; Lyu, D.; Li, Y.; Shi, G.; et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016, 7, 11215. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhu, L.; Lu, C.; Wang, C.; Wang, H.; Jin, H.; Ma, X.; Cheng, Z.; Yu, C.; Wang, S.; et al. circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat. Commun. 2021, 12, 295. [Google Scholar] [CrossRef] [PubMed]
- Du, W.W.; Yang, W.; Liu, E.; Yang, Z.; Dhaliwal, P.; Yang, B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016, 44, 2846–2858. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Yuan, Z.; Du, K.Y.; Fang, L.; Lyu, J.; Zhang, C.; He, A.; Eshaghi, E.; Zeng, K.; Ma, J.; et al. Translation of yes-associated protein (YAP) was antagonized by its circular RNA via suppressing the assembly of the translation initiation machinery. Cell Death Differ. 2019, 26, 2758–2773. [Google Scholar] [CrossRef]
- Abe, N.; Matsumoto, K.; Nishihara, M.; Nakano, Y.; Shibata, A.; Maruyama, H.; Shuto, S.; Matsuda, A.; Yoshida, M.; Ito, Y.; et al. Rolling Circle Translation of Circular RNA in Living Human Cells. Sci. Rep. 2015, 5, 16435. [Google Scholar] [CrossRef]
- Gao, X.; Xia, X.; Li, F.; Zhang, M.; Zhou, H.; Wu, X.; Zhong, J.; Zhao, Z.; Zhao, K.; Liu, D.; et al. Circular RNA-encoded oncogenic E-cadherin variant promotes glioblastoma tumorigenicity through activation of EGFR-STAT3 signalling. Nat. Cell. Biol. 2021, 23, 278–291. [Google Scholar] [CrossRef]
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.B.; Jaffrey, S.R. 5′ UTR m(6)A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Jakobsen, T.; Hager, H.; Kjems, J. The emerging roles of circRNAs in cancer and oncology. Nat. Rev. Clin. Oncol. 2022, 19, 188–206. [Google Scholar] [CrossRef]
- Vo, J.N.; Cieslik, M.; Zhang, Y.; Shukla, S.; Xiao, L.; Wu, Y.M.; Dhanasekaran, S.M.; Engelke, C.G.; Cao, X.; Robinson, D.R.; et al. The Landscape of Circular RNA in Cancer. Cell 2019, 176, 869–881.e813. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, K.; Tan, S.; Xin, J.; Yuan, Q.; Xu, H.; Xu, X.; Liang, Q.; Christiani, D.C.; Wang, M.; et al. Circular RNAs in body fluids as cancer biomarkers: The new frontier of liquid biopsies. Mol. Cancer 2021, 20, 13. [Google Scholar] [CrossRef] [PubMed]
- Kun-Peng, Z.; Chun-Lin, Z.; Jian-Ping, H.; Lei, Z. A novel circulating hsa_circ_0081001 act as a potential biomarker for diagnosis and prognosis of osteosarcoma. Int. J. Biol. Sci. 2018, 14, 1513–1520. [Google Scholar] [CrossRef]
- Zhu, K.; Niu, L.; Wang, J.; Wang, Y.; Zhou, J.; Wang, F.; Cheng, Y.; Zhang, Q.; Li, H. Circular RNA hsa_circ_0000885 Levels are Increased in Tissue and Serum Samples from Patients with Osteosarcoma. Med. Sci. Monit. 2019, 25, 1499–1505. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, J.; Ma, J.; Sun, T.; Zhou, Q.; Wang, W.; Wang, G.; Wu, P.; Wang, H.; Jiang, L.; et al. Exosomal circRNAs: Biogenesis, effect and application in human diseases. Mol. Cancer 2019, 18, 116. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Lin, Y.; Mi, C. Cisplatin-resistant osteosarcoma cell-derived exosomes confer cisplatin resistance to recipient cells in an exosomal circ_103801-dependent manner. Cell Biol. Int. 2021, 45, 858–868. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Pei, Y.; Wang, W.; Liu, F.; Zheng, K.; Zhang, X. Extracellular nanovesicles-transmitted circular RNA has_circ_0000190 suppresses osteosarcoma progression. J. Cell Mol. Med. 2020, 24, 2202–2214. [Google Scholar] [CrossRef]
- He, C.; Zheng, S.; Luo, Y.; Wang, B. Exosome Theranostics: Biology and Translational Medicine. Theranostics 2018, 8, 237–255. [Google Scholar] [CrossRef]
- Endzeliņš, E.; Berger, A.; Melne, V.; Bajo-Santos, C.; Soboļevska, K.; Ābols, A.; Rodriguez, M.; Šantare, D.; Rudņickiha, A.; Lietuvietis, V.; et al. Detection of circulating miRNAs: Comparative analysis of extracellular vesicle-incorporated miRNAs and cell-free miRNAs in whole plasma of prostate cancer patients. BMC Cancer 2017, 17, 730. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, H.; Yang, H.; Bai, M.; Ning, T.; Deng, T.; Liu, R.; Fan, Q.; Zhu, K.; Li, J.; et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol. Oncol. 2020, 14, 539–555. [Google Scholar] [CrossRef]
- Xin, R.; Gao, Y.; Wang, R.; Kadash-Edmondson, K.E.; Liu, B.; Wang, Y.; Lin, L.; Xing, Y. isoCirc catalogs full-length circular RNA isoforms in human transcriptomes. Nat. Commun. 2021, 12, 266. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Hou, L.; Zuo, Z.; Ji, P.; Zhang, X.; Xue, Y.; Zhao, F. Comprehensive profiling of circular RNAs with nanopore sequencing and CIRI-long. Nat. Biotechnol. 2021, 39, 836–845. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Kannisto, E.; Yu, G.; Yang, Y.; Reid, M.E.; Patnaik, S.K.; Wu, Y. Non-invasive Detection of Exosomal MicroRNAs via Tethered Cationic Lipoplex Nanoparticles (tCLN) Biochip for Lung Cancer Early Detection. Front. Genet. 2020, 11, 258. [Google Scholar] [CrossRef] [PubMed]
- Noren Hooten, N.; Byappanahalli, A.M.; Vannoy, M.; Omoniyi, V.; Evans, M.K. Influences of age, race, and sex on extracellular vesicle characteristics. Theranostics 2022, 12, 4459–4476. [Google Scholar] [CrossRef]
- Dluzen, D.F.; Noren Hooten, N.; Evans, M.K. Extracellular RNA in aging. Wiley Interdiscip. Rev. RNA 2017, 8, e1385. [Google Scholar] [CrossRef]
| miRNA Name | Expression Pattern | No. of Patients | Similar Expression Pattern in Tumor Tissue | Disease Monitoring | Method to Identify/ Quantify miRNA |
|---|---|---|---|---|---|
| Osteosarcoma | |||||
| miR-21 [23,26] | Upregulated (serum) | 65/69 | Not reported/yes | Not analyzed | RT-qPCR |
| miR-25-3p [27] | Upregulated (serum) | 10 | yes | Decrease after tumor resection or during chemotherapy | Microarray/ RT-qPCR |
| miR-196a/b [29] | Upregulated (serum) | 100 | yes | Not analyzed | RT-qPCR |
| miR-17 [30] | Upregulated (serum) | 46 | yes | Not analyzed | RT-qPCR |
| miR-191 [31] | Upregulated (serum) | 100 | yes | Not analyzed | RT-qPCR |
| miR-221 [32] | Upregulated (serum) | 108 | yes | Not analyzed | RT-qPCR |
| miR-300 [33] | Upregulated (serum) | 114 | yes | Decreased in patients with curative surgeries | RT-qPCR |
| miR-421 [34] | Upregulated (serum) | 40 | yes | Not analyzed | RT-qPCR |
| miR-9 [35] | Upregulated (serum) | 118 | Not reported | Not analyzed | RT-qPCR |
| miR-27a [36] | Upregulated (serum) | 166 | Not reported | Not analyzed | RT-qPCR |
| miR-26a-5p [37] | Upregulated (serum) | 243 | yes | Not analyzed | RT-qPCR |
| miR-29 family [38] | Upregulated (serum) | 80 | yes | Not analyzed | RT-qPCR |
| miR-215-5p [40] | Upregulated (serum) | 15 | Not reported | Not analyzed | miRNA Low Density Arrays (TLDAs)/ RT-qPCR |
| miR-542-3p [41] | Upregulated (serum) | 76 | Not reported | Not analyzed | RT-qPCR |
| miR-675 [49] | Upregulated (serum exosomes) | 10 | Not reported | Not analyzed | RNA sequencing/ RT-qPCR |
| miR-15a [50] | Upregulated (serum exosomes) | 31 | no | Not analyzed | RT-qPCR |
| miR-34b [53] | Downregulated (plasma) | 133 | yes | Not analyzed | RT-qPCR |
| miR-124 [54] | Downregulated (serum) | 114 | yes | Increased after chemotherapy treatment | RT-qPCR |
| miR-139-5p [55] | Downregulated (serum) | 98 | Not reported | Not analyzed | RT-qPCR |
| miR-101 [56] | Downregulated (serum) | 152 | Not reported | Increased after chemotherapy treatment | RT-qPCR |
| miR-375 [57] | Downregulated (serum) | 95 | Not reported | Not analyzed | RT-qPCR |
| miR-195 [58] | Downregulated (serum) | 195 | Not reported | Not analyzed | RT-qPCR |
| miR-214 [59] | Downregulated (plasma of metastatic patients) | 40 | Not reported | Not analyzed | RT-qPCR |
| miR-497 [60] | Downregulated (serum) | 185 | Not reported | Not analyzed | RT-qPCR |
| miR-223 [64] | Downregulated (serum) | 112 | Not reported | Not analyzed | RT-qPCR |
| miR-491 [65] | Downregulated (serum) | 102 | Not reported | Not analyzed | RT-qPCR |
| miR-194 [66] | Downregulated (serum) | 124 | Not reported | Increased after surgery | RT-qPCR |
| miR-133b [67] | Downregulated (serum) | 100 | yes | Not analyzed | RT-qPCR |
| miR-206 [68] | Downregulated (serum) | 100 | yes | Not analyzed | RT-qPCR |
| miR-125b [69] | Downregulated (plasma of unresectable OS patients) | 138 | Not reported | Not analyzed | RT-qPCR |
| Synovial sarcoma | |||||
| miR-92b-3p [42] | Upregulated (serum) | 12 | Not reported | Decreased after tumour resection and adjuvant chemotherapy | Microarray/ RT-qPCR |
| Kaposi’s sarcoma | |||||
| miR-375 [43] | Upregulated (serum) | 10 | Not reported | Decreased after cART-cell induced remission | Microarray/ RT-qPCR |
| Dedifferentiated liposarcoma | |||||
| miR-1246 [45] | Upregulated (serum) | 17 | yes | Not analyzed | Microarray/ RT-qPCR |
| miR-4532 [45] | Upregulated (serum) | 17 | yes | Not analyzed | Microarray/ RT-qPCR |
| miR-619-5p [45] | Upregulated (serum) | 17 | yes | Not analyzed | Microarray/ RT-qPCR |
| miR-3613-3p [46] | Upregulated (serum) | 6 | Not reported | Not analyzed | Affymetrix GeneChip Array/RT-qPCR |
| Rhabdomyosarcoma | |||||
| miR-206 [44] | Upregulated (serum) | 10 | yes | Decreased after treatment | RT-qPCR |
| miR-26a [71] | Downregulated (plasma) | 30 | Not reported | Not analyzed | ddPCR |
| miR-486-5p [51] | Upregulated (serum exosome) | 10 | Not reported | Decreased after chemotherapy | Microarray/ RT-qPCR |
| Ewing sarcoma | |||||
| miR-34a [47] | Upregulated (plasma of localized disease patient) | 31 | Not reported | Increased after chemotherapy | RT-qPCR |
| miR-125b [70] | Downregulated (serum) | 63 | Not reported | Decreased in case of poor response to chemotherapy | Microarray/ RT-qPCR |
| Liposarcoma | |||||
| miR-25-3p [52] | Upregulated (plasma vesicles) | 16 | no | Not analyzed | NanoString nCounter miRNA expression assay/ RT-qPCR |
| miR-92a-3p [52] | Upregulated (plasma vesicles) | 16 | no | Not analyzed | RT-qPCR |
| Myxofibrosarcoma | |||||
| miR-1260b [48] | Upregulated (serum) | 5 | no | Decreased postoperatively | Microarray/ RT-qPCR |
| Uterine sarcoma | |||||
| miR-24 [72] | Downregulated (serum) | 101 | Not reported | Not analyzed | RT-qPCR |
| miR-152 [72] | Downregulated (serum) | 101 | Not reported | Not analyzed | RT-qPCR |
| Circulating RNAs Detection Techniques | ||||
|---|---|---|---|---|
| Technique | Sensitivity | Specificity | Advantages | Disadvantages |
| RT-qPCR | low | Dependent to primer design | Quick and easy to perform | Analysis is limited to a restricted number of targets, normalization relative to housekeeping genes |
| Microarray | medium | Dependent to probe design and density | Quick and easy to perform | Probes can limit the detection of novel mutations |
| Droplet digital PCR | high | good | Absolute quantification, ideal for low input target concentration | Limited number of targets, prior knowledge of mutations is required |
| Next generation sequencing | high | for detection of genetic and epigenetic changes | Simultaneous profiling of several genes | Specialized bioinformatic approach |
| isoCirc | very high (for circRNAs) | reduced low-accuracy defects | 70-fold enrichment of circRNA reads compared with RNase R-treated short-read libraries High reproducibility among technical replicates | Specialized bioinformatic approach |
| CIRI-long | very high (for circRNAs) | reduced low-accuracy defects | mitochondrial circRNAs detection | Specialized bioinformatic approach |
| Cationic liposome nanoparticle biochips | very high | to distinguish cancer cell-derived exosomal miRNAs from normal cell-derived exosomal miRNAs | detection of tumor-derived exosomal microRNAs with high specificity and sensitivity | Specialized technical procedures and instruments |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chellini, L.; Palombo, R.; Riccioni, V.; Paronetto, M.P. Oncogenic Dysregulation of Circulating Noncoding RNAs: Novel Challenges and Opportunities in Sarcoma Diagnosis and Treatment. Cancers 2022, 14, 4677. https://doi.org/10.3390/cancers14194677
Chellini L, Palombo R, Riccioni V, Paronetto MP. Oncogenic Dysregulation of Circulating Noncoding RNAs: Novel Challenges and Opportunities in Sarcoma Diagnosis and Treatment. Cancers. 2022; 14(19):4677. https://doi.org/10.3390/cancers14194677
Chicago/Turabian StyleChellini, Lidia, Ramona Palombo, Veronica Riccioni, and Maria Paola Paronetto. 2022. "Oncogenic Dysregulation of Circulating Noncoding RNAs: Novel Challenges and Opportunities in Sarcoma Diagnosis and Treatment" Cancers 14, no. 19: 4677. https://doi.org/10.3390/cancers14194677
APA StyleChellini, L., Palombo, R., Riccioni, V., & Paronetto, M. P. (2022). Oncogenic Dysregulation of Circulating Noncoding RNAs: Novel Challenges and Opportunities in Sarcoma Diagnosis and Treatment. Cancers, 14(19), 4677. https://doi.org/10.3390/cancers14194677





