Simple Summary
The search for non-invasive biomarkers is a hot topic in modern oncology, since a tissue biopsy has significant limitations in terms of cost and invasiveness. The treatment perspectives have been significantly improved after the approval of immunotherapy for patients with hepatocellular carcinoma; therefore, the quick identification of responders is crucial to define the best therapeutic strategy. In this review, the current knowledge on the available non-invasive biomarkers of the response to immunotherapy is described.
Abstract
The treatment perspectives of advanced hepatocellular carcinoma (HCC) have deeply changed after the introduction of immunotherapy. The results in responders show improved survival compared with Sorafenib, but only one-third of patients achieve a significant benefit from treatment. As the tumor microenvironment exerts a central role in shaping the response to immunotherapy, the future goal of HCC treatment should be to identify a proxy of the hepatic tissue condition that is easy to use in clinical practice. Therefore, the search for biomarkers that are accurate in predicting prognosis will be the hot topic in the therapeutic management of HCC in the near future. Understanding the mechanisms of resistance to immunotherapy may expand the patient population that will benefit from it, and help researchers to find new combination regimens to improve patients’ outcomes. In this review, we describe the current knowledge on the prognostic non-invasive biomarkers related to treatment with immune checkpoint inhibitors, focusing on serological markers and gut microbiota.
1. Introduction
In recent years, the management of hepatocellular carcinoma (HCC) has dramatically changed. The recent update of the current guidelines emphasizes that the expansion of the drug armamentarium of tyrosine kinase inhibitors [1,2,3] (TKIs) and the introduction of immunotherapy as a first-line treatment has improved survival results, even in advanced stages [4]. Several trials have tested the efficacy of immune checkpoint inhibitors (ICIs), a class of drugs blocking the programmed cell death receptor 1 (PD-1), programmed cell death ligand 1 (PD-L1), cytotoxic T-lymphocyte antigen 4 (CTLA-4), lymphocyte activation gene 3 (LAG-3), and mucin domain molecule 3 (TIM-3) in monotherapy or in combination with other ICIs or TKIs [5]. The results of these studies have demonstrated the superiority of ICI-based therapies versus Sorafenib in terms of overall survival (OS) and progression free survival (PFS) [6,7]. The combination of Atezolizumab plus Bevacizumab or Durvalumab [8] plus Tremelimumab has been recently recommended as a first-line regimen in advanced HCC [4]. Other treatment regimens such as Nivolumab plus Ipilimumab [9] or Pembrolizumab [10] received Food and Drug Administration (FDA)-accelerated approval as second line therapies in patients previously treated with Sorafenib, even if the best sequential strategy for the administration of these drugs has yet to be well defined, and even though Pembrolizumab did not satisfy the pre-planned statistical threshold of effectiveness in spite of the improved OS and PFS vs. a placebo in a phase III trial involving patients progressing after Sorafenib [11]. Other ongoing studies evaluate the role of ICIs in the neoadjuvant or adjuvant setting in combination with locoregional treatments or surgical resection [12,13,14].
Despite the significant results obtained with immunotherapy, not all patients receive adequate benefits from this type of treatment [15,16]. Multiple factors may be associated with this variable effect, depending on the characteristics of the host and tumor microenvironment (TME), the latter being only marginally involved in the treatment allocation for HCC. This is the result of the biopsy-sparing diagnostic approach to this type of tumor, which has severely limited the progress in prognostic stratification in recent years, generating a gap between the clinical characteristics of patients and the histological counterpart of HCC [16]. Hence, it is crucial to identify non-invasive biomarkers to prognosticate the probability of response to ICIs, as this would allow patients to be directed to the most appropriate drug or combination of drugs. In light of these considerations, this review will address the current knowledge and future perspectives on the non-invasive biomarkers of a response to immunotherapy in patients with HCC.
2. Clinical Usefulness of Non-Invasive Biomarkers
Tumor biomarkers are cellular and molecular products linked directly or indirectly to the presence of cancer cells that are an expression of the tumor’s intrinsic characteristics and can be identified, measured, and analyzed by specific tests [17,18]. They can be used for multiple purposes, mainly for the early detection of a tumor, but also for defining its biological behavior and aggressiveness, and for assessing the response following a therapeutic intervention [19]. In the past few years, HCC biomarkers have been prevalently derived from histological analysis, but currently there is an increasing interest in developing novel techniques to extract information on tumor biology directly from patient’s body fluids in a non-invasive way [20,21,22]. Indeed, liver biopsy is invasive and requires the use of structural resources with consequent costs; furthermore, it renders a timely picture of the TME or genomic landscape and cannot capture dynamic changes over time. A liquid biopsy can overcome these limitations. It is a non-invasive approach of detecting tumor-derived products that can be performed at different time points, drawing a personalized and dynamic picture of HCC’s evolution and response to therapy [23]. There are various tumor by-products that can be measured in the bloodstream alone or in combination [24], some of which are well-known and traditionally linked to the mechanisms of carcinogenesis (e.g., cytokines and alpha-fetoprotein), while others have only recently been studied (e.g., circulating tumor cells, DNA, RNA, and exosomes).
3. Traditional Non-Invasive Biomarkers of Response to ICIs
3.1. Cytokines, Immune Checkpoints, and Immune Cells
Circulating soluble factors, such as cytokines, have been evaluated as biomarkers over the course of ICI therapy [25]. An evaluation of the baseline levels of 34 circulating serum cytokines and chemokines of patients receiving Atezolizumab plus Bevacizumab highlighted that only interleukin 6 (IL-6) and interferon alpha (IFN-alpha) were related to disease progression [26]. IL-6 is an inflammatory cytokine involved in liver regeneration, but also shows an oncogenic role [27,28], as it is involved in the recruitment of myeloid-derived suppressor cells, thereby favoring tumor immune escape [29]. It can be simply detected by an enzyme-linked immunosorbent assay (ELISA) [25,26].
A high serum level of IL-6 was also associated with worse PFS and OS and was more frequently found in females; in patients with high Aspartate Aminotransferase (AST), Alpha Fetoprotein (AFP), and Des-gamma-carboxy prothrombin (DCP) values; and those with reduced liver function. A limitation of this study is the small number of Asian patients enrolled. Moreover, IL-6 serum levels are influenced by inflammatory conditions, reducing their specificity [25,26,27,28,29].
In another study, performed to evaluate the correlation between circulating biomarkers and the response to the anti-PD-1 Pembrolizumab after 60–90 days of treatment in patients with advanced HCC, among several plasma biomarkers, only transforming growth factor (TGF)-beta serum levels significantly differed between the responders and non-responders (141.9 pg/mL vs. 1071.8 pg/mL) [30]. Plasma levels of TGF-beta higher than 200 pg/mL indicated a poor treatment response and a reduced PFS and OS (p = 0.003). Circulating biomarkers, assessed by ELISA, demonstrated that higher levels of PD-1 and PD-L1 were associated with the upregulation of IFN-gamma and IL-10 (p < 0.05); patients with a high tumor expression of PD-1 showed higher serum levels of IFN-gamma and IL-10, but plasma PD-L1 did not correlate with tumor PD-L1 expression. Changes in plasma biomarkers were also observed after treatment: only Chemokine (C-X-C motif) ligand 9 (CXCL9) increased regardless of the response (p = 0.008) [30].
The impact of PD-L1 expression in HCC TME and its correlation with prognosis has not been clarified yet. PD-L1 expression is not homogeneous in tumor tissue, varying over time depending on several stimuli; the different assays used to determine PD-L1 expression in different studies may also be partially responsible for these results [25,31,32].
PD-L1 expression on peripheral immune cells has been previously reported in patients with HCC [33]. Recent studies demonstrated that the response to ICIs may be influenced by the prevalence of PD-L1 expression on cluster differentiation (CD) 4 T lymphocytes before treatment [34]. Furthermore, immunotherapy may influence the expression of PD-1 and PD-L1 during systemic treatment on double positive CD4/CD8 T cells and on CD4 T cells in responder patients compared to non-responders, even in the absence of differences at the baseline [35,36]. Soluble PD-L1 (sPD-L1) has been tested in patients with renal cell carcinoma or melanoma prior to, and at two time points during, treatment with Nivolumab; a progressive or stable disease was associated with an increase in sPD-L1 [35]. The reliability of these results is affected by the absence of standardized reference levels for sPD-L1 and of pre-established cut-off levels for prognosis and response prediction. Moreover, ELISA results may vary depending on the assay kit [35]. In HCC patients, a meta-analysis demonstrated that a high sPD-L1 level correlates with a shorter survival (HR: 2.93; 95% CI: 2.20–3.91; p < 0.00001) [36]. In a study conducted in early HCC patients who underwent surgical liver resection, the persistence after treatment of sPD-L1 indicated a poor outcome, suggesting the possibility to apply sPD-L1 analysis for identifying patients eligible for adjuvant immunotherapy [37]. Overall, the predictive value of sPD-L1 during ICIs therapy remains controversial and further studies are needed [38].
Peripheral Blood Mononuclear Cells (PBMC) reflect the changing landscape of tumor-associated immune cells before and after immunotherapy. They can be easily identified and characterized using flow cytometry, with a subset of antibodies binding to different CD proteins [39]. In patients with HCC, the response to ICIs is influenced by the number of peripheral effector T cells, which are more prominent in the presence of CTLA-4 and the inducible Co-stimulator ICOS on PBMC surfaces, regardless of the etiology of liver disease. Moreover, anti-PD-1 agents upregulate cytotoxic T cell effectors with a memory phenotype, also reducing PD-1 expression on their surface, and cause the downregulation of B cells [40]. The presence of CD14+ CD16− Human Leukocyte Antigen-DR isotype (HLA-DRhi) monocytes in peripheral blood before ICIs treatment is another marker of a favorable outcome; monocytes presenting these receptors can favor the infiltration of T cells in tumor tissue, leading to the activation of T cell effectors against cancer [41]. Other studies demonstrated that baseline CD4+ PD-1+ cells predict a successful response to the anti-CTLA-4 Tremelimumab, and that a lower percentage of T regulatory cells as well as higher levels of double positive CD4/CD8 T cells are associated with the response to anti-PD-L1 inhibitors [40,42]. PBMCs obtained before and after 6 weeks of therapy with Pembrolizumab in patients with advanced HCC showed that the immune cell distribution was not homogeneous, with levels of CD8 T cells and CD4 T naive cells that differed among the samples, suggesting a role for this imbalance in response modulation [43]. After therapy, an increased number of activated CD4 and CD8 T cells and a reduction in CD4 and CD8 naive T cells was observed. Moreover, the responders had higher levels of cytotoxic CD8 T cells, while the poor responder patients expressed molecules associated with neutrophil-associated pathways. In another study, Nivolumab therapy was associated with an elevation of CD8 αβT cells after 4 weeks of therapy; these cells showed lower levels of PD-1 expression compared to that belonging to patients with disease progression. No significant alteration in regulatory T cells was observed, and the patients achieving disease control (DC) maintained a persistent expression of PD-L1 on their monocytes after 28 and 42 days of treatment. The post treatment changes in the PD-L1 positivity of the patients’ monocytes differed among responders and non-responders after 28 and 42 days. Finally, the low pretreatment expression of PD-1 on peripheral B cells was associated with DC [42]. In a study by Macek Jilkova et al., the composition and expression of CD in blood lymphocytes, natural killer (NK) T cells, and NK cells were evaluated and compared to T cells and NK cells within the liver in 21 patients with advanced HCC treated with Sorafenib [44]; the CD31+ and CD56+ T-cells found in the blood accounted for more than 60% of all CD45-high lymphocytes, whereas in the liver, their frequency decreased to 50%. Conversely, CD3+ CD56+ cells (NKT and CD3brightCD56+ T cells) and CD3- CD56+ NK cells represented a significantly smaller population in the blood (7% and 11%), but their frequency increased up to 21% and 21% in nontumor liver tissues and 19% and 17% in tumor liver tissues, respectively. Dividing the population according to their progression after 16 weeks, the elevation of IL-10 in the blood samples correlated with a higher CD4/CD8 ratio in tumor and non-tumor tissues and a poor prognosis. The expression of the immune checkpoint molecules LAG3 and TIM3 on circulating T cells was upregulated in non-responders and associated with poor survival [44].
3.2. Neutrophil to Lymphocyte Ratio, Platelet to Lymphocyte Ratio, Prognostic Nutritional Index, and Their Combined Prognostic Value
The Neutrophil to Lymphocytes Ratio (NLR) has been used to evaluate the risk of mortality in patients with liver disease [45,46], and has also been tested as a prognostic biomarker for immunotherapy in several studies. A decline in the NLR in patients with HCC receiving anti PD-1 therapy was associated with a better response to treatment and improved survival [46,47]. In patients receiving Atezolizumab plus Bevacizumab, the baseline NLR (cut-off 3.21) resulted to be an independent predictor of response, and was directly linked with PFS [48]; in another study, a low NLR was associated with a better OS [49]. Similarly, a high Platelet to Lymphocytes ratio (PLR), an inflammatory marker already correlated with prognosis in patients with HCC [46,50], has been associated with poor prognosis [46]. Finally, a high prognostic Nutritional Index (PNI), obtained by multiplying the serum albumin (g/dL) by the total lymphocyte count, was associated with a response to anti-PD-1 therapy and better survival [51].
Based on these results, a study combined NLR, PLR, and PNI to evaluate their roles as predictors of response to immunotherapy [52]. The blood samples of 362 HCC patients were collected; 74.3% of themhad advanced disease, and the most frequent liver disease etiology was the hepatitis C virus. Eighty percent of the patients received anti-PD-1 monotherapy, while the others received a combination of anti-PD-1 plus anti-CTLA-4 or TKIs. The median NLR value in the whole cohort was 3.55 (0.06–25.3), the median PLR value was 137.32 (0.17–1100), and the median PNI value was 40.29 (1.11–1270). The previously indicated thresholds for a high mortality risk (NLR ≥ 5, PLR ≥ 300, and PNI < 45) were associated with the presence of portal vein thrombosis, a worse performance status, a more advanced Barcelona Clinic Liver Cancer (BCLC) stage, and worse OS and PFS (limited to PLR and PNI). In the multivariate analysis, an NLR ≥ 5 and a PLR ≥ 300 remained independent prognostic factors for OS (HR 1.73, 95% CI 1.23–2.42, p = 0.002 and HR 1.60, 95% CI 1.6–2.40, p = 0.020, respectively), while the PLR was the only independent predictor of PFS. These results were partially confirmed by a Chinese study reporting that all the inflammation-based prognostic scores showed good discriminatory ability in OS, but only the PNI score was an independent predictor for OS in multivariate analysis [53].
Finally, albumin, lactate dehydrogenase (LDH), and the NLR were combined in a score named the Gustave Roussy immune score (GRIm-Score), which was able to stratify patients receiving ICIs into responders and non-responders with respect to several tumors [54]. To better mirror the characteristics of HCC, the GRIm-Score was implemented with other circulating markers that resulted independent prognostic factors of survival in a multivariate analysis; therefore, the HCC-GRIm-Score was obtained, including albumin (<35 g/L = 1), LDH (>245 U/L = 1), NLR (≥4.8 = 1), Aspartate Aminotransferase to Alanine Aminotransferase AST-to-ALT ratio (≥1.44 = 1), and total bilirubin (≥22.6 umol/L = 1). Lower scores (from 0 to 2 points) resulted in a better OS [55]. Despite the remarkable results highlighted by these studies on the utility of a combinatory approach of non-invasive biomarkers to improve sensitivity, further and larger clinical trials are needed to apply them in clinical practice.
3.3. Alpha-Fetoprotein
Serum alpha-fetoprotein is the HCC-related biomarker that is most used in clinical practice [56,57]. Studies showed that early AFP reduction is associated with a good prognosis in patients with HCC receiving ICIs [58,59,60,61]. Previous in vitro studies demonstrated a pro-oncogenic role of AFP in inducing resistance to tumor necrosis factor (TNF) cytotoxic activity, promoting protein kinase A activity and the expression of the pro-oncogenic proteins p53 and p21 [62,63]. In addition, NK cells’ activity has been shown to be impaired in the presence of AFP-treated dendritic cells (DCs), which decreases IL-12 production [64]. AFP also exerts a pro-oncogenic priming in both animal and human models of HCC by reducing the FAS/FAS-associated death domain protein (FADD) apoptotic pathway through the modulation of Human Antigen R RNA-binding protein, resulting in the promotion of tumor growth [65]. For these reasons, targeting AFP could be a new therapeutic strategy in the treatment of HCC, and an ongoing trial is evaluating the efficacy of T cells’ reprogramming against AFP in AFP-expressing HCC patients [66].
AFP has the advantage of being detectable via traditional methods, such as radio- and fluorescent-immunoassays, which are available in almost all laboratories [67]. However, current guidelines highlight the suboptimal performance of AFP for HCC screening due to several limitations [68,69]. In particular, when used as a diagnostic test, AFP shows good sensitivity but low specificity with a cut-off of 20 ng/mL, while the sensitivity decreases by increasing the cut-off to 200 ng/mL, although improving specificity [68]. Moreover, many HCCs do not express AFP; on the contrary, liver cirrhosis and other intrahepatic and extrahepatic non-HCC tumors may be associated with an increase in AFP serum levels [70]. Finally, when combined with ultrasonography, AFP increases the ability to detect previously unidentified nodules by 6–8% [68].
Models combining AFP with other biochemical parameters have reported promising results with respect to predicting ICIs response. One of them that includes AFP and C-reactive protein (CRP), named the CRAFITY score, has been recently evaluated in HCC patients receiving anti-PD-1 immunotherapy. The patients with a CRAFITY score of 0 showed a better radiological response and OS compared to patients with a CRAFITY score ≥ 1 [71,72].
Another study combined the CRAFITY score with AFP decline after 6 weeks of treatment with Atezolizumab plus Bevacizumab, generating a classification named CAR (CRAFITY score and AFP Response) [73]. The patients were divided into three classes: those with a low CRAFITY score and good AFP response (class I), either a high CRAFITY score or an unsatisfactory AFP response (class II), and a high CRAFITY score and an unsatisfactory AFP response (class III). The Median Objective Response Rate (ORR) was better in class I than in classes II or III (35% vs. 18.2% vs. 0%). The patients in class I had the best OS and PFS, followed by those in classes II and III (the median OS of class I did not reach the median time exceeding the 12 months of follow-up vs. 11.1 months vs. 4.3 months, p < 0.001; the median PFS was 7.9 months vs. 6.6 months vs. 2.6 months, p = 0.001).
Another study analyzed the prognostic ability of AFP plus prothrombin induced by vitamin K absence-II (PIVKA-II) in HCC patients treated with anti-PD-1 therapy [74]. Reductions in AFP and PIVKA-II of more than 50% from the baseline levels at 6 weeks of treatment were associated with a favorable ORR and a better OS and PFS. The combination of these results with the Albumin Bilirubin score (ALBI) was included in the AAP score; patients with an AAP score ≥ 2 points showed a significantly longer PFS and OS. Lower serum levels of AFP and soluble intercellular adhesion molecule 1 (sICAM-1)—a soluble factor derived from endothelial cells and involved in inflammatory responses, which is absent in normal hepatocytes [75,76]—were associated with a response to immunotherapy in a recent analysis of the Cancer Genome Atlas Liver Hepatocellular Carcinoma. AFP and ICAM-1 upregulation correlate with an immunosuppressive TME in which CD4 T cells, macrophages, and monocytes are predominant, and the expression of immune checkpoints such as T cell immunoglobulin and ITIM domain (TIGIT), Hepatitis A virus cellular receptor 2 (HAVCR2), PD-1, CTLA4, and LAG3 is enhanced [77].
In conclusion, AFP is a definitively cheap non-invasive biomarker, but its combination with other serum markers [71,72,73,74] deserves further investigation to improve diagnostic accuracy.
A summary of the main findings of the available studies on traditional non-invasive biomarkers described herein is reported in Table 1.

Table 1.
Traditional non-invasive biomarkers for immunotherapy in patients with hepatocellular carcinoma (HCC).
4. Novel Biomarkers
4.1. Circulating Tumor DNA
Circulanting Tumor DNA (ctDNA) are cell-free DNA products released by tumor cells in the bloodstream during apoptosis or necrosis that are rapidly cleared by macrophages [78]. ctDNA is detected using real-time PCR and digital PCR, which are able to recognize DNA aberrations in target genes in the presence of a specific probe; this limitation is overcome by Next Generation Sequencing (NGS), which allows for the identification of new genomic aberrations that may cause resistance to therapy [79]. Another method is the Sequenom’s (San Diego, CA, USA) MassARRAY Compact system, which is able to measure the direct mass of nucleic acids with high precision, in addition to quantifying gene expression and identifying genotypes and the methylation of ctDNAs [80]. The concentration of ctDNA in peripheral blood is higher in patients with HCC than in cirrhotic patients without a tumor and healthy controls. Interestingly, tumor size, extrahepatic spread, and vascular invasion are associated with higher ctDNA levels [81]. Tumor ctDNA fragments are longer when compared to circulating DNA derived from other apoptotic host cells; indeed, the ratio of ctDNA to the whole circulating DNA length, called DNA integrity, has been used as a marker for the early diagnosis of HCC [82].
A qualitative analysis of somatic gene mutations in HCC-derived ctDNA fragments showed that several oncogenes and tumor suppressor genes such as ARIDNA, tumor protein 53 (TP53), catenin (cadherin-associated protein), and beta 1 (CTNNB1) are involved, and the same mutations were found in tumor tissue in 63% of cases [82,83]. The presence of Telomerase reverse Transcriptase gene (TERT) mutations in ctDNAs are associated with vascular invasion [84]. Mutations in the PI3K/mTOR pathway correlate with a shorter PFS in patients treated with TKIs but not with ICIs [84], whereas mutations in MutL homolog 1 are linked to a worse OS [85].
The detection of a TP53 R249S mutation in ctDNA after HCC resection is a marker of a poor disease-free survival (DFS) [86]. ctDNA detectable after curative treatment has been related with microvascular invasion and may predict tumor recurrence and extrahepatic spread [87]. Accordingly, preoperative ctDNA detection was associated with larger HCCs, multiple lesions, microvascular invasion, and shorter PFS and OS [81]. A high tumor mutational burden (TMB) in the tumor genome was associated with an effective immune response against several tumors, due to the increased neoantigen load [88,89]. In addition to a TMB assessment on liver tissue, an emerging blood-based technique has been described and performed on ctDNA. Blood TMB (bTMB) accurately reflects tissue TMB, as described for several tumors. Recently, a commercial ctDNA platform for quantifying bTMB from blood has proven to be sensitive and reproducible, with an optimal ctDNA TMB cutoff of ≥20 mut/Mb that predicted positive results with Durvalumab plus Tremelimumab compared to chemotherapy in non-small cell lung cancer [90].
In a pilot study, the analysis of a cohort of patients with advanced solid tumors treated with immunotherapy demonstrated the correlation between bTMB and tTMB regardless of tumor histology [91]. However, the patients with higher levels of bTMB did not achieve a better OS, but the heterogeneity of the cohort may explain this result. Interestingly, an exploratory analysis from the same study demonstrated a reduction in the ctDNA mutant allele frequency (MAF) over the treatment period in the responders. Similarly, ctDNA MAF has been reported to change dynamically after surgery and to correlate with recurrence-free survival and OS [92]. Despite these promising results, at present, the application of ctDNA as a biomarker is limited, mainly due to the lack of standardized procedures for sample preparation and the difficulty of determining the assay when the ctDNA levels are very low [93].
4.2. Circulating Tumor Cells
Circulating Tumor Cells (CTCs) are rounded, nucleated cells released into the bloodstream from the primary tumor site or from metastatic sites [94]. CTCs express epithelial proteins such as Epithelial Cell Adhesion Molecules (EpCAM); cytokeratin 8, 18, or 19; and stem cells markers, while they lack the CD45 antigen. They can be found as single cells or in clusters, according to their mono- or oligo-clonal origin; CTC clusters are more prone to seeding, as they express transcription factors of genes that enhance proliferation.
CTCs can be isolated using different techniques, categorized as follows: (1) physical methods, according to size (filtration-based devices), electric charge (electrophoresis), density (Ficoll centrifugation), migratory capacity, and deformability; (2) biological methods, based on antigen–antibody binding, for a qualitative analysis of the proteins expressed on CTCs surfaces. Specifically, biological methods use antibodies against specific tumor biomarkers, such as EpCAM, which is not expressed in blood cells but only in cells of an epithelial origin. The EpCAM-based CellSearch platform (Veridex LLC, Raritan, NJ, USA) has been approved by the FDA for CTC detection. This method has a limitation because although EpCAM is expressed in most epithelial-derived cells but not in blood cells, tumor cells who undergo endothelial-mesenchymal transition are not detected; furthermore, the absence of standardization limits the diffusion of CTC detection [95]. To overcome this issue, CTCs can be detected with the Canpatrol platform, a multiplex RNA in situ hybridization against EpCAM and several epithelial cytokeratines [96]. More recently, a microscale technology was developed to increase sensitivity and identify lower levels of CTCs [97]. CTCs are increased in patients with HCC, and their levels correlate with the prolongation of the disease, tumor stage, and AFP serum levels [98]. The number of CD3, CD4, and CD8 T cells negatively influences the presence of CTCs, while the number of T regulatory cells is associated with a greater number of CTCs [78].
Despite their low concentration in peripheral blood (about 5–50 CTCs in 7.5 mL of blood) [99] and their short half-life (2.5 h) [100], several studies of different tumors showed that CTCs may be useful as prognostic markers after treatment. Indeed, it has been reported that after HCC surgical resection or locoregional treatment, the CTCs concentration drops [101], whereas any increase after treatment is associated with a higher risk of tumor recurrence [102]. Studies in several cancers showed that CTCs expose immune checkpoints such as PD-L1, PD-L2, and CTLA-4 on their membrane, so they have been evaluated as biomarkers to identify patients suitable for immunotherapy [103]. In a phase I trial, the total number of CTCs and the PD-L1 expression on CTCs was evaluated in patients with advanced gastrointestinal cancers, including HCC, at baseline and following anti-PD-1 therapy [104]. The patients were divided into four categories based on their PD-L1 expression at baseline (negative, low, medium, and high); 74% of the patients showed PD-L1-high CTCs, which correlated with a good treatment response, while the persistence of PD-L1-high CTCs after therapy was associated with a poor outcome. Reductions in total CTCs and in PD-L1 expression on CTCs from the baseline levels were also reported in patients with a stable disease. Even though a positive correlation between baseline PD-L1-high CTCs and disease status was not detected, the average percentage of PD-L1-high CTCs among the total number of CTCs in the patients with disease control was significantly higher compared to the patients with a lower expression of PD-L1-high CTCs; moreover, the PD-L1-high CTC/total CTCs ratio was higher in the responders. Further, the absence of PD-L1-high CTCs at baseline was associated with a higher risk of progression during anti-PD1 therapy. Taken together, these results suggest that the presence and distribution of PD-L1-high CTCs could be a better biomarker in predicting PD-1 therapy response compared to PD-L1 positive CTCs. Another study including only HCC patients confirmed that PD-L1+ CTCs identified responders to ICIs [105].
Despite the promising role of CTCs as a biomarker for HCC, their low concentration in the bloodstream and short half-life can limit the reliability of their respective assay. Moreover, the validated platform has a weak performance with respect to the recognition of CTCs with markers of endothelial-mesenchymal transition. The lack of standardized clinical trials with large cohorts of patients is another concern that should be overcome [106].
4.3. Extracellular Vesicles
Extracellular vesicles (EVs) are nanoparticles that have heterogeneous functions, including the ability to modulate inter-cellular communication, and could influence several processes, such as inflammation and tumorigenesis [107,108]. EVs can be classified in exosomes (size < 200 nm), derived from the internal budding of endocytic membranes, and microvesicles (MVs) (size > 200 nm), derived from the extroflession of activated cells’ membranes [109]. Exosomes express CD63, CD81, and CD9 on their surfaces [109] and can interact with target cells through exosomal proteins and cellular receptors, the direct fusion of the exosomal membrane with the cell membrane, or endocytosis [110].
Ultracentrifugation is the gold standard technique for exosomes’ isolation and extraction, alone or in combination with other methods, such as density gradient centrifugation for purification. Other physical methods include polymer precipitation, size exclusion chromatography, and ultrafiltration. Otherwise, exosomes can be detected by antibodies directed against specific markers exposed on their membranes, or by the combination of physical and biological methods for EV detection, such as immunoaffinity chromatography [111].
Through autocrine and paracrine mechanisms, exosomes enable the interaction between tumor cells and TME, with immune-modulatory effects and the stimulation of the epithelial to mesenchymal transition that favors vascular invasion and metastatization [112]. Moreover, increasing evidence suggests that the interaction between tumor-derived exosomes, tumor cells, and TME may play a significant role in the development of drug resistance to TKIs in patients with HCC [113,114,115,116,117,118]. The exposure of PD-L1 on exosomes causes the direct inhibition of T cells’ function. Exosomes expressing PD-L1 compete with cancer cells and peritumoral cells in binding ICIs; as a result, lower levels of a drug can target tumor cells, resulting in a mechanism of resistance against therapy [116]. Similar to exosomes, MVs express specific markers on their surface derived from the type of cell that generates them and contain nucleic acids and proteins that could influence several biologic processes [117]. MVs expressing Hepatocyte Paraffin 1 (HepPar1+) have been found to be higher in patients with HCC than in controls without cancer, and their lack of reduction 3 months after liver resection was observed in patients with tumor recurrence [118]. AnnexinV+ EpCAM+ Human Asialoglycoprotein Receptor 1 (ASGPR1+) MVs were able to distinguish patients with cirrhosis and liver cancer (HCC or cholangiocarcinoma) from those with no malignancy, and this was confirmed by the drop in the concentration 7 days after curative resection [119].
EVs contain a variety of proteins, lipids, DNAs, messenger RNAs (mRNAs), microRNAs (miRNAs), and other non-coding RNAs, such as circular RNAs (circRNAs) and long non-coding RNAs (lncRNA) [120,121]. These genetic products and proteins are similar to those expressed in the tumor tissue; thus, EVs not only reflect a cancer’s features and its dynamic changes [122] but can also regulate various cellular processes such as proliferation, survival, migration, and the inhibition of the anti-tumor response [123]. In particular, long non-coding RNAs (lncRNA) have shown a potential role in modulating immunotherapy responses via TME re-programming, leading to the exhaustion of CD8 cells, which is a well-known marker of a poor response to ICIs [124]. lncRNAs’ detection and amplification has been obtained using real-time PCR techniques or, lately, microarray technology [125]. Several signatures based on lncRNAs have been associated with a response to ICIs [126,127,128,129,130,131,132]. Furthermore, circular RNAs (circRNAs) can regulate gene expression and interfere with transcription and peptide translation through the modulation of microRNAs (miRNAs) [133]. Several methods exist to detect circRNAs, such as northern blotting and real-time PCR; droplet digital PCR, which divides a sample into 20 thousand droplets to obtain the PCR amplification of the template in each droplet, has also been applied to increase the sensitivity for the detection of low levels of circRNAs. Other methods that are used include isothermal exponential amplification, which can copy oligonucleotides within minutes with a high amplification ability [134], and rolling cycle amplification, which uses circular probes as templates to generate long single-stranded DNA or RNA of the target sequence [135]. The genotype analysis of circRNA is based predominantly on NGS, but this technique loses sensitivity with an increasing read length. Recently, a novel algorithm has been developed to overcome this problem: using full-length circular reverse transcription, a long complementary DNA strand including many copies of the circRNA target is obtained; then, the full length circRNAs are sequenced using nanopore technology. This algorithm is also able to quantify circRNA expression and the presence of mutations [136]. In HCC tissue, Circular ubiquitin-like with PHD and ring finger domain 1 (circUHRF1) RNA is upregulated. It decreases the activity and number of NK cells in tumor tissue and has been associated with a resistance to ICIs [137]. CircMET (hsa_circ_0082002), which is highly expressed in HCC tissue and exosomes compared to non-tumor cells, promotes HCC cells’ survival by acting on the miR-30-5p/Snail/dipeptidyl peptidase 4 (DPP4)/CXCL10 axis, with the consequent inhibition of CD8 T cell functions and a resistance to anti-PD-1 therapy [138]. Hsa-crc-0003288 has been demonstrated to increase PD-L1 expression in vitro by the activation of the PI3K/AKT signaling pathway, thereby promoting tumor proliferation and the epithelial to mesenchymal transition of cancer cells [139]. Hsa_circRNA_104348 is upregulated in HCC tumor cells and promotes tumor progression and invasion via the Wnt/beta catenin and miR-187–3p/RTKN2 pathways, leading to a poor response to ICIs [140]. CircRHBDD1 promotes a hypoxic environment by the upregulation of glycolysis; its expression has been analyzed in 18 patients with advanced HCC who received anti-PD-1 therapy: after 4 cycles of treatment, non-responders expressed significantly higher levels of circRHBDD1 compared to responders. To confirm the tumorigenic role of circRHBDD1, its inhibition has been studied in a xenograft model: mice with silenced circRHBDD1 cells presented a better response to anti-PD-1 therapy and an increased infiltration of CD8 cells in tumor tissue [141]. Finally, miRNAs located in EVs can derive from any type of cell, including HCC tumor cells and TME; they contribute to chronic hepatic inflammation and tumorigenesis by promoting tumor growth and immune tolerance and by influencing angiogenesis and extrahepatic spread [142]. Initially, miRNAs were isolated from cells using a phenol–chloroform extraction followed by RNA precipitation. Since this method causes a loss of RNA sequences full of GC base pairs, adding a column-based RNA absorption method reduces the presence of contaminants and stabilizes the miRNA sequences. The mirVana and miRNEasy kits are two of the available kits [143,144]. The detection of circulating miRNAs is based on quantitative PCR. Lately, the application of digital PCR, which detects the absolute level of miRNA expression with a high specificity and sensitivity even in fluids, has been proposed [145].
MiRNAs can target the 3′-UTR regions of PD-L1 mRNA; otherwise, they can act indirectly on PD-1, as observed in other non-HCC solid tumors [146,147,148]. Hsa-miR-329-3p inhibits lysine-specific dymethylase 1A (KDM1A), which increases the methylation of Myocyte Enhancer Factor 2D (MEF2D), reducing the expression of PD-L1 and blocking tumor growth, as demonstrated in a xenograft model of HCC [149]. Other miRNAs, such as miR-675-5p, can upregulate or, as in the case of miR-145, miR-194-5p, and MiR-200, downregulate PD-L1 expression in HCC TME [126,134,150,151]. MiR-34a can target the 3′-UTR regions of PD-L1, thereby reducing its ability to bind to PD-1, increasing the infiltration of CD8 T cells in tumor tissue, and activating DCs [152]. MiR-155 upregulates TIM-3, resulting in an enhanced degree of T cell exhaustion [153]. Conversely, the interaction between miR-155 and the lncRNA Nuclear-Enriched Abundant Transcript 1 (NEAT1) can interfere with tumor progression in mice, thus enhancing CD8 T cell cytolysis. Moreover, miR-449c-5p, which is expressed by NK cells, can bind to TIM-3 mRNA causing its degradation and boosting the immune response in HCC TME [132]. Despite the promising role of EVs as possible non-invasive biomarkers, no gold standard for exosome isolation exists; therefore, a remarkable difference in the terms of sensitivity among the different platforms has been reported. The costs of RNA extraction, sequencing, and characterization are other concerns [154].
4.4. Antidrug Antibodies
Following the administration of ICIs, patients may present a hyperstimulation of the immune response that leads to the production of antidrug antibodies (ADA). According to the Food and Drug Administration’s indications, ADA are usually tested in screening assays, followed by a characterization and titer quantification [155]. Among them, neutralizing antibodies (NAb) bind to the ICIs, forming an immunocomplex that is cleared by the circulation. This process may lead to a reduced efficacy of checkpoints’ blockade and to a higher sensitivity to immune adverse reactions (IRAEs) [156]. The appearance of ADA has been reported following anti-PD-1, anti-PD-L1, and anti-CTLA-4 therapy, with an incidence that differs between the ICIs classes: among anti-PD1 inhibitors, Nivolumab induced ADA expression in 11.2% of patients [157,158] compared to only 2.1% for Pembrolizumab [159]. The anti-CTLA-4 agent Ipilimumab induced ADA upregulation in 5.4% of patients [160], while the anti-PD-L1 Durvalumab induced such an upregulation in 2.9% [161] of patients; the higher levels of ADA (13–54% of patients, with NAb in 4–28% of the cases) have been reported in patients treated with Atezolizumab plus Bevacizumab at different doses and regardless of tumor histology [162]. Atezolizumab serum levels can be influenced by ADA exposure, with an average reduction of 22% in treatment activity in the ADA+ group compared with the ADA- group in vitro [163]. Atezolizumab’s efficacy is also reduced by its clearance promoted by ADA. ADA production was shown to be independent of tumor type, line of therapy, treatment dose, or administration as monotherapy or in combination with other drugs, while a male sex, Caucasian ethnicity, extended tumor burden, impaired liver function, a high level of serum C-reactive protein, NLR, and lactate dehydrogenase demonstrated a strong correlation with the development of ADA following ICIs therapy [163]. Despite these results, a meta-analysis that enrolled 7736 patients across 11 clinical trials showed no differences in terms of OS and PFS among ADA+ and ADA- patients or in ADA NAb+ versus ADA NAb-patients [164]. Another meta-analysis including 1086 patients treated with Nivolumab confirmed the absence of negative effects caused by ADA in terms of adverse reactions or a loss of efficacy [165]. According to these results, the assay of ADA as a biomarker affecting the response to immunotherapy appears to be controversial and without a demonstrated usefulness.
The main findings of the studies concerning the novel non-invasive biomarkers of the response to immunotherapy in HCC are reported in Table 2.

Table 2.
Novel non-invasive biomarkers for immunotherapy in patients with hepatocellular carcinoma (HCC).
4.5. Gut Microbiome
The liver and the gut interact in a bidirectional way. Via the portal circulation, there is a continuous passage of microorganisms, microbially derived proteins, and metabolic products, creating a functional interplay, known as the gut–liver axis. The gut microbiome plays a crucial role in maintaining the homeostasis of this system through the regulation of metabolic pathways, the immune system function, and intestinal barrier integrity [166,167]. Under normal conditions, the interaction between pathogen-associated molecular patterns (PAMPs) expressed by the gut microbiota and Toll-like receptors (TLRs) on the host cell’s surface induces a tolerogenic response, which is important for maintaining the proper functioning of the immune system and protecting the host from potentially harmful external aggressions [168,169]. Gut microbiome-derived metabolites, such as short-chain fatty acids (SCFAs) and bile acids, are also involved in the maintenance of this balance, favoring an immune-tolerogenic environment [170,171,172,173,174,175]. In patients with cirrhosis, qualitative and quantitative changes in the gut microbiota compositions have been described, with a relative decrease in beneficial bacteria and an increase in pathological ones [176,177]. These changes are associated with an inflammatory shift in metabolic and immune processes. Indeed, the intestinal barrier’s impairment and pathological bacterial translocation, which are hallmarks of chronic liver diseases, trigger a pro-inflammatory cascade that culminates in liver-focalized and systemic inflammation [178,179,180,181,182]. With the progression of liver cirrhosis, this chronic injury with persistent inflammation results in immune exhaustion, which favors HCC development [183,184,185].
In recent years, the scientific interest in the characterization of the gut microbiota has rapidly grown, followed by the development of two main techniques. The first to be adopted was the sequencing of the 16s rRNA gene [186], a highly conserved bacterial gene coding for the 30S ribosome, which is used for the identification of bacterial taxa down to the genus level [187]. Metagenomic sequencing, including the more accurate shotgun sequencing, is more recent and costly, but allows for the identification of species and even strains, as well as functional information [188].
Considering its profound impact on the immune system, several findings demonstrate that the gut microbiota influences the response to immunotherapy. Commensal bacteria are fundamental in orchestrating antitumor responses in TME, and distinct bacterial species modulate different immune responses [189]. In a study conducted on patients affected by advanced non-small cell lung cancer (NSCLC) and renal cell carcinoma receiving PD-1 blockade therapies, Routy et al. demonstrated an increased abundance of A. muciniphila in the stool specimens of responder patients versus the non-responders. Then, fecal microbiota transplantation (FMT) from the responders into germ-free (GF) or antibiotic-treated mice was performed. After tumor induction, the mice receiving an FMT from responders had a better response to the PD-1 blockade and showed an increased expression of CXCR3+ CD4+ T cells in TME. Intriguingly, an oral A. muciniphila supplementation in mice receiving an FMT from the non-responder patients was able to restore the efficacy of anti-PD-1 therapy [190]. In another study conducted on patients affected by metastatic melanoma receiving anti-PD-1 immunotherapy, a stool analysis of the responder patients showed a higher alpha diversity and increased abundance of Ruminococcaceae. Among Ruminococcaceae, an increased Faecalibacterium abundance was associated with a prolonged PFS and an improvement in CD4 and CD8 T cells, whereas Bacteroidales abundance correlated with a worse outcome and an immunosuppressive pathway. Likewise, an FMT from the responder patients into the GF mice was associated with a better response to therapy and greater anti-tumor activity [191]. These findings have been reinforced by the results of another phase 1 trial (NCT03353402), in which stool samples from two donors with a previously documented response to anti-PD-1 monotherapy were transplanted into a population of 10 patients affected by metastatic melanoma refractory to anti-PD-1 agents. Three patients obtained a complete or partial response, reverting the initial drug resistance, and showed a post-treatment increase of antigen-presenting cells infiltration in the gut, suggesting that the anti-tumor immune response may start in the intestine [192]. With respect to HCC patients, Zheng et al. [193] analyzed the gut microbial composition of eight patients affected by BCLC stage C HCC receiving Camrelizumab as a second-line treatment after Sorafenib; stool samples were collected before and after 3 to 12 weeks from the beginning of treatment. The baseline gut microbiota were mainly enriched with Bacteroidetes, followed by Firmicutes and Proteobacteria. After an ICI treatment, the patients who showed an objective tumor response presented an overgrowth of Proteobacteria, with a peak after 12 weeks. Among Proteobacteria, Klebsiella pneumoniae was the main species enriched in the responders, while in the non-responders an overabundance of Escherichia coli was reported. Furthermore, the responders presented an increased abundance of several probiotic bacteria such as Lactobacilli, Bifidobacterium dentium, and Streptococcus thermophilus, which are known to positively influence host immunity. In addition, an increase in Ruminococcaceae and A. muciniphila, which are involved in maintaining intestinal barrier integrity, was reported among responders [193]. Another small study collected stool samples from eight patients with HCC who received the anti-PD-1 agent Nivolumab [194]. The analysis of the gut microbiota demonstrated a higher concentration of Clostridia, Prevotella, and Ruminococcaceae in the responders, while Ruminococcus gnavus was predominant in the non-responder group. Citrobacter freundii, Azospirillum spp., and Enterococcus durans were correlated with a good prognosis in terms of OS and PFS, while Escherichia coli, Lactobacillus reuteri, Streptococcus mutans, and Enterococcus faecium predicted a negative outcome. The composition of the gut microbiota in HCC patients was analyzed at the phylum level, reporting an imbalance in the Firmicutes/Bacteroidetes ratio (below 0.5 or upper than 1.5) that occurred more prevalently in the non-responders than in the responders, while a higher mean ratio of Prevotella spp. to Bacteroides spp. (P/B ratio) was clearly identified in the responders; also, the presence of Akkermansia was detected only in the responders. Mao J. et al. [195] analyzed the fecal samples of 65 patients affected by advanced HCC or biliary tract cancer receiving anti-PD-1 therapies. The results showed that the patients with a clinical benefit response (CBR), considered as a partial or complete response to therapy or a stable disease for a minimum of 6 months, had a relative abundance of Lachnospiraceae bacterium-GAM79 and bacteria from Ruminococcaceae family, while the Veillonellales predominated among the patients without any clinical benefit (NCB). Moreover, a higher abundance of Lachnospiraceae bacterium-GAM79 was associated with a longer PFS and OS, while bacteria from the Veillonellaceae family were associated with a worse clinical outcome. A dynamic analysis of the gut microbiota composition also showed a decrease in bacterial diversity among the NCB group. The importance of Ruminococcaceae in predicting ICIs’ efficacy was confirmed by a retrospective Chinese study, in which the enrichment of Clostridiales/Ruminococcaceae was reported in the responders to anti-PD-1 therapy [196]. The study also showed a positive association between a high abundance of Faecalibacterium, belonging to the Ruminococcaceae family, and a longer PFS, while an increased abundance of Bacteroidales was associated with a worse prognosis.
A concern with respect to these results is that the majority of the studies included Asian patients, some of whom presented chronic hepatitis. However, a recent study including 11 Caucasian cirrhotic patients with HCC treated with Tremelimumab and/or Durvalumab demonstrated that those who achieved DCshowed a lower fecal calprotectin concentration and PD-L1 serum levels at baseline; also, the pre-treatment increased the abundance of Akkermansia observed in patients who achieved DC, in parallel with a reduction in Staphylococcus, Neisseria, and Enterobacteriaceae [197]. Dynamic analyses of the microbiota composition during treatment showed an inverse relationship between alpha diversity; Akkermansia to Enterobacteriaceae (AE) ratio, which was considered as a marker of dysbiosis; and calprotectin levels, reinforcing the hypothesis that intestinal inflammation plays a role in influencing clinical outcomes.
Metabolites derived from the microbiome can also contribute to modulating the response to ICIs, and can be used as biomarkers. A prospective study conducted on 52 patients with advanced solid tumors receiving Nivolumab or Pembrolizumab showed a higher fecal and plasma concentration of SCFAs among responders, and fecal propionic acid was identified as a marker of PFS [198]. A possible explanation for the immune-modulating activity of SCFAs is the inhibition of histone deacetylases (HDACs), which has been associated with a higher expression of PD-1 ligands and sustained PD-1 blockade in melanoma models [199,200,201,202]. Conversely, in patients affected by metastatic melanoma, high levels of butyrate and propionate seem to reduce the efficacy of anti-CTLA-4 therapy, with negative effects on DC maturation in mice, a reduced production of IL-2, and lower numbers of memory and ICOS+ T cells [202]. In patients with Nonalcoholic fatty liver disease (NAFLD)-related HCC, the overabundance of SCFA-producing bacteria was linked to an immunosuppressive condition, with a higher expression of T regs and a reduced cytotoxic CD8+ T cells response [203]. Accordingly, a recent study by Pfister et al. has demonstrated that, in preclinical models of nonalcoholic steatohepatitis (NASH)-induced HCC, the administration of anti-PD-1 agents induced the expansion of intratumoral CD8+ PD1+ T cells, but this phenomenon did not cause tumor regression, suggesting an impairment in the cytotoxic activity of these lymphocytes [204]. Moreover, the administration of an anti-PD-1 agent induced the development of NASH-HCC and led to an overexpression of exhausted T cells. These findings were followed by a meta-analysis of three major studies on the effect of immunotherapy on patients with non-viral HCC, which confirmed a poor prognosis in terms of the OS and PFS in these patients [11,204,205,206]. These results suggest an impaired and aberrant T cell activation in NASH patients that limits ICIs’ application, and that can be explained by a dysfunctional gut–liver axis [203,204].
In light of these findings, the gut microbiome’s modulation by antibiotics is a key factor to consider, because in various cancers it has been associated with a worse response to immunotherapy [207,208,209]. In particular, a recent study reported a worse survival in patients who received a prior antibiotic treatment, but not in those who had been undergoing a current antibiotic treatment, during ICI therapy [210]. However, in a study on murine models of HCC, a vancomycin administration was associated with a reduction of primary to secondary bile acid conversion, due to the depletion of Gram-positive bacteria in the gut. This study showed a positive correlation between the primary bile acid concentration and CXCR6+ NKT cells’ accumulation in the liver favoring tumor inhibition, whereas secondary bile acids had opposite effects [211]. Table 3 briefly summarizes the main findings of the studies on the role of the gut–liver axis in the response to immunotherapy.

Table 3.
Studies evaluating the gut microbiota as biomarkers in patients treated with immune checkpoints inhibitors (ICIs).
Currently, a limitation on the use of the gut microbiota as a biomarker of ICI response is the heterogeneity of the results obtained so far [193,194,195,196,197], making it impossible to identify a reliable signature or metabolic feature. Nevertheless, further studies based on human FMT are needed to confirm its efficacy as a co-adjuvant treatment for a successful immunotherapy. The influence of diet, ethnicity, lifestyle, and chronic therapies on the relationship between gut microbiota and the host [212,213,214] are other data that need to be taken into account to further stratify patients and refine gut microbiota’s prognostic significance and therapeutic usefulness.
The novel circulating biomarkers and the gut microbiome species whose changes are under evaluation as predictors of the response to immunotherapy in HCC patients are reported in Figure 1.
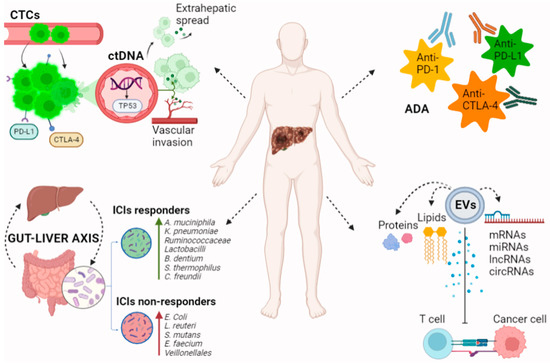
Figure 1.
Novel biomarkers for immunotherapy in patients with HCC. CtDNA and CTCs reflect tumor growth and invasiveness of HCC. Genome analysis of ctDNA allows for the detection of prognostic tumor mutations. CTC levels correlate with tumor extension and may have a role in predicting tumor recurrence after immunotherapy. EVs contain proteins, lipids, DNAs, mRNAs, miRNAs, and non-coding RNAs including circRNAs and lncRNAs. These products influence ICIs’ efficacy by down- or up-regulating PD-L1 expression in the tumor microenvironment. Immune system hyperstimulation by ICIs can lead to the production of ADA. ADA directed against ICIs promote immunocomplex formation and drug clearance, potentially affecting anti-tumor efficacy. The gut microbiota’s composition and products regulate several immune and metabolic pathways in the gut–liver axis, including the response to ICIs. Gut microbiota profiles differ among ICIs responders and non-responders, opening the field to studies testing microbiota-targeted therapies as a new strategy in immuno-oncology. In particular increased abundance of Akkermansia muciniphila, Klebsiella pneumoniae, Ruminococcaceae Lactobacilli, Bifidobacterium dentium, Streptococcus thermophilus, and Citrobacter freundii has been linked to response to immunotherapy; Firmicutes/Bacteroidetes ratio between 0.5 and 1.5 and a higher mean ratio of Prevotella spp. to Bacteroides spp. are also markers of improved survival, whereas increased abundance of Escherichia coli, Lactobacillus reuteri, Streptococcus mutans, Enterococcus faecium, and Veillonellales, and a reduction in bacterial diversity has been associated with non-response. ctDNA: circulating tumor DNA, CTCs: circulating tumor cells, EVs: extracellular vesicles, mRNAs: messenger RNAs, miRNAs: microRNAs, circRNAs: circular RNAs, lncRNAs: long non-coding RNAs, ICIs: immune checkpoint inhibitors, ADA: antidrug antibodies, and PD-L1: programmed death-ligand 1.
5. Conclusions and Future Perspectives
HCC often occurs in the setting of chronic inflammation and immune exhaustion [185,189]. Recently, immunotherapies have become part of the first-line treatment for advanced HCC [4], but several immune and phenotypical tumor features negatively influence a durable anti-tumor response. Nowadays, liver biopsy is not mandatory for the diagnosis of HCC in patients with liver cirrhosis and is mainly performed for the enrollment of patients in clinical trials; the liquid biopsy analysis of circulating tumor biproducts has emerged as a simple and reproducible tool for monitoring cancer progression and assessing pharmacological efficacy, with the possibility of multiple re-evaluations during treatments [23,117]. Studies aimed at identifying noninvasive biomarkers in easy-to-analyze body fluids such as saliva are being continuously published, but interesting results aside, the generalizability is limited by the small cohorts of patients [214,215]. Unraveling the characteristics of HCC and its response to ICIs through the analysis of samples derived from body fluids is fascinating. With the improvement of gene sequencing, the discovery of sensitive biomarkers has become widespread. In particular, ctDNA and CTCs have important prognostic implications, as they could identify mutational signatures that reflect the genomic landscape of the primary tumor [103,104,105,106]. In addition, exosomes, being able to modulate cellular communication directly and indirectly through the release of their cargos, provide important information on tumors’ mutational burden, invasiveness, and resistance during therapy to a degree surpassed by no other previous biomarker [112,113,114,115,116]. Compared with traditional AFP, it has been shown that ctDNA [81,82,83] might be a better prognostic marker of responses in patients with unresectable HCC, but these findings have not been confirmed in other studies. This could be due to the low levels of CTC and ctDNA that can be found in body fluids, which reduce their application in standard diagnostic and prognostic procedures, and to the lack of antibodies against membrane markers of these products [93,106,154]. High costs and the need for multiple platforms and technologies for the comprehensive analysis of tumor products should also be mentioned as limitations for the dissemination and application of noninvasive biomarkers [93,106,154]. The main limitation of liquid biopsy is related to the lack of standardized protocols and the limited data available. Further clinical studies are needed to define the most useful biomarkers obtained from different biological materials and to standardize the assay and characterization methods in order to provide reliable information driving therapeutic decisions [93,106,154].
Although the search for reliable non-invasive biomarkers of ICIs’ efficacy is a compelling clinical need, histologic evaluations still hold valuable and unique information regarding TMBs, oncogenic mutations, and TMEs, which closely influence responsiveness to ICIs [216]. The direct analysis of tumor tissue also suffers from pitfalls, as biopsy specimens do not always encode the full set of tumor characteristics; constructing multimodal predictive scores that include genomic, transcriptomic, proteomic, and immunophenotypic features of the tumor, taking into account the host’s characteristics and environmental modifiers, would probably be the most promising approach for identifying the optimal responders to immunotherapy. Growing evidence suggests that the use of prognostic scores based on the combination of multiple noninvasive biomarkers, or the association between invasive and noninvasive biomarkers, and the integration of new NGS and AI technologies in clinical research could overcome these concerns [55,72,73,217,218]. The application of bioinformatics to medical research has opened the field of hepato-oncology to innovative perspectives. Artificial intelligence (AI) can be used to extract complex information from visual data derived from digitized histological samples, and it is becoming a key tool for predicting prognoses and responses to treatment in gastrointestinal cancers, including HCC [217,218]. In this scenario, an important role can be played by radiomics, which allows for the analysis of tumor heterogeneity and characteristics derived from medical imaging in a multidimensional way using quantitative features. Radiomics can extract data related to TME and its cellularity, such as the infiltration of CD8 T cells, intratumoral lymphocytes, and macrophages; provide information about qualitative and quantitative immune checkpoints’ expression; and predict responses to immunotherapy [219,220,221,222]. Finally, the interplay between the liver and gut, as well as the influence of the gut microbiome on ICIs’ efficacy, are matters of fact in several tumor types, including HCC [191,194,195,198]. Microbiome profiling should be considered as the next frontier for an integrated evaluation of the candidates towards ICI treatment to be included in AI algorithms, whenever possible, as another piece in the puzzle. What biological specimen is the most informative and easy to use (i.e., saliva, stool, and blood) is yet to be defined, as well as the optimal timepoints for its harvesting. (After antibiotic treatment, at fixed intervals, after progression?) However, little is known about the correlation between the gut microbiome and its metabolic patterns and TME. The markers of gut barrier integrity could be another interesting field to explore in the near future; in fact, bacterial translocation is a hallmark of chronic liver disease and parallels immune dysfunction in its advanced stages, linking persistent gut-derived inflammation with the promotion of hepatocarcinogenesis [166,185,223].
In conclusion, the identification of sensitive and accurate prognostic biomarkers for the evaluation of responses to immunotherapy is a compelling and developing field in hepato-oncology. The combinatory analysis of tumor tissues’ intrinsic features, the peritumoral microenvironment, and the immunological and microbiological characteristics of the host is crucial for the development of a prognostic score capable of differentiating responders from non-responders. This dynamic characterization process better adapts to the continuous changes of tumor biology, and can prognosticate the responsiveness of HCC to ICIs in a personalized manner, tailored to the individual patient.
Author Contributions
M.P. (Maria Pallozzi): conceptualization, investigation, writing—original draft preparation, N.D.T.: Investigation, writing—original draft preparation, V.M.: investigation, writing—original draft preparation, F.S.: Conceptualization, review and editing, Supervision, A.G. Conceptualization, review and editing, Supervision, F.R.P.: conceptualization, review and editing, Supervision. M.P. (Maurizio Pompili): conceptualization, review and editing, Supervision. F.R.P. and M.P. (Maurizio Pompili) equally contributed to the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
Thanks to Fondazione Roma for the continuous support to our scientific research.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cerrito, L.; Santopaolo, F.; Monti, F.; Pompili, M.; Gasbarrini, A.; Ponziani, F.R. Advances in pharmacotherapeutics for hepatocellular carcinoma. Expert Opin. Pharmacother. 2021, 22, 1343–1354. [Google Scholar] [CrossRef] [PubMed]
- Ponziani, F.R.; Bhoori, S.; Germini, A.; Bongini, M.; Flores, M.; Sposito, C.; Facciorusso, A.; Gasbarrini, A.; Mazzaferro, V. Inducing tolerability of adverse events increases sorafenib exposure and optimizes patient’s outcome in advanced hepatocellular carcinoma. Liver Int. 2016, 36, 1033–1042. [Google Scholar] [CrossRef] [PubMed]
- Cerrito, L.; Annicchiarico, B.E.; Iezzi, R.; Gasbarrini, A.; Pompili, M.; Ponziani, F.R. Treatment of hepatocellular carcinoma in patients with portal vein tumor thrombosis: Beyond the known frontiers. World J. Gastroenterol. 2019, 25, 4360–4382. [Google Scholar] [CrossRef]
- Reig, M.; Forner, A.; Rimola, J.; Ferrer-Fàbrega, J.; Burrel, M.; Garcia-Criado, Á.; Kelley, R.K.; Galle, P.R.; Mazzaferro, V.; Salem, R.; et al. BCLC strategy for prognosis prediction and treatment recommendation: The 2022 update. J. Hepatol. 2022, 76, 681–693. [Google Scholar] [CrossRef]
- Buchbinder, E.I.; Desai, A. CTLA-4 and PD-1 Pathways: Similarities, Differences, and Implications of Their Inhibition. Am. J. Clin. Oncol. 2016, 39, 98–106. [Google Scholar] [CrossRef]
- Roche, H.-L. A Phase III, Open-Label, Randomized Study of Atezolizumab in Combination With Bevacizumab Compared with Sorafenib in Patients with Untreated Locally Advanced or Metastatic Hepatocellular Carcinoma. Available online: https://clinicaltrials.gov/ct2/show/NCT03434379 (accessed on 1 August 2022).
- Phase 3 Randomized, Open-Label, Multicenter Study of Tremelimumab (T) and Durvalumab (D) as First-Line Therapy in Patients (Pts) with Unresectable Hepatocellular Carcinoma (uHCC): HIMALAYA. Available online: https://ascopubs.org/doi/abs/10.1200/JCO.2022.40.4_suppl.379 (accessed on 1 August 2022).
- Maestri, M.; Pallozzi, M.; Santopaolo, F.; Cerrito, L.; Pompili, M.; Gasbarrini, A.; Ponziani, F.R. Durvalumab: An investigational agent for unresectable hepatocellular carcinoma. Expert Opin. Investig. Drugs 2021, 31, 347–360. [Google Scholar] [CrossRef] [PubMed]
- Yau, T.; Kang, Y.K.; Kim, T.Y.; El-Khoueiry, A.B.; Santoro, A.; Sangro, B.; Melero, I.; Kudo, M.; Hou, M.M.; Matilla, A.; et al. Efficacy and Safety of Nivolumab Plus Ipilimumab in Patients With Advanced Hepatocellular Carcinoma Previously Treated With Sorafenib: The CheckMate 040 Randomized Clinical Trial. JAMA Oncol. 2020, 6, e204564. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.X.; Finn, R.S.; Edeline, J.; Cattan, S.; Ogasawara, S.; Palmer, D.; Verslype, C.; Zagonel, V.; Fartoux, L.; Vogel, A.; et al. KEYNOTE-224 investigators. Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE-224): A non-randomised, open-label phase 2 trial. Lancet. Oncol. 2018, 19, 940–952. [Google Scholar] [CrossRef]
- Finn, R.S.; Ryoo, B.Y.; Merle, P.; Kudo, M.; Bouattour, M.; Lim, H.Y.; Breder, V.; Edeline, J.; Chao, Y.; Ogasawara, S.; et al. Pembrolizumab As Second-Line Therapy in Patients With Advanced Hepatocellular Carcinoma in KEYNOTE-240: A Randomized, Double-Blind, Phase III Trial. J. Clin. Oncol. 2020, 38, 193–202. [Google Scholar] [CrossRef]
- Giraud, J.; Chalopin, D.; Blanc, J.F.; Saleh, M. Hepatocellular Carcinoma Immune Landscape and the Potential of Immunotherapies. Front. Immunol. 2021, 12, 655697. [Google Scholar] [CrossRef]
- Ponziani, F.R.; Santopaolo, F.; Posa, A.; Pompili, M.; Tanzilli, A.; Maestri, M.; Pallozzi, M.; Ibba, F.; Manfredi, R.; Gasbarrini, A.; et al. SIRT in 2025. Cardiovasc. Interv. Radiol. 2022, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Cabibbo, G.; Aghemo, A.; Lai, Q.; Masarone, M.; Montagnese, S.; Ponziani, F.R.; Italian Association for the Study of the Liver (AISF). Optimizing systemic therapy for advanced hepatocellular carcinoma: The key role of liver function. Dig. Liver Dis. 2022, 54, 452–460. [Google Scholar] [CrossRef] [PubMed]
- Khemlina, G.; Ikeda, S.; Kurzrock, R. The biology of Hepatocellular carcinoma: Implications for genomic and immune therapies. Mol. Cancer. 2017, 16, 149. [Google Scholar] [CrossRef] [PubMed]
- Giannini, E.G.; Aglitti, A.; Borzio, M.; Gambato, M.; Guarino, M.; Iavarone, M.; Lai, Q.; Levi Sandri, G.B.; Melandro, F.; Morisco, F.; et al. Overview of Immune Checkpoint Inhibitors Therapy for Hepatocellular Carcinoma, and The ITA.LI.CA Cohort Derived Estimate of Amenability Rate to Immune Checkpoint Inhibitors in Clinical Practice. Cancers 2019, 11, 1689. [Google Scholar] [CrossRef] [PubMed]
- Robb, M.A.; McInnes, P.M.; Califf, R.M. Biomarkers and Surrogate Endpoints: Developing Common Terminology and Definitions. JAMA 2016, 315, 1107–1108. [Google Scholar] [CrossRef]
- Califf, R.M. Biomarker definitions and their applications. Exp. Biol. Med. 2018, 243, 213–221. [Google Scholar] [CrossRef]
- Febbo, P.G.; Ladanyi, M.; Aldape, K.D.; De Marzo, A.M.; Hammond, M.E.; Hayes, D.F.; Iafrate, A.J.; Kelley, R.K.; Marcucci, G.; Ogino, S.; et al. NCCN Task Force report: Evaluating the clinical utility of tumor markers in oncology. J. Natl. Compr. Cancer Netw. 2011, 9, S1–S32. [Google Scholar] [CrossRef]
- Guan, M.-C.; Ouyang, W.; Wang, M.-D.; Liang, L.; Li, N.; Fu, T.-T.; Shen, F.; Lau, W.-Y.; Xu, Q.-R.; Huang, D.-S.; et al. Biomarkers for hepatocellular carcinoma based on body fluids and feces. World J. Gastrointest. Oncol. 2021, 13, 351–365. [Google Scholar] [CrossRef]
- Trevisan, L.; de Lima, F.; Broszczak, D.; Zhang, X.; Bridle, K.; Crawford, D.; Punyadeera, C. The use of minimally invasive biomarkers for the diagnosis and prognosis of hepatocellular carcinoma. Biochim. Biophys. Acta 2020, 1874, 188451. [Google Scholar]
- Ao, H.; Xin, Z.; Jian, Z. Liquid biopsy to identify biomarkers for immunotherapy in hepatocellular carcinoma. Biomark. Res. 2021, 9, 91. [Google Scholar] [CrossRef]
- Maravelia, P.; Silva, D.N.; Rovesti, G.; Chrobok, M.; Stål, P.; Lu, Y.C.; Pasetto, A. Liquid Biopsy in Hepatocellular Carcinoma: Opportunities and Challenges for Immunotherapy. Cancers 2021, 13, 4334. [Google Scholar] [CrossRef] [PubMed]
- Shimagaki, T.; Yoshio, S.; Kawai, H.; Sakamoto, Y.; Doi, H.; Matsuda, M.; Mori, T.; Osawa, Y.; Fukai, M.; Yoshida, T.; et al. Serum milk fat globule-EGF factor 8 (MFG-E8) as a diagnostic and prognostic biomarker in patients with hepatocellular carcinoma. Sci. Rep. 2019, 9, 15788. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, A.; Brandi, G. Biochemical predictors of response to immune checkpoint inhibitors in unresectable hepatocellular carcinoma. Cancer Treat. Res. Commun. 2021, 27, 100328. [Google Scholar] [CrossRef]
- Myojin, Y.; Kodama, T.; Sakamori, R.; Maesaka, K.; Matsumae, T.; Sawai, Y.; Imai, Y.; Ohkawa, K.; Miyazaki, M.; Tanaka, S.; et al. Interleukin-6 Is a Circulating Prognostic Biomarker for Hepatocellular Carcinoma Patients Treated with Combined Immunotherapy. Cancers 2022, 14, 883. [Google Scholar] [CrossRef] [PubMed]
- Shakiba, E.; Ramezani, M.; Sadeghi, M. Evaluation of serum interleukin-6 levels in hepatocellular carcinoma patients: A systematic review and meta-analysis. Clin. Exp. Hepatol. 2018, 4, 182–190. [Google Scholar] [CrossRef]
- McLoughlin, R.M.; Jenkins, B.J.; Grail, D.; Williams, A.S.; Fielding, C.A.; Parker, C.R.; Ernst, M.; Topley, N.; Jones, S.A. IL-6 trans-signaling via STAT3 directs T cell infiltration in acute inflammation. Proc. Natl. Acad. Sci. USA 2005, 102, 9589–9594. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.F.; Kuan, F.C.; Yen, T.C.; Lu, M.S.; Lin, P.Y.; Chung, Y.H.; Chen, W.C.; Lee, K.D. IL-6-stimulated CD11b+ CD14+ HLA-DR- myeloid-derived suppressor cells, are associated with progression and poor prognosis in squamous cell carcinoma of the esophagus. Oncotarget 2014, 5, 8716–8728. [Google Scholar] [CrossRef] [PubMed]
- Feun, L.G.; Li, Y.; Wu, C.; Wangpaichitr, M.; Jones, P.D.; Richman, S.P.; Madrazo, B.; Kwon, D.; Garcia-Buitrago, M.; Martin, P.; et al. Phase 2 study of pembrolizumab and circulating biomarkers to predict anticancer response in advanced, unresectable hepatocellular carcinoma. Cancer 2019, 125, 3603–3614. [Google Scholar] [CrossRef]
- Patel, S.P.; Kurzrock, R. PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol. Cancer Ther. 2015, 14, 847–856. [Google Scholar] [CrossRef]
- Gibney, G.T.; Weiner, L.M.; Atkins, M.B. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol. 2016, 17, e542–e551. [Google Scholar] [CrossRef]
- Hung, Y.P.; Shao, Y.Y.; Hsu, C.; Hsu, C.H.; Lee, J.M.; Yang, M.H.; Chao, Y. The unique characteristic in peripheral immune cells in patients with advanced hepatocellular carcinoma. J. Formos. Med. Assoc. 2021, 120, 1581–1590. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.Y.; Kim, S.; Keam, B.; Kim, T.M.; Kim, D.W.; Heo, D.S. Soluble PD-L1 is a predictive and prognostic biomarker in advanced cancer patients who receive immune checkpoint blockade treatment. Sci. Rep. 2021, 11, 19712. [Google Scholar] [CrossRef] [PubMed]
- Mahoney, K.M.; Ross-Macdonald, P.; Yuan, L.; Song, L.; Veras, E.; Wind-Rotolo, M.; McDermott, D.F.; Hodi, F.S.; Choueiri, T.K.; Freeman, G.J. Soluble PD-L1 as an early marker of progressive disease on nivolumab. J. Immunother. Cancer 2020, 10, e003527. [Google Scholar] [CrossRef] [PubMed]
- Li, X.S.; Li, J.W.; Li, H.; Jiang, T. Prognostic value of programmed cell death ligand 1 (PD-L1) for hepatocellular carcinoma: A meta-analysis. Biosci. Rep. 2020, 40, BSR20200459. [Google Scholar] [CrossRef]
- Mocan, T.; Ilies, M.; Nenu, I.; Craciun, R.; Horhat, A.; Susa, R.; Minciuna, I.; Rusu, I.; Mocan, L.P.; Seicean, A.; et al. Serum levels of soluble programmed death-ligand 1 (sPD-L1): A possible biomarker in predicting post-treatment outcomes in patients with early hepatocellular carcinoma. Int. Immunopharmacol. 2021, 94, 107467. [Google Scholar] [CrossRef]
- Wang, T.; Denman, D.; Bacot, S.M.; Feldman, G.M. Challenges and the Evolving Landscape of Assessing Blood-Based PD-L1 Expression as a Biomarker for Anti-PD-(L)1 Immunotherapy. Biomedicines 2022, 10, 1181. [Google Scholar] [CrossRef]
- Park, L.M.; Lannigan, J.; Jaimes, M.C. OMIP-069: Forty-Color Full Spectrum Flow Cytometry Panel for Deep Immunophenotyping of Major Cell Subsets in Human Peripheral Blood. Citometry A 2020, 97, 1044–1051. [Google Scholar] [CrossRef]
- Agdashian, D.; ElGindi, M.; Xie, C.; Sandhu, M.; Pratt, D.; Kleiner, D.E.; Figg, W.D.; Rytlewski, J.A.; Sanders, C.; Yusko, E.C.; et al. The effect of anti-CTLA4 treatment on peripheral and intra-tumoral T cells in patients with hepatocellular carcinoma. Cancer Immunol. 2019, 68, 599–608. [Google Scholar] [CrossRef]
- Mengos, A.E.; Gastineau, D.A.; Gustafson, M.P. The CD14+HLA-DRlo/neg Monocyte: An Immunosuppressive Phenotype That Restrains Responses to Cancer Immunotherapy. Front. Immunol. 2019, 10, 1147. [Google Scholar] [CrossRef]
- Hung, Y.P.; Shao, Y.Y.; Lee, J.M.; Hsu, C.; Hsu, C.H.; Yang, M.H.; Chao, Y. Potential of circulating immune cells as biomarkers of nivolumab treatment efficacy for advanced hepatocellular carcinoma. J. Chin. Med. Assoc. 2021, 84, 144–150. [Google Scholar] [CrossRef]
- Hong, J.Y.; Cho, H.J.; Sa, J.K.; Liu, X.; Ha, S.Y.; Lee, T.; Kim, H.; Kang, W.; Sinn, D.H.; Gwak, G.-Y.; et al. Hepatocellular carcinoma patients with high circulating cytotoxic T cells and intra-tumoral immune signature benefit from pembrolizumab: Results from a single-arm phase 2 trial. Genome Med. 2022, 14, 1. [Google Scholar] [CrossRef] [PubMed]
- Macek Jilkova, Z.; Aspord, C.; Kurma, K.; Granon, A.; Sengel, C.; Sturm, N.; Marche, P.N.; Decaens, T. Immunologic Features of Patients With Advanced Hepatocellular Carcinoma Before and During Sorafenib or Anti-programmed Death-1/Programmed Death-L1 Treatment. Clin. Transl. Gastroenterol. 2019, 10, e00058. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Lu, M.; Che, J.; Chu, Q.; Zhang, P.; Chen, Y. Biomarkers and Future Perspectives for Hepatocellular Carcinoma Immunotherapy. Front. Oncol. 2021, 11, 716844. [Google Scholar] [CrossRef] [PubMed]
- Dharmapuri, S.; Özbek, U.; Lin, J.Y.; Sung, M.; Schwartz, M.; Branch, A.D.; Ang, C. Predictive value of neutrophil to lymphocyte ratio and platelet to lymphocyte ratio in advanced hepatocellular carcinoma patients treated with anti-PD-1 therapy. Cancer Med. 2020, 9, 4962–4970. [Google Scholar] [CrossRef] [PubMed]
- Hung, H.C.; Lee, J.C.; Wang, Y.C.; Cheng, C.H.; Wu, T.H.; Lee, C.F.; Wu, T.J.; Chou, H.S.; Chan, K.M.; Lee, W.C. Response Prediction in Immune Checkpoint Inhibitor Immunotherapy for Advanced Hepatocellular Carcinoma. Cancers 2020, 13, 1607. [Google Scholar] [CrossRef]
- Ogihara, K.; Kikuchi, E.; Shigeta, K.; Okabe, T.; Hattori, S.; Yamashita, R.; Yoshimine, S.; Shirotake, S.; Nakazawa, R.; Matsumoto, K.; et al. The pretreatment neutrophil-to-lymphocyte ratio is a novel biomarker for predicting clinical responses to pembrolizumab in platinum-resistant metastatic urothelial carcinoma patients. Urol. Oncol. 2020, 38, 602.e1–602.e10. [Google Scholar] [CrossRef]
- Tada, T.; Kumada, T.; Hiraoka, A.; Hirooka, M.; Kariyama, K.; Tani, J.; Atsukawa, M.; Takaguchi, K.; Itobayashi, E.; Fukunishi, S.; et al. Real-life Practice Experts for HCC (RELPEC) Study Group and the Hepatocellular Carcinoma Experts from 48 clinics in Japan (HCC 48) Group. Neutrophil-lymphocyte ratio predicts early outcomes in patients with unresectable hepatocellular carcinoma treated with atezolizumab plus bevacizumab: A multicenter analysis. Eur. J. Gastroenterol. Hepatol. 2022, 34, 698–706. [Google Scholar]
- Zheng, J.; Cai, J.; Li, H.; Zeng, K.; He, L.; Fu, H.; Zhang, J.; Chen, L.; Yao, J.; Zhang, Y.; et al. Neutrophil to Lymphocyte Ratio and Platelet to Lymphocyte Ratio as Prognostic Predictors for Hepatocellular Carcinoma Patients with Various Treatments: A Meta-Analysis and Systematic Review. Cell. Physiol. Biochem. 2017, 44, 967–981. [Google Scholar] [CrossRef]
- Jiang, Y.; Tu, X.; Zhang, X.; Liao, H.; Han, S.; Jiang, W.; Zheng, Y.; Zhao, P.; Tong, Z.; Fu, Q.; et al. Nutrition and metabolism status alteration in advanced hepatocellular carcinoma patients treated with anti-PD-1 immunotherapy. Support. Care Cancer 2020, 28, 5569–5579. [Google Scholar] [CrossRef]
- Muhammed, A.; Fulgenzi, C.A.M.; Dharmapuri, S.; Pinter, M.; Balcar, L.; Scheiner, B.; Marron, T.U.; Jun, T.; Saeed, A.; Hildebrand, H.; et al. The Systemic Inflammatory Response Identifies Patients with Adverse Clinical Outcome from Immunotherapy in Hepatocellular Carcinoma. Cancers 2021, 14, 186. [Google Scholar] [CrossRef]
- Mei, J.; Sun, X.Q.; Lin, W.P.; Li, S.H.; Lu, L.H.; Zou, J.W.; Wei, W.; Guo, R.P. Comparison of the Prognostic Value of Inflammation-Based Scores in Patients with Hepatocellular Carcinoma After Anti-PD-1 Therapy. J. Inflamm. Res. 2021, 14, 3879–3890. [Google Scholar] [CrossRef] [PubMed]
- Bigot, F.; Castanon, E.; Baldini, C.; Hollebecque, A.; Carmona, A.; Postel-Vinay, S.; Angevin, E.; Armand, J.-P.; Ribrag, V.; Aspeslagh, S.; et al. Prospective validation of a prognostic score for patients in immunotherapy phase I trials: The Gustave Roussy Immune Score (GRIm-Score). Eur. J. Cancer 1990, 84, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Pan, Y.; Lin, X.; Hou, J.; Hu, Z.; Xu, L.; Zhou, Z.; Zhang, Y.; Chen, M.; Hu, D. Development and Validation of a Prognostic Score for Hepatocellular Carcinoma Patients in Immune Checkpoint Inhibitors Therapies: The Hepatocellular Carcinoma Modified Gustave Roussy Immune Score. Front. Pharmacol. 2022, 12, 819985. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, G.; Zhang, P.; Zhang, J.; Li, X.; Gan, D.; Cao, X.; Han, M.; Du, H.; Ye, Y. The threshold of alpha-fetoprotein (AFP) for the diagnosis of hepatocellular carcinoma: A systematic review and meta-analysis. PLoS ONE 2020, 15, e0228857. [Google Scholar] [CrossRef] [PubMed]
- Mehta, N.; Dodge, J.L.; Roberts, J.P.; Hirose, R.; Yao, F.Y. Alpha-Fetoprotein Decrease from >1000 to <500 ng/mL in Patients with Hepatocellular Carcinoma Leads to Improved Posttransplant Outcomes. Hepatology 2019, 69, 1193–1205. [Google Scholar] [PubMed]
- Lee, P.C.; Chao, Y.; Chen, M.H.; Lan, K.H.; Lee, C.J.; Lee, I.C.; Chen, S.C.; Hou, M.C.; Huang, Y.H. Predictors of Response and Survival in Immune Checkpoint Inhibitor-Treated Unresectable Hepatocellular Carcinoma. Cancers 2020, 12, 182. [Google Scholar] [CrossRef]
- Shao, Y.Y.; Liu, T.H.; Hsu, C.; Lu, L.C.; Shen, Y.C.; Lin, Z.Z.; Cheng, A.L.; Hsu, C.H. Early alpha-foetoprotein response associated with treatment efficacy of immune checkpoint inhibitors for advanced hepatocellular carcinoma. Liver Int. 2019, 39, 2184–2189. [Google Scholar] [CrossRef] [PubMed]
- Hsu, W.F.; Wang, H.W.; Chen, C.K.; Lai, H.C.; Chuang, P.H.; Tsai, M.H.; Su, W.P.; Chen, H.Y.; Chu, C.S.; Chou, J.W.; et al. Alpha-fetoprotein response predicts treatment outcomes in patients with unresectable hepatocellular carcinoma receiving immune checkpoint inhibitors with or without tyrosine kinase inhibitors or locoregional therapies. Am. J. Cancer Res. 2021, 11, 6173–6187. [Google Scholar]
- Teng, W.; Lin, C.C.; Ho, M.M.; Lui, K.W.; Wang, S.F.; Hsu, C.W.; Lin, S.M. Alpha-fetoprotein response at different time-points is associated with efficacy of nivolumab monotherapy for unresectable hepatocellular carcinoma. Am. J. Cancer Res. 2021, 11, 2319–2330. [Google Scholar]
- Semenkova, L.N.; Dudich, E.I.; Dudich, I.V.; Shingarova, L.N.; Korobko, V.G. Alpha-fetoprotein as a TNF resistance factor for the human hepatocarcinoma cell line HepG2. Tumour Biol. 1997, 18, 30–40. [Google Scholar] [CrossRef]
- Li, M.S.; Li, P.F.; He, S.P.; Du, G.G.; Li, G. The promoting molecular mechanism of alpha-fetoprotein on the growth of human hepatoma Bel7402 cell line. World J. Gastroenterol. 2002, 8, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Tatsumi, T.; Miyagi, T.; Tsunematsu, H.; Aketa, H.; Hosui, A.; Kanto, T.; Hiramatsu, N.; Hayashi, N.; Takehara, T. α-Fetoprotein impairs activation of natural killer cells by inhibiting the function of dendritic cells. Clin. Exp. Immunol. 2011, 165, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Dai, X.; Dai, J.; Ding, C.; Zhang, Z.; Lin, Z.; Hu, J.; Lu, M.; Wang, Z.; Qi, Y.; et al. AFP promotes HCC progression by suppressing the HuR-mediated Fas/FADD apoptotic pathway. Cell Death Dis. 2020, 11, 822. [Google Scholar] [CrossRef] [PubMed]
- A Phase I Open Label Clinical Trial Evaluating the Safety and Anti-Tumor Activity of Autologous T Cells Expressing Enhanced TCRs Specific for Alpha Fetoprotein (AFPc332T) in HLA-A2 Positive Subjects With Advanced Hepatocellular Carcinoma (HCC) or Other AFP Expressing Tumor. Available online: https://clinicaltrials.gov/ct2/show/NCT03132792 (accessed on 1 August 2022).
- Wu, Y.; Wei, P.; Pengpumkiat, S.; Schumacher, E.A.; Remcho, V.T. Development of a carbon dot (C-Dot)-linked immunosorbent assay for the detection of human α-fetoprotein. Anal. Chem. 2015, 87, 8510–8516. [Google Scholar] [CrossRef]
- European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2018, 69, 182–236. [Google Scholar] [CrossRef]
- Heimbach, J.K.; Kulik, L.M.; Finn, R.S.; Sirlin, C.B.; Abecassis, M.M.; Roberts, L.R.; Zhu, A.X.; Murad, M.H.; Marrero, J.A. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology 2018, 67, 358–380. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.L.; Mo, F.; Johnson, P.J.; Siu, D.Y.; Chan, M.H.; Lau, W.Y.; Lai, P.B.; Lam, C.W.; Yeo, W.; Yu, S.C. Performance of serum α-fetoprotein levels in the diagnosis of hepatocellular carcinoma in patients with a hepatic mass. HPB 2014, 16, 366–372. [Google Scholar] [CrossRef] [PubMed]
- Scheiner, B.; Pomej, K.; Kirstein, M.M.; Hucke, F.; Finkelmeier, F.; Waidmann, O.; Himmelsbach, V.; Schulze, K.; von Felden, J.; Fründt, T.W.; et al. Prognosis of patients with hepatocellular carcinoma treated with immunotherapy—Development and validation of the CRAFITY score. J. Hepatol. 2022, 76, 353–363. [Google Scholar] [CrossRef]
- Hatanaka, T.; Kakizaki, S.; Hiraoka, A.; Tada, T.; Hirooka, M.; Kariyama, K.; Tani, J.; Atsukawa, M.; Takaguchi, K.; Itobayashi, E.; et al. Prognostic impact of C-reactive protein and alpha-fetoprotein in immunotherapy score in hepatocellular carcinoma patients treated with atezolizumab plus bevacizumab: A multicenter retrospective study. Hepatol. Int. 2022, 76, 353–363. [Google Scholar] [CrossRef]
- Teng, W.; Lin, C.C.; Su, C.W.; Lin, P.T.; Hsieh, Y.C.; Chen, W.T.; Ho, M.M.; Wang, C.T.; Chai, P.M.; Hsieh, J.C.; et al. Combination of CRAFITY score with Alpha-fetoprotein response predicts a favorable outcome of atezolizumab plus bevacizumab for unresectable hepatocellular carcinoma. Am. J. Cancer Res. 2022, 12, 1899–1911. [Google Scholar] [CrossRef]
- Sun, X.; Mei, J.; Lin, W.; Yang, Z.; Peng, W.; Chen, J.; Zhang, Y.; Xu, L.; Chen, M. Reductions in AFP and PIVKA-II can predict the efficiency of anti-PD-1 immunotherapy in HCC patients. BMC Cancer 2021, 21, 775. [Google Scholar] [CrossRef] [PubMed]
- Bui, T.M.; Wiesolek, H.L.; Sumagin, R. ICAM-1: A master regulator of cellular responses in inflammation, injury resolution, and tumorigenesis. J. Leukoc. Biol. 2020, 108, 787–799. [Google Scholar] [CrossRef] [PubMed]
- Witkowska, A.M.; Borawska, M.H. Soluble intercellular adhesion molecule-1 (sICAM-1): An overview. Eur. Cytokine. Netw. 2004, 15, 91–98. [Google Scholar] [PubMed]
- Cao, W.; Chen, Y.W.; Yuan, J.; Xie, W.; Liu, K.; Qiu, Y.; Wang, X.; Li, X. Potentiality of α-fetoprotein (AFP) and soluble intercellular adhesion molecule-1 (sICAM-1) in prognosis prediction and immunotherapy response for patients with hepatocellular carcinoma. Bioengineered 2021, 12, 9435–9451. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Zhang, F.; Li, H.; Yang, L.; Lv, T.; Gu, W.; Song, Y. Circulating Tumor Cells Were Associated with the Number of T Lymphocyte Subsets and NK Cells in Peripheral Blood in Advanced Non-Small-Cell Lung Cancer. Dis. Markers 2017, 2017, 5727815. [Google Scholar] [CrossRef]
- Postel, M.; Roosen, A.; Laurent-Puig, P.; Taly, V.; Wang-Renault, S.F. Droplet-based digital PCR and next generation sequencing for monitoring circulating tumor DNA: A cancer diagnostic perspective. Expert Rev. Mol. Diagn. 2018, 18, 7–17. [Google Scholar] [CrossRef]
- Cullinan, A.; Cantor, C. Sequenom, Inc. Pharmacogenomics 2008, 9, 1211–1215. [Google Scholar] [CrossRef]
- Wang, J.; Huang, A.; Wang, Y.P.; Yin, Y.; Fu, P.Y.; Zhang, X.; Zhou, J. Circulating tumor DNA correlates with microvascular invasion and predicts tumor recurrence of hepatocellular carcinoma. Ann. Transl. Med. 2020, 8, 237. [Google Scholar] [CrossRef]
- Wu, X.; Li, J.; Gassa, A.; Buchner, D.; Alakus, H.; Dong, Q.; Ren, N.; Liu, M.; Odenthal, M.; Stippel, D.; et al. Circulating tumor DNA as an emerging liquid biopsy biomarker for early diagnosis and therapeutic monitoring in hepatocellular carcinoma. Int. J. Biol. Sci. 2020, 16, 1551–1562. [Google Scholar] [CrossRef]
- Von Felden, J.; Craig, A.J.; Garcia-Lezana, T.; Labgaa, I.; Haber, P.K.; D’Avola, D.; Asgharpour, A.; Dieterich, D.; Bonaccorso, A.; Torres-Martin, M.; et al. Mutations in circulating tumor DNA predict primary resistance to systemic therapies in advanced hepatocellular carcinoma. Oncogene 2021, 40, 140–151. [Google Scholar] [CrossRef]
- Oversoe, S.K.; Clement, M.S.; Pedersen, M.H.; Weber, B.; Aagaard, N.K.; Villadsen, G.E.; Grønbæk, H.; Hamilton-Dutoit, S.J.; Sorensen, B.S.; Kelsen, J. TERT promoter mutated circulating tumor DNA as a biomarker for prognosis in hepatocellular carcinoma. Scand. J. Gastroenterol. 2020, 55, 1433–1440. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.S.; Eun, J.W.; Choi, J.H.; Woo, H.G.; Cho, H.J.; Ahn, H.R.; Suh, C.W.; Baek, G.O.; Cho, S.W.; Cheong, J.Y. MLH1 single-nucleotide variant in circulating tumor DNA predicts overall survival of patients with hepatocellular carcinoma. Sci. Rep. 2020, 10, 17862. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Li, S.; Wang, J.; Zhang, T.; Zhang, S.; Chen, H.; Xiao, Q.; Ren, W.; Liu, C.; Peng, B.; et al. TP53 R249S mutation detected in circulating tumour DNA is associated with Prognosis of hepatocellular carcinoma patients with or without hepatectomy. Liver Int. 2020, 40, 2834–2847. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wang, P.; Cheng, J.; Gong, Z.; Huang, A.; Zhou, K.; Hu, B.; Gao, P.; Cao, Y.; Qiu, S.; et al. Postoperative circulating tumor cells: An early predictor of extrahepatic metastases in patients with hepatocellular carcinoma undergoing curative surgical resection. Cancer Cytopathol. 2020, 128, 733–745. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Børresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Goodman, A.M.; Kato, S.; Bazhenova, L.; Patel, S.P.; Frampton, G.M.; Miller, V.; Stephens, P.J.; Daniels, G.A.; Kurzrock, R. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol. Cancer Ther. 2020, 16, 2598–2608. [Google Scholar] [CrossRef]
- Rizvi, N.A.; Cho, B.C.; Reinmuth, N.; Lee, K.H.; Luft, A.; Ahn, M.J.; van den Heuvel, M.M.; Cobo, M.; Vicente, D.; Smolin, A.; et al. MYSTIC Investigators. Durvalumab With or Without Tremelimumab vs Standard Chemotherapy in First-line Treatment of Metastatic Non-Small Cell Lung Cancer: The MYSTIC Phase 3 Randomized Clinical Trial. JAMA Oncol. 2020, 6, 661–674. [Google Scholar] [CrossRef]
- Araujo, D.V.; Wang, A.; Torti, D.; Leon, A.; Marsh, K.; McCarthy, A.; Berman, H.; Spreafico, A.; Hansen, A.R.; Razak, A.-A.; et al. Applications of Circulating Tumor DNA in a Cohort of Phase I Solid Tumor Patients Treated With Immunotherapy. JNCI Cancer Spectr. 2021, 5, pkaa122. [Google Scholar] [CrossRef]
- Zhu, G.; Liu, W.; Tang, Z.; Qu, W.; Fang, Y.; Jiang, X.; Song, S.; Wang, H.; Tao, C.; Zhou, P.; et al. Serial circulating tumor DNA to predict early recurrence in patients with hepatocellular carcinoma: A prospective study. Mol. Oncol. 2022, 16, 549–561. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, Y.; Wu, L.; Li, J.; Ji, J.; Yu, Q.; Dai, W.; Feng, J.; Wu, J.; Guo, C. Current status of ctDNA in precision oncology for hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2021, 40, 140. [Google Scholar] [CrossRef]
- Ahn, J.C.; Teng, P.C.; Chen, P.J.; Posadas, E.; Tseng, H.R.; Lu, S.C.; Yang, J.D. Detection of Circulating Tumor Cells and Their Implications as a Biomarker for Diagnosis, Prognostication, and Therapeutic Monitoring in Hepatocellular Carcinoma. Hepatology 2021, 73, 422–436. [Google Scholar] [CrossRef] [PubMed]
- Vona, G.; Estepa, L.; Béroud, C.; Damotte, D.; Capron, F.; Nalpas, B.; Mineur, A.; Franco, D.; Lacour, B.; Pol, S.; et al. Impact of cytomorphological detection of circulating tumor cells in patients with liver cancer. Hepatology 2014, 39, 792–797. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.N.; Xiang, B.D.; Wu, F.X.; Ye, J.Z.; Zhong, J.H.; Wang, Y.Y.; Chen, Y.Y.; Chen, Z.S.; Ma, L.; Chen, J.; et al. Circulating Tumor Cells Undergoing EMT Provide a Metric for Diagnosis and Prognosis of Patients with Hepatocellular Carcinoma. Cancer Res. 2018, 78, 4731–4744. [Google Scholar] [CrossRef] [PubMed]
- Sierra-Agudelo, J.; Rodriguez-Trujillo, R.; Samitier, J. Microfluidics for the Isolation and Detection of Circulating Tumor Cells. Adv. Exp. Med. Biol. 2022, 1379, 389–412. [Google Scholar] [PubMed]
- Chen, J.; Cao, S.W.; Cai, Z.; Zheng, L.; Wang, Q. Epithelial-mesenchymal transition phenotypes of circulating tumor cells correlate with the clinical stages and cancer metastasis in hepatocellular carcinoma patients. Cancer Biomark. 2017, 20, 487–498. [Google Scholar] [CrossRef]
- Okegawa, T.; Itaya, N.; Hara, H.; Tambo, M.; Nutahara, K. Circulating tumor cells as a biomarker predictive of sensitivity to docetaxel chemotherapy in patients with castration-resistant prostate cancer. Anticancer. Res. 2014, 34, 6705–6710. [Google Scholar]
- Chen, L.; Bode, A.M.; Dong, Z. Circulating Tumor Cells: Moving Biological Insights into Detection. Theranostics 2017, 7, 2606–2619. [Google Scholar] [CrossRef]
- Yu, J.-J.; Xiao, W.; Dong, S.-L.; Liang, H.-F.; Zhang, Z.-W.; Zhang, B.-X.; Huang, Z.-Y.; Chen, Y.-F.; Zhang, W.-G.; Luo, H.-P.; et al. Effect of surgical liver resection on circulating tumor cells in patients with hepatocellular carcinoma. BMC cancer 2018, 18, 835. [Google Scholar] [CrossRef]
- Ye, X.; Li, G.; Han, C.; Han, Q.; Shang, L.; Su, H.; Han, B.; Gong, Y.; Lu, G.; Peng, T. Circulating tumor cells as a potential biomarker for postoperative clinical outcome in HBV-related hepatocellular carcinoma. Cancer Manag. Res. 2018, 10, 5639–5647. [Google Scholar] [CrossRef]
- Winograd, P.; Hou, S.; Court, C.M.; Lee, Y.-T.; Chen, P.-J.; Zhu, Y.; Sadeghi, S.; Finn, R.S.; Teng, P.-C.; Wang, J.J.; et al. Hepatocellular Carcinoma-Circulating Tumor Cells Expressing PD-L1 Are Prognostic and Potentially Associated With Response to Checkpoint Inhibitors. Hepatol. Commun. 2020, 4, 1527–1540. [Google Scholar] [CrossRef]
- Yue, C.; Jiang, Y.; Li, P.; Wang, Y.; Xue, J.; Li, N.; Li, D.; Wang, R.; Dang, Y.; Hu, Z.; et al. Dynamic change of PD-L1 expression on circulating tumor cells in advanced solid tumor patients undergoing PD-1 blockade therapy. Oncoimmunology 2018, 7, e1438111. [Google Scholar] [CrossRef] [PubMed]
- Winograd, P.; Hou, S.; Court, C.; Sadeghi, S.; Finn, R.; DiPardo, B.; Li, Q.; Kaldas, F.; Busuttil, R.; Tomlinson, J.; et al. Evaluation of hepatocellular carcinoma circulating tumor cells expressing programmed death-ligand 1. HPB 2018, 20, S2–S3. [Google Scholar] [CrossRef]
- Kowalik, A.; Kowalewska, M.; Góźdź, S. Current approaches for avoiding the limitations of circulating tumor cells detection methods-implications for diagnosis and treatment of patients with solid tumors. Transl. Res. 2017, 185, 58–84.e15. [Google Scholar] [CrossRef] [PubMed]
- Bao, Q.; Huang, Q.; Chen, Y.; Wang, Q.; Sang, R.; Wang, L.; Xie, Y.; Chen, W. Tumor-Derived Extracellular Vesicles Regulate Cancer Progression in the Tumor Microenvironment. Front. Mol. Biosci. 2022, 8, 796385. [Google Scholar] [CrossRef] [PubMed]
- Maacha, S.; Bhat, A.A.; Jimenez, L.; Raza, A.; Haris, M.; Uddin, S.; Grivel, J.C. Extracellular vesicles-mediated intercellular communication: Roles in the tumor microenvironment and anti-cancer drug resistance. Mol. Cancer 2019, 18, 55. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Mashouri, L.; Yousefi, H.; Aref, A.R.; Ahadi, A.M.; Molaei, F.; Alahari, S.K. Exosomes: Composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol. Cancer 2019, 18, 75. [Google Scholar] [CrossRef]
- Zhang, Y.; Bi, J.; Huang, J.; Tang, Y.; Du, S.; Li, P. Exosome: A Review of Its Classification, Isolation Techniques, Storage, Diagnostic and Targeted Therapy Applications. Int. J. Nanomed. 2020, 15, 6917–6934. [Google Scholar] [CrossRef]
- Wortzel, I.; Dror, S.; Kenific, C.M.; Lyden, D. Exosome-Mediated Metastasis: Communication from a Distance. Dev. Cell 2019, 49, 347–360. [Google Scholar] [CrossRef]
- Li, I.; Nabet, B.Y. Exosomes in the tumor microenvironment as mediators of cancer therapy resistance. Mol. Cancer. 2019, 18, 32. [Google Scholar] [CrossRef]
- Li, S.; Chen, L. Exosomes in Pathogenesis, Diagnosis, and Treatment of Hepatocellular Carcinoma. Front. Oncol. 2022, 12, 793432. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yu, L.; Huang, P.; Zhou, Y.; Zheng, W.; Meng, N.; He, R.; Xu, Y.; Keong, T.S.; Cui, Y. Tumor-associated Exosomes Are Involved in Hepatocellular Carcinoma Tumorigenesis, Diagnosis, and Treatment. J. Clin. Transl. Hepatol. 2022, 10, 496–508. [Google Scholar] [CrossRef] [PubMed]
- Xue, D.; Han, J.; Liang, Z.; Jia, L.; Liu, Y.; Tuo, H.; Peng, Y. Current Perspectives on the Unique Roles of Exosomes in Drug Resistance of Hepatocellular Carcinoma. J. Hepatocell. Carcinoma 2022, 9, 99–112. [Google Scholar] [CrossRef] [PubMed]
- Moldogazieva, N.T.; Zavadskiy, S.P.; Terentiev, A.A. Genomic Landscape of Liquid Biopsy for Hepatocellular Carcinoma Personalized Medicine. Cancer Genom. Proteom. 2021, 18, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Abbate, V.; Marcantoni, M.; Giuliante, F.; Vecchio, F.M.; Gatto, I.; Mele, C.; Saviano, A.; Arciuolo, D.; Gaetani, E.; Ferrari, M.C.; et al. HepPar1-Positive Circulating Microparticles Are Increased in Subjects with Hepatocellular Carcinoma and Predict Early Recurrence after Liver Resection. Int. J. Mol. Sci. 2017, 18, 1043. [Google Scholar] [CrossRef]
- Julich-Haertel, H.; Urban, S.K.; Krawczyk, M.; Willms, A.; Jankowski, K.; Patkowski, W.; Kruk, B.; Krasnodębski, M.; Ligocka, J.; Schwab, R.; et al. Cancer-associated circulating large extracellular vesicles in cholangiocarcinoma and hepatocellular carcinoma. J. Hepatol. 2017, 67, 282–292. [Google Scholar] [CrossRef]
- Abels, E.R.; Breakefield, X.O. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell Mol. Neurobiol. 2016, 36, 301–312. [Google Scholar] [CrossRef]
- Henderson, M.C.; Azorsa, D.O. The genomic and proteomic content of cancer cell-derived exosomes. Front. Oncol. 2020, 2, 38. [Google Scholar] [CrossRef]
- D’Agnano, I.; Berardi, A.C. Extracellular Vesicles, A Possible Theranostic Platform Strategy for Hepatocellular Carcinoma-An Overview. Cancers 2020, 12, 261. [Google Scholar] [CrossRef]
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Ji, J.; Yin, Y.; Ju, H.; Xu, X.; Liu, W.; Fu, Q.; Hu, J.; Zhang, X.; Sun, B. Long non-coding RNA Lnc-Tim3 exacerbates CD8 T cell exhaustion via binding to Tim-3 and inducing nuclear translocation of Bat3 in HCC. Cell Death Dis. 2018, 9, 478. [Google Scholar] [CrossRef] [PubMed]
- Chowdhary, A.; Satagopam, V.; Schneider, R. Long Non-coding RNAs: Mechanisms, Experimental, and Computational Approaches in Identification, Characterization, and Their Biomarker Potential in Cancer. Front. Genet. 2021, 12, 649619. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xie, R.; Lu, G. Identification of m6A methyltransferase-related lncRNA signature for predicting immunotherapy and prognosis in patients with hepatocellular carcinoma. Biosci. Rep. 2021, 41, BSR20210760. [Google Scholar] [CrossRef]
- Xu, Q.; Wang, Y.; Huang, W. Identification of immune-related lncRNA signature for predicting immune checkpoint blockade and prognosis in hepatocellular carcinoma. Int. Immunopharmacol. 2021, 92, 107333. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Lu, Y.; Zhang, Y.; Wang, L. Construction of an Immune-Related Six-lncRNA Signature to Predict the Outcomes, Immune Cell Infiltration, and Immunotherapy Response in Patients With Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 661758. [Google Scholar] [CrossRef] [PubMed]
- Tang, P.; Qu, W.; Wang, T.; Liu, M.; Wu, D.; Tan, L.; Zhou, H. Identifying a Hypoxia-Related Long Non-Coding RNAs Signature to Improve the Prediction of Prognosis and Immunotherapy Response in Hepatocellular Carcinoma. Front. Genet. 2021, 12, 785185. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, L.; Xu, Y.; Wu, X.; Zhou, Y.; Mo, J. Immune-related long noncoding RNA signature for predicting survival and immune checkpoint blockade in hepatocellular carcinoma. J. Cell. Physiol. 2020, 235, 9304–9316. [Google Scholar] [CrossRef]
- Fan, F.; Chen, K.; Lu, X.; Li, A.; Liu, C.; Wu, B. Dual targeting of PD-L1 and PD-L2 by PCED1B-AS1 via sponging hsa-miR-194-5p induces immunosuppression in hepatocellular carcinoma. Hepatol. Int. 2021, 15, 444–458. [Google Scholar] [CrossRef]
- Peng, L.; Chen, Z.; Chen, Y.; Wang, X.; Tang, N. MIR155HG is a prognostic biomarker and associated with immune infiltration and immune checkpoint molecules expression in multiple cancers. Cancer Med. 2019, 8, 7161–7173. [Google Scholar] [CrossRef]
- Shen, H.; Liu, B.; Xu, J.; Zhang, B.; Wang, Y.; Shi, L.; Cai, X. Circular RNAs: Characteristics, biogenesis, mechanisms and functions in liver cancer. J. Hematol. Oncol. 2021, 14, 134. [Google Scholar] [CrossRef]
- Mi, Z.; Zhongqiang, C.; Caiyun, J.; Yanan, L.; Jianhua, W.; Liang, L. Circular RNA detection methods: A minireview. Talanta 2022, 238, 123066. [Google Scholar] [CrossRef] [PubMed]
- Mok, E.; Wee, E.; Wang, Y.; Trau, M. Comprehensive evaluation of molecular enhancers of the isothermal exponential amplification reaction. Sci. Rep. 2016, 6, 37837. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Duan, J.; Chen, J.; Ding, S.; Cheng, W. Recent advances in rolling circle amplification-based biosensing strategies—A review. Anal. Chim. Acta 2021, 1148, 238187. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.F.; Gao, C.; Huang, X.Y.; Lu, J.C.; Guo, X.J.; Shi, G.M.; Cai, J.B.; Ke, A.W. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol. Cancer 2020, 19, 110. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.-Y.; Zhang, P.-F.; Wei, C.-Y.; Peng, R.; Lu, J.-C.; Gao, C.; Cai, J.-B.; Yang, X.; Fan, J.; Ke, A.-W.; et al. Circular RNA circMET drives immunosuppression and anti-PD1 therapy resistance in hepatocellular carcinoma via the miR-30-5p/snail/DPP4 axis. Mol. Cancer 2020, 19, 92. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Zhang, P.; Liang, H.; Xu, Y.; Shen, J.; Wang, W.; Li, M.; Huang, J.; Ni, C.; Zhang, X.; et al. Circular RNA hsa_circ_0003288 induces EMT and invasion by regulating hsa_circ_0003288/miR-145/PD-L1 axis in hepatocellular carcinoma. Cancer Cell Int. 2021, 21, 212. [Google Scholar] [CrossRef]
- Huang, G.; Liang, M.; Liu, H.; Huang, J.; Li, P.; Wang, C.; Zhang, Y.; Lin, Y.; Jiang, X. CircRNA hsa_circRNA_104348 promotes hepatocellular carcinoma progression through modulating miR-187-3p/RTKN2 axis and activating Wnt/β-catenin pathway. Cell Death Dis. 2020, 11, 1065. [Google Scholar] [CrossRef]
- Cai, J.; Chen, Z.; Zhang, Y.; Wang, J.; Zhang, Z.; Wu, J.; Mao, J.; Zuo, X. CircRHBDD1 augments metabolic rewiring and restricts immunotherapy efficacy via m6A modification in hepatocellular carcinoma. Mol. Ther. Oncolytics 2022, 24, 755–771. [Google Scholar] [CrossRef]
- Pan, J.H.; Zhou, H.; Zhao, X.X.; Ding, H.; Li, W.; Qin, L.; Pan, Y.L. Role of exosomes and exosomal microRNAs in hepatocellular carcinoma: Potential in diagnosis and antitumour treatments (Review). Int. J. Mol. Med. 2018, 41, 1809–1816. [Google Scholar] [CrossRef]
- Chugh, P.; Tamburro, K.; Dittmer, D.P. Profiling of pre-micro RNAs and microRNAs using quantitative real-time PCR (qPCR) arrays. J. Vis. Exp. 2010, 46, 2210. [Google Scholar] [CrossRef]
- Lu, T.X.; Rothenberg, M.E. MicroRNA. J. Allergy Clin. Immunol. 2018, 141, 1202–1207. [Google Scholar] [CrossRef] [PubMed]
- Sekovanić, A.; Dorotić, A.; Jurasović, J.; Pašalić, D.; Kovačić, J.; Stasenko, S.; Mioč, T.; Piasek, M.; Orct, T. Pre-amplification as a method for improvement of quantitative RT-PCR analysis of circulating miRNAs. Biochem. Med. 2021, 31, 010901. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Tao, Z.; Hai, B.; Liang, H.; Shi, Y.; Wang, T.; Song, W.; Chen, Y.; OuYang, J.; Chen, J.; et al. miR-424(322) reverses chemoresistance via T-cell immune response activation by blocking the PD-L1 immune checkpoint. Nat. Commun. 2016, 7, 11406. [Google Scholar] [CrossRef] [PubMed]
- Guyon, N.; Garnier, D.; Briand, J.; Nadaradjane, A.; Bougras-Cartron, G.; Raimbourg, J.; Campone, M.; Heymann, D.; Vallette, F.M.; Frenel, J.-S.; et al. Anti-PD1 therapy induces lymphocyte-derived exosomal miRNA-4315 release inhibiting Bim-mediated apoptosis of tumor cells. Cell Death Dis. 2020, 11, 1048. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Nduom, E.K.; Kong, L.Y.; Hashimoto, Y.; Xu, S.; Gabrusiewicz, K.; Ling, X.; Huang, N.; Qiao, W.; Zhou, S.; et al. MiR-138 exerts anti-glioma efficacy by targeting immune checkpoints. Neuro-Oncology 2016, 18, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cao, K. KDM1A Promotes Immunosuppression in Hepatocellular Carcinoma by Regulating PD-L1 through Demethylating MEF2D. J. Immunol. Res. 2021, 2021, 9965099. [Google Scholar] [CrossRef]
- Liu, Z.; Ning, F.; Cai, Y.; Sheng, H.; Zheng, R.; Yin, X.; Lu, Z.; Su, L.; Chen, X.; Zeng, C.; et al. The EGFR-P38 MAPK axis up-regulates PD-L1 through miR-675-5p and down-regulates HLA-ABC via hexokinase-2 in hepatocellular carcinoma cells. Cancer Commun. 2021, 41, 62–78. [Google Scholar] [CrossRef]
- Guangshun, S.; Guoqiang, S.; Xin, C.; Xiangyi, K.; Wubin, Z.; Zhitao, L.; Zhiying, Z.; Hongyong, C.; Chengyu, L.; Yongxiang, X.; et al. Meloxicam Inhibits Hepatocellular Carcinoma Progression and Enhances the Sensitivity of Immunotherapy via the MicroRNA-200/PD-L1 Pathway. J. Oncol. 2022, 2022, 4598573. [Google Scholar] [CrossRef]
- Cortez, M.A.; Ivan, C.; Valdecanas, D.; Wang, X.; Peltier, H.J.; Ye, Y.; Araujo, L.; Carbone, D.P.; Shilo, K.; Giri, D.K.; et al. PDL1 Regulation by p53 via miR-34. J. Natl. Cancer Inst. 2015, 108, djv303. [Google Scholar] [CrossRef]
- Yan, K.; Fu, Y.; Zhu, N.; Wang, Z.; Hong, J.L.; Li, Y.; Li, W.J.; Zhang, H.B.; Song, J.H. Repression of lncRNA NEAT1 enhances the antitumor activity of CD8+T cells against hepatocellular carcinoma via regulating miR-155/Tim-3. Int. J. Biochem. Cell Biol. 2019, 110, 1–8. [Google Scholar] [CrossRef]
- Ludwig, N.; Whiteside, T.L.; Reichert, T.E. Challenges in Exosome Isolation and Analysis in Health and Disease. Int. J. Mol. Sci. 2019, 20, 4684. [Google Scholar] [CrossRef] [PubMed]
- US Food and Drug Administration (FDA). 2019: Immunogenicity Testing of Therapeutic Protein Products—Developing and Validating Assays for Anti-Drug Antibody Detection. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/immunogenicity-testing-therapeutic-protein-products-developing-and-validating-assays-anti-drug. (accessed on 1 August 2022).
- Van Brummelen, E.M.; Ros, W.; Wolbink, G.; Beijnen, J.H.; Schellens, J.H. Antidrug Antibody Formation in Oncology: Clinical Relevance and Challenges. Oncoligist 2016, 21, 1260–1268. [Google Scholar] [CrossRef] [PubMed]
- FDA Grants Accelerated Approval to Nivolumab and Ipilimumab Combination for Hepatocellular Carcinoma. Available online: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-grants-accelerated-approval-nivolumab-and-ipilimumab-combination-hepatocellular-carcinoma (accessed on 1 August 2022).
- Food and Drug Administration. Opdivo Prescribing Information. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2019/125554s070lbl.pdf (accessed on 1 August 2022).
- Food and Drug Administration. Keytruda Prescribing Information. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2022/125514s110lbl.pdf (accessed on 1 August 2022).
- Food and Drug Administration. Yervoy Prescribing Information. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2015/125377s073lbl.pdf (accessed on 1 August 2022).
- Food and Drug Administration. Imfinzi Prescribing Information. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2020/761069s018lbl.pdf (accessed on 1 August 2022).
- Enrico, D.; Paci, A.; Chaput, N.; Karamouza, E.; Besse, B. Antidrug Antibodies Against Immune Checkpoint Blockers: Impairment of Drug Efficacy or Indication of Immune Activation? Clin. Cancer Res. 2020, 26, 787–792. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Sternheim, N.; Agarwal, P.; Suchomel, J.; Vadhavkar, S.; Bruno, R.; Ballinger, M.; Bernaards, C.A.; Chan, P.; Ruppel, J.; et al. Evaluation of atezolizumab immunogenicity: Clinical pharmacology (part 1). Clin. Transl. Sci. 2022, 15, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Peters, S.; Galle, P.R.; Bernaards, C.A.; Ballinger, M.; Bruno, R.; Quarmby, V.; Ruppel, J.; Vilimovskij, A.; Wu, B.; Sternheim, N.; et al. Evaluation of atezolizumab immunogenicity: Efficacy and safety (Part 2). Clin. Transl. Sci. 2022, 15, 141–157. [Google Scholar] [CrossRef]
- Agrawal, S.; Statkevich, P.; Bajaj, G.; Feng, Y.; Saeger, S.; Desai, D.D.; Park, J.S.; Waxman, I.M.; Roy, A.; Gupta, M. Evaluation of Immunogenicity of Nivolumab Monotherapy and Its Clinical Relevance in Patients With Metastatic Solid Tumors. J. Clin. Pharmacol. 2017, 57, 394–400. [Google Scholar] [CrossRef] [PubMed]
- Albillos, A.; de Gottardi, A.; Rescigno, M. The gut-liver axis in liver disease: Pathophysiological basis for therapy. J. Hepatol. 2020, 72, 558–577. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.; Debelius, J.; Brenner, D.A.; Karin, M.; Loomba, R.; Schnabl, B.; Knight, R. The gut-liver axis and the intersection with the microbiome. Nature reviews. Gastroenterol. Hepatol. 2018, 15, 397–411. [Google Scholar]
- Rakoff-Nahoum, S.; Paglino, J.; Eslami-Varzaneh, F.; Edberg, S.; Medzhitov, R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 2004, 118, 229–241. [Google Scholar] [CrossRef]
- Mazmanian, S.K.; Round, J.L.; Kasper, D.L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 2008, 453, 620–625. [Google Scholar] [CrossRef]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Qie, Y.; Park, J.; Kim, C.H. Gut Microbial Metabolites Fuel Host Antibody Responses. Cell Host Microbe 2016, 20, 202–214. [Google Scholar] [CrossRef] [PubMed]
- Hao, H.; Cao, L.; Jiang, C.; Che, Y.; Zhang, S.; Takahashi, S.; Wang, G.; Gonzalez, F.J. Farnesoid X Receptor Regulation of the NLRP3 Inflammasome Underlies Cholestasis-Associated Sepsis. Cell Metab. 2017, 25, 856–867.e5. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.D.; Chen, W.D.; Wang, M.; Yu, D.; Forman, B.M.; Huang, W. Farnesoid X receptor antagonizes nuclear factor kappaB in hepatic inflammatory response. Hepatology 2008, 48, 1632–1643. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.D.; Chen, W.D.; Yu, D.; Forman, B.M.; Huang, W. The G-protein-coupled bile acid receptor, Gpbar1 (TGR5), negatively regulates hepatic inflammatory response through antagonizing nuclear factor κ light-chain enhancer of activated B cells (NF-κB) in mice. Hepatology 2011, 54, 1421–1432. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, C.D.; Trauner, M. Role of bile acids and their receptors in gastrointestinal and hepatic pathophysiology. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 432–450. [Google Scholar] [CrossRef]
- Bajaj, J.S.; Heuman, D.M.; Hylemon, P.B.; Sanyal, A.J.; White, M.B.; Monteith, P.; Noble, N.A.; Unser, A.B.; Daita, K.; Fisher, A.R.; et al. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J. Hepatol. 2014, 60, 940–947. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, F.; Lu, H.; Wang, B.; Chen, Y.; Lei, D.; Wang, Y.; Zhu, B.; Li, L. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology 2011, 54, 562–572. [Google Scholar] [CrossRef]
- Yu, L.X.; Schwabe, R.F. The gut microbiome and liver cancer: Mechanisms and clinical translation. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 527–539. [Google Scholar] [CrossRef]
- Tsiaoussis, G.I.; Assimakopoulos, S.F.; Tsamandas, A.C.; Triantos, C.K.; Thomopoulos, K.C. Intestinal barrier dysfunction in cirrhosis: Current concepts in pathophysiology and clinical implications. World J. Hepatol. 2017, 7, 2058–2068. [Google Scholar] [CrossRef]
- Wies, R.; Lawson, M.; Geuking, M. Pathological bacterial translocation in liver cirrhosis. J. Hepatol. 2014, 60, 197–209. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Baker, S.S.; Gill, C.; Liu, W.; Alkhouri, R.; Baker, R.D.; Gill, S.R. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: A connection between endogenous alcohol and NASH. Hepatology 2013, 57, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Zheng, J.; Zhang, S.; Wang, B.; Wu, C.; Guo, X. Advances in the Involvement of Gut Microbiota in Pathophysiology of NAFLD. Front. Med. 2020, 7, 361. [Google Scholar] [CrossRef]
- Albillos, A.; Lario, M.; Álvarez-Mon, M. Cirrhosis-associated immune dysfunction: Distinctive features and clinical relevance. J. Hepatol. 2014, 61, 1385–1396. [Google Scholar] [CrossRef]
- Yu, L.X.; Ling, Y.; Wang, H.Y. Role of nonresolving inflammation in hepatocellular carcinoma development and progression. NPJ Precis Oncol. 2018, 2, 6. [Google Scholar] [CrossRef] [PubMed]
- Ponziani, F.R.; Nicoletti, A.; Gasbarrini, A.; Pompili, M. Diagnostic and therapeutic potential of the gut microbiota in patients with early hepatocellular carcinoma. Ther. Adv. Med. Oncol. 2019, 11, 1758835919848184. [Google Scholar] [CrossRef]
- Shahi, S.K.; Zarei, K.; Guseva, N.V.; Mangalam, A.K. Microbiota Analysis Using Two-step PCR and Next-generation 16S rRNA Gene Sequencing. J. Vis. Exp. 2019, 152. [Google Scholar] [CrossRef]
- Petti, C.A. Detection and identification of microorganisms by gene amplification and sequencing. Clin. Infect. Dis. 2007, 44, 1108–1114. [Google Scholar]
- Durazzi, F.; Sala, C.; Castellani, G.; Manfreda, G.; Remondini, D.; De Cesare, A. Comparison between 16S rRNA and shotgun sequencing data for the taxonomic characterization of the gut microbiota. Sci. Rep. 2021, 11, 3030. [Google Scholar] [CrossRef]
- Iida, N.; Dzutsev, A.; Stewart, C.A.; Smith, L.; Bouladoux, N.; Weingarten, R.A.; Molina, D.A.; Salcedo, R.; Back, T.; Cramer, S.; et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013, 342, 967–970. [Google Scholar] [CrossRef]
- Routy, B.; le Chatelier, E.; DeRosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Baruch, E.N.; Youngster, I.; Ben-Betzalel, G.; Ortenberg, R.; Lahat, A.; Katz, L.; Adler, K.; Dick-Necula, D.; Raskin, S.; Bloch, N.; et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 2021, 371, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wang, T.; Tu, X.; Huang, Y.; Zhang, H.; Tan, D.; Jiang, W.; Cai, S.; Zhao, P.; Song, R.; et al. Gut microbiome affects the response to anti-PD-1 immunotherapy in patients with hepatocellular carcinoma. J. Immunother. Cancer 2019, 7, 193. [Google Scholar] [CrossRef]
- Chung, M.-W.; Kim, M.-J.; Won, E.J.; Lee, Y.J.; Yun, Y.-W.; Cho, S.B.; Joo, Y.-E.; Hwang, J.-E.; Bae, W.K.; Chung, I.-J.; et al. Gut microbiome composition can predict the response to nivolumab in advanced hepatocellular carcinoma patients. World J. Gastroenterol. 2021, 27, 7340–7349. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.; Wang, D.; Long, J.; Yang, X.; Lin, J.; Song, Y.; Xie, F.; Xun, Z.; Wang, Y.; Wang, Y.; et al. Gut microbiome is associated with the clinical response to anti-PD-1 based immunotherapy in hepatobiliary cancers. J. Immunother. Cancer 2021, 9, e003334. [Google Scholar] [CrossRef]
- Li, L.; Ye, J. Characterization of gut microbiota in patients with primary hepatocellular carcinoma received immune checkpoint inhibitors: A Chinese population-based study. Medicine 2020, 2020, e21788. [Google Scholar] [CrossRef]
- Ponziani, F.R.; De Luca, A.; Picca, A.; Marzetti, E.; Petito, V.; Del Chierico, F.; Reddel, S.; Sterbini, F.P.; Sanguinetti, M.; Putignani, L.; et al. Gut Dysbiosis and Fecal Calprotectin Predict Response to Immune Checkpoint Inhibitors in Patients With Hepatocellular Carcinoma. Hepatol. Commun. 2022, 6, 1492–1501. [Google Scholar] [CrossRef]
- Nomura, M.; Nagatomo, R.; Doi, K.; Shimizu, J.; Baba, K.; Saito, T.; Matsumoto, S.; Inoue, K.; Muto, M. Association of Short-Chain Fatty Acids in the Gut Microbiome With Clinical Response to Treatment With Nivolumab or Pembrolizumab in Patients With Solid Cancer Tumors. JAMA Netw. Open 2020, 3, e202895. [Google Scholar] [CrossRef]
- Li, M.; van Esch, B.; Henricks, P.; Folkerts, G.; Garssen, J. The Anti-inflammatory Effects of Short Chain Fatty Acids on Lipopolysaccharide- or Tumor Necrosis Factor α-Stimulated Endothelial Cells via Activation of GPR41/43 and Inhibition of HDACs. Front. Pharmacol. 2018, 9, 533. [Google Scholar] [CrossRef]
- Licciardi, P.V.; Ververis, K.; Karagiannis, T.C. Histone deacetylase inhibition and dietary short-chain Fatty acids. ISRN Allergy 2011, 2011, 869647. [Google Scholar] [CrossRef] [PubMed]
- Woods, D.M.; Sodré, A.L.; Villagra, A.; Sarnaik, A.; Sotomayor, E.M.; Weber, J. HDAC Inhibition Upregulates PD-1 Ligands in Melanoma and Augments Immunotherapy with PD-1 Blockade. Cancer Immunol. Res. 2015, 3, 1375–1385. [Google Scholar] [CrossRef] [PubMed]
- Coutzac, C.; Jouniaux, J.M.; Paci, A.; Schmidt, J.; Mallardo, D.; Seck, A.; Asvatourian, V.; Cassard, L.; Saulnier, P.; Lacroix, L.; et al. Systemic short chain fatty acids limit antitumor effect of CTLA-4 blockade in hosts with cancer. Nat. Commun. 2020, 11, 2168. [Google Scholar] [CrossRef] [PubMed]
- Behary, J.; Amorim, N.; Jiang, X.T.; Raposo, A.; Gong, L.; McGovern, E.; Ibrahim, R.; Chu, F.; Stephens, C.; Jebeili, H.; et al. Gut microbiota impact on the peripheral immune response in non-alcoholic fatty liver disease related hepatocellular carcinoma. Nat. Commun. 2021, 12, 187. [Google Scholar] [CrossRef] [PubMed]
- Pfister, D.; Núñez, N.G.; Pinyol, R.; Govaere, O.; Pinter, M.; Szydlowska, M.; Gupta, R.; Qiu, M.; Deczkowska, A.; Weiner, A.; et al. NASH limits anti-tumour surveillance in immunotherapy-treated HCC. Nature 2021, 592, 450–456. [Google Scholar] [CrossRef] [PubMed]
- CheckMate 459: A Randomized, Multi-Center Phase III Study of Nivolumab (NIVO) vs Sorafenib (SOR) as First-Line (1L) Treatment in Patients (pts) with Advanced Hepatocellular Carcinoma (aHCC). Available online: https://www.sciencedirect.com/science/article/pii/S0923753419603893 (accessed on 1 August 2022).
- Derosa, L.; Hellmann, M.D.; Spaziano, M.; Halpenny, D.; Fidelle, M.; Rizvi, H.; Long, N.; Plodkowski, A.J.; Arbour, K.C.; Chaft, J.E.; et al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Ann. Oncol. 2018, 29, 1437–1444. [Google Scholar] [CrossRef]
- Tinsley, N.; Zhou, C.; Tan, G.; Rack, S.; Lorigan, P.; Blackhall, F.; Krebs, M.; Carter, L.; Thistlethwaite, F.; Graham, D.; et al. Cumulative Antibiotic Use Significantly Decreases Efficacy of Checkpoint Inhibitors in Patients with Advanced Cancer. Oncologist 2020, 25, 55–63. [Google Scholar] [CrossRef]
- Hakozaki, T.; Okuma, Y.; Omori, M.; Hosomi, Y. Impact of prior antibiotic use on the efficacy of nivolumab for non-small cell lung cancer. Oncol. Lett. 2019, 17, 2946–2952. [Google Scholar] [CrossRef]
- Pinato, D.J.; Howlett, S.; Ottaviani, D.; Urus, H.; Patel, A.; Mineo, T.; Brock, C.; Power, D.; Hatcher, O.; Falconer, A.; et al. Association of Prior Antibiotic Treatment With Survival and Response to Immune Checkpoint Inhibitor Therapy in Patients With Cancer. JAMA Oncol. 2019, 5, 1774–1778. [Google Scholar] [CrossRef]
- Ma, C.; Han, M.; Heinrich, B.; Fu, Q.; Zhang, Q.; Sandhu, M.; Agdashian, D.; Terabe, M.; Berzofsky, J.A.; Fako, V.; et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 2018, 360, eaan5931. [Google Scholar] [CrossRef]
- Dwiyanto, J.; Hussain, M.H.; Reidpath, D.; Ong, K.S.; Qasim, A.; Lee, S.; Lee, S.M.; Foo, S.C.; Chong, C.W.; Rahman, S. Ethnicity influences the gut microbiota of individuals sharing a geographical location: A cross-sectional study from a middle-income country. Sci. Rep. 2021, 11, 2618. [Google Scholar] [CrossRef] [PubMed]
- Redondo-Useros, N.; Nova, E.; González-Zancada, N.; Díaz, L.E.; Gómez-Martínez, S.; Marcos, A. Microbiota and Lifestyle: A Special Focus on Diet. Nutrients 2020, 12, 1776. [Google Scholar] [CrossRef] [PubMed]
- Weersma, R.K.; Zhernakova, A.; Fu, J. Interaction between drugs and the gut microbiome. Gut 2020, 69, 1510–1519. [Google Scholar] [CrossRef] [PubMed]
- Mariam, A.; Miller-Atkins, G.; Moro, A.; Rodarte, A.I.; Siddiqi, S.; Acevedo-Moreno, L.A.; Brown, J.M.; Allende, D.S.; Aucejo, F.; Rotroff, D.M. Salivary miRNAs as non-invasive biomarkers of hepatocellular carcinoma: A pilot study. PeerJ 2022, 10, e12715. [Google Scholar] [CrossRef] [PubMed]
- Hershberger, C.E.; Rodarte, A.I.; Siddiqi, S.; Moro, A.; Acevedo-Moreno, L.A.; Brown, J.M.; Allende, D.S.; Aucejo, F.; Rotroff, D.M. Salivary Metabolites are Promising Non-Invasive Biomarkers of Hepatocellular Carcinoma and Chronic Liver Disease. Liver Cancer Int. 2021, 2, 33–44. [Google Scholar] [CrossRef]
- Xie, F.; Bai, Y.; Yang, X.; Long, J.; Mao, J.; Lin, J.; Wang, D.; Song, Y.; Xun, Z.; Huang, H.; et al. Comprehensive analysis of tumour mutation burden and the immune microenvironment in hepatocellular carcinoma. Int. Immunopharmacol. 2020, 89, 107135. [Google Scholar] [CrossRef]
- Kather, J.N.; Calderaro, J. Development of AI-based pathology biomarkers in gastrointestinal and liver cancer. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 591–592. [Google Scholar] [CrossRef]
- Zeng, Q.; Klein, C.; Caruso, S.; Maille, P.; Laleh, N.G.; Sommacale, D.; Laurent, A.; Amaddeo, G.; Gentien, D.; Rapinat, A.; et al. Artificial intelligence predicts immune and inflammatory gene signatures directly from hepatocellular carcinoma histology. J. Hepatol. 2022, 77, 116–127. [Google Scholar] [CrossRef]
- Tian, Y.; Komolafe, T.E.; Zheng, J.; Zhou, G.; Chen, T.; Zhou, B.; Yang, X. Assessing PD-L1 Expression Level via Preoperative MRI in HCC Based on Integrating Deep Learning and Radiomics Features. Diagnostics 2021, 11, 1875. [Google Scholar] [CrossRef]
- Chen, S.; Feng, S.; Wei, J.; Liu, F.; Li, B.; Li, X.; Hou, Y.; Gu, D.; Tang, M.; Xiao, H.; et al. Pretreatment prediction of immunoscore in hepatocellular cancer: A radiomics-based clinical model based on Gd-EOB-DTPA-enhanced MRI imaging. Eur. Radiol. 2019, 29, 4177–4187. [Google Scholar] [CrossRef]
- Liao, H.; Zhang, Z.; Chen, J.; Liao, M.; Xu, L.; Wu, Z.; Yuan, K.; Song, B.; Zeng, Y. Preoperative Radiomic Approach to Evaluate Tumor-Infiltrating CD8+ T Cells in Hepatocellular Carcinoma Patients Using Contrast-Enhanced Computed Tomography. Ann. Surg. Oncol. 2019, 26, 4537–4547. [Google Scholar] [CrossRef] [PubMed]
- Hectors, S.J.; Lewis, S.; Besa, C.; King, M.J.; Said, D.; Putra, J.; Ward, S.; Higashi, T.; Thung, S.; Yao, S.; et al. MRI radiomics features predict immuno-oncological characteristics of hepatocellular carcinoma. Eur. Radiol. 2020, 30, 3759–3769. [Google Scholar] [CrossRef] [PubMed]
- Di Tommaso, N.; Gasbarrini, A.; Ponziani, F.R. Intestinal Barrier in Human Health and Disease. Int. J. Environ. Res. Public Health 2021, 18, 12836. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).