Low Expression of RGS2 Promotes Poor Prognosis in High-Grade Serous Ovarian Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Patients and Methods
2.1. Clinical Cohort
2.2. Tissue-Microarrays and Immunohistochemistry
2.3. Digital Image Analysis
2.4. TCGA HGSOC Data Set Gene Expression Analysis
2.5. Single-Cell Gene Expression Analysis in Three Publicly Available HGSOC Datasets
2.6. Statistical Analysis
3. Results
3.1. Staining Pattern of RGS2 in HGSOC
3.2. Clinical Characteristics
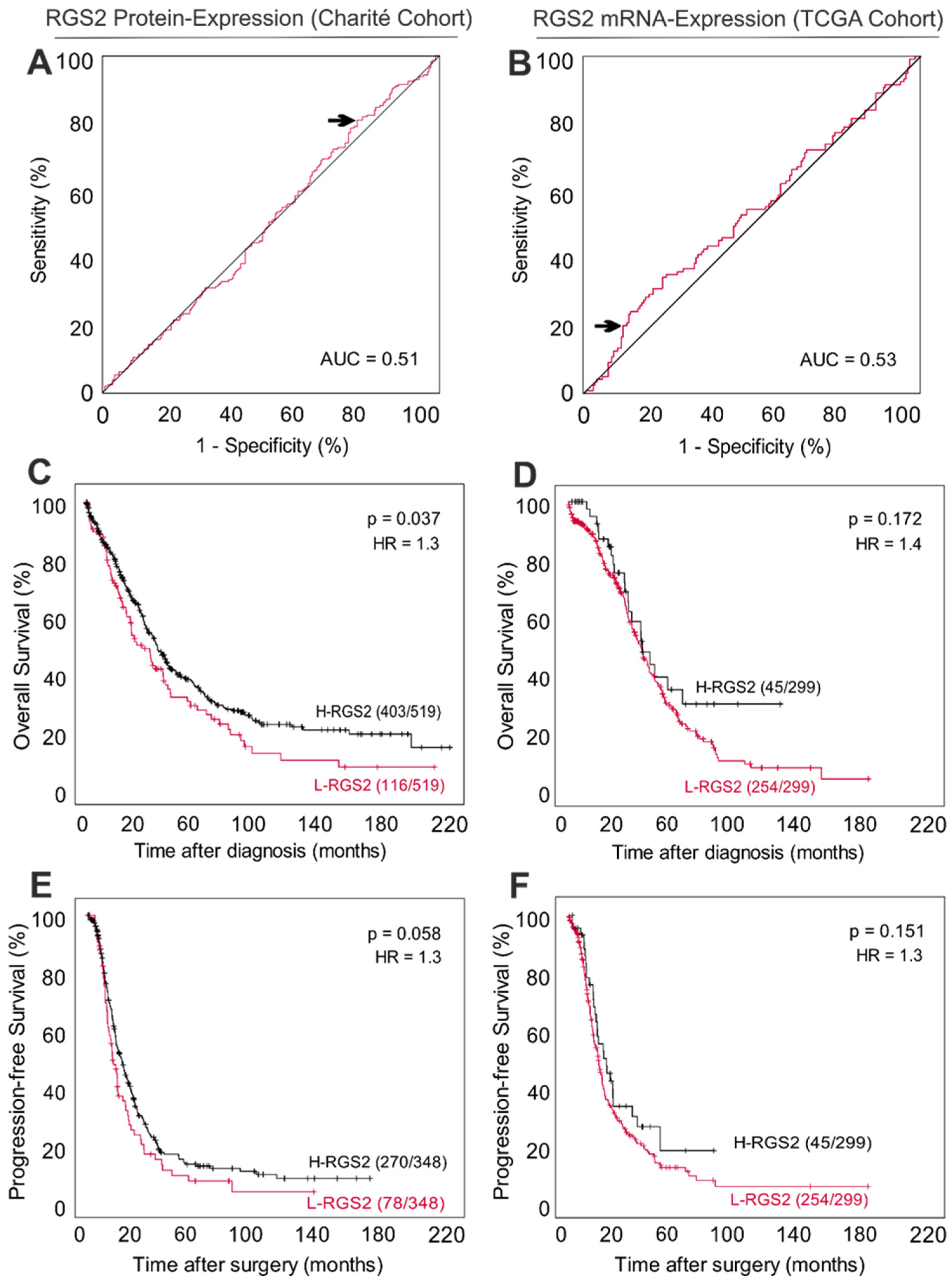
3.3. Low RGS2 Protein Expression Is Partially Associated with a Poor Long-Term Survival in HGSOC
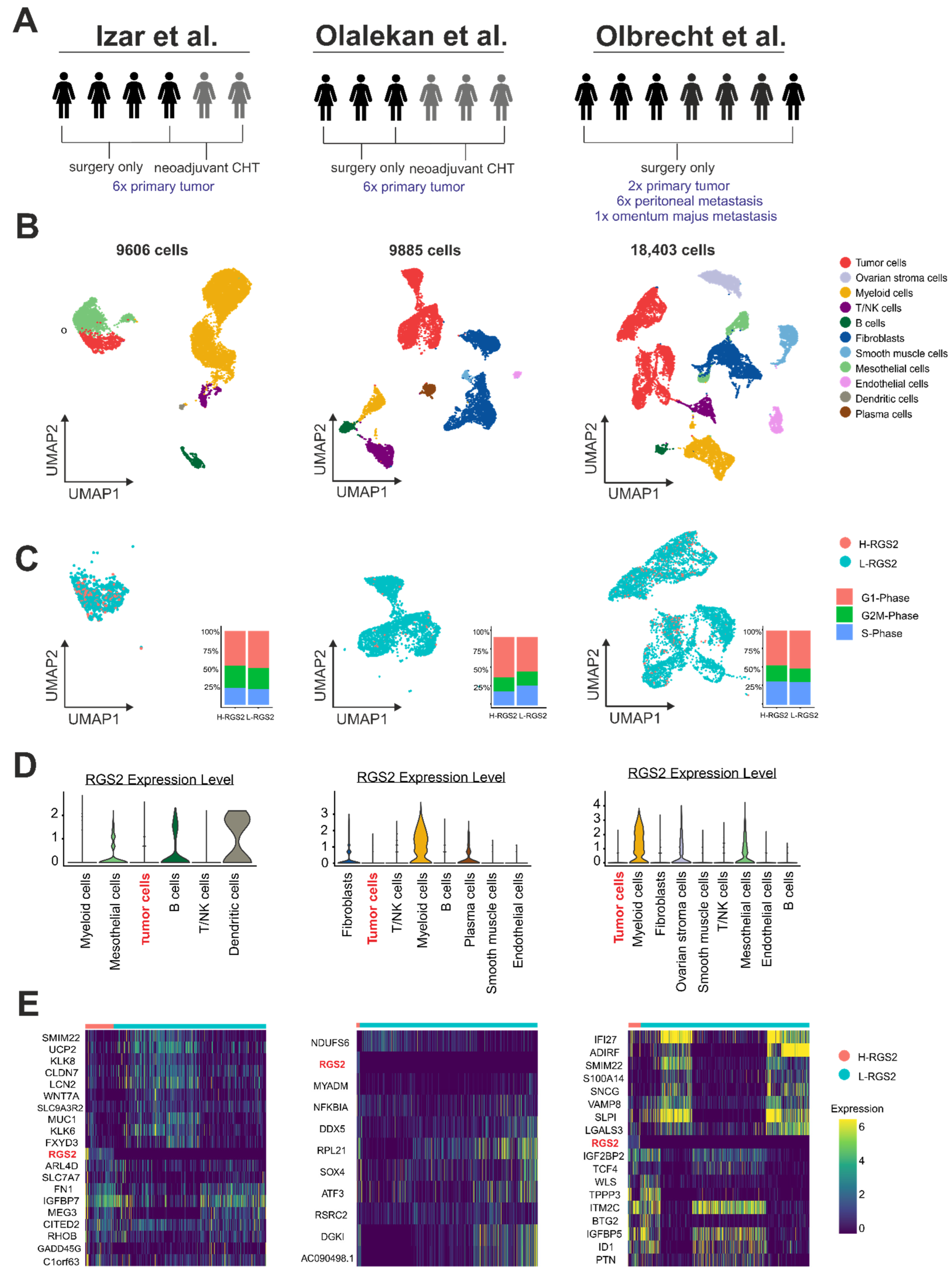
3.4. RGS2 mRNA Expression in Ovarian Cancer Cells Is Weak on the Single-Cell Level
3.5. RGS2 mRNA-Suppression Is Linked to Tumor Cell Plasticity on a Single-Cell Level
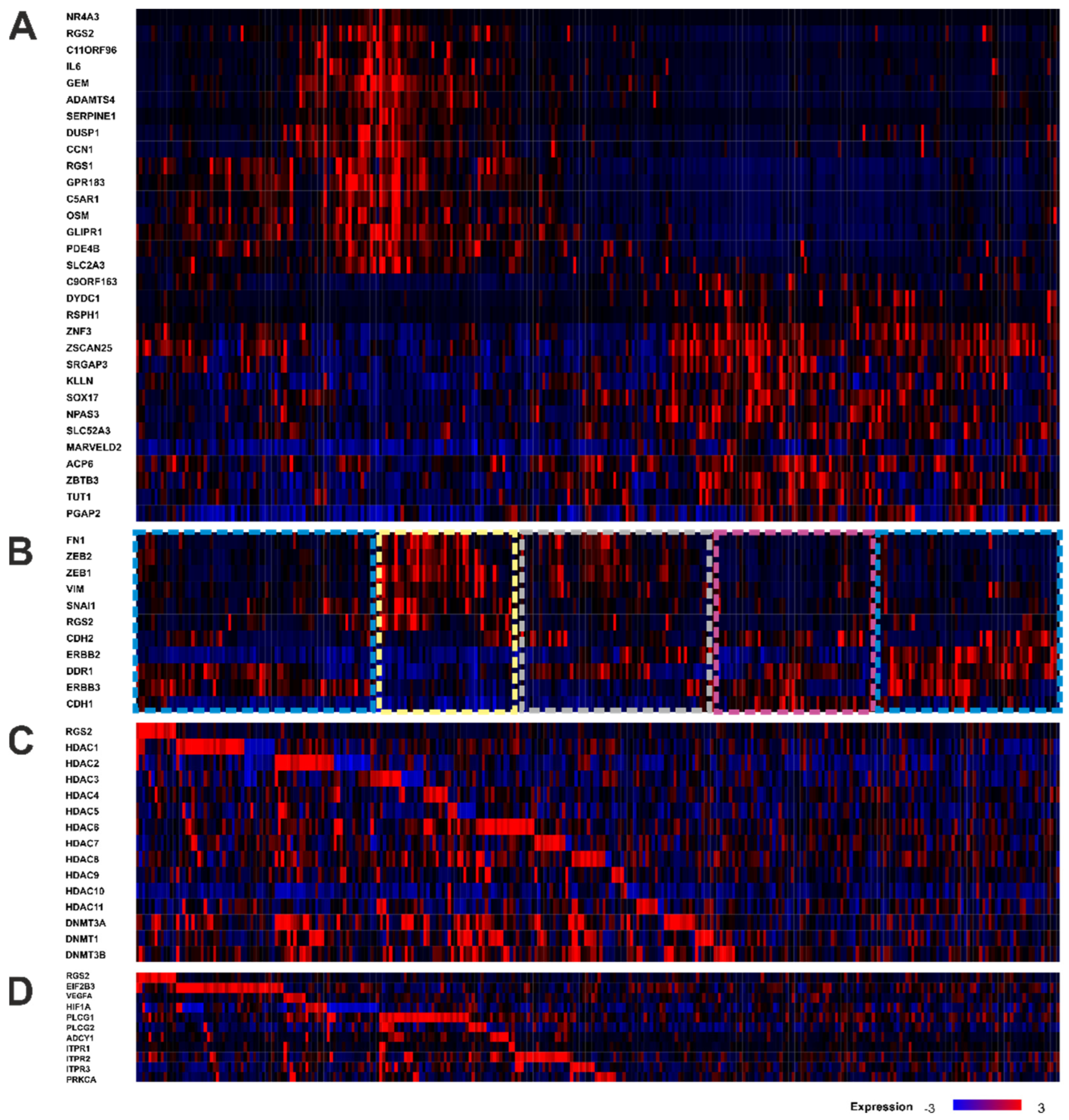
3.6. TCGA Gene Expression Analysis Reveals an Association between RGS2 Expression and Protein Synthesis and Co-Dependence on Methylation
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Heximer, S.P.; Watson, N.; Linder, M.E.; Blumer, K.J.; Hepler, J.R. RGS2/G0S8 is a selective inhibitor of Gqalpha function. Proc. Natl. Acad. Sci. USA 1997, 94, 14389–14393. [Google Scholar] [CrossRef] [PubMed]
- Sinnarajah, S.; Dessauer, C.; Srikumar, D.; Chen, J.; Yuen, J.; Yilma, S.; Dennis, J.C.; Morrison, E.E.; Vodyanoy, V.; Kehrl, J. RGS2 regulates signal transduction in olfactory neurons by attenuating activation of adenylyl cyclase III. Nature 2001, 409, 1051–1055. [Google Scholar] [CrossRef] [PubMed]
- Salim, S.; Sinnarajah, S.; Kehrl, J.H.; Dessauer, C.W. Identification of RGS2 and Type V Adenylyl Cyclase Interaction Sites. J. Biol. Chem. 2003, 278, 15842–15849. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, L.S.; Ramineni, S.; Hague, C.; Cladman, W.; Chidiac, P.; Levey, A.I.; Hepler, J.R. RGS2 Binds Directly and Selectively to the M1 Muscarinic Acetylcholine Receptor Third Intracellular Loop to Modulate Gq/11α Signaling. J. Biol. Chem. 2004, 279, 21248–21256. [Google Scholar] [CrossRef]
- Nunn, C.; Zou, M.X.; Sobiesiak, A.J.; Roy, A.A.; Kirshenbaum, L.A.; Chidiac, P. RGS2 inhibits beta-adrenergic receptor-induced cardiomyocyte hypertrophy. Cell. Signal. 2010, 22, 1231–1239. [Google Scholar] [CrossRef]
- Druey, K.M.; Blumer, K.J.; Kang, V.H.; Kehrl, J.H. Inhibition of G-protein-mediated MAP kinase activation by a new mammalian gene family. Nature 1996, 379, 726–742. [Google Scholar] [CrossRef]
- Masuho, I.; Balaji, S.; Muntean, B.S.; Skamangas, N.K.; Chavali, S.; Tesmer, J.J.; Babu, M.M.; Martemyanov, K.A. A Global Map of G Protein Signaling Regulation by RGS Proteins. Cell 2020, 183, 503–521.e19. [Google Scholar] [CrossRef]
- Yang, J.; Kamide, K.; Kokubo, Y.; Takiuchi, S.; Tanaka, C.; Banno, M.; Miwa, Y.; Yoshii, M.; Horio, T.; Okayama, A.; et al. Genetic variations of regulator of G-protein signaling 2 in hypertensive patients and in the general population. J. Hypertens. 2005, 23, 1497–1505. [Google Scholar] [CrossRef]
- Calò, L.A.; Pagnin, E.; Davis, P.A.; Sartori, M.; Ceolotto, G.; Pessina, A.C.; Semplicini, A. Increased expression of regulator of G protein signaling-2 (RGS-2) in Bartter’s/Gitelman’s syndrome. A role in the control of vascular tone and implication for hypertension. J. Clin. Endocrinol. Metabol. 2004, 89, 4153–4157. [Google Scholar] [CrossRef]
- Semplicini, A.; Strapazzon, G.; Papparella, I.; Sartori, M.; Realdi, A.; Macchini, L.; A Calò, L.; Ceolotto, G. RGS2 expression and aldosterone: Renin ratio modulate response to drug therapy in hypertensive patients. J. Hypertens. 2010, 28, 1104–1108. [Google Scholar] [CrossRef]
- Dong, H.; Zhang, Y.; Wang, J.; Kim, D.S.; Wu, H.; Sjögren, B.; Gao, W.; Luttrell, L.; Wang, H. Regulator of G protein signaling 2 is a key regulator of pancreatic β-cell mass and function. Cell Death Dis. 2017, 8, e2821. [Google Scholar] [CrossRef] [PubMed]
- Lifschytz, T.; Broner, E.C.; Zozulinsky, P.; Slonimsky, A.; Eitan, R.; Greenbaum, L.; Lerer, B. Relationship between Rgs2 gene expression level and anxiety and depression-like behaviour in a mutant mouse model: Serotonergic involvement. Intern. J. Neuropsychopharmacol. 2012, 15, 1307–1318. [Google Scholar] [CrossRef] [PubMed]
- Dusonchet, J.; Li, H.; Guillily, M.; Liu, M.; Stafa, K.; Troletti, C.D.; Boon, J.Y.; Saha, S.; Glauser, L.; Mamais, A.; et al. A Parkinson’s disease gene regulatory network identifies the signaling protein RGS2 as a modulator of LRRK2 activity and neuronal toxicity. Hum. Mol. Genet. 2014, 23, 4887–4905. [Google Scholar] [CrossRef] [PubMed]
- Hadar, A.; Milanesi, E.; Squassina, A.; Niola, P.; Chillotti, C.; Pasmanik-Chor, M.; Yaron, O.; Martásek, P.; Rehavi, M.; Weissglas-Volkov, D.; et al. RGS2 expression predicts amyloid-β sensitivity, MCI and Alzheimer’s disease: Genome-wide transcriptomic profiling and bioinformatics data mining. Transl. Psych. 2016, 6, e909. [Google Scholar] [CrossRef] [PubMed]
- Schwäble, J.; Choudhary, C.; Thiede, C.; Tickenbrock, L.; Sargin, B.; Steur, C.; Rehage, M.; Rudat, A.; Brandts, C.; Berdel, W.E.; et al. RGS2 is an important target gene of Flt3-ITD mutations in AML and functions in myeloid differentiation and leukemic transformation. Blood 2005, 105, 2107–2114. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.K.; Heng, H.H.; Shi, X.M.; Forsdyke, D.R.; Tsui, L.C.; Mak, T.W.; Minden, M.D.; Siderovski, D.P. Differential expression of a basic helix-loop-helix phosphoprotein gene, G0S8, in acute leukemia and localization to human chromosome 1q31. Leukemia 1995, 9, 1291–1298. [Google Scholar]
- Wang, C.; Ye, Q.; Cao, Y.; Tan, J.; Wang, F.; Jiang, J.; Cao, Y. Downregulation of regulator of G protein signaling 2 expression in breast invasive carcinoma of no special type: Clinicopathological associations and prognostic relevance. Oncol. Lett. 2018, 15, 213–220. [Google Scholar] [CrossRef]
- Cao, X.; Qin, J.; Xie, Y.; Khan, O.; Dowd, F.; Scofield, M.; Lin, M.-F.; Tu, Y. Regulator of G-protein signaling 2 (RGS2) inhibits androgen-independent activation of androgen receptor in prostate cancer cells. Oncogene 2006, 25, 3719–3734. [Google Scholar] [CrossRef]
- Wolff, D.W.; Xie, Y.; Deng, C.; Gatalica, Z.; Yang, M.; Wang, B.; Wang, J.; Lin, M.-F.; Abel, P.; Tu, Y. Epigenetic repression of regulator of G-protein signaling 2 promotes androgen-independent prostate cancer cell growth. Intern. J. Cancer 2012, 130, 1521–1531. [Google Scholar] [CrossRef]
- Linder, A.; Hagberg Thulin, M.; Damber, J.-E.; Welén, K. Analysis of regulator of G-protein signalling 2 (RGS2) expression and function during prostate cancer progression. Sci. Rep. 2018, 8, 17259. [Google Scholar] [CrossRef]
- Hurst, J.H.; Mendpara, N.; Hooks, S.B. Regulator of G-protein signalling expression and function in ovarian cancer cell lines. Cell. Mol. Biol. Lett. 2009, 14, 153–174. [Google Scholar] [CrossRef] [PubMed]
- Cacan, E. Epigenetic regulation of RGS2 (Regulator of G-protein signaling 2) in chemoresistant ovarian cancer cells. J. Chemother. 2017, 29, 173–178. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.B.; Wilkinson, J.C.; Roman, D.L. Regulator of G-protein signaling (RGS) proteins as drug targets: Progress and future potentials. J. Biol. Chem. 2019, 294, 18571–18585. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, S.; Coward, J.I.; Bast, R.C., Jr.; Berchuck, A.; Berek, J.S.; Brenton, J.D.; Coukos, G.; Crum, C.C.; Drapkin, R.; Etemadmoghadam, D.; et al. Rethinking ovarian cancer: Recommendations for improving outcomes. Nat. Rev. Cancer 2011, 11, 719–725. [Google Scholar] [CrossRef] [PubMed]
- Dorsam, R.T.; Gutkind, J.S. G-protein-coupled receptors and cancer. Nat. Rev. Cancer 2007, 7, 79–94. [Google Scholar] [CrossRef]
- Lappano, R.; Maggiolini, M. G protein-coupled receptors: Novel targets for drug discovery in cancer. Nat. Rev. Drug Discov. 2011, 10, 47–60. [Google Scholar] [CrossRef]
- O’Hayre, M.; Degese, M.S.; Gutkind, J.S. Novel insights into G protein and G protein-coupled receptor signaling in cancer. Curr. Opin. Bell. Biol. 2014, 27, 126–135. [Google Scholar] [CrossRef]
- Kurman, R.J.; Carcangiu, M.L.; Young, R.H.; Herrington, C.S. WHO Classification of Tumours of Femal Reproductive Organs. In WHO Classification of Tumours, 4th ed.; International Agency for Research on Cancer: Lyon, France, 2014; Volume 6, ISBN 13:978-92-832-2435-8. [Google Scholar]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.E.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Izar, B.; Tirosh, I.; Stover, E.H.; Wakiro, I.; Cuoco, M.S.; Alter, I.; Rodman, C.; Leeson, R.; Su, M.-J.; Shah, P.; et al. A single-cell landscape of high-grade serous ovarian cancer. Nat. Med. 2020, 26, 1271–1279. [Google Scholar] [CrossRef] [PubMed]
- Olalekan, S.; Xie, B.; Back, R.; Eckart, H.; Basu, A. Characterizing the tumor microenvironment of metastatic ovarian cancer by single-cell transcriptomics. Cell Rep. 2021, 35, 109165. [Google Scholar] [CrossRef]
- Olbrecht, S.; Busschaert, P.; Qian, J.; Vanderstichele, A.; Loverix, L.; Van Gorp, T.; Van Nieuwenhuysen, E.; Han, S.; Van den Broeck, A.; Coosemans, A.; et al. High-grade serous tubo-ovarian cancer refined with single-cell RNA sequencing: Specific cell subtypes influence survival and determine molecular subtype classification. Genom. Med. 2021, 13, 111. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M., 3rd; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573–3587.e29. [Google Scholar] [CrossRef]
- Budczies, J.; Klauschen, F.; Sinn, B.V.; Gyorffy, B.; Schmitt, W.D.; Darb-Esfahani, S.; Denkert, C. Cutoff Finder: A Comprehensive and Straightforward Web Application Enabling Rapid Biomarker Cutoff Optimization. PLoS ONE 2012, 7, e51862. [Google Scholar] [CrossRef] [PubMed]
- Schemper, M.; Smith, T.L. A note on quantifying follow-up in studies of failure time. Control Clin. Trials 1996, 17, 343–346. [Google Scholar] [CrossRef]
- Yin, H.; Wang, Y.; Chen, W.; Zhong, S.; Liu, Z.; Zhao, J. Drug-resistant CXCR4-positive cells have the molecular characteristics of EMT in NSCLC. Gene 2016, 594, 23–29. [Google Scholar] [CrossRef]
- Wang, C.-H.; Chidiac, P. RGS2 promotes the translation of stress-associated proteins ATF4 and CHOP via its eIF2B-inhibitory domain. Cell. Signal. 2019, 59, 163–170. [Google Scholar] [CrossRef]
- Ying, L.; Lin, J.; Qiu, F.; Cao, M.; Chen, H.; Liu, Z.; Huang, Y. Epigenetic repression of regulator of G-protein signaling 2 by ubiquitin-like with PHD and ring-finger domain 1 promotes bladder cancer progression. FEBS J. 2015, 282, 174–182. [Google Scholar] [CrossRef]
- Jiang, Z.; Wang, Z.; Xu, Y.; Wang, B.; Huang, W.; Cai, S. Analysis of RGS2 expression and prognostic significance in stage II and III colorectal cancer. Biosci. Rep. 2010, 30, 383–390. [Google Scholar] [CrossRef]
- Gu, S.; Anton, A.; Salim, S.; Blumer, K.J.; Dessauer, C.W.; Heximer, S.P. Alternative Translation Initiation of Human Regulators of G-Protein Signaling-2 Yields a Set of Functionally Distinct Proteins. Mol. Pharmacol. 2008, 73, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Raveh, A.; Schultz, P.J.; Aschermann, L.; Carpenter, C.; Tamayo-Castillo, G.; Cao, S.; Clardy, J.; Neubig, R.R.; Sherman, D.H.; Sjögren, B. Identification of PKC Activation as a Novel Mechanism for RGS2 Protein Up-regulation Through Phenotypic Screening of Natural Product Extracts. Mol. Pharmacol. 2014, 86, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Sjögren, B.; Parra, S.; Heath, L.J.; Atkins, K.B.; Xie, Z.-J.; Neubig, R.R. Cardiotonic Steroids Stabilize Regulator of G Protein Signaling 2 Protein Levels. Mol. Pharmacol. 2012, 82, 500. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, M.L.; Waldo, G.L.; Hollinger, S.; Hepler, J.R.; Harden, T.K. Protein kinase C phosphorylates RGS2 and modulates its capacity for negative regulation of Galpha 11 signaling. J. Biol. Chem. 2001, 276, 5438–5444. [Google Scholar] [CrossRef] [PubMed]
- Sjögren, B.; Parra, S.; Atkins, K.B.; Karaj, B.; Neubig, R.R. Digoxin-Mediated Upregulation of RGS2 Protein Protects against Cardiac Injury. J. Pharmacol. Exp. Ther. 2016, 357, 311–319. [Google Scholar] [CrossRef]
- Nguyen, C.H.; Ming, H.; Zhao, P.; Hugendubler, L.; Gros, R.; Kimball, S.R.; Chidiac, P. Translational control by RGS2. J. Cell Biol. 2009, 186, 755–765. [Google Scholar] [CrossRef]
- Nguyen, C.H.; Zhao, P.; Sobiesiak, A.J.; Chidiac, P. RGS2 is a component of the cellular stress response. Biochem. Biophys. Res. Commun. 2012, 426, 129–134. [Google Scholar] [CrossRef]
- Song, L.; Jope, R.S. Cellular stress increases RGS2 mRNA and decreases RGS4 mRNA levels in SH-SY5Y cells. Neurosci. Lett. 2006, 402, 205–209. [Google Scholar] [CrossRef]
- Yin, J.; Yan, X.; Yao, X.; Zhang, Y.; Shan, Y.; Mao, N.; Yang, Y.; Pan, L. Secretion of annexin A3 from ovarian cancer cells and its association with platinum resistance in ovarian cancer patients. J. Cell. Mol. Med. 2012, 16, 337–348. [Google Scholar] [CrossRef]
- Lucidi, A.; Buca, D.; Ronsini, C.; Tinari, S.; Bologna, G.; Buca, D.; Leombroni, M.; Liberati, M.; D’Antonio, F.; Scambia, G.; et al. Role of Extracellular Vesicles in Epithelial Ovarian Cancer: A Systematic Review. Int. J. Mol. Sci. 2020, 21, 8762. [Google Scholar] [CrossRef]
- Yan, X.D.; Pan, L.Y.; Yuan, Y.; Lang, J.H.; Mao, N. Identification of platinum-resistance associated proteins through proteomic analysis of human ovarian cancer cells and their platinum-resistant sublines. J. Proteome. Res. 2007, 6, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xu, Q.-H.; Dong, Y.-H.; Li, G.-X.; Yang, L.; Wang, L.-W.; Li, H.-Y. MiR-181a upregulation is associated with epithelial-to-mesenchymal transition (EMT) and multidrug resistance (MDR) of ovarian cancer cells. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 2004–2010. [Google Scholar] [PubMed]
- Tsuyoshi, H.; Inoue, D.; Kurokawa, T.; Yoshida, Y. Hyperthermic intraperitoneal chemotherapy (HIPEC) for gynecological cancer. J. Obstet. Gynecol. Res. 2020, 46, 1661–1671. [Google Scholar] [CrossRef]
- Ghirardi, V.; Ronsini, C.; Trozzi, R.; Di Ilio, C.; Di Giorgio, A.; Cianci, S.; Draisci, G.; Scambia, G.; Fagotti, A. Hyperthermic intraperitoneal chemotherapy in interval debulking surgery for advanced epithelial ovarian cancer: A single-center, real-life experience. Cancer 2020, 126, 5256–5262. [Google Scholar] [CrossRef]
- Khetan, R.; Dharmayanti, C.; Gillam, T.A.; Kübler, E.; Klingler-Hoffmann, M.; Ricciardelli, C.; Oehler, M.K.; Blencowe, A.; Garg, S.; Albrecht, H. Using GPCRs as Molecular Beacons to Target Ovarian Cancer with Nanomedicines. Cancers 2022, 14, 2362. [Google Scholar] [CrossRef] [PubMed]
- Keunecke, C.; Kulbe, H.; Dreher, F.; Taube, E.T.; Chekerov, R.; Horst, D.; Hummel, M.; Kessler, T.; Pietzner, K.; Kassuhn, W.; et al. Predictive biomarker for surgical outcome in patients with advanced primary high-grade serous ovarian cancer. Are we there yet? An analysis of the prospective biobank for ovarian cancer. Gynecol. Oncol. 2022, 166, 334–343. [Google Scholar] [CrossRef] [PubMed]
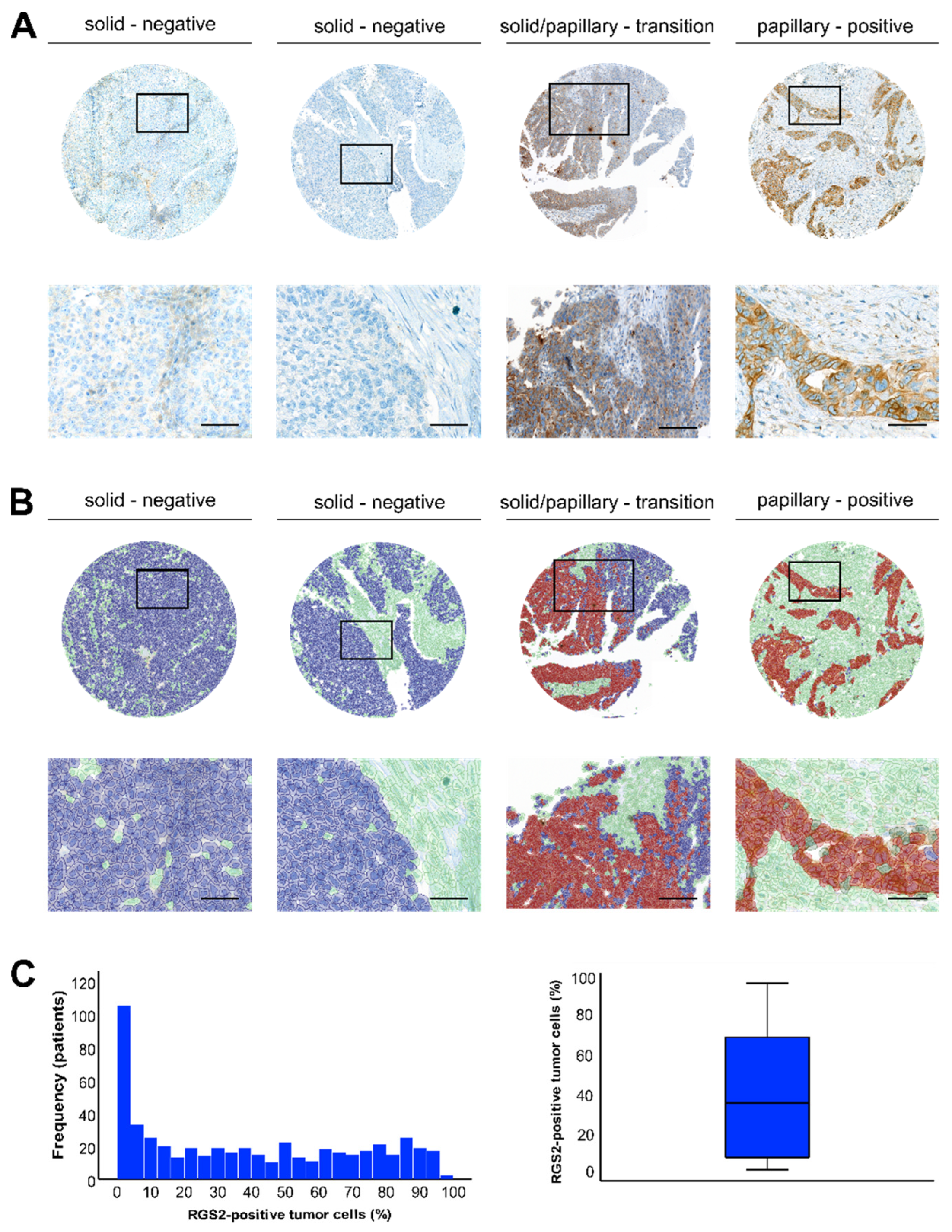
| Characteristics | Entire Cohort | L-RGS2 | H-RGS2 | p-Value |
|---|---|---|---|---|
| n (%) | 519 | 116 (22) | 403 (78) | |
| Age (years), median (IQR) | 61.5 (53–69) | 62 (55–68) | 61 (53–70) | 0.926 |
| RGS2-positive tumor cells, median, % (IQR) | 34.5 (6.4–68.4) | 1.1 (0.3–2.4) | 49.2 (24.2–75.4) | <0.001 |
| Primary tumor stage (pT) | 0.092 | |||
| - pT1, n (%) | 31 (6) | 5 (4) | 26 (6) | |
| - pT2, n (%) | 32 (6) | 3 (3) | 29 (7) | |
| - pT3, n (%) | 456 (88) | 108 (93) | 348 (87) | |
| Lymph node stage (pN) | 0.140 | |||
| - pN0, n (%) | 127 (24) | 23 (20) | 74 (18) | |
| - pN1, n (%) | 290 (56) | 74 (64) | 216 (54) | |
| - pNX, n (%) | 102 (20) | 19 (16) | 290 (72) | |
| Distant metastasis (pM) | 0.104 | |||
| - pM0, n (%) | 211 (41) | 43 (37) | 168 (42) | |
| - pM1, n (%) | 105 (20) | 30 (26) | 75 (18) | |
| - pMX, n (%) | 203 (39) | 43 (37) | 160 (40) | |
| FIGO stage | 0.185 | |||
| - FIGO I, n (%) | 24 (5) | 5 (4) | 19 (5) | |
| - FIGO II, n (%) | 22 (4) | 2 (2) | 20 (5) | |
| - FIGO III, n (%) | 373 (72) | 80 (69) | 293 (73) | |
| - FIGO IV, n (%) | 100 (19) | 29 (25) | 71 (17) | |
| Residual tumor burden, n (%) | 0.699 | |||
| - present, n (%) | 133 (26) | 32 (27.5) | 101 (25) | |
| - not present, n (%) | 236 (45) | 52 (45) | 184 (46) | |
| - n.A., n (%) | 150 (29) | 32 (27.5) | 118 (29) | |
| OS, median (95% CI) | 41.2 (36.1–46.3) | 30.6 (21.0–40.1) | 43.0 (37.3–48.7) | 0.037 |
| PFS (n= 348 pts.), median (95% CI) | 19.3 (16.9–21.7) | 15.9 (12.3–19.6) | 19.3 (16.9–21.7) | 0.058 |
| OS (n = 519) | PFS (n = 348) | |||||
|---|---|---|---|---|---|---|
| Variable | HR | 95% CI | p | HR | 95% CI | p |
| Age > 60 | 1.16 | 0.96–1.51 | 0.233 | 1.02 | 0.78–1.33 | 0.912 |
| FIGO > II | 2.08 | 1.02–4.24 | 0.045 | 1.90 | 0.94–3.90 | 0.076 |
| Residual tumor | 2.10 | 1.61–2.72 | <0.001 | 1.64 | 1.22–2.20 | 0.001 |
| L-RGS2 | 1.24 | 0.84–1.51 | 0.440 | 1.24 | 0.90–1.71 | 0.193 |
| Gene | Localization | Spearman’s Correlation | p-Value |
|---|---|---|---|
| A | |||
| C5AR1 | 19q13.32 | 0.591 | 1.24 × 10−29 |
| RGS1 | 1q31.2 | 0.590 | 1.44 × 10−29 |
| NR4A3 | 9q22 | 0.587 | 3.64 × 10−29 |
| GPR183 | 13q32.3 | 0.584 | 8.27 × 10−29 |
| DUSP1 | 5q35.1 | 0.576 | 6.98 × 10−28 |
| OSM | 22q12.2 | 0.574 | 1.24 × 10−27 |
| IL6 | 7p15.3 | 0.564 | 1.28 × 10−26 |
| GEM | 8q22.1 | 0.563 | 1.55 × 10−26 |
| ADAMTS4 | 1q23.3 | 0.550 | 4.03 × 10−25 |
| C11ORF96 | 11p11.2 | 0.547 | 7.71 × 10−25 |
| SERPINE1 | 7q22.1 | 0.547 | 7.82 × 10−25 |
| CCN1 | 1p22.3 | 0.538 | 7.01 × 10−24 |
| GLIPR1 | 12q21.2 | 0.521 | 3.10 × 10−22 |
| PDE4B | 1p31.3 | 0.520 | 3.79 × 10−22 |
| SLC2A3 | 12p13.31 | 0.514 | 1.38 × 10−21 |
| B | |||
| ZNF3 | 7q22.1 | −0.329 | 5.15 × 10−9 |
| C9ORF163 | 9q34.3 | −0.315 | 2.40 × 10−8 |
| ZBTB3 | 11q12.3 | −0.309 | 4.83 × 10−8 |
| ZSCAN25 | 7q22.1 | −0.301 | 1.11 × 10−7 |
| MARVELD2 | 5q13.2 | −0.300 | 1.15 × 10−7 |
| NPAS3 | 14q13.1 | −0.292 | 2.63 × 10−7 |
| RSPH1 | 21q22.3 | −0.284 | 5.88 × 10−7 |
| SLC52A3 | 20p13 | −0.283 | 6.28 × 10−7 |
| DYDC1 | 10q23.1 | −0.280 | 8.02 × 10−7 |
| KLLN | 10q23 | −0.278 | 9.90 × 10−7 |
| PGAP2 | 11p15.4 | −0.276 | 1.12 × 10−3 |
| SOX17 | 8q11.23 | −0.274 | 1.48 × 10−3 |
| SRGAP3 | 3p25.3 | −0.273 | 1.57 × 10−3 |
| TUT1 | 11q12.3 | −0.269 | 2.18 × 10−3 |
| ACP6 | 1q21.2 | −0.269 | 2.19 × 10−3 |
| C | |||
| VIM | 10p13 | 0.293 | 2.47 × 10−7 |
| ZEB2 | 2q22.3 | 0.424 | 1.55 × 10−14 |
| ZEB1 | 10p11.22 | 0.409 | 1.59 × 10−13 |
| FN1 | 2q35 | 0.402 | 4.54 × 10−13 |
| SNAI2 | 8q11.21 | 0.343 | 1.05 × 10−9 |
| TWIST1 | 7p21.1 | 0.389 | 2.65 × 10−12 |
| SNAI1 | 20q13.13 | 0.427 | 1.06 × 10−14 |
| CDH2 | 18q12.1 | −0.019 | 0.749 |
| ERBB2 | 17q12 | −0.055 | 0.343 |
| DDR1 | 6p21.33 | −0.090 | 0.118 |
| ERBB3 | 12q13.2 | −0.153 | 7.92 × 10−3 |
| CDH1 | 16q22.1 | −0.104 | 0.072 |
| D | |||
| HDAC6 | Xp11.23 | −0.142 | 0.014 |
| HDAC11 | 3p25.1 | −0.135 | 0.019 |
| HDAC7 | 12q13.11 | −0.135 | 0.020 |
| HDAC4 | 2q37.3 | −0.111 | 0.054 |
| HDAC1 | 1p35.2-p35.1 | −0.083 | 0.154 |
| HDAC3 | 5q31.3 | −0.078 | 0.179 |
| HDAC5 | 17q21.31 | 0.077 | 0.186 |
| HDAC10 | 22q13.33 | 0.054 | 0.355 |
| HDAC2 | 6q21 | 0.051 | 0.379 |
| HDAC9 | 7p21.1 | −0.015 | 0.796 |
| HDAC8 | Xq13.1 | −0.007 | 0.900 |
| DNMT1 | 19p13.2 | −0.010 | 0.085 |
| DNMT3A | 2p23.3 | 0.048 | 0.410 |
| DNMT3B | 20q11.21 | −0.028 | 0.625 |
| E | |||
| EIF2B3 | 1p34.1 | 0.137 | 0.018 |
| VEGFA | 6p21.1 | 0.109 | 0.061 |
| HIF1A | 14q23.2 | 0.039 | 0.503 |
| PLCG2 | 16q24.1 | 0.081 | 0.164 |
| PLCG1 | 20q12 | 9.46 × 10−3 | 0.870 |
| ITPR1 | 3p26.1 | 0.112 | 0.053 |
| ITPR3 | 6p21.31 | −0.085 | 0.142 |
| ITPR2 | 12p11.23 | 0.049 | 0.398 |
| ADCY1 | 7p12.3 | −0.028 | 0.634 |
| PRKCA | 17q24.2 | 0.116 | 0.148 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ihlow, J.; Monjé, N.; Hoffmann, I.; Bischoff, P.; Sinn, B.V.; Schmitt, W.D.; Kunze, C.A.; Darb-Esfahani, S.; Kulbe, H.; Braicu, E.I.; et al. Low Expression of RGS2 Promotes Poor Prognosis in High-Grade Serous Ovarian Cancer. Cancers 2022, 14, 4620. https://doi.org/10.3390/cancers14194620
Ihlow J, Monjé N, Hoffmann I, Bischoff P, Sinn BV, Schmitt WD, Kunze CA, Darb-Esfahani S, Kulbe H, Braicu EI, et al. Low Expression of RGS2 Promotes Poor Prognosis in High-Grade Serous Ovarian Cancer. Cancers. 2022; 14(19):4620. https://doi.org/10.3390/cancers14194620
Chicago/Turabian StyleIhlow, Jana, Nanna Monjé, Inga Hoffmann, Philip Bischoff, Bruno Valentin Sinn, Wolfgang Daniel Schmitt, Catarina Alisa Kunze, Sylvia Darb-Esfahani, Hagen Kulbe, Elena Ioana Braicu, and et al. 2022. "Low Expression of RGS2 Promotes Poor Prognosis in High-Grade Serous Ovarian Cancer" Cancers 14, no. 19: 4620. https://doi.org/10.3390/cancers14194620
APA StyleIhlow, J., Monjé, N., Hoffmann, I., Bischoff, P., Sinn, B. V., Schmitt, W. D., Kunze, C. A., Darb-Esfahani, S., Kulbe, H., Braicu, E. I., Sehouli, J., Denkert, C., Horst, D., & Taube, E. T. (2022). Low Expression of RGS2 Promotes Poor Prognosis in High-Grade Serous Ovarian Cancer. Cancers, 14(19), 4620. https://doi.org/10.3390/cancers14194620







