Feasibility of Leukemia-Derived Exosome Enrichment and Co-isolated dsDNA Sequencing in Acute Myeloid Leukemia Patients: A Proof of Concept for New Leukemia Biomarkers Detection
Abstract
Simple Summary
Abstract
1. Introduction
1.1. Extracellular Vesicles (EVs) and Liquid Biopsy
1.2. Acute Myeloid Leukemias (AMLs)
1.3. A proof of Concept for the Study of AML Markers by EVs
2. Materials and Methods
2.1. Patients
2.2. Blood Sampling and Plasma and Cells Isolation
2.3. Cellular DNA Extraction
2.4. Exosomal dsDNA Isolation
2.5. Exosomal dsDNA Amplification by Whole Genome Amplification (WGA)
2.6. Next Generation Sequencing (NGS) Analysis
2.7. Statistical Analysis
3. Results
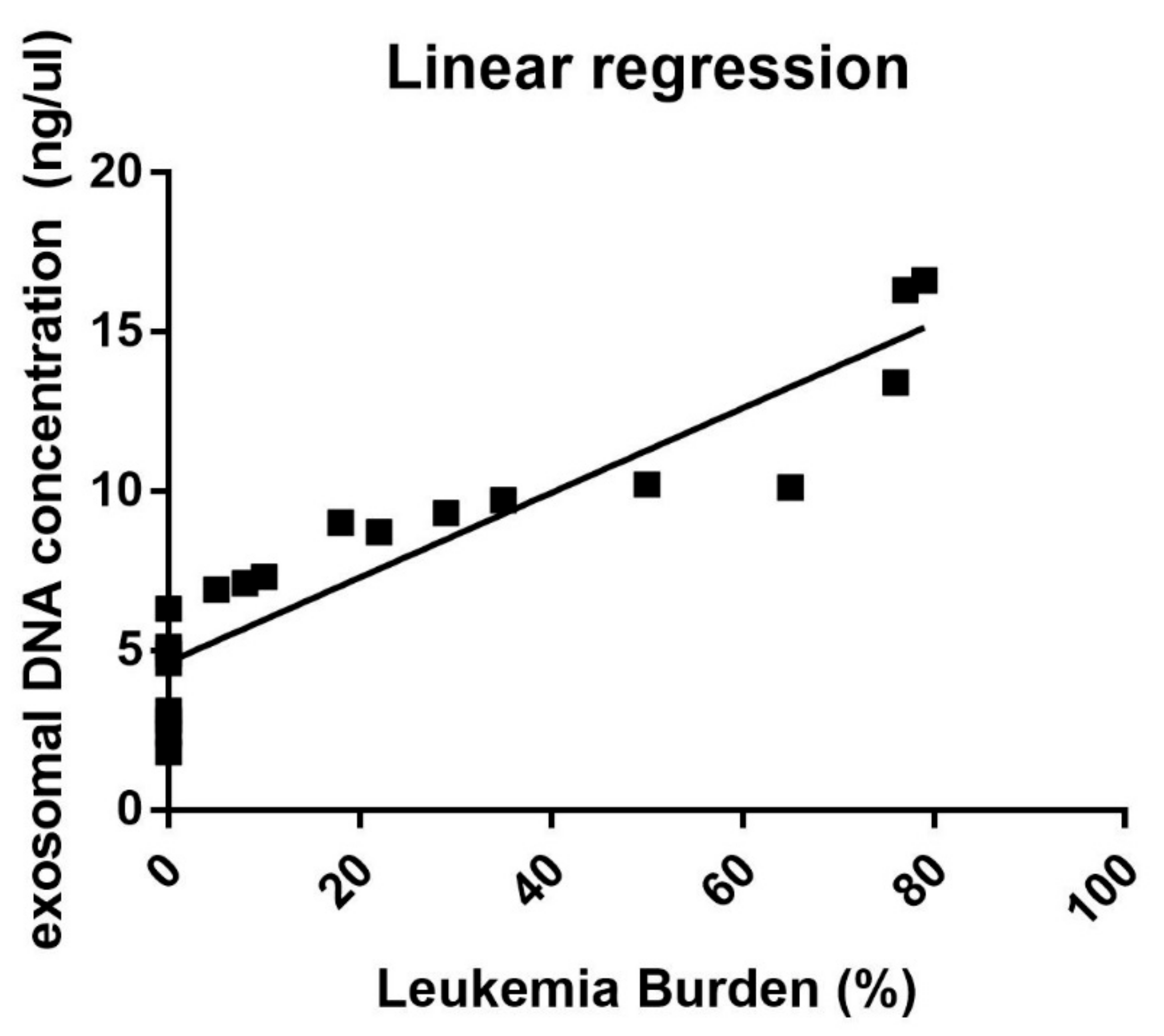
3.1. Correlation between Exosomal dsDNA and Leukemia Burden
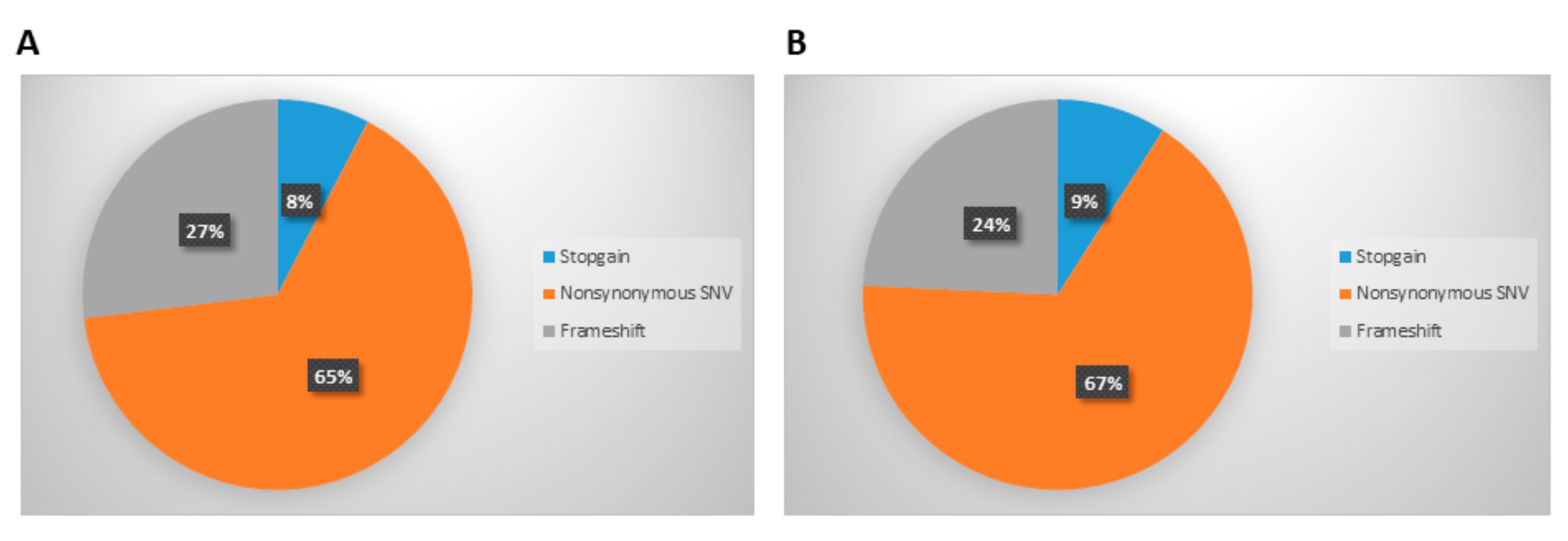
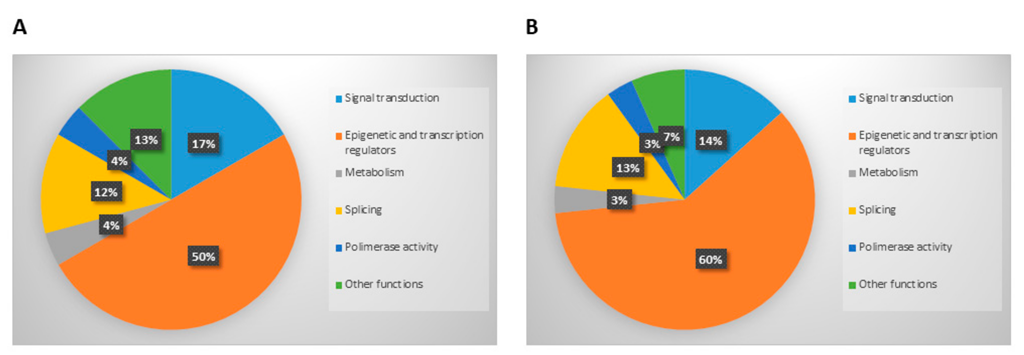
3.2. Analysis of Mutations Detected in Cellular and Exosomal dsDNA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Urabe, F.; Kosaka, N.; Ito, K.; Kimura, T.; Egawa, S.; Ochiya, T. Extracellular vesicles as biomarkers and therapeutic targets for cancer. Am. J. Physiol. Cell Physiol. 2020, 318, C29–C39. [Google Scholar] [CrossRef]
- Bernardi, S.; Balbi, C. Extracellular Vesicles: From Biomarkers to Therapeutic Tools. Biology 2020, 9, 258. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Gargiulo, E.; Paggetti, J.; Moussay, E. Hematological Malignancy-Derived Small Extracellular Vesicles and Tumor Microenvironment: The Art of Turning Foes into Friends. Cells 2019, 8, 511. [Google Scholar] [CrossRef]
- Galimberti, S.; Genuardi, E.; Mazziotta, F.; Iovino, L.; Morabito, F.; Grassi, S.; Ciabatti, E.; Guerrini, F.; Petrini, M. The minimal residual disease in non-hodgkin’s lymphomas: From the laboratory to the clinical practice. Front. Oncol. 2019, 9, 528. [Google Scholar] [CrossRef]
- Bernardi, S.; Farina, M. Exosomes and Extracellular Vesicles in Myeloid Neoplasia: The Multiple and Complex Roles Played by These “Magic Bullets”. Biology 2021, 10, 105. [Google Scholar] [CrossRef]
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef]
- Döhner, H.; Weisdorf, D.J.; Bloomfield, C.D. Acute Myeloid Leukemia. N. Engl. J. Med. 2015, 373, 1136–1152. [Google Scholar] [CrossRef]
- Farina, M.; Rossi, G.; Bellotti, D.; Marchina, E.; Gale, R.P. Is Having Clonal Cytogenetic Abnormalities the Same as Having Leukaemia? Acta Haematol. 2016, 135, 39–42. [Google Scholar] [CrossRef]
- Bernasconi, P.; Farina, M.; Boni, M.; Dambruoso, I.; Calvello, C. Therapeutically targeting SELF-reinforcing leukemic niches in acute myeloid leukemia: A worthy endeavor? Am. J. Hematol. 2016, 91, 507–517. [Google Scholar] [CrossRef]
- Simonetti, G.; Padella, A.; do Valle, I.F.; Fontana, M.C.; Fonzi, E.; Bruno, S.; Baldazzi, C.; Guadagnuolo, V.; Manfrini, M.; Ferrari, A.; et al. Aneuploid acute myeloid leukemia exhibits a signature of genomic alterations in the cell cycle and protein degradation machinery. Cancer 2019, 125, 712–725. [Google Scholar] [CrossRef]
- Churpek, J.E.; Pyrtel, K.; Kanchi, K.L.; Shao, J.; Koboldt, D.; Miller, C.A.; Shen, D.; Fulton, R.; O’Laughlin, M.; Fronick, C.; et al. Genomic analysis of germ line and somatic variants in familial myelodysplasia/acute myeloid leukemia. Blood 2015, 126, 2484–2490. [Google Scholar] [CrossRef]
- Tawana, K.; Drazer, M.W.; Churpek, J.E. Universal genetic testing for inherited susceptibility in children and adults with myelodysplastic syndrome and acute myeloid leukemia: Are we there yet? Leukemia 2018, 32, 1482–1492. [Google Scholar] [CrossRef]
- Bernardi, S.; Farina, M.; Zanaglio, C.; Cattina, F.; Polverelli, N.; Schieppati, F.; Re, F.; Foroni, C.; Malagola, M.; Dunbar, A.J.; et al. ETV6: A Candidate Gene for Predisposition to “Blend Pedigrees”? A Case Report from the NEXT-Famly Clinical Trial. Case Rep. Hematol. 2020, 2020, 2795656. [Google Scholar] [CrossRef]
- Goldin, L.R.; Kristinsson, S.Y.; Liang, X.S.; Derolf, A.R.; Landgren, O.; Björkholm, M. Familial aggregation of acute myeloid leukemia and myelodysplastic syndromes. J. Clin. Oncol. 2012, 30, 179–183. [Google Scholar] [CrossRef]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef]
- Borlenghi, E.; Cattaneo, C.; Cerqui, E.; Archetti, S.; Bertoli, D.; Bellotti, D.; Gramegna, D.; Soverini, G.; Oberti, M.; Schieppati, F.; et al. Postremission therapy with repeated courses of high-dose cytarabine, idarubicin, and limited autologous stem cell support achieves a very good long-term outcome in European leukemia net favorable and intermediate-risk acute myeloid leukemia. Hematol. Oncol. 2020, 38, 754–762. [Google Scholar] [CrossRef]
- Malagola, M.; Polverelli, N.; Cancelli, V.; Morello, E.; Turra, A.; Borlenghi, E.; Cattina, F.; Rambaldi, B.; Bernardi, S.; Zanaglio, C.; et al. Biological versus Clinical Risk Factors in Acute Myeloid Leukemia: Is There a Winner? Case Rep. Hematol. 2019, 2019, 3914828. [Google Scholar] [CrossRef]
- Malagola, M.; Skert, C.; Ruggeri, G.; Turra, A.; Ribolla, R.; Cancelli, V.; Cattina, F.; Alghisi, E.; Bernardi, S.; Perucca, S.; et al. Peripheral blood WT1 expression predicts relapse in AML patients undergoing allogeneic stem cell transplantation. BioMed Res. Int. 2014, 2014, 123079. [Google Scholar] [CrossRef]
- Malagola, M.; Skert, C.; Borlenghi, E.; Chiarini, M.; Cattaneo, C.; Morello, E.; Cancelli, V.; Cattina, F.; Cerqui, E.; Pagani, C.; et al. Postremission sequential monitoring of minimal residual disease by WT1 Q-PCR and multiparametric flow cytometry assessment predicts relapse and may help to address risk-adapted therapy in acute myeloid leukemia patients. Cancer Med. 2016, 5, 265–274. [Google Scholar] [CrossRef]
- Jeyakumar, D.; O’Brien, S. Minimal Residual Disease in Acute Myeloid Leukemia. JAMA Oncol. 2020, 6, 1899–1900. [Google Scholar] [CrossRef]
- Richard-Carpentier, G.; DiNardo, C.D. Single-agent and combination biologics in acute myeloid leukemia. Hematology 2019, 2019, 548–556. [Google Scholar] [CrossRef]
- Walker, A.R. How to approach shared decision making when determining consolidation, maintenance therapy, and transplantation in acute myeloid leukemia. Hematology 2020, 2020, 51–56. [Google Scholar] [CrossRef]
- Kantarjian, H. Acute myeloid leukemia-Major progress over four decades and glimpses into the future. Am. J. Hematol. 2016, 91, 131–145. [Google Scholar] [CrossRef]
- Bernardi, S.; Zanaglio, C.; Farina, M.; Polverelli, N.; Malagola, M.; Russo, D. dsDNA from extracellular vesicles (EVs) in adult AML. Ann. Hematol. 2021, 100, 1355–1356. [Google Scholar] [CrossRef]
- Rücker, F.G.; Agrawal, M.; Corbacioglu, A.; Weber, D.; Kapp-Schwoerer, S.; Gaidzik, V.I.; Jahn, N.; Schroeder, T.; Wattad, M.; Lübbert, M.; et al. Measurable residual disease monitoring in acute myeloid leukemia with t(8;21)(q22;q22.1): Results from the AML Study Group. Blood 2019, 134, 1608–1618. [Google Scholar] [CrossRef]
- Zocco, D.; Bernardi, S.; Novelli, M.; Astrua, C.; Fava, P.; Zarovni, N.; Carpi, F.M.; Bianciardi, L.; Malavenda, O.; Quaglino, P.; et al. Isolation of extracellular vesicles improves the detection of mutant DNA from plasma of metastatic melanoma patients. Sci. Rep. 2020, 10, 15745. [Google Scholar] [CrossRef]
- Liu, H.E.; Triboulet, M.; Zia, A.; Vuppalapaty, M.; Kidess-Sigal, E.; Coller, J.; Natu, V.S.; Shokoohi, V.; Che, J.; Renier, C.; et al. Workflow optimization of whole genome amplification and targeted panel sequencing for CTC mutation detection. NPJ Genomic Med. 2017, 2, 34. [Google Scholar] [CrossRef]
- Yang, H.; Wang, K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat. Protoc. 2015, 10, 1556–1566. [Google Scholar] [CrossRef]
- Colosini, A.; Bernardi, S.; Foroni, C.; Pasinetti, N.; Guerini, A.E.; Russo, D.; Bresciani, R.; Tomasi, C.; Magrini, S.M.; Bardoscia, L.; et al. Stratification of Oligometastatic Prostate Cancer Patients by Liquid Biopsy: Clinical Insights from a Pilot Study. Biomedicines 2022, 10, 1321. [Google Scholar] [CrossRef]
- Mishra, P.; Pandey, C.M.; Singh, U.; Gupta, A.; Sahu, C.; Keshri, A. Descriptive Statistics and Normality Tests for Statistical Data. Ann. Card. Anaesth. 2019, 22, 67. [Google Scholar] [CrossRef] [PubMed]
- Kontopoulou, E.; Strachan, S.; Reinhardt, K.; Kunz, F.; Walter, C.; Walkenfort, B.; Jastrow, H.; Hasenberg, M.; Giebel, B.; von Neuhoff, N.; et al. Evaluation of dsDNA from extracellular vesicles (EVs) in pediatric AML diagnostics. Ann. Hematol. 2020, 99, 459–475. [Google Scholar] [CrossRef] [PubMed]
- Taylor, C.; Chacko, S.; Davey, M.; Lacroix, J.; Macpherson, A.; Finn, N.; Wajnberg, G.; Ghosh, A.; Crapoulet, N.; Lewis, S.M.; et al. Peptide-affinity precipitation of extracellular vesicles and cell-free dna improves sequencing performance for the detection of pathogenic mutations in lung cancer patient plasma. Int. J. Mol. Sci. 2020, 21, 9083. [Google Scholar] [CrossRef] [PubMed]
- Castillo, J.; Bernard, V.; San Lucas, F.A.; Allenson, K.; Capello, M.; Kim, D.U.; Gascoyne, P.; Mulu, F.C.; Stephens, B.M.; Huang, J.; et al. Surfaceome profiling enables isolation of cancerspecific exosomal cargo in liquid biopsies from pancreatic cancer patients. Ann. Oncol. 2018, 29, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Möhrmann, L.; Huang, H.J.; Hong, D.S.; Tsimberidou, A.M.; Fu, S.; Piha-Paul, S.A.; Subbiah, V.; Karp, D.D.; Naing, A.; Krug, A.; et al. Liquid biopsies using plasma exosomal nucleic acids and plasma cell-free DNA compared with clinical outcomes of patients with advanced cancers. Clin. Cancer Res. 2018, 24, 181–188. [Google Scholar] [CrossRef]
- Kim, Y.; Shin, S.; Lee, K.A. Exosome-based detection of EGFR T790M in plasma and pleural fluid of prospectively enrolled non-small cell lung cancer patients after first-line tyrosine kinase inhibitor therapy. Cancer Cell Int. 2021, 21, 50. [Google Scholar] [CrossRef]
- Tang, S.; Cheng, J.; Yao, Y.; Lou, C.; Wang, L.; Huang, X.; Zhang, Y. Combination of Four Serum Exosomal MiRNAs as Novel Diagnostic Biomarkers for Early-Stage Gastric Cancer. Front. Genet. 2020, 11, 237. [Google Scholar] [CrossRef]
- Theodoraki, M.N.; Yerneni, S.; Gooding, W.E.; Ohr, J.; Clump, D.A.; Bauman, J.E.; Ferris, R.L.; Whiteside, T.L. Circulating exosomes measure responses to therapy in head and neck cancer patients treated with cetuximab, ipilimumab, and IMRT. Oncoimmunology 2019, 8, e1593805. [Google Scholar] [CrossRef]
- Zare, N.; Haghjooy Javanmard, S.; Mehrzad, V.; Eskandari, N.; Kefayat, A. Evaluation of exosomal miR-155, let-7g and let-7i levels as a potential noninvasive biomarker among refractory/relapsed patients, responsive patients and patients receiving R-CHOP. Leuk. Lymphoma 2019, 60, 1877–1889. [Google Scholar] [CrossRef]
- Van Eijndhoven, M.A.J.; Zijlstra, J.M.; Groenewegen, N.J.; Drees, E.E.E.; van Niele, S.; Baglio, S.R.; Koppers-Lalic, D.; van der Voorn, H.; Libregts, S.F.W.M.; Wauben, M.H.M.; et al. Plasma vesicle miRNAs for therapy response monitoring in Hodgkin lymphoma patients. JCI Insight 2016, 1, e89631. [Google Scholar] [CrossRef]
- De Luca, L.; D’Arena, G.; Simeon, V.; Trino, S.; Laurenzana, I.; Caivano, A.; La Rocca, F.; Villani, O.; Mansueto, G.; Deaglio, S.; et al. Characterization and prognostic relevance of circulating microvesicles in chronic lymphocytic leukemia. Leuk. Lymphoma 2017, 58, 1424–1432. [Google Scholar] [CrossRef]
- Boyiadzis, M.; Whiteside, T.L. Plasma-derived exosomes in acute myeloid leukemia for detection of minimal residual disease: Are we ready? Expert Rev. Mol. Diagn. 2016, 16, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Yoon, H.; Park, S.; Kim, J.S.; Ahn, Y.H.; Kwon, K.; Lee, D.; Kim, K.H. Urinary Exosomal and cell-free DNA Detects Somatic Mutation and Copy Number Alteration in Urothelial Carcinoma of Bladder. Sci. Rep. 2018, 8, 14707. [Google Scholar] [CrossRef] [PubMed]
- Longjohn, M.N.; Hudson, J.A.B.J.; Smith, N.C.; Rise, M.L.; Moorehead, P.C.; Christian, S.L. Deciphering the messages carried by extracellular vesicles in hematological malignancies. Blood Rev. 2021, 46, 100734. [Google Scholar] [CrossRef] [PubMed]
- Malagola, M.; Bernardi, S.; Polverelli, N.; Russo, D. Minimal Residual Disease Monitoring in Acute Myeloid Leukaemia: Are We Ready to Move from Bone Marrow to Peripheral Blood? Br. J. Hematol. 2020, 190, 135–136. [Google Scholar] [CrossRef]
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef]
- Miyamoto, K.N.; Bonatto, D. Circulating cells and exosomes in acute myelogenous leukemia and their role in disease progression and survival. Clin. Immunol. 2020, 217, 108489. [Google Scholar] [CrossRef]
- Rykova, E.; Sizikov, A.; Roggenbuck, D.; Antonenko, O.; Bryzgalov, L.; Morozkin, E.; Skvortsova, K.; Vlassov, V.; Laktionov, P.; Kozlov, V. Circulating DNA in rheumatoid arthritis: Pathological changes and association with clinically used serological markers. Arthritis Res. Ther. 2017, 19, 85. [Google Scholar] [CrossRef]
- Rossi, D.; Spina, V.; Bruscaggin, A.; Gaidano, G. Liquid biopsy in lymphoma. Haematologica 2019, 104, 648–652. [Google Scholar] [CrossRef]
- Galbiati, S.; Damin, F.; Brambilla, D.; Ferraro, L.; Soriani, N.; Ferretti, A.M.; Burgio, V.; Ronzoni, M.; Vago, R.; Sola, L.; et al. Small EVs-Associated DNA as Complementary Biomarker to Circulating Tumor DNA in Plasma of Metastatic Colorectal Cancer Patients. Pharmaceuticals 2021, 14, 128. [Google Scholar] [CrossRef]
- San Lucas, F.A.; Allenson, K.; Bernard, V.; Castillo, J.; Kim, D.U.; Ellis, K.; Ehli, E.A.; Davies, G.E.; Petersen, J.L.; Li, D.; et al. Minimally invasive genomic and transcriptomic profiling of visceral cancers by next-generation sequencing of circulating exosomes. Ann. Oncol. 2016, 27, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Lázaro-Ibáñez, E.; Sanz-Garcia, A.; Visakorpi, T.; Escobedo-Lucea, C.; Siljander, P.; Ayuso-Sacido, Á.; Yliperttula, M. Different gDNA content in the subpopulations of prostate cancer extracellular vesicles: Apoptotic bodies, microvesicles, and exosomes. Prostate 2014, 74, 1379–1390. [Google Scholar] [CrossRef] [PubMed]
- Ravandi, F.; Walter, R.B.; Freeman, S.D. Evaluating measurable residual disease in acute myeloid leukemia. Blood Adv. 2018, 2, 1356–1366. [Google Scholar] [CrossRef] [PubMed]
- Foroni, C.; Zarovni, N.; Bianciardi, L.; Bernardi, S.; Triggiani, L.; Zocco, D.; Venturella, M.; Chiesi, A.; Valcamonico, F.; Berruti, A. When less is more: Specific capture and analysis of tumor exosomes in plasma increases the sensitivity of liquid biopsy for comprehensive detection of multiple androgen receptor phenotypes in advanced prostate cancer patients. Biomedicines 2020, 8, 131. [Google Scholar] [CrossRef] [PubMed]
- Kunz, F.; Kontopoulou, E.; Reinhardt, K.; Soldierer, M.; Strachan, S.; Reinhardt, D.; Thakur, B.K. Detection of AML-specific mutations in pediatric patient plasma using extracellular vesicle–derived RNA. Ann. Hematol. 2019, 98, 595–603. [Google Scholar] [CrossRef]
- Bernardi, S.; Foroni, C.; Zanaglio, C.; Re, F.; Polverelli, N.; Turra, A.; Morello, E.; Farina, M.; Cattina, F.; Gandolfi, L.; et al. Feasibility of tumor-derived exosome enrichment in the onco-hematology leukemic model of chronic myeloid leukemia. Int. J. Mol. Med. 2019, 44, 2133–2144. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, S.; Malagola, M.; Polverelli, N.; Russo, D. Exosomes in Chronic Myeloid Leukemia: Are We Reading a New Reliable Message? Acta Haematol. 2020, 143, 509–510. [Google Scholar] [CrossRef]
- Skert, C.; Perucca, S.; Chiarini, M.; Giustini, V.; Sottini, A.; Ghidini, C.; Martellos, S.; Cattina, F.; Rambaldi, B.; Cancelli, V.; et al. Sequential monitoring of lymphocyte subsets and of T-and-B cell neogenesis indexes to identify time-varying immunologic profiles in relation to graft-versus-host disease and relapse after allogeneic stem cell transplantation. PLoS ONE 2017, 12, e0175337. [Google Scholar] [CrossRef]
- Kröger, N.M.; Deeg, J.H.; Olavarria, E.; Niederwieser, D.; Bacigalupo, A.; Barbui, T.; Rambaldi, A.; Mesa, R.; Tefferi, A.; Griesshammer, M.; et al. Indication and management of allogeneic stem cell transplantation in primary myelofibrosis: A consensus process by an EBMT/ELN international working group. Leukemia 2015, 29, 2126–2133. [Google Scholar] [CrossRef]
- Morello, E.; Malagola, M.; Bernardi, S.; Pristipino, C.; Russo, D. The role of allogeneic hematopoietic stem cell transplantation in the four P medicine era. Blood Res. 2018, 53, 3–6. [Google Scholar] [CrossRef]

| Case | Sex | Age | Disease Status at Enrollment | Known Mutations/Alteration | Disease Status at Sampling and Corresponding Specimens Number |
|---|---|---|---|---|---|
| 1 | M | 61 y | Relapse post-alloSCT | WT1 overexpression | Relapse post-alloSCT (1) |
| 2 | F | 47 y | Relapse | WT1 overexpression FLT3 D835Y NPM1 * | Relapse (2) |
| 3 | F | 30 y | CR post-alloSCT | WT1 overexpression DNMT3A | CR post-alloSCT (3) CR 3 months post-alloSCT (4) |
| 4 | M | 71 y | Diagnosis of AML secondary to MDS | WT1 overexpression TP53 | Diagnosis of AML secondary to MDS (5) |
| 5 | F | 44 y | Relapse | WT1 overexpression | Relapse (6) Relapse post- second alloSCT (7) CR post-therapy Ven-Aza (8) |
| 6 | F | 64 y | Relapse post-alloSCT | Complex Karyotype | Relapse post-alloSCT (9) |
| 7 | M | 44 y | Relapse post-alloSCT | WT1 overexpression | Relapse post-alloSCT (10) CR post- therapy (11) |
| 8 | M | 70 y | CR post relapse post-alloSCT | WT1 overexpression | CR post relapse post-alloSCT (12) |
| 9 | M | 41 y | AML MRD+ pre-alloSCT | FLT3-ITD | AML MRD+ pre-alloSCT (13) CR post-alloSCT (14) |
| 10 | F | 44 y | Pre-alloSCT | WT1 overexpression DNMT3A*IDH1 | Pre-alloSCT (15) CR post-alloSCT (16) Relapse 7 months post-alloSCT (17) Relapse 10 months post-alloSCT (18) Post therapy Azacitidine (19) Post therapy Azacitidine (20) |
| 11 | M | 77 y | Diagnosis of AML secondary to MDS | Diagnosis of AML secondary to MDS (21) CR post-alloSCT (22) Relapse post-alloSCT (23) | |
| 12 | M | 43 y | Pre-alloSCT | Pre-alloSCT (24) CR post-alloSCT (25) Relapse post-alloSCT (26) | |
| 13 | F | 49 y | Relapse 2 months post-alloSCT | FLT3-ITD | Relapse 2 months post-alloSCT (27) Relapse 4 months post-alloSCT under target therapy (28) Relapse 6 months post-alloSCT under target therapy (29) |
| 14 | F | 71 y | Pre-alloSCT | Pre-alloSCT (30) CR post-alloSCT (31) |
| Material | BM Cells | PB Cells | Exo dsDNA | Disease Status | |
|---|---|---|---|---|---|
| Specimen 1 (Case 1) | 4 | 3 | 4 | Relapse post-alloSCT | |
| Specimen 2 (Case 2) | 3 | 2 | 3 | Relapse | |
| Specimen 3 (Case 3) | 0 | 0 | 0 | CR post-alloSCT | |
| Specimen 4 (Case 3) | 1 | 0 | 1 | CR 3 months post-alloSCT | |
| Specimen 5 (Case 4) | 3 | 3 | 3 | Diagnosis of AML secondary to MDS | |
| Specimen 6 (Case 5) | 3 | 3 | 4 | Relapse | |
| Specimen 7 (Case 5) | 5 | 4 | 4 | Relapse post- second alloSCT | |
| Specimen 8 (Case 5) | 1 | 0 | 1 | CR post-therapy Ven-Aza | |
| Specimen 9 (Case 6) | 4 | 4 | 3 | Relapse post-alloSCT | |
| Specimen 10 (Case 7) | 3 | 2 | 3 | Relapse post-alloSCT | |
| Specimen 11 (Case 7) | 0 | 0 | 1 | CR post- therapy | |
| Specimen 12 (Case 8) | 0 | 0 | 0 | CR post relapse post-alloSCT | |
| Specimen 13 (Case 9) | 2 | 1 | 2 | AML MRD+ pre-alloSCT | |
| Specimen 14 (Case 9) | 0 | 0 | 0 | CR post-alloSCT | |
| Specimen 15 (Case 10) | 0 | 0 | 0 | Pre-alloSCT | |
| Specimen 16 (Case 10) | 0 | 0 | 3 | CR post-alloSCT | |
| Specimen 17 (Case 10) | 2 | 1 | 3 | Relapse 7 months post-alloSCT | |
| Specimen 18 (Case 10) | 2 | 2 | 3 | Relapse 10 months post-alloSCT | |
| Specimen 19 (Case 10) | 3 | 3 | 4 | Post therapy Azacitidine | |
| Specimen 20 (Case 10) | 3 | 3 | 3 | Post therapy Azacitidine | |
| Specimen 21 (Case 11) | 5 | 5 | 6 | Diagnosis of AML secondary to MDS | |
| Specimen 22 (Case 11) | 1 | 0 | 2 | CR post-alloSCT | |
| Specimen 23 (Case 11) | 3 | 1 | 3 | Relapse post-alloSCT | |
| Specimen 24 (Case 12) | 1 | 0 | 1 | Pre-alloSCT | |
| Specimen 25 (Case 12) | 1 | 0 | 1 | CR post-alloSCT | |
| Specimen 26 (Case 12) | 2 | 1 | 2 | Relapse post-alloSCT | |
| Specimen 27 (Case 13) | 3 | 3 | 4 | Relapse 2 months post-alloSCT | |
| Specimen 28 (Case 13) | 3 | 3 | 3 | Relapse 4 months post-alloSCT under target therapy | |
| Specimen 29 (Case 13) | 2 | 1 | 2 | Relapse 6 months post-alloSCT under target therapy | |
| Specimen 30 (Case 14) | 0 | 0 | 0 | Pre-alloSCT | |
| Specimen 31 (Case 14) | 0 | 0 | 0 | CR post-alloSCT | |
| Specimen 32 (Healthy Control) | n.d. | 0 | 0 | Healthy status | |
| Specimen 33 (Healthy Control) | n.d. | 0 | 0 | Healthy status | |
| Specimen 34 (Healthy Control) | n.d. | 0 | 0 | Healthy status |
| BM Cells | PB Cells | Exo dsDNA | |
|---|---|---|---|
| Case 1 | |||
| Specimen 1 | DNMT3A (c.327dupG; p.Q110Afs*13) ASXL1 (c.1927delG; p.G645Vfs*57) ASXL1 (c.1927dupG; p.G646Wfs*10) RUNX1 (c.667_668insA; p.R223Qfs*10) | DNMT3A (c.327dupG; p.Q110Afs*13) ASXL1 (c.1927delG; p.G645Vfs*57) ASXL1 (c.1927dupG; p.G646Wfs*10) | DNMT3A (c.327dupG; p.Q110Afs*13) ASXL1 (c.1927delG; p.G645Vfs*57) ASXL1 (c.1927dupG; p.G646Wfs*10) RUNX1 (c.667_668insA; p.R223Qfs*10) |
| Case 2 | |||
| Specimen 2 | DNMT3A (c.G2189A; p.R730H) FLT3 (c.G2503T; p.D835Y) RUNX1 (c.337dupT; p.Y113Lfs*3) | DNMT3A (c.G2189A; p.R730H) FLT3 (c.G2503T; p.D835Y) | DNMT3A (c.G2189A; p.R730H) FLT3 (c.G2503T; p.D835Y) RUNX1 (c.337dupT; p.Y113Lfs*3) |
| Case 3 | |||
| Specimen 3 | n.d. | n.d. | n.d. |
| Specimen 4 | ETV6 (c.C1198G; p.H400D) | n.d. | ETV6 (c.C1198G; p.H400D) |
| Case 4 | |||
| Specimen 5 | EZH2 (c.G1522A; p.G508R) ASXL1 (c.G4265C; p.S1422T) RUNX1 (c.G364C; p.A122P) | EZH2 (c.G1522A; p.G508R) ASXL1 (c.G4265C; p.S1422T) RUNX1 (c.G364C; p.A122P) | EZH2 (c.G1522A; p.G508R) ASXL1 (c.G4265C; p.S1422T) RUNX1 (c.G364C; p.A122P) TP53 (c.T194G; p.V65G) |
| Case 5 | |||
| Specimen 6 | TET2 (c.A1532C; p.H511P) BCOR (c.A1589T; p.K530M) TP53 (c.C472A; p.R158S) | TET2 (c.A1532C; p.H511P) BCOR (c.A1589T; p.K530M) TP53 (c.C472A; p.R158S) | EZH2 (c.T748G; p.C250G) TET2 (c.A1532C; p.H511P) BCOR (c.A1589T; p.K530M) TP53 (c.C472A; p.R158S) |
| Specimen 7 | EZH2 (c.T748G; p.C250G) TET2 (c.A1532C; p.H511P) BCOR (c.A1589T; p.K530M) TP53 (c C472A; p.R158S) TP53 (c.A1T; p.E2_M40del) | EZH2 (c.T748G; p.C250G) TET2 (c.A1532C; p.H511P) BCOR (c.A1589T; p.K530M) TP53 (c. C472A; p.R158S) | EZH2(c.T748G; p.C250G) TET2 (c.A1532C; p.H511P) BCOR (c.A1589T; p.K530M) TP53(c. C472A; p.R158S) |
| Specimen 8 | TET2 (c.A1532C; p.H511P) | n.d. | TET2 (c.A1532C; p.H511P) |
| Case 6 | |||
| Specimen 9 | TET2 (c.C4144A; p.H1382Y) SF3B1 (c.A856T; p.I286F) TP53 (c.T262A; p.S88T) RUNX1 (c.G364C; p.A122P) | TET2 (c. C4144A; p.H1382Y) SF3B1 (c.A856T; p.I286F) TP53 (c.T262A; p.S88T) | TET2 (c.C4144A; p.H1382Y) |
| Case 7 | |||
| Specimen 10 | SRP72 (c.A926C; p.E309A) ETV6 (:c.G1167C; p.M389I) ASXL1 (c.C1260T; p.A420A) | ETV6 (:c.G1167C; p.M389I) ASXL1 (c.C1260T; p.A420A) | SRP72 (c.A926C; p.E309A) ETV6 (:c.G1167C; p.M389I) ASXL1 (c.C1260T; p.A420A) |
| Specimen 11 | n.d. | n.d. | ASXL1 (c.C1260T; p.A420A) |
| Case 8 | |||
| Specimen 12 | n.d. | n.d. | n.d. |
| Case 9 | |||
| Specimen 13 | BCOR (c.G1306A; p.V436I) TP53 (c.C472A; p.R158S) | BCOR (c.G1306A; p.V436I) | BCOR (c.G1306A; p.V436I) TP53 (c.C472A; p.R158S) |
| Specimen 14 | n.d. | n.d. | n.d. |
| Case 10 | |||
| Specimen 15 | n.d. | n.d. | n.d. |
| Specimen 16 | n.d. | n.d. | TET2 (c.C4144T; p.H1382Y) ASXL1 (c.A4501T; p.S1501C) RUNX1 (c.G1183T; p.E395X) |
| Specimen 17 | ASXL1 (c.A4501T; p.S1501C) RUNX1 (c.G1183T; p.E395X) | ASXL1 (c.A4501T; p.S1501C) | ASXL1 (c.A4501T; p.S1501C) RUNX1 (c.G1183T; p.E395X) TET2 (c.C4144T; p.H1382Y) |
| Specimen 18 | ASXL1 (c.A4501T; p.S1501C) RUNX1 (c.G1183T; p.E395X) | ASXL1 (c.A4501T; p.S1501C) RUNX1 (c.G1183T; p.E395X) | ASXL1 (c.A4501T; p.S1501C) RUNX1 (c.G1183T; p.E395X) TET2 (c.C4144T; p.H1382Y) |
| Specimen 19 | TET2 (c.C4144T; p.H1382Y) ASXL1 (c.A4501T; p.S1501C) RUNX1 (c.G1183T; p.E395X) | TET2 (c.C4144T; p.H1382Y) ASXL1 (c.A4501T; p.S1501C) RUNX1 (c.G1183T; p.E395X) | TET2(c.C4144T; p.H1382Y) ASXL1 (c.A4501T; p.S1501C) RUNX1 (c.G1183T; p.E395X) IDH1 (c.A643T:p.I215F) |
| Specimen 20 | TET2 (c.C4144T; p.H1382Y) ASXL1 (c.A4501T; p.S1501C) RUNX1 (c.G1183T; p.E395X) | TET2 (c.C4144T; p.H1382Y) ASXL1 (c.A4501T; p.S1501C) RUNX1 (c.G1183T; p.E395X) | TET2 (c.C4144T; p.H1382Y) ASXL1 (c.A4501T; p.S1501C) RUNX1 (c.G1183T; p.E395X) |
| Case 11 | |||
| Specimen 21 | ASXL1 (c.1786dupG; p.R596P) ASXL1 (c.A2957G; p.N986S) EZH2 (c.T748G; p.C250G) IDH2 (c.435dupG; p.T146Dfs172) U2AF1 (c.A476G; p.E159G) | ASXL1 (c.1786dupG; p.R596P) ASXL1 (c.A2957G; p.N986S) EZH2 (c.T748G; p.C250G) IDH2 (c.435dupG; p.T146Dfs) U2AF1 (c.A476G; p.E159G) | ASXL1 (c.1786dupG; p.R596P) ASXL1 (c.A2957G; p.N986S) EZH2(c.T748G; p.C250) IDH2 (c.435dupG; p.T146Dfs) U2AF1 (c.A476G; p.E159G) TP53 (c.C523T; p.R175C) |
| Specimen 22 | EZH2 (c.T748G; p.C250G) | n.d. | EZH2 (c.T748G; p.C250G) TP53 (c.C523T; p.R175C) |
| Specimen 23 | EZH2 (c.T748G; p.C250G) ASXL1 (c.A2957G; p.N986S) TP53 (c.C523T; p.R175C) | EZH2 (c.T748G; p.C250G) | EZH2 (c.T748G; p.C250G) ASXL1 (c.A2957G; p.N986S) TP53 (c.C523T; p.R175C) |
| Case 12 | |||
| Specimen 24 | ETV6 (c.G1167C; p.M389I) | n.d. | ETV6 (c.G1167C; p.M389I) |
| Specimen 25 | ETV6 (c.G1167C; p.M389I) | n.d. | ETV6 (c.G1167C; p.M389I) |
| Specimen 26 | ETV6 (c.G1167C; p.M389I) CEBPalpha (c.564_566del; p.P189del) | ETV6 (c.G1167C; p.M389I) | ETV6 (c.G1167C; p.M389I) CEBPalpha (c.564_566del; p.P189del) |
| Case 13 | |||
| Specimen 27 | SRSF2 (c. 287dupC; p. P97Gfs27) KRAS (c.A9T; p. E3D)* DNMT3A (c.G2189A; p.R730H) | SRSF2 (c. 287dupC; p. P97Gfs27) KRAS (c.A9T; p. E3D)* DNMT3A (c.G2189A; p.R730H) | SRSF2 (c. 287dupC; p. P97Gfs27) KRAS (c.A9T; p. E3D)* DNMT3A (c.G2189A; p.R730H) BCOR (c.A1589T; p.K530M) |
| Specimen 28 | KRAS (c.A9T; p. E3D)* DNMT3A (c.G2189A; p.R730H) BCOR (c.A1589T; p.K530M) | KRAS (c.A9T; p. E3D)* DNMT3A (c.G2189A; p.R730H) SRSF2 (c. 287dupC; p. P97Gfs27) | KRAS (c.A9T; p. E3D)* DNMT3A (c.G2189A; p.R730H) BCOR (c.A1589T; p.K530M) |
| Specimen 29 | KRAS (c.A9T; p. E3D)* BCOR (c.A1589T; p.K530M) | KRAS (c.A9T; p. E3D)* | KRAS (c.A9T; p. E3D)* BCOR (c.A1589T; p.K530M) |
| Case 14 | |||
| Specimen 30 | n.d. | n.d. | n.d. |
| Specimen 31 | n.d. | n.d. | n.d. |
| Healthy Subject 1 | |||
| Specimen 32 | n.a. | n.d. | n.d. |
| Healthy Subject 2 | |||
| Specimen 33 | n.a. | n.d. | n.d. |
| Healthy Subject 3 | |||
| Specimen 34 | n.a. | n.d. | n.d. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bernardi, S.; Farina, M.; Bosio, K.; Di Lucanardo, A.; Leoni, A.; Re, F.; Polverelli, N.; Turra, A.; Morello, E.; Accorsi Buttini, E.; et al. Feasibility of Leukemia-Derived Exosome Enrichment and Co-isolated dsDNA Sequencing in Acute Myeloid Leukemia Patients: A Proof of Concept for New Leukemia Biomarkers Detection. Cancers 2022, 14, 4504. https://doi.org/10.3390/cancers14184504
Bernardi S, Farina M, Bosio K, Di Lucanardo A, Leoni A, Re F, Polverelli N, Turra A, Morello E, Accorsi Buttini E, et al. Feasibility of Leukemia-Derived Exosome Enrichment and Co-isolated dsDNA Sequencing in Acute Myeloid Leukemia Patients: A Proof of Concept for New Leukemia Biomarkers Detection. Cancers. 2022; 14(18):4504. https://doi.org/10.3390/cancers14184504
Chicago/Turabian StyleBernardi, Simona, Mirko Farina, Katia Bosio, Anna Di Lucanardo, Alessandro Leoni, Federica Re, Nicola Polverelli, Alessandro Turra, Enrico Morello, Eugenia Accorsi Buttini, and et al. 2022. "Feasibility of Leukemia-Derived Exosome Enrichment and Co-isolated dsDNA Sequencing in Acute Myeloid Leukemia Patients: A Proof of Concept for New Leukemia Biomarkers Detection" Cancers 14, no. 18: 4504. https://doi.org/10.3390/cancers14184504
APA StyleBernardi, S., Farina, M., Bosio, K., Di Lucanardo, A., Leoni, A., Re, F., Polverelli, N., Turra, A., Morello, E., Accorsi Buttini, E., Zollner, T., Bonvicini, C., Malagola, M., & Russo, D. (2022). Feasibility of Leukemia-Derived Exosome Enrichment and Co-isolated dsDNA Sequencing in Acute Myeloid Leukemia Patients: A Proof of Concept for New Leukemia Biomarkers Detection. Cancers, 14(18), 4504. https://doi.org/10.3390/cancers14184504









