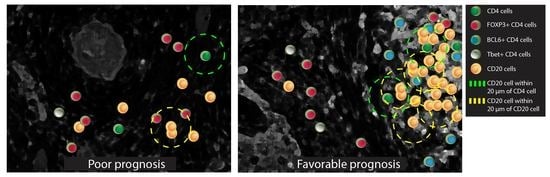
Intratumoral Niches of B Cells and Follicular Helper T Cells, and the Absence of Regulatory T Cells, Associate with Longer Survival in Early-Stage Oral Tongue Cancer Patients
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Cohort
2.2. Histopathological Analysis
2.3. Immunofluorescence Staining
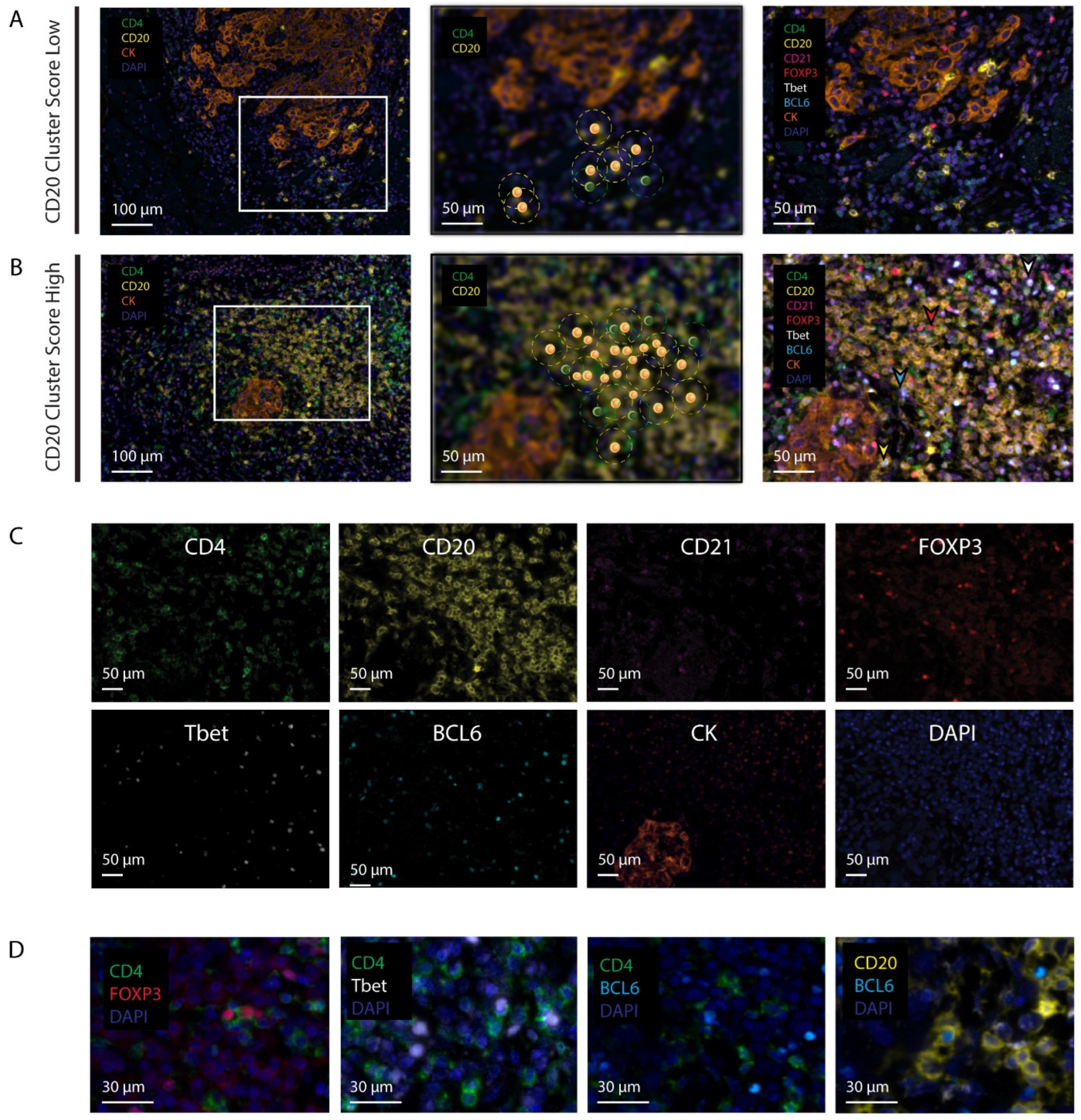
2.4. Digital Image Analysis
2.5. Assessment of Cellular Densities and Networks
2.6. The CD20 Cluster Score
2.7. Quantification of Tertiary Lymphoid Structures
2.8. Statistical Analysis
3. Results
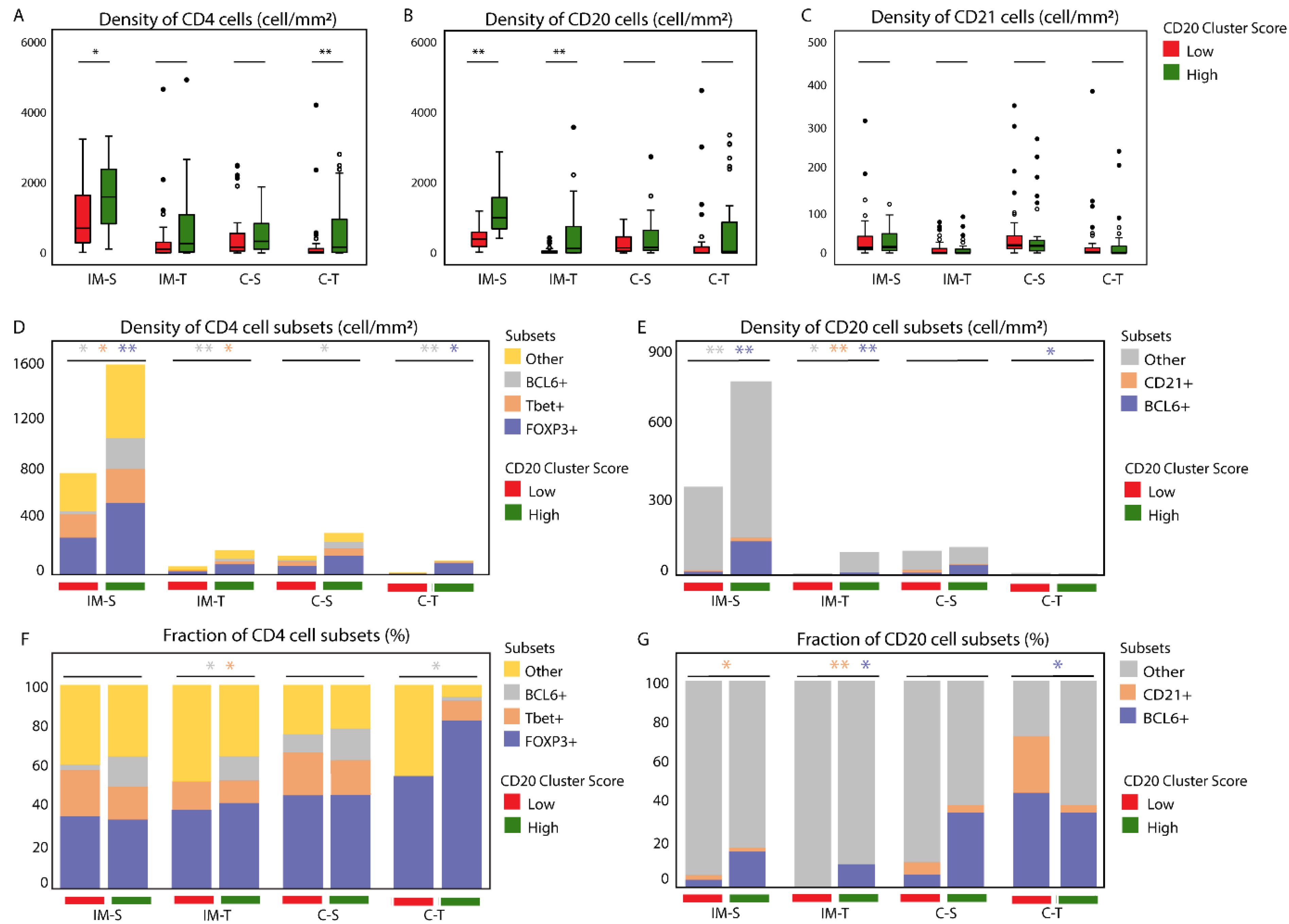
3.1. The CD20 Cluster Score Predicts Longer OS in Early OSCC and Associates with High Abundance of CD4 T Cells, CD20 B Cells, and TLSs, but Not with CD21 Cells in the IM-S Region
3.2. Patients with High CD20 Cluster Scores Demonstrate High Abundance of Stromal Treg, Th1, or Tfh, as well as GC B Cells
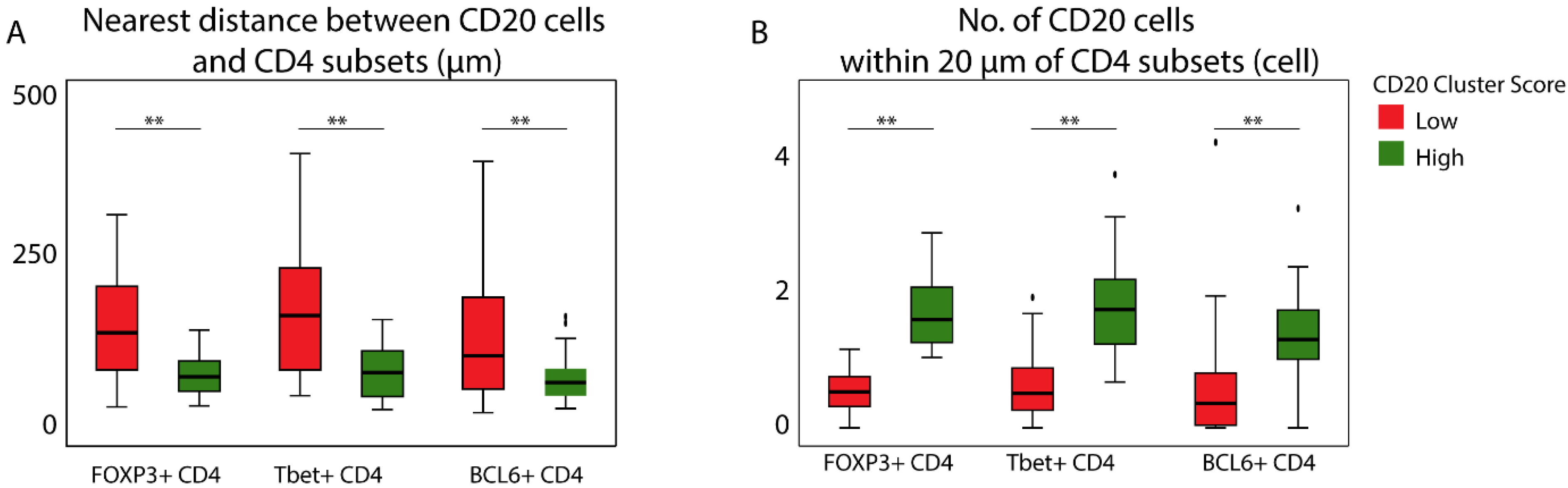
3.3. Patients with a High CD20 Cluster Score Demonstrate Co-Clustering of GC B Cells and Tfh Cells
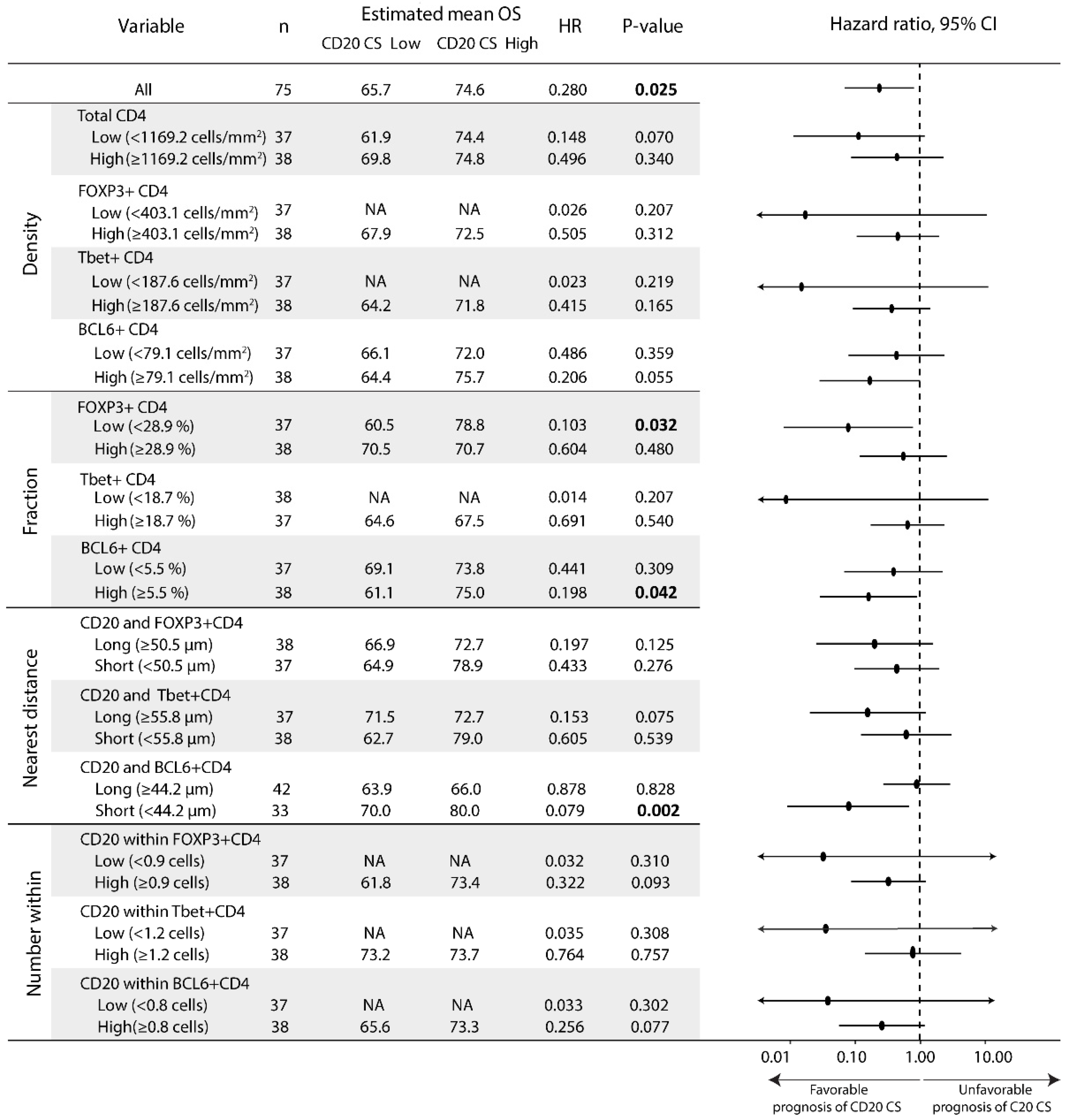
3.4. The Prognostic Value of the CD20 Cluster Score Is Impacted by Enriched Abundance of Tfh Relative to Treg and Short Distance between Tfh Cells and B Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- D’Cruz, A.K.; Vaish, R.; Kapre, N.; Dandekar, M.; Gupta, S.; Hawaldar, R.; Agarwal, J.P.; Pantvaidya, G.; Chaukar, D.; Deshmukh, A.; et al. Elective versus Therapeutic Neck Dissection in Node-Negative Oral Cancer. N. Engl. J. Med. 2015, 373, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Shim, S.J.; Cha, J.; Koom, W.S.; Kim, G.E.; Lee, C.G.; Choi, E.C.; Keum, K.C. Clinical outcomes for T1-2N0-1 oral tongue cancer patients underwent surgery with and without postoperative radiotherapy. Radiat. Oncol. 2010, 5, 43. [Google Scholar] [CrossRef] [PubMed]
- Tam, S.; Amit, M.; Zafereo, M.; Bell, D.; Weber, R.S. Depth of invasion as a predictor of nodal disease and survival in patients with oral tongue squamous cell carcinoma. Head Neck 2019, 41, 177–184. [Google Scholar] [CrossRef]
- Luryi, A.L.; Chen, M.M.; Mehra, S.; Roman, S.A.; Sosa, J.A.; Judson, B.L. Treatment Factors Associated with Survival in Early-Stage Oral Cavity Cancer: Analysis of 6830 Cases From the National Cancer Data Base. JAMA Otolaryngol. Head Neck Surg. 2015, 141, 593–598. [Google Scholar] [CrossRef]
- Mandal, R.; Senbabaoglu, Y.; Desrichard, A.; Havel, J.J.; Dalin, M.G.; Riaz, N.; Lee, K.W.; Ganly, I.; Hakimi, A.A.; Chan, T.A.; et al. The head and neck cancer immune landscape and its immunotherapeutic implications. JCI Insight 2016, 1, e89829. [Google Scholar] [CrossRef]
- Hammerman, P.S.; Hayes, D.N.; Grandis, J.R. Therapeutic insights from genomic studies of head and neck squamous cell carcinomas. Cancer Discov. 2015, 5, 239–244. [Google Scholar] [CrossRef]
- Borsetto, D.; Tomasoni, M.; Payne, K.; Polesel, J.; Deganello, A.; Bossi, P.; Tysome, J.R.; Masterson, L.; Tirelli, G.; Tofanelli, M.; et al. Prognostic Significance of CD4+ and CD8+ Tumor-Infiltrating Lymphocytes in Head and Neck Squamous Cell Carcinoma: A Meta-Analysis. Cancers 2021, 13, 781. [Google Scholar] [CrossRef]
- de Ruiter, E.J.; Ooft, M.L.; Devriese, L.A.; Willems, S.M. The prognostic role of tumor infiltrating T-lymphocytes in squamous cell carcinoma of the head and neck: A systematic review and meta-analysis. Oncoimmunology 2017, 6, e1356148. [Google Scholar] [CrossRef]
- Galon, J.; Costes, A.; Sanchez-Cabo, F.; Kirilovsky, A.; Mlecnik, B.; Lagorce-Pages, C.; Tosolini, M.; Camus, M.; Berger, A.; Wind, P.; et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006, 313, 1960–1964. [Google Scholar] [CrossRef]
- Tsujikawa, T.; Mitsuda, J.; Ogi, H.; Miyagawa-Hayashino, A.; Konishi, E.; Itoh, K.; Hirano, S. Prognostic significance of spatial immune profiles in human solid cancers. Cancer Sci. 2020, 111, 3426–3434. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Cantor, H. CD4 T-cell subsets and tumor immunity: The helpful and the not-so-helpful. Cancer Immunol. Res. 2014, 2, 91–98. [Google Scholar] [CrossRef]
- Hadler-Olsen, E.; Wirsing, A.M. Tissue-infiltrating immune cells as prognostic markers in oral squamous cell carcinoma: A systematic review and meta-analysis. Br. J. Cancer 2019, 120, 714–727. [Google Scholar] [CrossRef] [PubMed]
- Tay, R.E.; Richardson, E.K.; Toh, H.C. Revisiting the role of CD4(+) T cells in cancer immunotherapy-new insights into old paradigms. Cancer Gene Ther. 2021, 28, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lozano-Ruiz, B.; Yang, F.M.; Fan, D.D.; Shen, L.; Gonzalez-Navajas, J.M. The Multifaceted Role of Th1, Th9, and Th17 Cells in Immune Checkpoint Inhibition Therapy. Front. Immunol. 2021, 12, 625667. [Google Scholar] [CrossRef] [PubMed]
- Charoentong, P.; Finotello, F.; Angelova, M.; Mayer, C.; Efremova, M.; Rieder, D.; Hackl, H.; Trajanoski, Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017, 18, 248–262. [Google Scholar] [CrossRef]
- Yuen, G.J.; Demissie, E.; Pillai, S. B lymphocytes and cancer: A love-hate relationship. Trends Cancer 2016, 2, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Phanthunane, C.; Wijers, R.; de Herdt, M.; Langeveld, T.P.M.; Koljenovic, S.; Dasgupta, S.; Sleijfer, S.; Baatenburg de Jong, R.J.; Hardillo, J.; Balcioglu, H.E.; et al. B-cell clusters at the invasive margin associate with longer survival in early-stage oral-tongue cancer patients. Oncoimmunology 2021, 10, 1882743. [Google Scholar] [CrossRef]
- Petitprez, F.; de Reynies, A.; Keung, E.Z.; Chen, T.W.; Sun, C.M.; Calderaro, J.; Jeng, Y.M.; Hsiao, L.P.; Lacroix, L.; Bougouin, A.; et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature 2020, 577, 556–560. [Google Scholar] [CrossRef]
- Helmink, B.A.; Reddy, S.M.; Gao, J.; Zhang, S.; Basar, R.; Thakur, R.; Yizhak, K.; Sade-Feldman, M.; Blando, J.; Han, G.; et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature 2020, 577, 549–555. [Google Scholar] [CrossRef]
- Dieu-Nosjean, M.C.; Giraldo, N.A.; Kaplon, H.; Germain, C.; Fridman, W.H.; Sautes-Fridman, C. Tertiary lymphoid structures, drivers of the anti-tumor responses in human cancers. Immunol. Rev. 2016, 271, 260–275. [Google Scholar] [CrossRef] [PubMed]
- Cabrita, R.; Lauss, M.; Sanna, A.; Donia, M.; Skaarup Larsen, M.; Mitra, S.; Johansson, I.; Phung, B.; Harbst, K.; Vallon-Christersson, J.; et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature 2020, 577, 561–565. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.W.; Hill, D.G.; Jones, S.A. Understanding Immune Cells in Tertiary Lymphoid Organ Development: It Is All Starting to Come Together. Front. Immunol. 2016, 7, 401. [Google Scholar] [CrossRef] [PubMed]
- Pipi, E.; Nayar, S.; Gardner, D.H.; Colafrancesco, S.; Smith, C.; Barone, F. Tertiary Lymphoid Structures: Autoimmunity Goes Local. Front. Immunol. 2018, 9, 1952. [Google Scholar] [CrossRef]
- Noel, G.; Fontsa, M.L.; Garaud, S.; De Silva, P.; de Wind, A.; Van den Eynden, G.G.; Salgado, R.; Boisson, A.; Locy, H.; Thomas, N.; et al. Functional Th1-oriented T follicular helper cells that infiltrate human breast cancer promote effective adaptive immunity. J. Clin. Investig. 2021, 131, e139905. [Google Scholar] [CrossRef]
- Joshi, N.S.; Akama-Garren, E.H.; Lu, Y.; Lee, D.Y.; Chang, G.P.; Li, A.; DuPage, M.; Tammela, T.; Kerper, N.R.; Farago, A.F.; et al. Regulatory T Cells in Tumor-Associated Tertiary Lymphoid Structures Suppress Anti-tumor T Cell Responses. Immunity 2015, 43, 579–590. [Google Scholar] [CrossRef]
- Colbeck, E.J.; Ager, A.; Gallimore, A.; Jones, G.W. Tertiary Lymphoid Structures in Cancer: Drivers of Antitumor Immunity, Immunosuppression, or Bystander Sentinels in Disease? Front. Immunol. 2017, 8, 1830. [Google Scholar] [CrossRef]
- Wirsing, A.M.; Rikardsen, O.G.; Steigen, S.E.; Uhlin-Hansen, L.; Hadler-Olsen, E. Presence of tumour high-endothelial venules is an independent positive prognostic factor and stratifies patients with advanced-stage oral squamous cell carcinoma. Tumour Biol. 2016, 37, 2449–2459. [Google Scholar] [CrossRef]
- Wouters, M.C.A.; Nelson, B.H. Prognostic Significance of Tumor-Infiltrating B Cells and Plasma Cells in Human Cancer. Clin. Cancer Res. 2018, 24, 6125–6135. [Google Scholar] [CrossRef]
- Suarez-Sanchez, F.J.; Lequerica-Fernandez, P.; Rodrigo, J.P.; Hermida-Prado, F.; Suarez-Canto, J.; Rodriguez-Santamarta, T.; Dominguez-Iglesias, F.; Garcia-Pedrero, J.M.; de Vicente, J.C. Tumor-Infiltrating CD20(+) B Lymphocytes: Significance and Prognostic Implications in Oral Cancer Microenvironment. Cancers 2021, 13, 395. [Google Scholar] [CrossRef]
- Laumont, C.M.; Banville, A.C.; Gilardi, M.; Hollern, D.P.; Nelson, B.H. Tumour-infiltrating B cells: Immunological mechanisms, clinical impact and therapeutic opportunities. Nat. Rev. Cancer 2022, 22, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Pinto, D. B cells as antigen presenting cells. Cell Immunol. 2005, 238, 67–75. [Google Scholar] [CrossRef]
- Sharonov, G.V.; Serebrovskaya, E.O.; Yuzhakova, D.V.; Britanova, O.V.; Chudakov, D.M. B cells, plasma cells and antibody repertoires in the tumour microenvironment. Nat. Rev. Immunol. 2020, 20, 294–307. [Google Scholar] [CrossRef] [PubMed]
- Lund, F.E.; Randall, T.D. Effector and regulatory B cells: Modulators of CD4+ T cell immunity. Nat. Rev. Immunol. 2010, 10, 236–247. [Google Scholar] [CrossRef] [PubMed]
- Harris, D.P.; Goodrich, S.; Gerth, A.J.; Peng, S.L.; Lund, F.E. Regulation of IFN-gamma production by B effector 1 cells: Essential roles for T-bet and the IFN-gamma receptor. J. Immunol. 2005, 174, 6781–6790. [Google Scholar] [CrossRef]
- Lechner, A.; Schlosser, H.A.; Thelen, M.; Wennhold, K.; Rothschild, S.I.; Gilles, R.; Quaas, A.; Siefer, O.G.; Huebbers, C.U.; Cukuroglu, E.; et al. Tumor-associated B cells and humoral immune response in head and neck squamous cell carcinoma. Oncoimmunology 2019, 8, 1535293. [Google Scholar] [CrossRef]
- Harris, R.J.; Cheung, A.; Ng, J.C.F.; Laddach, R.; Chenoweth, A.M.; Crescioli, S.; Fittall, M.; Dominguez-Rodriguez, D.; Roberts, J.; Levi, D.; et al. Tumor-Infiltrating B Lymphocyte Profiling Identifies IgG-Biased, Clonally Expanded Prognostic Phenotypes in Triple-Negative Breast Cancer. Cancer Res. 2021, 81, 4290–4304. [Google Scholar] [CrossRef]
- Garaud, S.; Buisseret, L.; Solinas, C.; Gu-Trantien, C.; de Wind, A.; Van den Eynden, G.; Naveaux, C.; Lodewyckx, J.N.; Boisson, A.; Duvillier, H.; et al. Tumor infiltrating B-cells signal functional humoral immune responses in breast cancer. JCI Insight 2019, 5, e129641. [Google Scholar] [CrossRef]
- Qi, H. T follicular helper cells in space-time. Nat. Rev. Immunol. 2016, 16, 612–625. [Google Scholar] [CrossRef]
- Silina, K.; Soltermann, A.; Attar, F.M.; Casanova, R.; Uckeley, Z.M.; Thut, H.; Wandres, M.; Isajevs, S.; Cheng, P.; Curioni-Fontecedro, A.; et al. Germinal Centers Determine the Prognostic Relevance of Tertiary Lymphoid Structures and Are Impaired by Corticosteroids in Lung Squamous Cell Carcinoma. Cancer Res. 2018, 78, 1308–1320. [Google Scholar] [CrossRef]
- Thommen, D.S.; Koelzer, V.H.; Herzig, P.; Roller, A.; Trefny, M.; Dimeloe, S.; Kiialainen, A.; Hanhart, J.; Schill, C.; Hess, C.; et al. A transcriptionally and functionally distinct PD-1(+) CD8(+) T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat. Med. 2018, 24, 994–1004. [Google Scholar] [CrossRef] [PubMed]
- Crotty, S. T Follicular Helper Cell Biology: A Decade of Discovery and Diseases. Immunity 2019, 50, 1132–1148. [Google Scholar] [CrossRef]
- Hagn, M.; Jahrsdorfer, B. Why do human B cells secrete granzyme B? Insights into a novel B-cell differentiation pathway. Oncoimmunology 2012, 1, 1368–1375. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; He, Y.; Cao, Y.; Wang, Y.; Jia, A.; Yang, Q.; Li, W.; Bi, Y.; Liu, G. Functional differentiation and regulation of follicular T helper cells in inflammation and autoimmunity. Immunology 2021, 163, 19–32. [Google Scholar] [CrossRef]
- Song, W.; Craft, J. T follicular helper cell heterogeneity: Time, space, and function. Immunol. Rev. 2019, 288, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Wing, J.B.; Tanaka, A.; Sakaguchi, S. Human FOXP3. Immunity 2019, 50, 302–316. [Google Scholar] [CrossRef]
- León, B.; Bradley, J.E.; Lund, F.E.; Randall, T.D.; Ballesteros-Tato, A. FoxP3+ regulatory T cells promote influenza-specific Tfh responses by controlling IL-2 availability. Nat. Commun. 2014, 5, 3495. [Google Scholar] [CrossRef]
- Germain, C.; Devi-Marulkar, P.; Knockaert, S.; Biton, J.; Kaplon, H.; Letaïef, L.; Goc, J.; Seguin-Givelet, A.; Gossot, D.; Girard, N.; et al. Tertiary Lymphoid Structure-B Cells Narrow Regulatory T Cells Impact in Lung Cancer Patients. Front. Immunol. 2021, 12, 626776. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, Z.; Wang, H.; Ba, X.; Shen, P.; Lin, W.; Wang, Y.; Qin, K.; Tu, S. Follicular regulatory T cells: A novel target for immunotherapy? Clin. Transl. Immunol. 2020, 9, e1106. [Google Scholar] [CrossRef]
- Wirsing, A.M.; Ervik, I.K.; Seppola, M.; Uhlin-Hansen, L.; Steigen, S.E.; Hadler-Olsen, E. Presence of high-endothelial venules correlates with a favorable immune microenvironment in oral squamous cell carcinoma. Mod. Pathol. 2018, 31, 910–922. [Google Scholar] [CrossRef]
- Germain, C.; Gnjatic, S.; Dieu-Nosjean, M.C. Tertiary Lymphoid Structure-Associated B Cells are Key Players in Anti-Tumor Immunity. Front. Immunol. 2015, 6, 67. [Google Scholar] [CrossRef] [PubMed]
- Engelhard, V.H.; Rodriguez, A.B.; Mauldin, I.S.; Woods, A.N.; Peske, J.D.; Slingluff, C.L., Jr. Immune Cell Infiltration and Tertiary Lymphoid Structures as Determinants of Antitumor Immunity. J. Immunol. 2018, 200, 432–442. [Google Scholar] [CrossRef]
- Wirsing, A.M.; Rikardsen, O.G.; Steigen, S.E.; Uhlin-Hansen, L.; Hadler-Olsen, E. Characterisation and prognostic value of tertiary lymphoid structures in oral squamous cell carcinoma. BMC Clin. Pathol. 2014, 14, 38. [Google Scholar] [CrossRef] [PubMed]
- Cillo, A.R.; Kurten, C.H.L.; Tabib, T.; Qi, Z.; Onkar, S.; Wang, T.; Liu, A.; Duvvuri, U.; Kim, S.; Soose, R.J.; et al. Immune Landscape of Viral- and Carcinogen-Driven Head and Neck Cancer. Immunity 2020, 52, 183–199.e189. [Google Scholar] [CrossRef]
- Li, K.; Guo, Q.; Zhang, X.; Dong, X.; Liu, W.; Zhang, A.; Li, Y.; Yan, J.; Jia, G.; Zheng, Z.; et al. Oral cancer-associated tertiary lymphoid structures: Gene expression profile and prognostic value. Clin. Exp. Immunol. 2020, 199, 172–181. [Google Scholar] [CrossRef]
- Hetta, H.F.; Elkady, A.; Yahia, R.; Meshall, A.K.; Saad, M.M.; Mekky, M.A.; Al-Kadmy, I.M.S. T follicular helper and T follicular regulatory cells in colorectal cancer: A complex interplay. J. Immunol. Methods 2020, 480, 112753. [Google Scholar] [CrossRef] [PubMed]
- Cardenas, M.G.; Oswald, E.; Yu, W.; Xue, F.; MacKerell, A.D.; Melnick, A.M. The Expanding Role of the BCL6 Oncoprotein as a Cancer Therapeutic Target. Clin. Cancer Res. 2017, 23, 885–893. [Google Scholar] [CrossRef]
- Frohwitter, G.; Buerger, H.; Korsching, E.; van Diest, P.J.; Kleinheinz, J.; Fillies, T. Site-specific gene expression patterns in oral cancer. Head Face Med. 2017, 13, 6. [Google Scholar] [CrossRef]
- Baitsch, L.; Baumgaertner, P.; Devêvre, E.; Raghav, S.K.; Legat, A.; Barba, L.; Wieckowski, S.; Bouzourene, H.; Deplancke, B.; Romero, P.; et al. Exhaustion of tumor-specific CD8⁺ T cells in metastases from melanoma patients. J Clin Investig. 2011, 121, 2350–2360. [Google Scholar] [CrossRef]
- Li, H.; van der Leun, A.M.; Yofe, I.; Lubling, Y.; Gelbard-Solodkin, D.; van Akkooi, A.C.J.; van den Braber, M.; Rozeman, E.A.; Haanen, J.B.A.G.; Blank, C.U.; et al. Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. Cell 2019, 176, 775–789.e718. [Google Scholar] [CrossRef]
- Zheng, C.; Zheng, L.; Yoo, J.K.; Guo, H.; Zhang, Y.; Guo, X.; Kang, B.; Hu, R.; Huang, J.Y.; Zhang, Q.; et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell 2017, 169, 1342–1356.e16. [Google Scholar] [CrossRef] [PubMed]
- Workel, H.H.; Lubbers, J.M.; Arnold, R.; Prins, T.M.; van der Vlies, P.; de Lange, K.; Bosse, T.; van Gool, I.C.; Eggink, F.A.; Wouters, M.C.A.; et al. A Transcriptionally Distinct CXCL13. Cancer Immunol. Res. 2019, 7, 784–796. [Google Scholar] [CrossRef] [PubMed]
- Im, S.J.; Hashimoto, M.; Gerner, M.Y.; Lee, J.; Kissick, H.T.; Burger, M.C.; Shan, Q.; Hale, J.S.; Nasti, T.H.; Sharpe, A.H.; et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 2016, 537, 417–421. [Google Scholar] [CrossRef]
- He, R.; Hou, S.; Liu, C.; Zhang, A.; Bai, Q.; Han, M.; Yang, Y.; Wei, G.; Shen, T.; Yang, X.; et al. Follicular CXCR5-expressing CD8(+) T cells curtail chronic viral infection. Nature 2016, 537, 412–428. [Google Scholar] [CrossRef] [PubMed]

| Variables | CD20 Cluster Score Low (n = 43) | CD20 Cluster Score High (n = 32) | p-Value |
|---|---|---|---|
| Median (Interquartile range) | |||
| Age (years) | 63 (50–71) | 62 (51–73) | 0.945 |
| Number (%) | |||
| Gender | 0.481 | ||
| Male | 26 (60%) | 16 (50%) | |
| Female | 17 (40%) | 16 (50%) | |
| Tumor (T) stage | 0.420 | ||
| pT1 | 19 (44%) | 19 (59%) | |
| pT2 | 20 (47%) | 10 (31%) | |
| pT3 | 4 (9%) | 3 (10%) | |
| Nodal (N) stage | 0.487 | ||
| pN0 | 39 (91%) | 30 (94%) | |
| pN1 | 4 (9%) | 2 (6%) | |
| Stage | 0.449 | ||
| pStage 1 | 18 (42%) | 18 (56%) | |
| pStage2 | 17 (40%) | 9 (28%) | |
| pStage3 | 8 (18%) | 5 (16%) | |
| Differentiation grade | 0.159 | ||
| Well differentiated | 6 (14%) | 10 (31%) | |
| Moderately differentiated | 32 (74%) | 18 (56%) | |
| Poorly differentiated | 5 (12%) | 4 (13%) | |
| Lymphovascular invasion | 0.991 | ||
| Absence | 39 (91%) | 29 (91%) | |
| Presence | 4 (9%) | 3 (9%) | |
| 5-year survival | 0.034 | ||
| Alive | 28 (65%) | 28 (88%) | |
| Dead | 15 (35%) | 4 (12%) | |
| Locoregional recurrence | 0.568 | ||
| No recurrence | 37 (86%) | 28 (88%) | |
| Recurrence | 6 (14%) | 4 (12%) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phanthunane, C.; Wijers, R.; De Herdt, M.J.; Koljenović, S.; Sleijfer, S.; Baatenburg de Jong, R.J.; Hardillo, J.A.U.; Debets, R.; Balcioglu, H.E. Intratumoral Niches of B Cells and Follicular Helper T Cells, and the Absence of Regulatory T Cells, Associate with Longer Survival in Early-Stage Oral Tongue Cancer Patients. Cancers 2022, 14, 4298. https://doi.org/10.3390/cancers14174298
Phanthunane C, Wijers R, De Herdt MJ, Koljenović S, Sleijfer S, Baatenburg de Jong RJ, Hardillo JAU, Debets R, Balcioglu HE. Intratumoral Niches of B Cells and Follicular Helper T Cells, and the Absence of Regulatory T Cells, Associate with Longer Survival in Early-Stage Oral Tongue Cancer Patients. Cancers. 2022; 14(17):4298. https://doi.org/10.3390/cancers14174298
Chicago/Turabian StylePhanthunane, Chumut, Rebecca Wijers, Maria J. De Herdt, Senada Koljenović, Stefan Sleijfer, Robert J. Baatenburg de Jong, José Angelito U. Hardillo, Reno Debets, and Hayri E. Balcioglu. 2022. "Intratumoral Niches of B Cells and Follicular Helper T Cells, and the Absence of Regulatory T Cells, Associate with Longer Survival in Early-Stage Oral Tongue Cancer Patients" Cancers 14, no. 17: 4298. https://doi.org/10.3390/cancers14174298
APA StylePhanthunane, C., Wijers, R., De Herdt, M. J., Koljenović, S., Sleijfer, S., Baatenburg de Jong, R. J., Hardillo, J. A. U., Debets, R., & Balcioglu, H. E. (2022). Intratumoral Niches of B Cells and Follicular Helper T Cells, and the Absence of Regulatory T Cells, Associate with Longer Survival in Early-Stage Oral Tongue Cancer Patients. Cancers, 14(17), 4298. https://doi.org/10.3390/cancers14174298