Circulating Tumour Cells, Cell Free DNA and Tumour-Educated Platelets as Reliable Prognostic and Management Biomarkers for the Liquid Biopsy in Multiple Myeloma
Abstract
Simple Summary
Abstract
1. Introduction
2. Liquid Biopsy and Circulating Tumour Cells
2.1. Methods of Detection of CTCs
2.2. CTC, Progression and Prognosis of Monoclonal Gammopathies
2.3. CTCs and Prognosis of Multiple Myeloma
3. Liquid Biopsy and Cell Free DNA
3.1. Origin of Cell Free DNA
3.2. Methods of Detection of cfDNA
3.3. cfDNA Disease Advancement and Relapse
3.4. cfDNA and MRD Evaluation
4. Liquid Biopsy and MiRNAs
Methods of Detection and Biological Significance of miRNAs
5. Liquid Biopsy and Exosomes
5.1. Origin and Detection of Exosomes
5.2. Biological and Clinical Significance of Exosomes
6. Tumor-Educated Platelets (TEP) and Liquid Biopsy

| Methods | Targets | Results | Refs. | |
|---|---|---|---|---|
| Prognostic significance | Small RNA sequencing analysis and quantitative reverse transcription polymerase chain reaction array | Circulating exosomal microRNAs | miRNAs, let-7b and miR-18a, were significantly associated with PFS and OS | [112] |
| TaqMan Low Density miRNA Arrays | Expression of 667 miRNAs | Lower levels of miR-744 and let-7e were associated with shorter overall survival and remission of myeloma patients | [126] | |
| Flow cytometric analysis | Analysis of circulating large extracellular vesicles | Elevated levels of P-glycoprotein and phosphatidylserine correlate with disease progression and treatment unresponsiveness | [131] | |
| Diagnostic significance | RT2 lncRNA PCR Array—Human lncRNA Finder | Long noncoding RNA molecules | Dysregulation of exosomal lncRNA PRINS in MM vs. controls | [113] |
| Quantitative RT-PCR | Exosome miR155 levels | Exosome miR155 levels were significantly lower in multiple myeloma vs. controls | [125] | |
| Real-time quantitative PCR | Expression of let-7c-5p, let-7d-5p, miR-140-3p, miR-185-5p, and miR-425-5p | Expression of exosomal miRNA is related to the expression levels of a clinical feature-related factor | [127] | |
| Multiple myeloma educated platelets | ||||
| Prognostic significance | Hematological analyzer | Evaluation of mean platelet volume in patients with multiple myeloma | Low mean platelet volume is correlated with poor prognosis in MM patients | [135,136,137] |
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Statistics of N. Cancer Survival by Stage at Diagnosis for England. 2019. Available online: https://www.ons.gov.uk/peoplepopulationandcommunity/healthandsocialcare/conditionsanddiseases/datasets/cancersurvivalratescancersurvivalinenglandadultsdiagnosed (accessed on 17 June 2022).
- Davies, F.E.; Pawlyn, C.; Usmani, S.Z.; San-Miguel, J.F.; Einsele, H.; Boyle, E.M.; Corre, J.; Auclair, D.; Cho, H.J.; Lonial, S.; et al. Perspectives on the Risk-Stratified Treatment of Multiple Myeloma. Blood Cancer Discov. 2022, 3, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Allegra, A.; Innao, V.; Gerace, D.; Vaddinelli, D.; Musolino, C. Adoptive immunotherapy for hematological malignancies: Current status and new insights in chimeric antigen receptor T cells. Blood Cells Mol. Dis. 2016, 62, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Touzeau, C.; Moreau, P. How I treat extramedullary myeloma. Blood 2015, 127, 971. [Google Scholar] [CrossRef] [PubMed]
- João, C.; Coelho, I.; Costa, C.; Esteves, S.; Lucio, P. Efficacy and safety of lenalidomide in relapse/refractory multiple myeloma--real life experience of a tertiary cancer center. Ann. Hematol. 2015, 94, 97–105. [Google Scholar] [CrossRef]
- Kumar, S.K.; Dispenzieri, A.; Lacy, M.Q.; Gertz, M.A.; Buadi, F.K.; Pandey, S.; Kapoor, P.; Dingli, D.; Hayman, S.R.; Leung, N.; et al. Continued improvement in survival in multiple myeloma: Changes in early mortality and outcomes in older patients. Leukemia 2014, 28, 1122–1128. [Google Scholar] [CrossRef]
- Rasche, L.; Chavan, S.S.; Stephens, O.W.; Patel, P.H.; Tytarenko, R.; Ashby, C.; Bauer, M.; Stein, C.; Deshpande, S.; Wardell, C.; et al. Spatial genomic heterogeneity in multiple myeloma revealed by multi-region sequencing. Nat. Commun. 2017, 8, 268. [Google Scholar] [CrossRef] [PubMed]
- Ponti, G.; Manfredini, M.; Tomasi, A. Non-blood sources of cell-free DNA for cancer molecular profiling in clinical pathology and oncology. Crit. Rev. Oncol. Hematol. 2019, 141, 36–42. [Google Scholar] [CrossRef]
- Colmenares, R.; Álvarez, N.; Barrio, S.; Martínez-López, J.; Ayala, R. The Minimal Residual Disease Using Liquid Biopsies in Hematological Malignancies. Cancers 2022, 14, 1310. [Google Scholar] [CrossRef]
- Charalampous, C.; Kourelis, T. Minimal Residual Disease Assessment in Multiple Myeloma Patients: Minimal Disease With Maximal Implications. Front. Oncol. 2022, 11, 801851. [Google Scholar] [CrossRef]
- Wallington-Beddoe, C.T.; Mynott, R.L. Prognostic and predictive biomarker developments in multiple myeloma. J. Hematol. Oncol. 2021, 14, 151. [Google Scholar] [CrossRef]
- Soliman, A.M.; Das, S.; Teoh, S.L. Next-Generation Biomarkers in Multiple Myeloma: Understanding the Molecular Basis for Potential Use in Diagnosis and Prognosis. Int. J. Mol. Sci. 2021, 22, 7470. [Google Scholar] [CrossRef] [PubMed]
- Oberle, A.; Brandt, A.; Voigtlaender, M.; Thiele, B.; Radloff, J.; Schulenkorf, A.; Alawi, M.; Akyuz, N.; Marz, M.; Ford, C.T.; et al. Monitoring multiple myeloma by next-generation sequencing of V(D)J rearrangements from circulating myeloma cells and cell-free myeloma DNA. Haematologica 2017, 102, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Levin, A.; Hari, P.; Dhakal, B. Novel biomarkers in multiple myeloma. Transl. Res. 2018, 201, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Sundling, K.E.; Lowe, A.C. Circulating tumor cells: Overview and opportunities in cytology. Adv. Anat. Pathol. 2019, 26, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Krebs, M.G.; Metcalf, R.L.; Carter, L.; Brady, G.; Blackhall, F.H.; Dive, C. Molecular analysis of circulating tumour cells–biology and biomarkers. Nat. Rev. Clin. Oncol. 2014, 11, 129–144. [Google Scholar] [CrossRef] [PubMed]
- Potaro, P.; Lotey, N. Role of circulating tumor cells in future diagnosis and therapy of cancer. J. Cancer Metastasis Treat 2015, 1, 44. [Google Scholar]
- Ghobrial, I.M. Myeloma as a model for the process of metastasis: Implications for therapy. Blood 2012, 120, 20–30. [Google Scholar] [CrossRef]
- Krebs, M.G.; Hou, J.-M.; Ward, T.H.; Blackhall, F.H.; Dive, C. Circulating tumour cells: Their utility in cancer management and predicting outcomes. Ther. Adv. Med. Oncol. 2010, 2, 351–365. [Google Scholar] [CrossRef]
- Paiva, B.; Paino, T.; Sayagues, J.-M.; Garayoa, M.; San-Segundo, L.; Martín, M.; Mota, I.; Sanchez, M.-L.; Bárcena, P.; Aires-Mejia, I.; et al. Detailed characterization of multiple myeloma circulating tumor cells shows unique phenotypic, cytogenetic, functional, and circadian distribution profile. Blood 2013, 122, 3591–3598. [Google Scholar] [CrossRef]
- Kraj, M.; Kopec-Szlezak, J.; Pogłód, R.; Kruk, B. Flow cytometric immunophenotypic characteristics of 36 cases of plasma cell leukemia. Leuk. Res. 2011, 35, 169–176. [Google Scholar] [CrossRef]
- Klimiene, I.; Radzevicius, M.; Matuzeviciene, R.; Sinkevic-Belliot, K.; Kucinskiene, Z.A.; Peceliunas, V. Adhesion molecule immunophenotype of bone marrow multiple myeloma plasma cells impacts the presence of malignant circulating plasma cells in peripheral blood. Int. J. Lab. Hematol. 2021, 43, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Alunni-Fabbroni, M.; Sandri, M.T. Circulating tumour cells in clinical practice: Methods of detection and possible characterization. Methods 2010, 50, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Pantel, K.; Alix-Panabières, C.; Riethdorf, S. Cancer micrometastases. Nat. Rev. Clin. Oncol. 2009, 6, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Paterlini-Brechot, P.; Benali, N.L. Circulating tumor cells (CTC) detection: Clinical impact and future directions. Cancer Lett. 2007, 253, 180–204. [Google Scholar] [CrossRef]
- Farace, F.; Massard, C.; Vimond, N.; Drusch, F.; Jacques, N.; Billiot, F.; Laplanche, A.; Chauchereau, A.; Lacroix, L.; Planchard, D.; et al. A direct comparison of Cell Search and ISET for circulating tumour-cell detection in patients with metastatic carcinomas. Br. J. Cancer 2011, 105, 847. [Google Scholar] [CrossRef]
- Miller, M.C.; Doyle, G.V.; Terstappen, L.W.M.M. Significance of circulating tumor cells detected by the CellSearch system in patients with metastatic breast colorectal and prostate cancer. J. Oncol. 2010, 2010, 617421. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef]
- Cohen, S.J.; Punt, C.J.A.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.; Mitchell, E.; Miller, M.C.; et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J. Clin. Oncol. 2008, 26, 3213–3221. [Google Scholar] [CrossRef]
- de Bono, J.S.; Scher, H.I.; Montgomery, R.B.; Parker, C.; Miller, M.C.; Tissing, H.; Doyle, G.V.; Terstappen, L.W.; Pienta, K.J.; Raghavan, D. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin. Cancer Res. 2008, 14, 6302–6309. [Google Scholar] [CrossRef]
- Sun, Y.-F.; Yang, X.-R.; Zhou, J.; Qiu, S.-J.; Fan, J.; Xu, Y. Circulating tumor cells: Advances in detection methods, biological issues, and clinical relevance. J. Cancer Res. Clin. Oncol. 2011, 137, 1151–1173. [Google Scholar] [CrossRef]
- Williams, A.L.; Fitzgerald, J.E.; Ivich, F.; Sontag, E.D.; Niedre, M. Short-Term Circulating Tumor Cell Dynamics in Mouse Xenograft Models and Implications for Liquid Biopsy. Front. Oncol. 2020, 10, 601085. [Google Scholar] [CrossRef] [PubMed]
- Ndacayisaba, L.J.; Rappard, K.E.; Shishido, S.N.; Ruiz Velasco, C.; Matsumoto, N.; Navarez, R.; Tang, G.; Lin, P.; Setayesh, S.M.; Naghdloo, A.; et al. Enrichment-Free Single-Cell Detection and Morphogenomic Profiling of Myeloma Patient Samples to Delineate Circulating Rare Plasma Cell Clones. Curr. Oncol. 2022, 29, 2954–2972. [Google Scholar] [CrossRef] [PubMed]
- Foulk, B.; Schaffer, M.; Gross, S.; Rao, C.; Smirnov, D.; Connelly, M.C.; Chaturvedi, S.; Reddy, M.; Brittingham, G.; Mata, M.; et al. Enumeration and characterization of circulating multiple myeloma cells in patients with plasma cell disorders. Br. J. Hematol. 2018, 180, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Beasley, S.; Prigozhina, N.L.; Higgins, R.; Ikeda, S.; Lee, F.Y.; Marrinucci, D.; Jia, S. Detection and characterization of circulating tumour cells in multiple myeloma. J. Circ. Biomark. 2016, 5, 1–8. [Google Scholar] [CrossRef][Green Version]
- Garcés, J.J.; Bretones, G.; Burgos, L.; Valdes-Mas, R.; Puig, N.; Cedena, M.T.; Alignani, D.; Rodriguez, I.; Puente, D.Á.; Álvarez, M.G.; et al. Circulating tumor cells for comprehensive and multiregional non-invasive genetic characterization of multiple myeloma. Leukemia 2020, 34, 3007–3018. [Google Scholar] [CrossRef]
- Wang, C.; Xu, Y.; Li, S.; Zhou, Y.; Qian, Q.; Liu, Y.; Mi, X. Designer tetrahedral DNA framework-based microfluidic technology for multivalent capture and release of circulating tumor cells. Mater. Today Bio 2022, 16, 100346. [Google Scholar] [CrossRef]
- Xu, X.; Lin, J.; Guo, Y.; Wu, X.; Xu, Y.; Zhang, D.; Zhang, X.; Yujiao, X.; Wang, J.; Yao, C.; et al. TiO2-based Surface-Enhanced Raman Scattering bio-probe for efficient circulating tumor cell detection on microfilter. Biosens. Bioelectron. 2022, 210, 114305. [Google Scholar] [CrossRef]
- Lohr, J.G.; Kim, S.; Gould, J.; Knoechel, B.; Drier, Y.; Cotton, M.J.; Gray, D.; Birrer, N.; Wong, B.; Ha, G.; et al. Genetic interrogation of circulating multiple myeloma cells at single-cell resolution. Sci. Transl. Med. 2016, 8, 363ra147. [Google Scholar] [CrossRef]
- Mishima, Y.; Paiva, B.D.L.; Shi, J.; Park, J.; Manier, S.; Takagi, S.; Massoud, M.; Perilla-Glen, A.; Aljawai, Y.; Huynh, D.; et al. The Mutational Landscape of Circulating Tumor Cells in Multiple Myeloma. Cell Rep. 2017, 19, 218–224. [Google Scholar] [CrossRef]
- Zhan, F.; Huang, Y.; Colla, S.; Stewart, J.P.; Hanamura, I.; Gupta, S.; Epstein, J.; Yaccoby, S.; Sawyer, J.; Burington, B.; et al. The molecular classification of multiple myeloma. Blood 2006, 108, 2020–2028. [Google Scholar] [CrossRef]
- Ledergor, G.; Weiner, A.; Zada, M.; Wang, S.-Y.; Cohen, Y.C.; Gatt, M.E.; Snir, N.; Magen, H.; Koren-Michowitz, M.; Herzog-Tzarfati, K.; et al. Single cell dissection of plasma cell heterogeneity in symptomatic and asymptomatic myeloma. Nat. Med. 2018, 24, 1867–1876. [Google Scholar] [CrossRef] [PubMed]
- Fokkema, C.; de Jong, M.M.E.; Tahri, S.; Kellermayer, Z.; den Hollander, C.; Vermeulen, M.; Papzian, N.; van Duin, M.; Wevers, M.J.W.; Sanders, M.A.; et al. Abstract #1566: High Levels of Circulating Tumor Cells Are Associated with Increased Bone Marrow Proliferation in Newly Diagnosed Multiple Myeloma Patients. In Proceedings of the 63rd ASH Annual Meeting & Exposition, Atlanta, GA, USA, 11–14 December 2021. [Google Scholar]
- Sanoja-Flores, L.; Flores-Montero, J.; Garcés, J.J.; Paiva, B.; Puig, N.; García-Mateo, A.; García-Sánchez, O.; Corral-Mateos, A.; Burgos, L.; Blanco, E.; et al. Next generation flow for minimally invasive blood characterization of MGUS and multiple myeloma at diagnosis based on circulating tumor plasma cells (CTPC). Blood Cancer J. 2018, 8, 117. [Google Scholar] [CrossRef] [PubMed]
- Bataille, R.; Jégo, G.; Robillard, N.; Barille-Nion, S.; Harousseau, J.-L.; Moreau, P.; Amiot, M.; Pellat-Deceunynck, C. The phenotype of normal, reactive and malignant plasma cells. Identification of “many and multiple myelomas” and of new targets for myeloma therapy. Haematologica 2006, 91, 1234–1240. [Google Scholar] [PubMed]
- Deceunynck, C.; Barille-Nion, S.; Jego, G.; Puthier, D.; Robillard, N.; Pineau, D.; Rapp, M.-J.; Harousseau, J.-L.; Amiot, M.; Bataille, R. The absence of CD56 (NCAM) on malignant plasma cells is a hallmark of plasma cell leukemia and of a special subset of multiple myeloma. Leukemia 1998, 12, 1977–1982. [Google Scholar] [CrossRef]
- Lonial, S.; Jacobus, S.; Fonseca, R.; Weiss, M.; Kumar, S.; Orlowski, R.Z.; Kaufman, J.L.; Yacoub, A.M.; Buadi, F.K.; O’Brien, T.; et al. Randomized trial of lenalidomide versus observation in smoldering multiple myeloma. J. Clin. Oncol. 2020, 38, 1126–1137. [Google Scholar] [CrossRef]
- Garcés, J.-J.; Puig, N.; Termini, R.; Cedena, M.-T.; Moreno, C.; Pérez, J.J.; Alignani, D.; Sarvide, S.; Oriol, A.; González-García, E.; et al. Abstract #76: Circulating Tumor Cells (CTCs) in Smoldering and Active Multiple Myeloma (MM): Mechanism of Egression, Clinical Significance and Therapeutic Endpoints. In Proceedings of the 63rd ASH Annual Meeting & Exposition, Atlanta, GA, USA, 11–14 December 2021. [Google Scholar]
- Vasco-Mogorrón, M.A.; Campillo, J.A.; Periago, A.; Cabañas, V.; Berenguer, M.; García-Garay, M.C.; Gimeno, L.; Soto-Ramírez, M.F.; Martínez-Hernández, M.D.; Muro, M.; et al. Blood-based risk stratification for pre-malignant and symptomatic plasma cell neoplasms to improve patient management. Am. J. Cancer Res. 2021, 11, 2736–2753. [Google Scholar]
- Garcés, J.J.; Cedena, M.T.; Puig, N.; Burgos, L.; Perez, J.J.; Cordon, L.; Flores-Montero, J.; Sanoja-Flores, L.; Calasanz, M.J.; Ortiol, A.; et al. Circulating Tumor Cells for the Staging of Patients with Newly Diagnosed Transplant-Eligible Multiple Myeloma. J. Clin. Oncol. 2022, JCO2101365. [Google Scholar] [CrossRef]
- Manier, S.; Park, J.; Capelletti, M.; Bustoros, M.; Freeman, S.S.; Ha, G.; Rhoades, J.; Liu, C.J.; Huynh, D.; Reed, S.C.; et al. Whole-exome sequencing of cell free DNA and circulating tumor cells in multiple myeloma. Nat. Commun. 2018, 9, 1691. [Google Scholar] [CrossRef]
- Jelinek, T.; Bezdekova, R.; Zatopkova, M.; Burgos, L.; Simicek, M.; Sevcikova, T.; Paiva, B.; Hajek, R. Current applications of multiparameter fow cytometry in plasma cell disorders. Blood Cancer J. 2017, 7, e617. [Google Scholar] [CrossRef]
- Huhn, S.; Weinhold, N.; Nickel, J.; Pritsch, M.; Hielscher, T.; Hummel, M.; Bertsch, U.; Huegle-Doerr, B.; Vogel, M.; Angermund, R.; et al. Circulating tumor cells as a biomarker for response to therapy in multiple myeloma patients treated within the GMMG-MM5 trial. Bone Marrow Transplant. 2017, 52, 1194–1198. [Google Scholar] [CrossRef]
- Li, J.; Wang, N.; Tesfaluul, N.; Gao, X.; Liu, S.; Yue, B. Prognostic value of circulating plasma cells in patients with multiple myeloma: A meta-analysis. PLoS ONE 2019, 12, e0181447. [Google Scholar] [CrossRef] [PubMed]
- Nowakowski, G.S.; Witzig, T.E.; Dingli, D.; Tracz, M.J.; Gertz, M.A.; Lacy, M.Q.; Lust, J.A.; Dispenzieri, A.; Greipp, P.R.; Kyle, R.A.; et al. Circulating plasma cells detected by flow cytometry as a predictor of survival in 302 patients with newly diagnosed multiple myeloma. Blood 2005, 106, 2276–2279. [Google Scholar] [CrossRef] [PubMed]
- Gonsalves, W.I.; Morice, W.G.; Rajkumar, V.; Gupta, V.; Timm, M.M.; Dispenzieri, A.; Buadi, F.K.; Lacy, M.Q.; Singh, P.P.; Kapoor, P.; et al. Quantification of clonal circulating plasma cells in relapsed multiple myeloma. Br. J. Haematol. 2014, 167, 500–505. [Google Scholar] [CrossRef]
- Bianchi, G.; Richardson, P.G.; Anderson, K.C. Promising therapies in multiple myeloma. Blood 2015, 126, 300–310. [Google Scholar] [CrossRef] [PubMed]
- Flores-Montero, J.; Sanoja-Flores, L.; Paiva, B.; Puig, N.; García Sánchez, O.; Böttcher, S.; van der Velden, V.H.J.; Pérez-Morán, J.J.; Vidriales, M.B.; García-Sanz, R.; et al. Next generation flow for highly sensitive and standardized detection of minimal residual disease in multiple myeloma. Leukemia 2017, 31, 2094. [Google Scholar] [CrossRef] [PubMed]
- Giuliano, M.; Shaikh, A.; Lo, H.C.; Arpino, G.; De Placido, S.; Zhang, X.H.; Cristofanilli, M.; Schiff, R.; Trivedi, M.V. Perspective on Circulating Tumor Cell Clusters: Why It Takes a Village to Metastasize. Cancer Res. 2018, 78, 845–852. [Google Scholar] [CrossRef]
- Hou, J.M.; Krebs, M.G.; Lancashire, L.; Sloane, R.; Backen, A.; Swain, R.K.; Priest, L.J.; Greystoke, A.; Zhou, C.; Morris, K.; et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J. Clin. Oncol. 2012, 30, 525–532. [Google Scholar] [CrossRef]
- Wang, C.; Mu, Z.; Chervoneva, I.; Austin, L.; Ye, Z.; Rossi, G.; Palazzo, J.P.; Sun, C.; Abu-Khalaf, M.; Myers, R.E.; et al. Longitudinally collected CTCs and CTC-clusters and clinical outcomes of metastatic breast cancer. Breast Cancer Res. Treat. 2017, 161, 83–94. [Google Scholar] [CrossRef]
- Macaraniag, C.; Luan, Q.; Zhou, J.; Papautsky, I. Microfluidic techniques for isolation, formation, and characterization of circulating tumor cells and clusters. APL Bioeng. 2022, 6, 31501. [Google Scholar] [CrossRef]
- Snyder, M.W.; Kircher, M.; Hill, A.J.; Daza, R.M.; Shendure, J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 2016, 164, 57–68. [Google Scholar] [CrossRef]
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Kis, O.; Kaedbey, R.; Chow, S.; Danesh, A.; Dowar, M.; Li, T.; Li, Z.; Liu, J.; Mansour, M.; Masih-Khan, E.; et al. Circulating tumour DNA sequence analysis as an alternative to multiple myeloma bone marrow aspirates. Nat Commun. 2017, 8, 15086. [Google Scholar] [CrossRef] [PubMed]
- Mithraprabhu, S.; Khong, T.; Ramachandran, M.; Chow, A.; Klarica, D.; Mai, L.; Walsh, S.; Broemeling, D.; Marziali, A.; Wiggin, M.; et al. Circulating tumour DNA analysis demonstrates spatial mutational heterogeneity that coincides with disease relapse in myeloma. Leukemia 2017, 31, 1695–1705. [Google Scholar] [CrossRef] [PubMed]
- Bolli, N.; Avet-Loiseau, H.; Wedge, D.C.; Van Loo, P.; Alexandrov, L.B.; Martincorena, I.; Dawson, K.J.; Iorio, F.; Nik-Zainal, S.; Bignell, G.R.; et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat. Commun. 2014, 5, 2997. [Google Scholar] [CrossRef]
- Kumar, S.K.; Rajkumar, S.V. The multiple myelomas—current concepts in cytogenetic classification and therapy. Nat. Rev. Clin. Oncol. 2018, 15, 409–421. [Google Scholar] [CrossRef]
- Walker, B.A.; Mavrommatis, K.; Wardell, C.P.; Cody Ashby, T.; Bauer, M.; Davies, F.E. Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. Blood 2018, 132, 587–597. [Google Scholar] [CrossRef]
- Guo, G.; Raje, N.S.; Seifer, C.; Kloeber, J.; Isenhart, R.; Ha, G.; Yee, A.J.; O’Donnell, E.K.; Tai, Y.T.; Richardson, P.G.; et al. Genomic discovery and clonal tracking in multiple myeloma by cell-free DNA sequencing. Leukemia 2018, 32, 1838–1841. [Google Scholar] [CrossRef]
- Gahan, P.B.; Swaminathan, R. Circulating nucleic acids in plasma and serum. Recent developments. Ann. N. Y. Acad. Sci. 2008, 1137, 1–6. [Google Scholar] [CrossRef]
- Mandel, P.; Metais, P. Les acides nucleiques du plasma sanguin chez l’homme. Seances Soc. Biol. Ses. Fil. 1948, 142, 241–243. [Google Scholar]
- Heitzer, E.; Auer, M.; Hoffmann, E.M.; Pichler, M.; Gasch, C.; Ulz, P.; Lax, S.; Waldispuehl-Geigl, J.; Mauermann, O.; Mohan, S.; et al. Establishment of tumor-specific copy number alterations from plasma DNA of patients with cancer. Int. J. Cancer 2013, 133, 346–356. [Google Scholar] [CrossRef]
- Murtaza, M.; Dawson, S.J.; Tsui, D.W.; Gale, D.; Forshew, T.; Piskorz, A.M.; Parkinson, C.; Chin, S.F.; Kingsbury, Z.; Wong, A.S.; et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013, 497, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.H.; Jiang, P.; Sun, K.; Cheng, Y.K.; Chan, K.C.; Leung, T.Y.; Chiu, R.W.; Lo, Y.M. Noninvasive prenatal testing by nanopore sequencing of maternal plasma DNA: Feasibility assessment. Clin. Chem. 2015, 61, 1305–1306. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.D.; Zhang, J.; Leung, T.N.; Lau, T.K.; Chang, A.M.; Hjelm, N.M. Rapid clearance of fetal DNA from maternal plasma. Am. J. Hum. Genet. 1999, 64, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F.O.; Hesch, R.D.; Knippers, R. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001, 61, 1659–1665. [Google Scholar]
- Dwivedi, D.J.; Toltl, L.J.; Swystun, L.L.; Pogue, J.; Liaw, K.L.; Weitz, J.I.; Cook, D.J.; Fox-Robichaud, A.E.; Liaw, P.C. Prognostic utility and characterization of cell-free DNA in patients with severe sepsis. Crit. Care 2012, 16, R151. [Google Scholar] [CrossRef]
- Lim, J.K.; Kuss, B.; Talaulikar, D. Role of cell-free DNA in haematological malignancies. Pathology 2021, 53, 416–426. [Google Scholar] [CrossRef]
- Gerber, B.; Manzoni, M.; Spina, V.; Bruscaggin, A.; Lionetti, M.; Fabris, S.; Barbieri, M.; Ciceri, G.; Pompa, A.; Forestieri, G.; et al. Circulating tumor DNA as a liquid biopsy in plasma cell dyscrasias. Haematologica 2018, 103, e245–e248. [Google Scholar] [CrossRef]
- Rustad, E.H.; Coward, E.; Skytøen, E.R.; Misund, K.; Holien, T.; Standal, T.; Børset, M.; Beisvag, V.; Myklebost, O.; Meza-Zepeda, L.A.; et al. Monitoring multiple myeloma by quantification of recurrent mutations in serum. Haematologica 2017, 102, 1266–1272. [Google Scholar] [CrossRef]
- Weinhold, N.; Ashby, C.; Rasche, L.; Chavan, S.S.; Stein, C.; Stephens, O.W.; Tytarenko, R.; Bauer, M.A.; Meissner, T.; Deshpande, S.; et al. Clonal selection and double-hit events involving tumor suppressor genes underlie relapse in myeloma. Blood 2016, 128, 1735–1744. [Google Scholar] [CrossRef]
- Hohaus, S.; Giachelia, M.; Massini, G.; Mansueto, G.; Vannata, B.; Bozzoli, V.; Criscuolo, M.; D’Alò, F.; Martini, M.; Larocca, L.M.; et al. Cell-free circulating DNA in Hodgkin’s and non-Hodgkin’s lymphomas. Ann. Oncol. 2009, 20, 1408–1413. [Google Scholar] [CrossRef]
- Schwarz, A.K.; Stanulla, M.; Cario, G.; Flohr, T.; Sutton, R.; Möricke, A.; Anker, P.; Stroun, M.; Welte, K.; Bartram, C.R.; et al. Quantification of free total plasma DNA and minimal residual disease detection in the plasma of children with acute lymphoblastic leukemia. Ann. Hematol. 2009, 88, 897–905. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Biancon, G.; Gimondi, S.; Vendramin, A.; Carniti, C.; Corradini, P. Noninvasive Molecular Monitoring in Multiple Myeloma Patients Using Cell-Free Tumor DNA: A Pilot Study. J. Mol. Diagn. JMD 2018, 20, 859–870. [Google Scholar] [CrossRef] [PubMed]
- Manzoni, M.; Pompa, A.; Fabris, S.; Pelizzoni, F.; Ciceri, G.; Seia, M.; Ziccheddu, B.; Bolli, N.; Corradini, P.; Baldini, L.; et al. Limits and Applications of Genomic Analysis of Circulating Tumor DNA as a Liquid Biopsy in Asymptomatic Forms of Multiple Myeloma. Hemasphere 2020, 4, e402. [Google Scholar] [CrossRef]
- Alidousty, C.; Brandes, D.; Heydt, C.; Wagener, S.; Wittersheim, M.; Schäfer, S.C.; Holz, B.; Merkelbach-Bruse, S.; Büttner, R.; Fassunke, J.; et al. Comparison of Blood Collection Tubes from Three Different Manufacturers for the Collection of Cell-Free DNA for Liquid Biopsy Mutation Testing. J. Mol. Diagn. JMD 2017, 19, 801–804. [Google Scholar] [CrossRef] [PubMed]
- Mithraprabhu, S.; Spencer, A. Analysis of Circulating Tumor DNA. Methods Mol. Biol. Clifton NJ 2018, 1792, 129–145. [Google Scholar] [CrossRef]
- Long, X.; Xu, Q.; Lou, Y.; Li, C.; Gu, J.; Cai, H.; Wang, D.; Xu, J.; Li, T.; Zhou, X.; et al. The utility of non-invasive liquid biopsy for mutational analysis and minimal residual disease assessment in extramedullary multiple myeloma. Br. J. Haematol. 2020, 189, e45–e48. [Google Scholar] [CrossRef]
- Rajkumar, S.V.; Landgren, O.; Mateos, M.V. Smoldering multiple myeloma. Blood 2015, 125, 3069–3075. [Google Scholar] [CrossRef]
- Bolli, N.; Maura, F.; Minvielle, S.; Gloznik, D.; Szalat, R.; Fullam, A.; Martincorena, I.; Dawson, K.J.; Samur, M.K.; Zamora, J.; et al. Genomic patterns of progression in smoldering multiple myeloma. Nat. Commun. 2018, 9, 3363. [Google Scholar] [CrossRef]
- Bustoros, M.; Sklavenitis-Pistofidis, R.; Park, J.; Redd, R.; Zhitomirsky, B.; Dunford, A.J.; Salem, K.; Tai, Y.T.; Anand, S.; Mouhieddine, T.H.; et al. Genomic Profiling of Smoldering Multiple Myeloma Identifies Patients at a High Risk of Disease Progression. J. Clin. Oncol. 2020, 38, 2380–2389. [Google Scholar] [CrossRef]
- Deshpande, S.; Tytarenko, R.G.; Wang, Y.; Boyle, E.M.; Ashby, C.; Schinke, C.D.; Thanendrarajan, S.; Zangari, M.; Zhan, F.; Davies, F.E.; et al. Monitoring treatment response and disease progression in myeloma with circulating cell-free DNA. Eur. J. Haematol. 2021, 106, 230–240. [Google Scholar] [CrossRef]
- Yasui, H.; Kobayashi, M.; Sato, K.; Kondoh, K.; Ishida, T.; Kaito, Y.; Tamura, H.; Handa, H.; Tsukune, Y.; Sasaki, M.; et al. Circulating cell-free DNA in the peripheral blood plasma of patients is an informative biomarker for multiple myeloma relapse. Int. J. Clin. Oncol. 2021, 26, 2142–2150. [Google Scholar] [CrossRef] [PubMed]
- Vrabel, D.; Sedlarikova, L.; Besse, L.; Rihova, L.; Bezdekova, R.; Almasi, M.; Kubaczkova, V.; Brožová, L.; Jarkovsky, J.; Plonkova, H.; et al. Dynamics of tumor-specific cfDNA in response to therapy in multiple myeloma patients. Eur. J. Haematol. 2020, 104, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Mithraprabhu, S.; Hocking, J.; Ramachandran, M.; Choi, K.; Klarica, D.; Khong, T.; Reynolds, J.; Spencer, A. DNA-Repair Gene Mutations Are Highly Prevalent in Circulating Tumour DNA from Multiple Myeloma Patients. Cancers 2019, 11, 917. [Google Scholar] [CrossRef] [PubMed]
- Waldschmidt, J.M.; Vijaykumar, T.; Knoechel, B.; Lohr, J.G. Tracking myeloma tumor DNA in peripheral blood. Best Pract. Res. Clin. Haematol. 2020, 33, 101146. [Google Scholar] [CrossRef] [PubMed]
- Pugh, T.J. Circulating Tumour DNA for Detecting Minimal Residual Disease in Multiple Myeloma. Semin. Hematol. 2018, 55, 38–40. [Google Scholar] [CrossRef]
- Mazzotti, C.; Buisson, L.; Maheo, S.; Perrot, A.; Chretien, M.-L.; Leleu, X.; Hulin, C.; Manier, S.; Hébraud, B.; Roussel, M.; et al. Myeloma MRD by Deep Sequencing from Circulating Tumor DNA Does Not Correlate with Results Obtained in the Bone Marrow. Blood Adv. 2018, 2, 2811–2813. [Google Scholar] [CrossRef]
- Ntanasis-Stathopoulos, I.; Gavriatopoulou, M.; Terpos, E.; Fotiou, D.; Kastritis, E.; Dimopoulos, M.A. Monitoring Plasma Cell Dyscrasias with Cell-Free DNA Analysis. Clin. Lymphoma Myeloma Leuk. 2020, 20, e905–e909. [Google Scholar] [CrossRef]
- Chawla, S.S.; Kumar, S.K.; Dispenzieri, A.; Greenberg, A.J.; Larson, D.R.; Kyle, R.A.; Lacy, M.Q.; Gertz, M.A.; Rajkumar, S.V. Clinical Course and Prognosis of Non-Secretory Multiple Myeloma. Eur. J. Haematol 2015, 95, 57–64. [Google Scholar] [CrossRef]
- Dupuis, M.M.; Tuchman, S.A. Non-secretory multiple myeloma: From biology to clinical management. OncoTargets Ther. 2016, 9, 7583–7590. [Google Scholar] [CrossRef]
- Allegra, A.; Cicero, N.; Tonacci, A.; Musolino, C.; Gangemi, S. Circular RNA as a Novel Biomarker for Diagnosis and Prognosis and Potential Therapeutic Targets in Multiple Myeloma. Cancers 2022, 14, 1700. [Google Scholar] [CrossRef]
- Allegra, A.; Ettari, R.; Innao, V.; Bitto, A. Potential Role of microRNAs in inducing Drug Resistance in Patients with Multiple Myeloma. Cells 2021, 10, 448. [Google Scholar] [CrossRef]
- Avenoso, A.; Campo, S.; Scuruchi, M.; Mania, M.; Innao, V.; D’Ascola, A.; Mandraffino, G.; Allegra, A.G.; Musolino, C.; Allegra, A. Quantitative polymerase Chain reaction profiling of microRNAs in peripheral lymph-monocytes from MGUS subjects. Pathol. Res. Pract. 2021, 218, 153317. [Google Scholar] [CrossRef] [PubMed]
- Musolino, C.; Oteri, G.; Allegra, A.; Mania, M.; D’Ascola, A.; Avenoso, A.; Innao, V.; Allegra, A.G.; Campo, S. Altered microRNA expression profile in the peripheral lymphoid compartment of multiple myeloma patients with bisphosphonate-induced osteonecrosis of the jaw. Ann. Hematol. 2018, 97, 1259–1269. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Xu, M.; Tian, X.; Cai, S.; Zeng, S. Research advances in the detection of miRNA. J. Pharm. Anal. 2019, 9, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Kshirsagar, P.; Seshacharyulu, P.; Muniyan, S.; Rachagani, S.; Smith, L.M.; Thompson, C.; Shah, A.; Mallya, K.; Kumar, S.; Jain, M.; et al. DNA-gold nanoprobe-based integrated biosensing technology for non-invasive liquid biopsy of serum miRNA: A new frontier in prostate cancer diagnosis. Nanomedicine 2022, 43, 102566. [Google Scholar] [CrossRef]
- Chen, M.; Mithraprabhu, S.; Ramachandran, M.; Choi, K.; Khong, T.; Spencer, A. Utility of Circulating Cell-Free RNA Analysis for the Characterization of Global Transcriptome Profiles of Multiple Myeloma Patients. Cancers 2019, 11, 887. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.J.; Liu, Y.; Ding, Z.; Zhang, L.; Han, C.; Yan, C.; Amador, E.; Yuan, L.; Wu, Y.; Song, C.; et al. The exploration of quantum dot-molecular beacon based MoS2 fluorescence probing for myeloma-related Mirnas detection. Bioact. Mater. 2022, 17, 360–368. [Google Scholar] [CrossRef]
- Xu, R.; Rai, A.; Chen, M.; Suwakulsiri, W.; Greening, D.W.; Simpson, R.J. Extracellular vesicles in cancer—Implications for future improvements in cancer care. Nat. Rev. Clin. Oncol. 2018, 15, 617–638. [Google Scholar] [CrossRef]
- Manier, S.; Liu, C.J.; Avet-Loiseau, H.; Park, J.; Shi, J.; Campigotto, F.; Salem, K.Z.; Huynh, D.; Glavey, S.V.; Rivotto, B.; et al. Prognostic role of circulating exosomal miRNAs in multiple myeloma. Blood 2017, 129, 2429–2436. [Google Scholar] [CrossRef]
- Sedlarikova, L.; Bollova, B.; Radova, L.; Brozova, L.; Jarkovsky, J.; Almasi, M.; Penka, M.; Kuglik, P.; Sandecka, V.; Stork, M.; et al. Circulating exosomal long noncoding RNA PRINS-First findings in monoclonal gammopathies. Hematol. Oncol. 2018, 36, 786–791. [Google Scholar] [CrossRef]
- Allegra, A.; Petrarca, C.; Di Gioacchino, M.; Casciaro, M.; Musolino, C.; Gangemi, S. Exosome-Mediated Therapeutic Strategies for Management of Solid and Hematological Malignancies. Cells 2022, 11, 1128. [Google Scholar] [CrossRef] [PubMed]
- Allegra, A.; Di Gioacchino, M.; Tonacci, A.; Petrarca, C.; Musolino, C.; Gangemi, S. Multiple Myeloma Cell-Derived Exosomes: Implications on Tumorigenesis, Diagnosis, Prognosis and Therapeutic Strategies. Cells 2021, 10, 2865. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour Exosome Integrins Determine Organotropic Metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef]
- Peinado, H.; Alečković, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; García-Santos, G.; Ghajar, C.M.; et al. Melanoma Exosomes Educate Bone Marrow Progenitor Cells Toward a Pro-Metastatic Phenotype Through MET. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef]
- Tutrone, R.; Donovan, M.J.; Torkler, P.; Tadigotla, V.; McLain, T.; Noerholm, M.; Skog, J.; McKiernan, J. Clinical Utility of the Exosome Based ExoDx Prostate (IntelliScore) EPI Test in Men Presenting for Initial Biopsy with a PSA 2–10 Ng/Ml. Prostate Cancer Prostatic Dis. 2020, 23, 607–614. [Google Scholar] [CrossRef] [PubMed]
- Reale, A.; Carmichael, I.; Xu, R.; Mithraprabhu, S.; Khong, T.; Chen, M.; Fang, H.; Savvidou, I.; Ramachandran, M.; Bingham, N.; et al. Human Myeloma Cell- and Plasma-Derived Extracellular Vesicles Contribute to Functional Regulation of Stromal Cells. Proteomics 2021, 21, e2000119. [Google Scholar] [CrossRef] [PubMed]
- Reale, A.; Khong, T.; Mithraprabhu, S.; Spencer, A. Translational Potential of RNA Derived From Extracellular Vesicles in Multiple Myeloma. Front. Oncol. 2021, 11, 718502. [Google Scholar] [CrossRef]
- Fan, W.; Han, P.; Feng, Q.; Sun, Y.; Ren, W.; Lawson, T.; Liu, C. Nucleic Acid Substrate-Independent DNA Polymerization on the Exosome Membrane: A Mechanism Study and Application in Exosome Analysis. Anal. Chem. 2022, 94, 2172–2179. [Google Scholar] [CrossRef]
- Fattahi, Z.; Khosroushahi, A.Y.; Hasanzadeh, M. Recent progress on developing of plasmon biosensing of tumor biomarkers: Efficient method towards early stage recognition of cancer. Biomed. Pharmacother. 2020, 132, 110850. [Google Scholar] [CrossRef]
- Farhana, F.Z.; Umer, M.; Saeed, A.; Pannu, A.S.; Shahbazi, M.; Jabur, A.; Nam, H.J.; Ostrikov, K.; Sonar, P.; Firoz, S.H.; et al. Isolation and detection of exosomes using Fe2O3 nanoparticles. ACS Appl. Nano Mater. 2021, 4, 1175–1186. [Google Scholar] [CrossRef]
- Li, S.; Ma, Q. Electrochemical nano-sensing interface for exosomes analysis and cancer diagnosis. Biosens. Bioelectron. 2022, 214, 114554. [Google Scholar] [CrossRef] [PubMed]
- Caivano, A.; La Rocca, F.; Simeon, V.; Girasole, M.; Dinarelli, S.; Laurenzana, I.; De Stradis, A.; De Luca, L.; Trino, S.; Traficante, A.; et al. MicroRNA-155 in Serum-Derived Extracellular Vesicles as a Potential Biomarker for Hematologic Malignancies–A Short Report. Cell Oncol. 2017, 40, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Kubiczkova, L.; Kryukov, F.; Slaby, O.; Dementyeva, E.; Jarkovsky, J.; Nekvindova, J.; Radova, L.; Greslikova, H.; Kuglik, P.; Vetesnikova, E.; et al. Circulating Serum microRNAs as Novel Diagnostic and Prognostic Biomarkers for Multiple Myeloma and Monoclonal Gammopathy of Undetermined Significance. Haematologica 2014, 99, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Y.; Li, Y.C.; Geng, C.Y.; Wang, H.J.; Chen, W.M. Potential Relationship Between Clinical Significance and Serum Exosomal miRNAs in Patients with Multiple Myeloma. BioMed Res. Int. 2019, 2019, 1575468. [Google Scholar] [CrossRef]
- Gong, J.; Jaiswal, R.; Mathys, J.M.; Combes, V.; Grau, G.E.; Bebawy, M. Microparticles and their emerging role in cancer multidrug resistance. Cancer Treat. Rev. 2012, 38, 226–234. [Google Scholar] [CrossRef]
- Innao, V.; Rizzo, V.; Allegra, A.G.; Musolino, C.; Allegra, A. Promising Anti-Mitochondrial Agents for Overcoming Acquired Drug Resistance in Multiple Myeloma. Cells 2021, 10, 439. [Google Scholar] [CrossRef]
- Allegra, A.; Casciaro, M.; Barone, P.; Musolino, C.; Gangemi, S. Epigenetic Crosstalk between Malignant Plasma Cells and the Tumour Microenvironment in Multiple Myeloma. Cancers 2022, 14, 2597. [Google Scholar] [CrossRef]
- Rajeev Krishnan, S.; De Rubis, G.; Suen, H.; Joshua, D.; Lam Kwan, Y.; Bebawy, M. A liquid biopsy to detect multidrug resistance and disease burden in multiple myeloma. Blood Cancer J. 2020, 10, 37. [Google Scholar] [CrossRef]
- Leslie, M. Cell Biology. Beyond Clotting: The Powers of Platelets. Science 2010, 328, 562–564. [Google Scholar] [CrossRef]
- Nilsson, R.J.A.; Balaj, L.; Hulleman, E.; van Rijn, S.; Pegtel, D.M.; Walraven, M.; Widmark, A.; Gerritsen, W.R.; Verheul, H.M.; Vandertop, W.P.; et al. Blood Platelets Contain Tumor-Derived RNA Biomarkers. Blood 2011, 118, 3680–3683. [Google Scholar] [CrossRef]
- Meng, Y.; Sun, J.; Zheng, Y.; Zhang, G.; Yu, T.; Piao, H. Platelets: The Emerging Clinical Diagnostics and Therapy Selection of Cancer Liquid Biopsies. Oncotargets Ther. 2021, 14, 3417–3428. [Google Scholar] [CrossRef] [PubMed]
- Shah, U.J.; Alsulimani, A.; Ahmad, F.; Mathkor, D.M.; Alsaieedi, A.; Harakeh, S.; Nasiruddin, M.; Haque, S. Bioplatforms in liquid biopsy: Advances in the techniques for isolation, characterization and clinical applications. Biotechnol. Genet. Eng. Rev. 2022, 13, 3303. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, X.; Guo, E.; Mao, X.; Miao, S. Emerging roles of platelets in cancer biology and their potential as therapeutic targets. Front. Oncol. 2022, 12, 939089. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Xiao, Z.P.; Fu, K.; Han, M. Clinical significance of mean platelet volume determination in multiple myeloma. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2017, 25, 827–831. [Google Scholar] [CrossRef]
- Zhuang, Q.; Xiang, L.; Xu, H.; Fang, F.; Xing, C.; Liang, B.; Yu, K.; Feng, J. The independent association of mean platelet volume with overall survival in multiple myeloma. Oncotarget 2016, 7, 62640–62646. [Google Scholar] [CrossRef]
- Flach, J.; Shumilov, E.; Joncourt, R.; Porret, N.; Novak, U.; Pabst, T.; Bacher, U. Current concepts and future directions for hemato-oncologic diagnostics. Crit. Rev. Oncol. Hematol. 2020, 151, 102977. [Google Scholar] [CrossRef]
- Cavo, M.; Terpos, E.; Nanni, C.; Moreau, P.; Lentzsch, S.; Zweegman, S.; Hillengass, J.; Engelhardt, M.; Usmani, S.Z.; Vesole, D.H.; et al. Role of 18F-FDG PET/CT in the diagnosis and management of multiple myeloma and other plasma cell disorders: A consensus statement by the International Myeloma Working Group. Lancet Oncol. 2017, 18, e217. [Google Scholar] [CrossRef]
- López-Anglada, L.; Gutiérrez, N.C.; García, J.L.; Mateos, M.V.; Flores, T.; San Miguel, J.F. P53 deletion may drive the clinical evolution and treatment response in multiple myeloma. Eur. J. Haematol. 2010, 84, 359–361. [Google Scholar] [CrossRef]
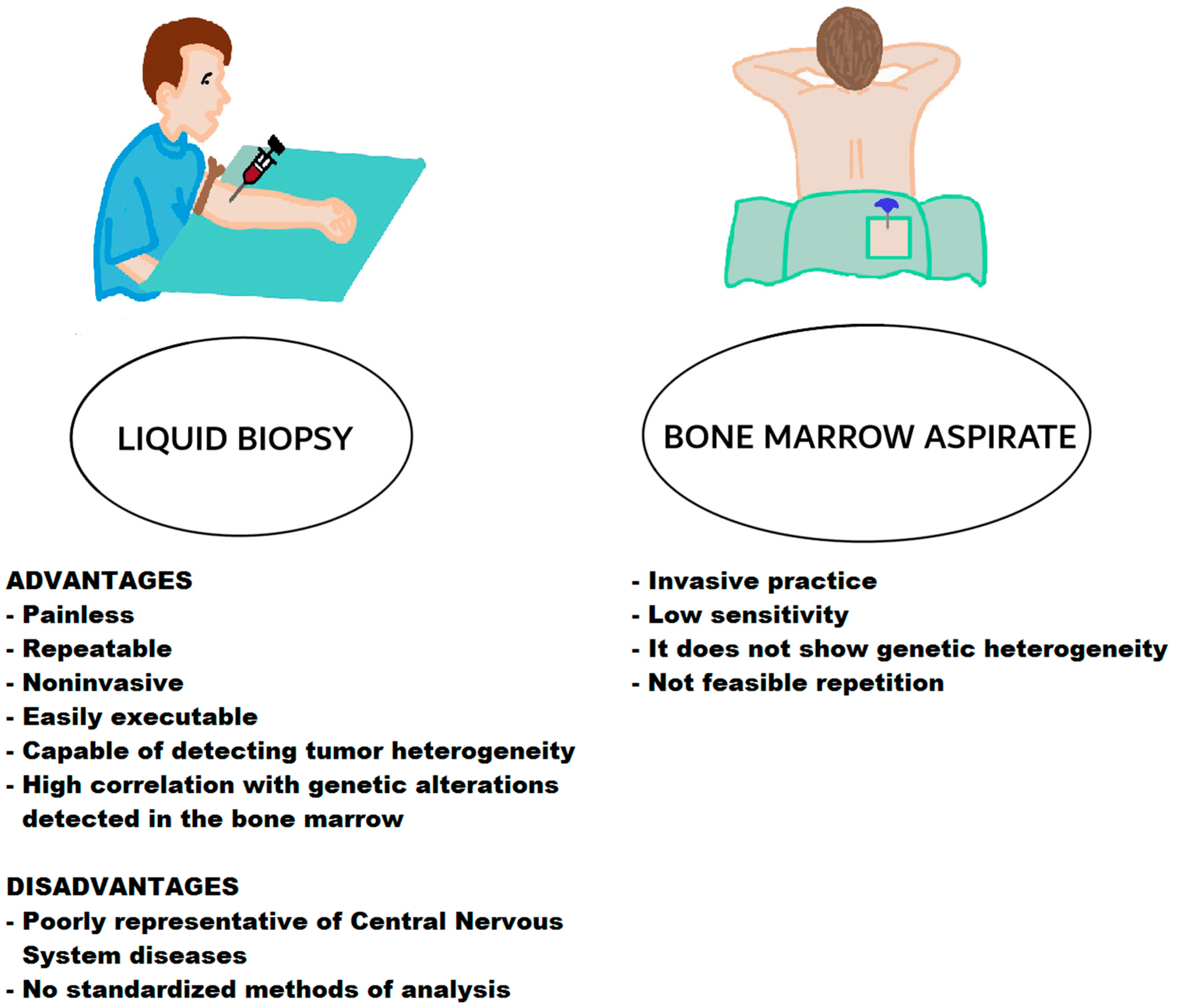
| Technique | Target | Marker | Features | Refs. |
|---|---|---|---|---|
| DiFc | To evaluate short time modifications | Green fluorescent protein-expressing CTCs | Small samples are unlikely to yield a quantitatively accurate estimate of mean CTC numbers. | [32] |
| Immunofluorescence technique and single cell DNA sequencing | Identification and genomic classification | CD138, CD56, CD45, DAPI | Detection of rare cell populations | [33] |
| FISH and NGS | Correlation between CTCs and clinical parameters | CD38, CD38/138, CD45, CD19 | It can be used to study disease biology and monitor clinical disease progression | [34] |
| Immunofluorescence test | To differentiate normal and clonal cells | CD138, CD45, phospho-ribosomal protein S6 | The assay is highly repeatable and reproducible | [35] |
| NGF cytometry | Evaluation of EMM | Patient-specific aberrant phenotypes identified with NGF were used for highly purified fluorescence-activated cell sorting of CTCs | Concordance between BM tumor cells and CTCs was high for chromosome arm-level copy number alterations (≥95%) though not for translocations (39%). | [36] |
| WES | Correlation with genetic alterations found in BM samples | CD138 | 100% of clonal mutations in patient BM were detected in CTCs and 99% of clonal mutations in CTCs were present in BM MM. | [40] |
| Diagnosis | Techniques | Target | Features | Refs. |
|---|---|---|---|---|
| Extramedullary MM | CAPP-seq ultra-deep targeted next-generation sequencing | Identification of cancer-gene somatic mutations | Greater amounts of cfDNA with respect to subjects without EMMM | [80] |
| Multiple myeloma | Digital droplet polymerase chain reaction analysis | Evaluation of recurrent mutations, mainly in mitogen activated protein kinase pathway genes NRAS, KRAS and BRAF | Correlation between the number of mutated alleles in the PB and the proportion of BM plasma cells | [81] |
| Smoldering MM | Gene expression profile | Analysis of 70 gene expression | Reduced amounts of cfDNA with respect to overt MM | [93] |
| Prognosis | ||||
| Relapsed MM | Next-generation sequencing | Identify driver mutations | Percentage of ras/raf and TP53 mutations was remarkably greater in RR subjects with a reduced OS | [94] |
| MRD assessment | ||||
| Extramedullary MM | Targeted deep sequencing and ddPCR assays | Mutational analysis | The concordance of plasma ctDNA to extramedullary tumor concordance was 0873, which is much higher than that of BM aspirates | [89] |
| Multiple myeloma | Ultra-low-pass whole genome sequencing | Detectability of cfDNA and CTCs | Correlation with disease progression is higher for CTCs | [51] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Allegra, A.; Cancemi, G.; Mirabile, G.; Tonacci, A.; Musolino, C.; Gangemi, S. Circulating Tumour Cells, Cell Free DNA and Tumour-Educated Platelets as Reliable Prognostic and Management Biomarkers for the Liquid Biopsy in Multiple Myeloma. Cancers 2022, 14, 4136. https://doi.org/10.3390/cancers14174136
Allegra A, Cancemi G, Mirabile G, Tonacci A, Musolino C, Gangemi S. Circulating Tumour Cells, Cell Free DNA and Tumour-Educated Platelets as Reliable Prognostic and Management Biomarkers for the Liquid Biopsy in Multiple Myeloma. Cancers. 2022; 14(17):4136. https://doi.org/10.3390/cancers14174136
Chicago/Turabian StyleAllegra, Alessandro, Gabriella Cancemi, Giuseppe Mirabile, Alessandro Tonacci, Caterina Musolino, and Sebastiano Gangemi. 2022. "Circulating Tumour Cells, Cell Free DNA and Tumour-Educated Platelets as Reliable Prognostic and Management Biomarkers for the Liquid Biopsy in Multiple Myeloma" Cancers 14, no. 17: 4136. https://doi.org/10.3390/cancers14174136
APA StyleAllegra, A., Cancemi, G., Mirabile, G., Tonacci, A., Musolino, C., & Gangemi, S. (2022). Circulating Tumour Cells, Cell Free DNA and Tumour-Educated Platelets as Reliable Prognostic and Management Biomarkers for the Liquid Biopsy in Multiple Myeloma. Cancers, 14(17), 4136. https://doi.org/10.3390/cancers14174136







