DNA Polymerase Theta Plays a Critical Role in Pancreatic Cancer Development and Metastasis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. DNA Sequencing
2.3. Lentiviral Production and Infection
2.4. Microarray Analysis
2.5. Quantitative PCR (qPCR)
2.6. Western Blotting
2.7. Cell Cycle Analysis
2.8. MTT Assay
2.9. The Traffic Light Reporter
2.10. Animal Models
2.11. Patient Sample Collection
2.12. Histology, Immunohistochemistry, and Alcian Blue
2.13. Statistical Analysis
3. Results
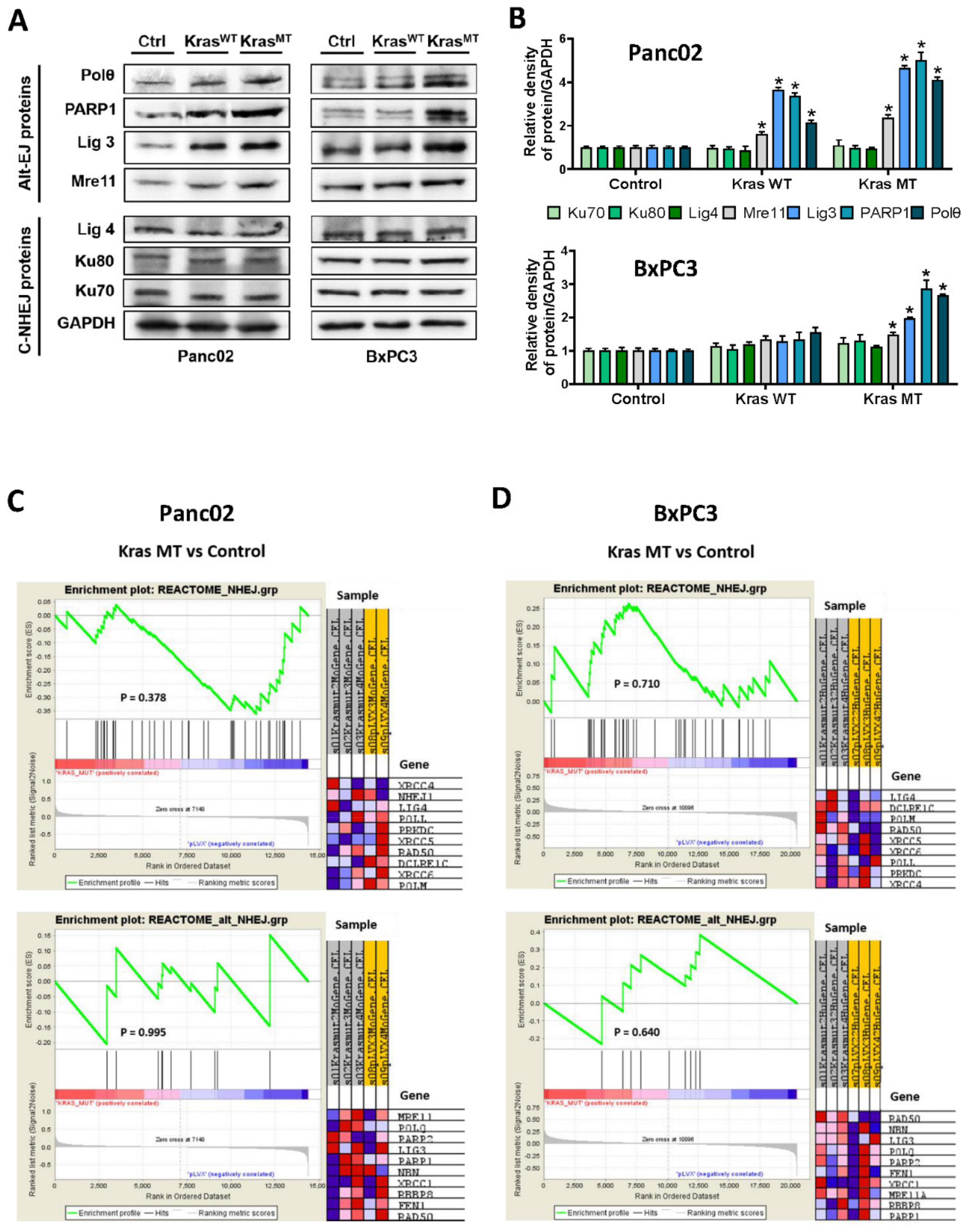
3.1. Oncogenic KRASG12D and Its Impact on the Protein Expression of the Alt-EJ Repair Pathway
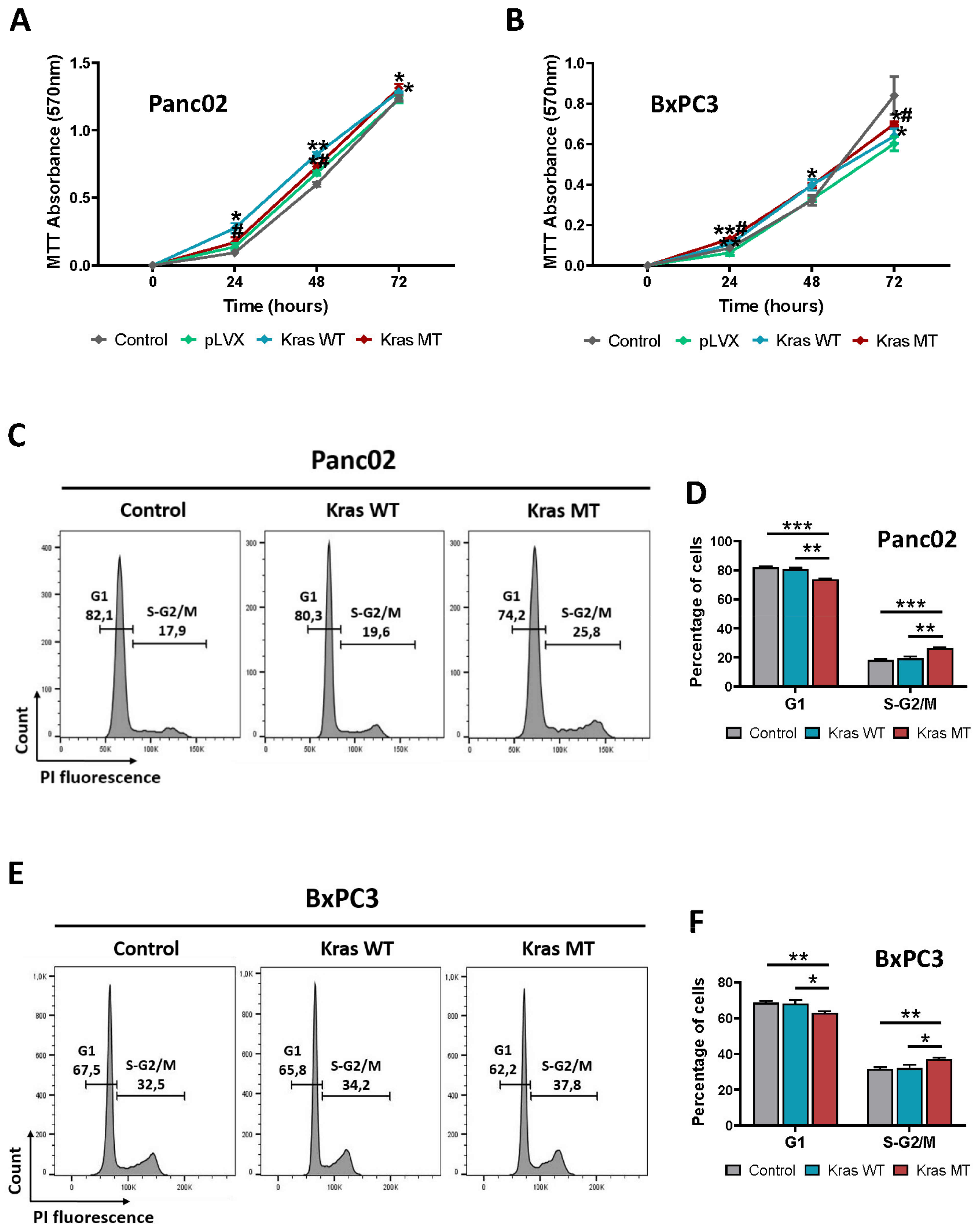
3.2. KRAS Overexpression Promotes Proliferation and Arrest in S/G2-M Phase of the Cell Cycle in Mouse and Human Pancreatic Cancer Cells In Vitro
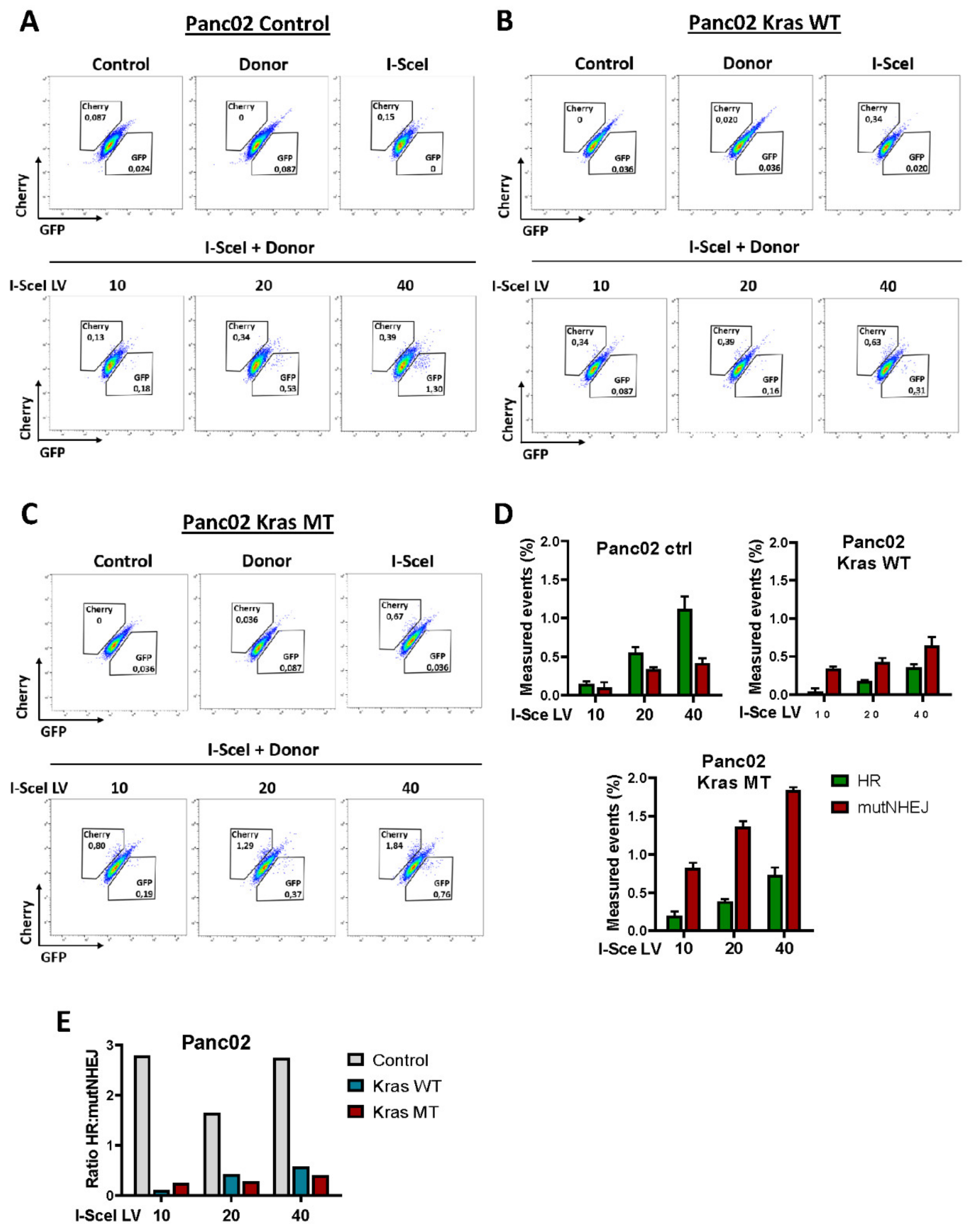
3.3. Exogenous KRASG12D Activates alt-EJ in Pancreatic Cancer Cells
3.4. Polθ Ablation Affects Disease Progression in Animal Models of Pancreatic Ductal Adenocarcinoma
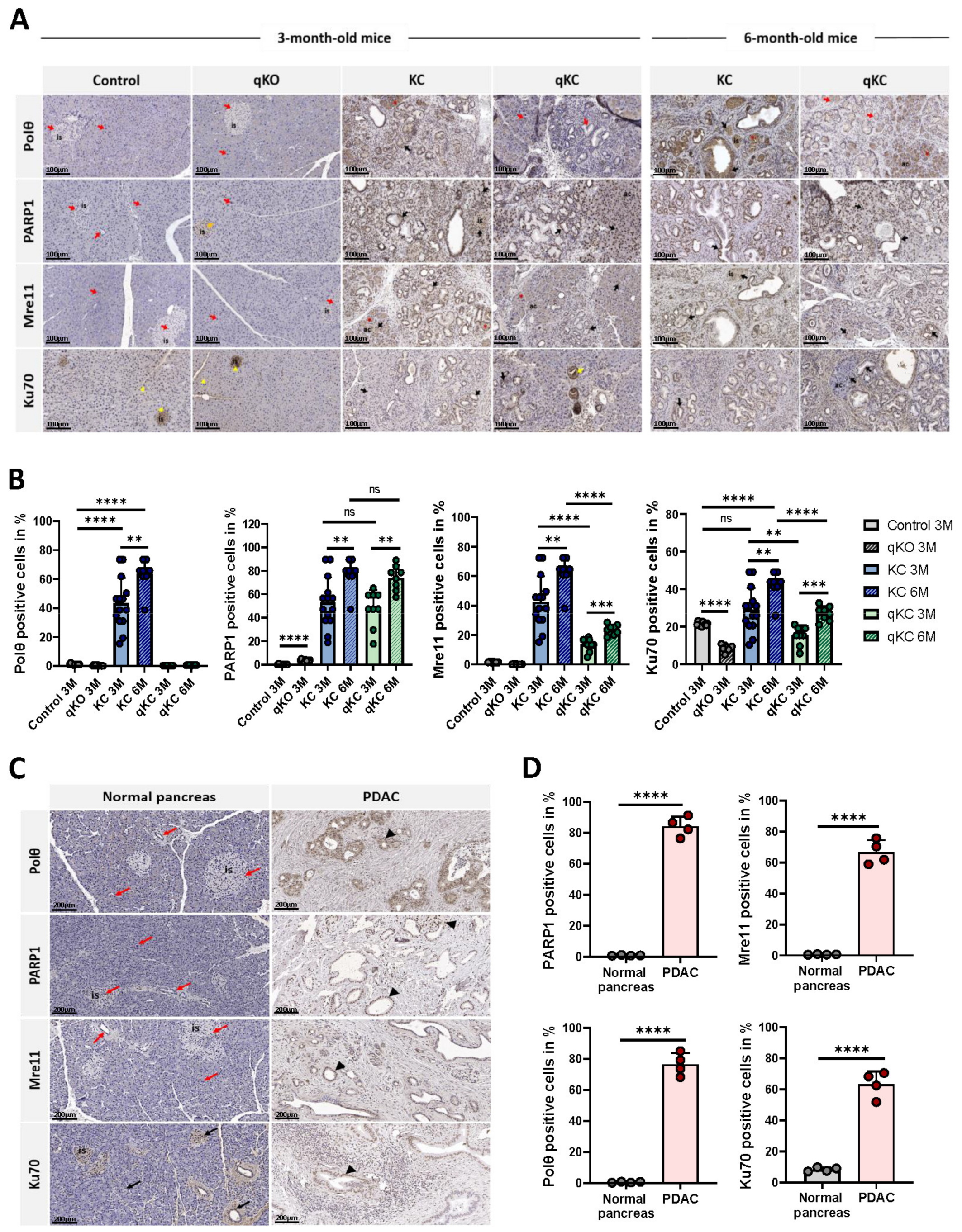
3.5. Oncogenic KRASG12D Promotes Expression of NHEJ Proteins in Pancreatic Ductal Adenocarcinoma
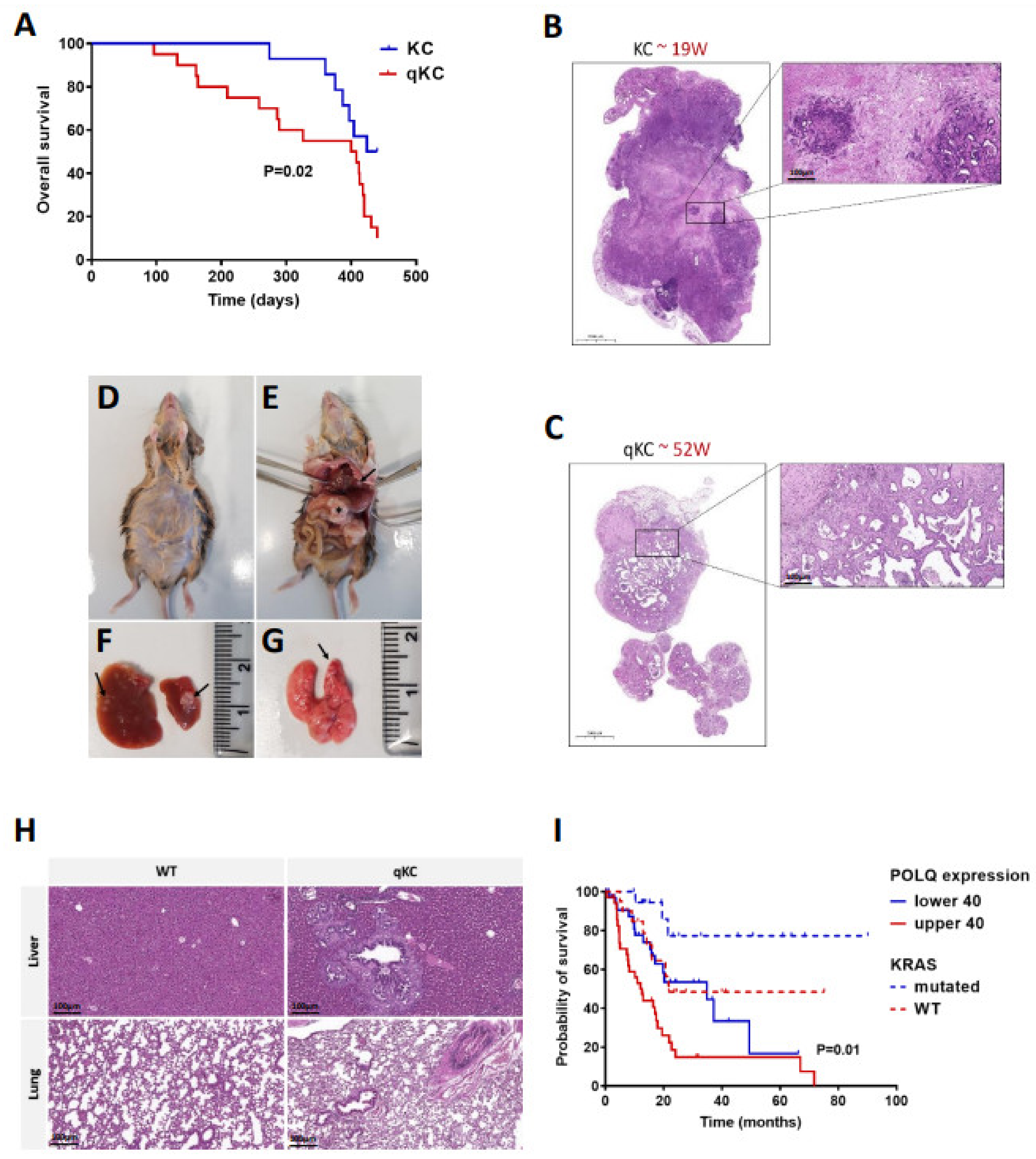
3.6. Polθ Is Essential for PDAC Progression and Overall Survival
3.7. Effect of Polθ Deficiency on Signaling Pathways in Oncogenic KRAS-Driven Mouse Models
4. Discussion
4.1. The alt-EJ Pathway Proteins Are Upregulated in Pancreatic Cancer Cells Expressing Oncogenic KRASG12D
4.2. Repair of DNA Double-Strand Breaks by alt-EJ Pathway in Pancreatic Cancer Cells Harboring Oncogenic KRASG12D
4.3. Depletion of Polymerase Theta Delays Pancreatic Cancer Progression in KrasG12D-Driven Mouse Model
4.4. Polθ Deficiency Dampens Cell Proliferation, Migration, and Invasion in KrasG12D-Driven Mouse Model of PDAC
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Basturk, O.; Hong, S.-M.; Wood, L.D.; Adsay, V.; Albores-Saavedra, J.; Biankin, A.; Brosens, L.A.; Fukushima, N.; Goggins, M.; Hruban, R.H.; et al. A Revised Classification System and Recommendations from the Baltimore Consensus Meeting for Neoplastic Precursor Lesions in the Pancreas. Am. J. Surg. Pathol. 2015, 39, 1730–1741. [Google Scholar] [CrossRef] [PubMed]
- Hruban, R.H.; Maitra, A.; Goggins, M. Update on Pancreatic Intraepithelial Neoplasia. Int. J. Clin. Exp. Pathol. 2008, 1, 306–316. [Google Scholar] [PubMed]
- Hunter, J.C.; Manandhar, A.; Carrasco, M.A.; Gurbani, D.; Gondi, S.; Westover, K.D. Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations. Mol. Cancer Res. 2015, 13, 1325–1335. [Google Scholar] [CrossRef]
- Zafra, M.P.; Parsons, M.J.; Kim, J.; Alonso-Curbelo, D.; Goswami, S.; Schatoff, E.M.; Han, T.; Katti, A.; Fernandez, M.T.C.; Wilkinson, J.E.; et al. An In Vivo Kras Allelic Series Reveals Distinct Phenotypes of Common Oncogenic Variants. Cancer Discov. 2020, 10, 1654–1671. [Google Scholar] [CrossRef]
- Winters, I.P.; Chiou, S.H.; Paulk, N.K.; McFarland, C.D.; Lalgudi, P.V.; Ma, R.K.; Lisowski, L.; Connolly, A.J.; Petrov, D.A.; Kay, M.A.; et al. Multiplexed in vivo homology-directed repair and tumor barcoding enables parallel quantification of Kras variant oncogenicity. Nat. Commun. 2017, 8, 2053. [Google Scholar] [CrossRef]
- Hingorani, S.R.; Petricoin, E.F.; Maitra, A.; Rajapakse, V.; King, C.; Jacobetz, M.A.; Ross, S.; Conrads, T.P.; Veenstra, T.D.; Hitt, B.A.; et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 2003, 4, 437–450. [Google Scholar] [CrossRef]
- Hingorani, S.R.; Wang, L.; Multani, A.S.; Combs, C.; Deramaudt, T.B.; Hruban, R.H.; Rustgi, A.K.; Chang, S.; Tuveson, D.A. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 2005, 7, 469–483. [Google Scholar] [CrossRef]
- Buscail, L.; Bournet, B.; Cordelier, P. Role of oncogenic Kras in the diagnosis, prognosis and treatment of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 153–168. [Google Scholar] [CrossRef]
- Schrempf, A.; Slyskova, J.; Loizou, J.I. Targeting the DNA Repair Enzyme Polymerase θ in Cancer Therapy. Trends Cancer 2021, 7, 98–111. [Google Scholar] [CrossRef]
- Brambati, A.; Barry, R.M.; Sfeir, A. DNA polymerase theta (Polθ)—An error-prone polymerase necessary for genome stability. Curr. Opin. Genet. Dev. 2020, 60, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Dueva, R.; Iliakis, G. Alternative pathways of non-homologous end joining (NHEJ) in genomic instability and cancer. Transl. Cancer Res. 2013, 2, 163–177. [Google Scholar]
- Wang, H.; Rosidi, B.; Perrault, R.; Wang, M.; Zhang, L.; Windhofer, F.; Iliakis, G. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 2005, 65, 4020–4030. [Google Scholar] [CrossRef] [PubMed]
- Mladenov, E.; Iliakis, G. Induction and repair of DNA double strand breaks: The increasing spectrum of non-homologous end joining pathways. Mutat. Res./Fundam. Mol. Mech. Mutagenesis 2011, 711, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Thyme, S.B.; Schier, A.F. Polq-mediated end joining is essential for surviving DNA double-strand breaks during early zebrafish development. Cell Rep. 2016, 15, 707–714. [Google Scholar] [CrossRef]
- Corneo, B.; Wendland, R.L.; Deriano, L.; Cui, X.; Klein, I.A.; Wong, S.-Y.; Arnal, S.; Holub, A.J.; Weller, G.R.; Pancake, B.A.; et al. Rag mutations reveal robust alternative end joining. Nature 2007, 449, 483486. [Google Scholar] [CrossRef]
- Sallmyr, A.; Tomkinson, A.E. Repair of DNA double-strand breaks by mammalian alternative end-joining pathways. J. Biol. Chem. 2018, 293, 10536–10546. [Google Scholar] [CrossRef]
- Wood, R.D.; Doublié, S. DNA polymerase θ (POLQ), double-strand break repair, and cancer. DNA Repair 2016, 44, 22–32. [Google Scholar] [CrossRef]
- Kawamura, K.; Bahar, R.; Seimiya, M.; Chiyo, M.; Wada, A.; Okada, S.; Hatano, M.; Tokuhisa, T.; Kimura, H.; Watanabe, S.; et al. DNA polymerase theta is preferentially expressed in lymphoid tissues and upregulated in human cancers. Int. J. Cancer 2004, 109, 9–16. [Google Scholar] [CrossRef]
- Ceccaldi, R.; Liu, J.C.; Amunugama, R.; Hajdu, I.; Primack, B.; Petalcorin, M.; O’Connor, K.; Konstantinopoulos, P.A.; Elledge, S.J.; Boulton, S.J.; et al. Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature 2015, 518, 258–262. [Google Scholar] [CrossRef]
- Mateos-Gomez, P.A.; Gong, F.; Nair, N.; Miller, K.M.; Denchi, E.L.; Sfeir, A. Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature 2015, 518, 254–257. [Google Scholar] [CrossRef] [PubMed]
- Lemée, F.; Bergoglio, V.; Fernandez-Vidal, A.; Machado-Silva, A.; Pillaire, M.J.; Bieth, A.; Gentil, C.; Baker, L.; Martin, A.L.; Leduc, C.; et al. DNA polymerase theta up-regulation is associated with poor survival in breast cancer, perturbs DNA replication, and promotes genetic instability. Proc. Natl. Acad. Sci. USA 2010, 107, 13390–13395. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2015, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Waters, A.M.; Der, C.J. KRAS: The Critical Driver and Therapeutic Target for Pancreatic Cancer. Cold Spring Harb. Perspect. Med. 2018, 8, a031435. [Google Scholar] [CrossRef]
- Collins, M.A.; Pasca di Magliano, M. Kras as a key oncogene and therapeutic target in pancreatic cancer. Front. Physiol. 2014, 4, 407. [Google Scholar] [CrossRef]
- Ozaki, T.; Nakagawara, A. Role of p53 in Cell Death and Human Cancers. Cancers 2011, 3, 994–1013. [Google Scholar] [CrossRef]
- Powell, E.; Piwnica-Worms, D.; Piwnica-Worms, H. Contribution of p53 to metastasis. Cancer Discov. 2014, 4, 405–414. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Yang, J.; Ni, X.; Liu, S.; Li, Z.; EHodges, S.; EFisher, W.; CBrunicardi, F.; AGibbs, R.; et al. Genomic sequencing of key genes in mouse pancreatic cancer cells. Curr. Mol. Med. 2012, 12, 331–341. [Google Scholar] [CrossRef]
- Jinesh, G.G.; Sambandam, V.; Vijayaraghavan, S.; Balaji, K.; Mukherjee, S. Molecular genetics and cellular events of K-Ras-driven tumorigenesis. Oncogene 2018, 37, 839–846. [Google Scholar] [CrossRef]
- Evan, G.; Vousden, K. Proliferation, cell cycle and apoptosis in cancer. Nature 2001, 411, 342348. [Google Scholar] [CrossRef]
- Feitelson, M.A.; Arzumanyan, A.; Kulathinal, R.J.; Blain, S.W.; Holcombe, R.F.; Mahajna, J.; Marino, M.; Martinez-Chantar, M.L.; Nawroth, R.; Sanchez-Garcia, I.; et al. Sustained proliferation in cancer: Mechanisms and novel therapeutic targets. Semin. Cancer Biol. 2015, 35, 25–54. [Google Scholar]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-homologous DNA end joining and alternative pathways to doublestrand break repair. Nat. Rev. Mol. Cell Biol. 2017, 18, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Carvajal-Garcia, J.; Cho, J.-E.; Carvajal-Garcia, P.; Feng, W.; Wood, R.D.; Sekelsky, J.; Gupta, G.P.; Roberts, S.A.; Ramsden, D.A. Mechanistic basis for microhomology identification and genome scarring by polymerase theta. Proc. Natl. Acad. Sci. USA 2020, 117, 8476–8485. [Google Scholar] [CrossRef] [PubMed]
- Certo, M.T.; Ryu, B.Y.; E Annis, J.; Garibov, M.; Jarjour, J.; Rawlings, D.J.; Scharenberg, A.M. Tracking genome engineering outcome at individual DNA breakpoints. Nat. Methods 2011, 8, 671–676. [Google Scholar] [CrossRef] [PubMed]
- Herreros-Villanueva, M.; Hijona, E.; Cosme, A.; Bujanda, L. Mouse models of pancreatic cancer. World J. Gastroenterol. 2012, 18, 1286–1294. [Google Scholar] [CrossRef] [PubMed]
- Allera-Moreau, C.; Rouquette, I.; Lepage, B.A.; Oumouhou, N.; Walschaerts, M.; Leconte, E.; Schilling, V.; Gordien, K.; Brouchet, L.; Delisle, M.B.; et al. DNA replication stress response involving PLK1, CDC6, POLQ, RAD51 and CLASPIN upregula-tion prognoses the outcome of early/mid-stage non-small cell lung cancer patients. Oncogenesis 2012, 1, e30. [Google Scholar] [CrossRef] [PubMed]
- Higgins, G.S.; Harris, A.L.; Prevo, R.; Helleday, T.; McKenna, W.G.; Buffa, F.M. Overexpression of POLQ confers a poor prognosis in early breast cancer patients. Oncotarget 2010, 1, 175–184. [Google Scholar] [CrossRef]
- Shima, N.; Munroe, R.J.; Schimenti, J.C. The Mouse Genomic Instability Mutation chaos1 Is an Allele of Polq That Exhibits Genetic Interaction with Atm. Mol. Cell. Biol. 2004, 24, 10381–10389. [Google Scholar] [CrossRef]
- Goff, J.P.; Shields, D.S.; Seki, M.; Choi, S.; Epperly, M.W.; Dixon, T.; Wang, H.; Bakkenist, C.J.; Dertinger, S.D.; Torous, D.K.; et al. Lack of DNA polymerase Q radiosensitizes bone marrow stromal cells in vitro and increases reticulocyte micronuclei after total-body irradiaton. Radiat. Res. 2009, 172, 165–174. [Google Scholar] [CrossRef]
- Hruban, R.H.; Adsay, N.V.; Albores-Saavedra, J.; Compton, C.; Garrett, E.S.; Goodman, S.N.; Kern, S.E.; Klimstra, D.S.; Klöppel, G.; Longnecker, D.S.; et al. Pancreatic intraepithelial neoplasia: A new nomenclature and classification system for pancreatic duct lesions. Am. J. Surg. Pathol. 2001, 25, 579–586. [Google Scholar] [CrossRef]
- Jonckheere, N.; Vasseur, R.; van Seuningen, I. The cornerstone K-RAS mutation in pancreatic adenocarcinoma: From cell signaling network, target genes, biological processes to therapeutic targeting. Crit. Rev. Oncol. Hembuscailatol. 2017, 111, 7–19. [Google Scholar] [CrossRef] [PubMed]
- Scholzen, T.; Gerdes, J. The Ki-67 protein: From the known and the unknown. J. Cell. Physiol. 2000, 182, 311–322. [Google Scholar] [CrossRef]
- Wang, S.C. PCNA: A silent housekeeper or a potential therapeutic target? Trends Pharmacol. Sci. 2014, 35, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Maga, G.; Hubscher, U. Proliferating cell nuclear antigen (PCNA): A dancer with many partners. J. Cell Sci. 2003, 116, 3051–3060. [Google Scholar] [CrossRef]
- Zinczuk, J.; Zareba, K.; Guzinska-Ustymowicz, K.; Kedra, B.; Kemona, A.; Pryczynicz, A. Expression of chosen cell cycle and proliferation markers in pancreatic intraepithelial neoplasia. Prz. Gastroenterol. 2018, 13, 118–126. [Google Scholar] [CrossRef]
- Fusté, N.P.; Castelblanco, E.; Felip, I.; Santacana, M.; Fernández-Hernández, R.; Gatius, S.; Pedraza, N.; Pallarés, J.; Cemeli, T.; Valls, J.; et al. Characterization of cytoplasmic cyclin D1 as a marker of invasiveness in cancer. Oncotarget 2016, 7, 26979–26991. [Google Scholar] [CrossRef]
- Principe, D.; Diaz, A.M.; Torres, C.; Mangan, R.J.; DeCant, B.; McKinney, R.; Tsao, M.S.; Lowy, A.; Munshi, H.G.; Jung, B.; et al. TGFβ engages MEK/ERK to differentially regulate benign and malignant pancreas cell function. Oncogene 2017, 36, 4336–4348. [Google Scholar] [CrossRef]
- Cagnol, S.; Chambard, J.C. ERK and cell death: Mechanisms of ERK-induced cell death-apoptosis, autophagy and senescence. FEBS J. 2010, 277, 2–21. [Google Scholar] [CrossRef]
- Dubois, R.N.; Abramson, S.B.; Crofford, L.; Gupta, R.A.; Simon, L.S.; van de Putte, L.B.; Lipsky, P.E. Cyclooxygenase in biology and disease. FASEB J. 2010, 12, 1063–1073. [Google Scholar] [CrossRef]
- Maitra, A.; Ashfaq, R.; Gunn, C.R.; Rahman, A.; Yeo, C.J.; Sohn, T.A.; Cameron, J.L.; Hruban, R.H.; Wilentz, R.E. Cyclooxygenase 2 expression in pancreatic adenocarcinoma and pancreatic intraepithelial neoplasia: An immunohistochemical analysis with automated cellular imaging. Am. J. Clin. Pathol. 2002, 118, 194–201. [Google Scholar] [CrossRef]
- Mueller, S.; Engleitner, T.; Maresch, R.; Zukowska, M.; Lange, S.; Kaltenbacher, T.; Konukiewitz, B.; Öllinger, R.; Zwiebel, M.; Strong, A.; et al. Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. Nature 2018, 554, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Waddell, N.; Pajic, M.; Patch, A.M.; Chang, D.K.; Kassahn, K.S.; Bailey, P.; Johns, A.L.; Miller, D.; Nones, K.; Quek, K.; et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015, 518, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Rowley, M.; Ohashi, A.; Mondal, G.; Mills, L.; Yang, L.; Zhang, L.; Sundsbak, R.; Shapiro, V.; Muders, M.H.; Smyrk, T.; et al. Inactivation of BRCA2 promotes Trp53-associated but inhibits KrasG12D-dependent pancreatic cancer development in mice. Gastroenterology 2011, 140, 1303–1313. [Google Scholar] [CrossRef] [PubMed]
- Sebesta, M.; Krejci, L. Mechanism of homologous recombination. In DNA Replication, Recombination, and Repair; Hanaoka, F., Sugasawa, K., Eds.; Springer: Tokyo, Japan, 2016. [Google Scholar]
- Moynahan, M.E.; Jasin, M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol. 2010, 11, 196–207. [Google Scholar] [CrossRef]
- Hähnel, P.S.; Enders, B.; Sasca, D.; Roos, W.P.; Kaina, B.; Bullinger, L.; Theobald, M.; Kindler, T. Targeting components of the alternative NHEJ pathway sensitizes KRAS mutant leukemic cells to chemotherapy. Blood 2014, 123, 2355–2366. [Google Scholar] [CrossRef]
- Sallmyr, A.; Tomkinson, A.E.; Rassool, F.V. Up-regulation of WRN and DNA ligase IIIalpha in chronic myeloid leukemia: Consequences for the repair of DNA double-strand breaks. Blood 2008, 112, 1413–1423. [Google Scholar] [CrossRef]
- Pulciani, S.; Santos, E.; Long, L.K.; Sorrentino, V.; Barbacid, M. Ras gene amplification and malignant transformation. Mol. Cell. Biol. 1985, 5, 2836–2841. [Google Scholar]
- Wong, G.S.; Zhou, J.; Liu, J.B.; Wu, Z.; Xu, X.; Li, T.; Xu, D.; Schumacher, S.E.; Puschhof, J.; McFarland, J.; et al. Targeting wild-type KRAS-amplified gastroesophageal cancer through combined MEK and SHP2 inhibition. Nat. Med. 2018, 24, 968–977. [Google Scholar] [CrossRef]
- Chang, E.H.; Furth, M.E.; Scolnick, E.M.; Lowy, D.R. Tumorigenic transformation of mammalian cells induced by a normal human gene homologous to the oncogene of Harvey murine sarcoma virus. Nature 1982, 297, 479–483. [Google Scholar] [CrossRef]
- Spandidos, D.A.; Wilkie, N.M. Malignant transformation of early passage rodent cells by single mutated human oncogene. Nature 1984, 310, 469–475. [Google Scholar] [CrossRef]
- Taylor, R.; Webb Robertson, B.-J.M.; Markillie, L.M.; Serres, M.H.; Linggi, B.E.; Aldrich, J.T.; Hill, E.A.; Romine, M.F.; Lipton, M.S.; Wiley, H.S. Steady State Analysis of Integrated Proteomics and Transcriptomics Data Shows Changes in Translational Efficiency a Dominant Regulatory Mechanism in the Environmental Response of Bacteria. Integr. Biol. 2013, 5, 1393–1406. [Google Scholar] [CrossRef] [PubMed]
- Oliveto, S.; Mancino, M.; Manfrini, N.; Biffo, S. Role of microRNAs in translation regulation and cancer. World J. Biol. Chem. 2017, 8, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Harvey, R.F.; Smith, T.S.; Mulroney, T.; Queiroz, R.M.L.; Pizzinga, M.; Dezi, V.; Villenueva, E.; Ramakrishna, M.; Lilley, K.S.; Willis, A.E. Trans-acting translational regulatory RNA binding proteins. Wiley Interdiscip. Rev. RNA 2018, 9, e1465. [Google Scholar] [CrossRef]
- Vogel, C.; Marcotte, E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012, 13, 227–232. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Li, G.; Schätzlein, A.G.; Humphrey, P.A.; Weiss, R.M.; Uchegbu, I.F.; Martin, D.T. Down-regulation of GP130 signaling sensitizes bladder cancer to cisplatin by impairing Ku70 DNA repair signaling and promoting apoptosis. Cell. Signal. 2021, 81, 109931. [Google Scholar] [CrossRef] [PubMed]
- Deriano, L.; Roth, D.B. Modernizing the Nonhomologous End-Joining Repertoire: Alternative and Classical NHEJ Share the Stage. Annu. Rev. Genet. 2013, 47, 433–455. [Google Scholar] [CrossRef] [PubMed]
- Hanscom, T.; McVey, M. Regulation of Error-Prone DNA Double-Strand Break Repair and Its Impact on Genome Evolution. Cells 2020, 9, 1657. [Google Scholar] [CrossRef]
- Truong, L.N.; Li, Y.; Shi, L.Z.; Hwang, P.Y.; He, J.; Wang, H.; Razavian, N.; Berns, M.W.; Wu, X. Microhomology-mediated end joining and homologous recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc. Natl. Acad. Sci. USA 2013, 110, 7720–7725. [Google Scholar] [CrossRef]
- Han, J.; Huang, J. DNA double-strand break repair pathway choice: The fork in the road. Genome Instab. Dis. 2020, 1, 10–19. [Google Scholar] [CrossRef]
- Agbunag, C.; Bar-Sagi, D. Oncogenic K-ras drives cell cycle progression and phenotypic conversion of primary pancreatic duct epithelial cells. Cancer Res. 2004, 64, 5659–5663. [Google Scholar] [CrossRef]
- Bennardo, N.; Gunn, A.; Cheng, A.; Hasty, P.; Stark, J.M. Limiting the persistence of a chromosome break diminishes its mutagenic potential. PLoS Genet. 2009, 5, e1000683. [Google Scholar] [CrossRef] [PubMed]
- Bentley, J.; Diggle, C.P.; Harnden, P.; Knowles, M.A.; Kiltie, A.E. DNA double strand break repair in human bladder cancer is error prone and involves microhomology-associated end-joining. Nucleic Acids Res. 2004, 32, 5249–5259. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ceccaldi, R.; Rondinelli, B.; D’Andrea, A.D. Repair Pathway Choices and Consequences at the Double-Strand Break. Trends Cell Biol. 2016, 26, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Andreassen, P.R.; Ho, G.P.; D’Andrea, A.D. DNA damage responses and their many interactions with the replication fork. Carcinogenesis 2006, 27, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Sonoda, E.; Sasaki, M.S.; Morrison, C.; Yamaguchi-Iwai, Y.; Takata, M.; Takeda, S. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol. Cell. Biol. 1999, 19, 5166–5169. [Google Scholar] [CrossRef]
- Bunting, S.F.; Callén, E.; Wong, N.; Chen, H.T.; Polato, F.; Gunn, A.; Bothmer, A.; Feldhahn, N.; Fernandez-Capetillo, O.; Cao, L. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 2010, 141, 243–254. [Google Scholar] [CrossRef]
- Jimeno, S.; Mejías-Navarro, F.; Prados-Carvajal, R.; Huertas, P. Controlling the balance between chromosome break repair pathways. Adv. Protein Chem. Struct. Biol. 2019, 115, 95–134. [Google Scholar]
- Wyatt, D.W.; Feng, W.; Conlin, M.P.; Yousefzadeh, M.J.; Roberts, S.A.; Mieczkowski, P.; Wood, R.D.; Gupta, G.P.; Ramsden, D.A. Essential Roles for Polymerase θ-Mediated End Joining in the Repair of Chromosome Breaks. Mol. Cell 2016, 63, 662–673. [Google Scholar] [CrossRef]
- Kent, T.; Chandramouly, G.; McDevitt, S.M.; Ozdemir, A.Y.; Pomerantz, R.T. Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase theta. Nat. Struct. Mol. Biol. 2015, 22, 230–237. [Google Scholar] [CrossRef]
- Krah, N.M.; de la O, J.P.; Swift, G.H.; Hoang, C.Q.; Willet, S.G.; Chen Pan, F.; Cash, G.M.; Bronner, M.P.; Wright, C.V.; MacDonald, R.J.; et al. The acinar differentiation determinant PTF1A inhibits initiation of pancreatic ductal adenocarcinoma. Elife 2015, 4, e07125. [Google Scholar] [CrossRef]
- Lee, A.Y.L.; Dubois, C.L.; Sarai, K.; Zarei, S.; Schaeffer, D.F.; Sander, M.; Kopp, J.L. Cell of origin affects tumour development and phenotype in pancreatic ductal adenocarcinoma. Gut 2019, 68, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.L.; d’Alincourt Salazar, M.; Mackenzie-Dyck, S.; D’Apuzzo, M.; Shih, H.P.; Manuel, E.R.; Diamond, D.J. Desmoplasia and oncogene driven acinar-to-ductal metaplasia are concurrent events during acinar cell-derived pancreatic cancer initiation in young adult mice. PLoS ONE. 2019, 14, e0221810. [Google Scholar] [CrossRef] [PubMed]
- King, T.C. Neoplasia. In Elsevier’s Integrated Pathology; Elsevier: Amsterdam, The Netherlands, 2007; Volume 1, pp. 111–143. [Google Scholar]
- Klein, W.M.; Hruban, R.H.; Klein-Szanto, A.J.; Wilentz, R.E. Direct correlation between proliferative activity and dysplasia in pancreatic intraepithelial neoplasia (PanIN): Additional evidence for a recently proposed model of progression. Mod. Pathol. 2002, 15, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, R.P.; Mortimer, G.; Muircheartaigh, I.O. Cell proliferation in breast tumours: Analysis of histological parameters Ki67 and PCNA expression. Ir. J. Med. Sci. 1993, 162, 343–347. [Google Scholar] [CrossRef]
- Qiu, X.; Mei, J.; Yin, J.; Wang, H.; Wang, J.; Xie, M. Correlation analysis between expression of PCNA, Ki67 and COX-2 and Xray features in mammography in breast cancer. Oncol. Lett. 2017, 14, 2912–2918. [Google Scholar] [CrossRef]
- Biankin, A.V.; Kench, J.G.; Biankin, S.A.; Lee, C.S.; Morey, A.L.; Dijkman, F.P.; Coleman, M.J.; Sutherland, R.L.; Henshall, S.M. Pancreatic intraepithelial neoplasia in association with intraductal papillary mucinous neoplasms of the pancreas: Implications for disease progression and recurrence. Am. J. Surg. Pathol. 2004, 28, 1184–1192. [Google Scholar] [CrossRef]
- Hitomi, M.; Stacey, D.W. Cyclin D1 production in cycling cells depends on ras in a cellcycle-specific manner. Curr. Biol. 1999, 9, 1075–1084. [Google Scholar] [CrossRef]
- Javle, M.M.; Gibbs, J.F.; Iwata, K.K.; Pak, Y.; Rutledge, P.; Yu, J.; Black, J.D.; Tan, D.; Khoury, T. Epithelial-mesenchymal transition (EMT) and activated extracellular signal-regulated kinase (p-Erk) in surgically resected pancreatic cancer. Ann. Surg. Oncol. 2007, 14, 3527–3533. [Google Scholar] [CrossRef]
- Yan, Z.; Ohuchida, K.; Fei, S.; Zheng, B.; Guan, W.; Feng, H.; Kibe, S.; Ando, Y.; Koikawa, K.; Abe, T.; et al. Inhibition of ERK1/2 in cancer-associated pancreatic stellate cells suppresses cancer–stromal interaction and metastasis. J. Exp. Clin. Cancer Res. 2019, 38, 221. [Google Scholar] [CrossRef]
- Wolff, H.; Saukkonen, K.; Anttila, S.; Karjalainen, A.; Vainio, H.; Ristimäki, A. Expression of cyclooxygenase-2 in human lung carcinoma. Cancer Res. 1998, 58, 4997–5001. [Google Scholar]
- Morris, C.D.; Armstrong, G.R.; Bigley, G.; Green, H.; Attwood, S.E. Cyclooxygenase-2 expression in the Barrett’s metaplasia-dysplasia adenocarcinoma sequence. Am. J. Gastroenterol. 2001, 96, 990–996. [Google Scholar] [CrossRef]
- Hwang, D.; Scollard, D.; Byrne, J.; Levine, E. Expression of cyclooxygenase-1 and cyclooxygenase-2 in human breast cancer. J. Natl. Cancer Inst. 1998, 90, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Komhoff, M.; Guan, Y.; Shappell, H.W.; Davis, L.; Jack, G.; Shyr, Y.; Koch, M.O.; Shappell, S.B.; Breyer, M.D. Enhanced expression of cyclooxygenase-2 in high grade human transitional cell bladder carcinomas. Am. J. Pathol. 2000, 157, 29–35. [Google Scholar] [CrossRef]
- Lee, L.M.; Pan, C.C.; Cheng, C.J.; Chi, C.W.; Liu, T.Y. Expression of cyclooxygenase-2 in prostate adenocarcinoma and benign prostatic hyperplasia. Anticancer Res. 2001, 21, 1291–1294. [Google Scholar] [PubMed]
- Colby, J.K.; Klein, R.D.; McArthur, M.J.; Conti, C.J.; Kiguchi, K.; Kawamoto, T.; Riggs, P.K.; Pavone, A.I.; Sawicki, J.; Fischer, S.M. Progressive metaplastic and dysplastic changes in mouse pancreas induced by cyclooxygenase-2 overexpression. Neoplasia 2008, 10, 782–796. [Google Scholar] [CrossRef] [PubMed]
- Hill, R.; Li, Y.; Tran, L.M.; Dry, S.; Calvopina, J.H.; Garcia, A.; Kim, C.; Wang, Y.; Donahue, T.R.; Herschman, H.R.; et al. Cell intrinsic role of COX-2 in pancreatic cancer development. Mol. Cancer Ther. 2012, 11, 2127–2137. [Google Scholar] [CrossRef]
- Wang, D.; Dubois, R.N. Prostaglandins and cancer. Gut 2006, 55, 115–122. [Google Scholar] [CrossRef]
- Gupta, G.P.; Nguyen, D.X.; Chiang, A.C.; Bos, P.D.; Kim, J.Y.; Nadal, C.; Gomis, R.R.; Manova-Todorova, K.; Massagué, J. Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature 2007, 446, 765–770. [Google Scholar] [CrossRef]
- Greenhough, A.; Smartt, H.J.; Moore, A.E.; Roberts, H.R.; Williams, A.C.; Paraskeva, C.; Kaidi, A. The COX-2/PGE2 pathway: Key roles in the hallmarks of cancer and adaptation to the tumour microenvironment. Carcinogenesis 2009, 30, 377–386. [Google Scholar] [CrossRef]
- Yu, J.R.; Wu, Y.J.; Qin, Q.; Lu, K.Z.; Yan, S.; Liu, X.S.; Zheng, S.S. Expression of cyclooxygenase-2 in gastric cancer and its relation to liver metastasis and long-term prognosis. World J. Gastroenterol. 2005, 11, 4908–4911. [Google Scholar] [CrossRef]
- Ghasemi, M.; Afshar, P.; Sheidaei, S.; Moeini, Y.; Vahedi Larijani, L. The role of immunohistochemistry expression of COX-2 in differentiating pigmented benign and malignant skin neoplasms. Med. J. Islamic Repub. Iran 2019, 33, 75. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smolinska, A.; Singer, K.; Golchert, J.; Smyczynska, U.; Fendler, W.; Sendler, M.; van den Brandt, J.; Singer, S.; Homuth, G.; Lerch, M.M.; et al. DNA Polymerase Theta Plays a Critical Role in Pancreatic Cancer Development and Metastasis. Cancers 2022, 14, 4077. https://doi.org/10.3390/cancers14174077
Smolinska A, Singer K, Golchert J, Smyczynska U, Fendler W, Sendler M, van den Brandt J, Singer S, Homuth G, Lerch MM, et al. DNA Polymerase Theta Plays a Critical Role in Pancreatic Cancer Development and Metastasis. Cancers. 2022; 14(17):4077. https://doi.org/10.3390/cancers14174077
Chicago/Turabian StyleSmolinska, Agnieszka, Kerstin Singer, Janine Golchert, Urszula Smyczynska, Wojciech Fendler, Matthias Sendler, Jens van den Brandt, Stephan Singer, Georg Homuth, Markus M. Lerch, and et al. 2022. "DNA Polymerase Theta Plays a Critical Role in Pancreatic Cancer Development and Metastasis" Cancers 14, no. 17: 4077. https://doi.org/10.3390/cancers14174077
APA StyleSmolinska, A., Singer, K., Golchert, J., Smyczynska, U., Fendler, W., Sendler, M., van den Brandt, J., Singer, S., Homuth, G., Lerch, M. M., & Moskwa, P. (2022). DNA Polymerase Theta Plays a Critical Role in Pancreatic Cancer Development and Metastasis. Cancers, 14(17), 4077. https://doi.org/10.3390/cancers14174077





