Brain Tumor Characterization Using Radiogenomics in Artificial Intelligence Framework
Abstract
Simple Summary
Abstract
1. Introduction
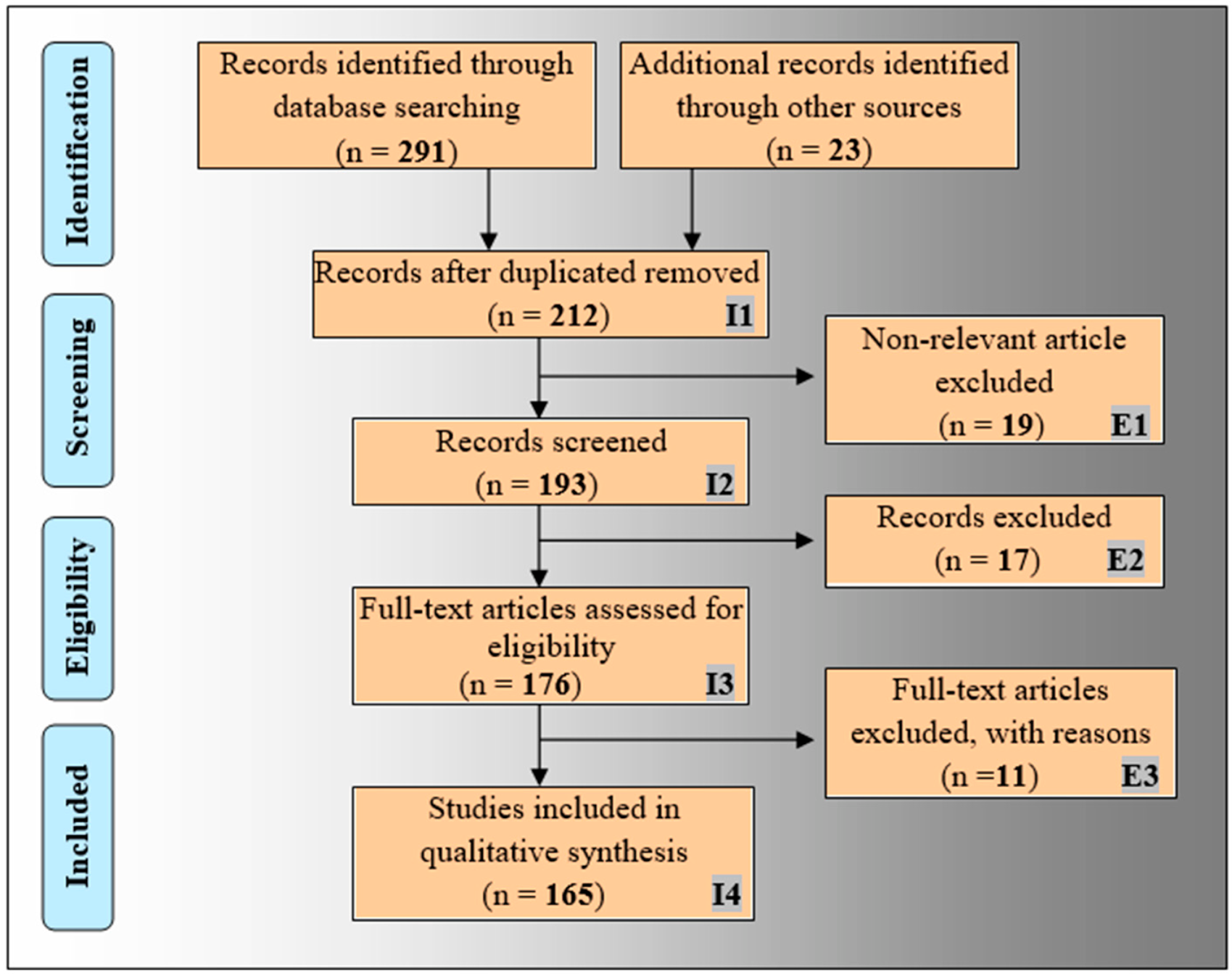
2. PRISMA Model and Search Strategy
2.1. The PRISMA Model
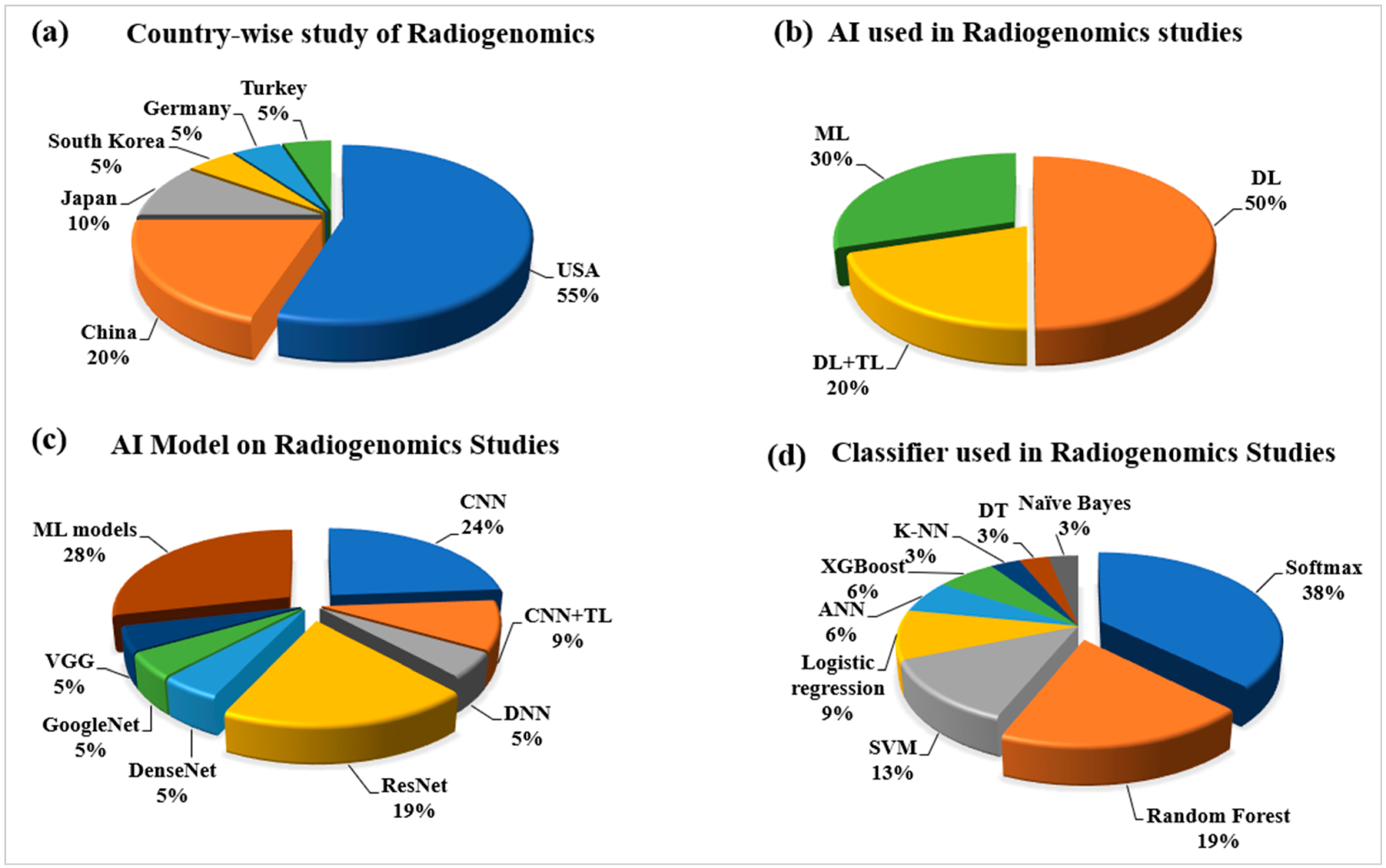
2.2. Statistical Distributions of AI Attributes for Radiogenomics Studies
2.2.1. Statistical Distribution of Country-Wise Study of Radiogenomics Using AI
2.2.2. Statistical Distribution of AI Used in the Radiogenomics’ System
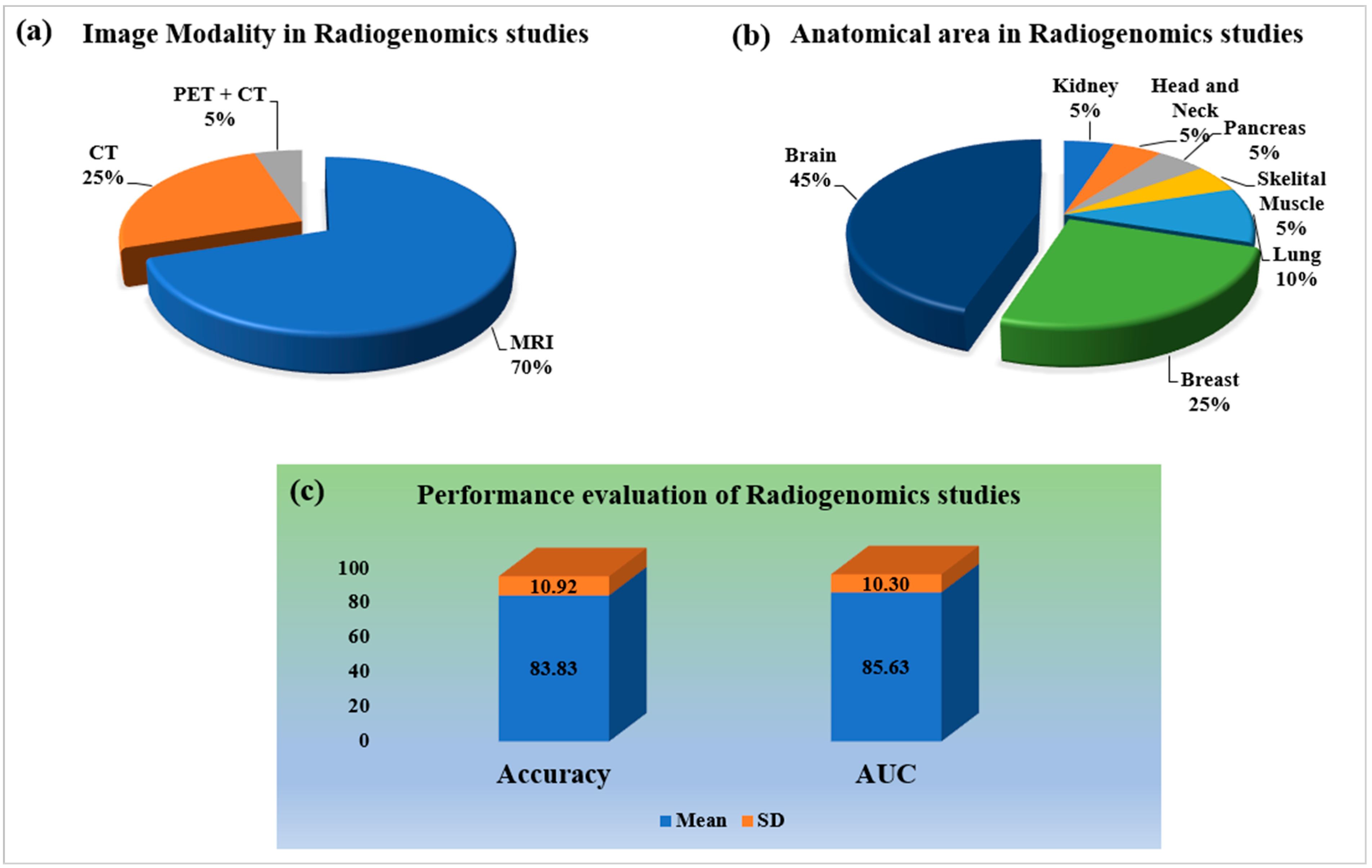
2.2.3. Statistical Distribution by Image Modality and Anatomical Area of Radiogenomics’ System
2.2.4. Statistical Distribution of Performance Evaluation on Radiogenomics System
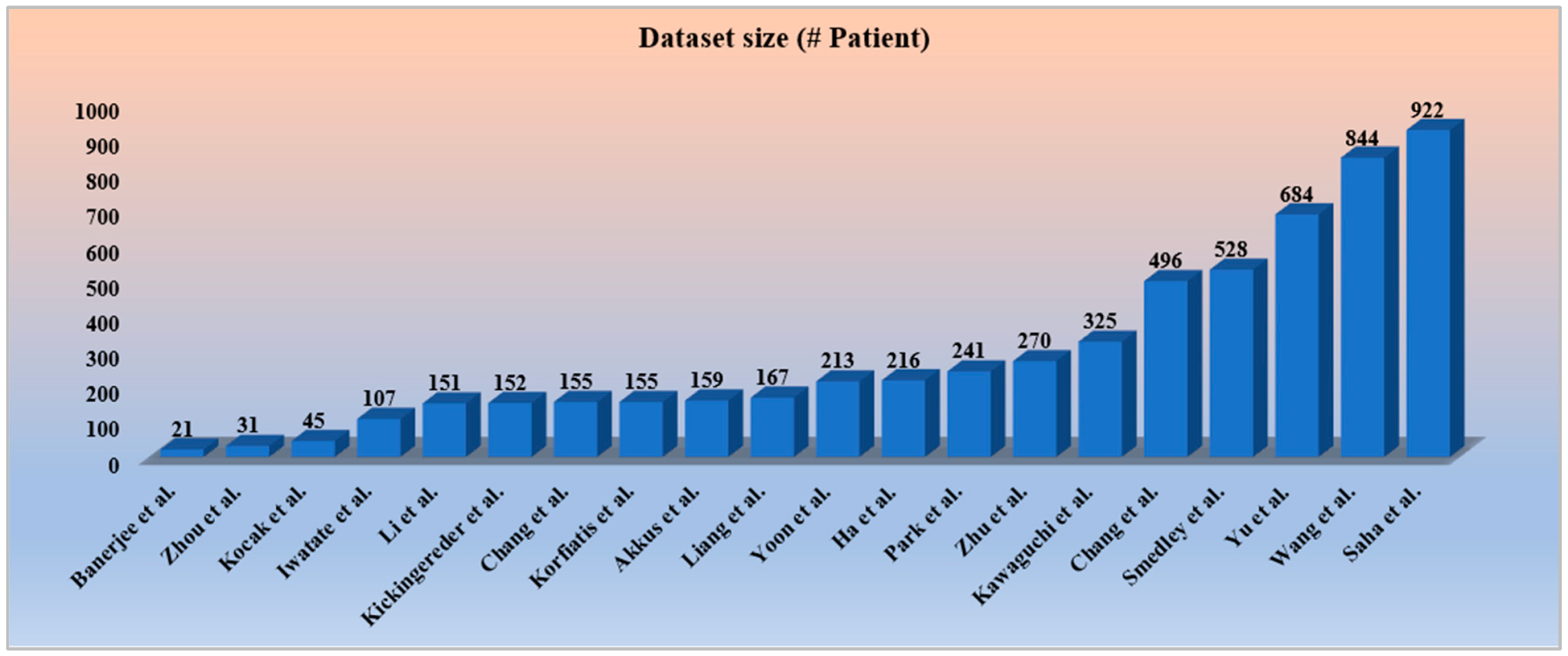
2.2.5. Statistical Distribution by Dataset Size Covering all the Objectives and Modalities
3. Biology of Brain Tumor
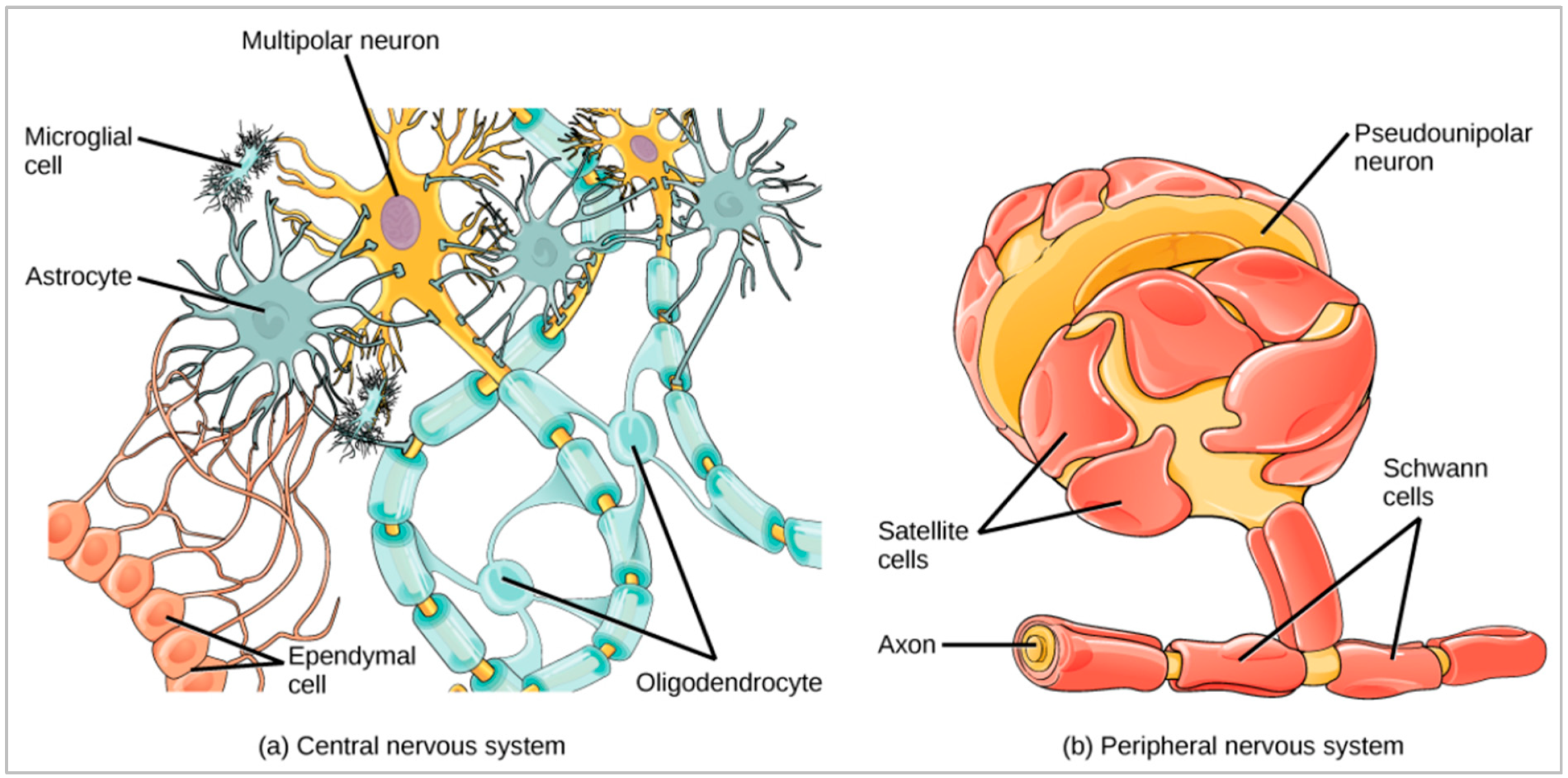
3.1. Brain Glia Cell and Deoxyribonucleic Acid
3.2. Mutation and Its Process
3.3. Central Nervous System and Brain Tumor Types’ Classification and Grading
4. Genetics of Brain Tumor
4.1. Genomics Types’ Information of Brain Tumor
4.2. Brain Tumor Types and Their Associated Genes
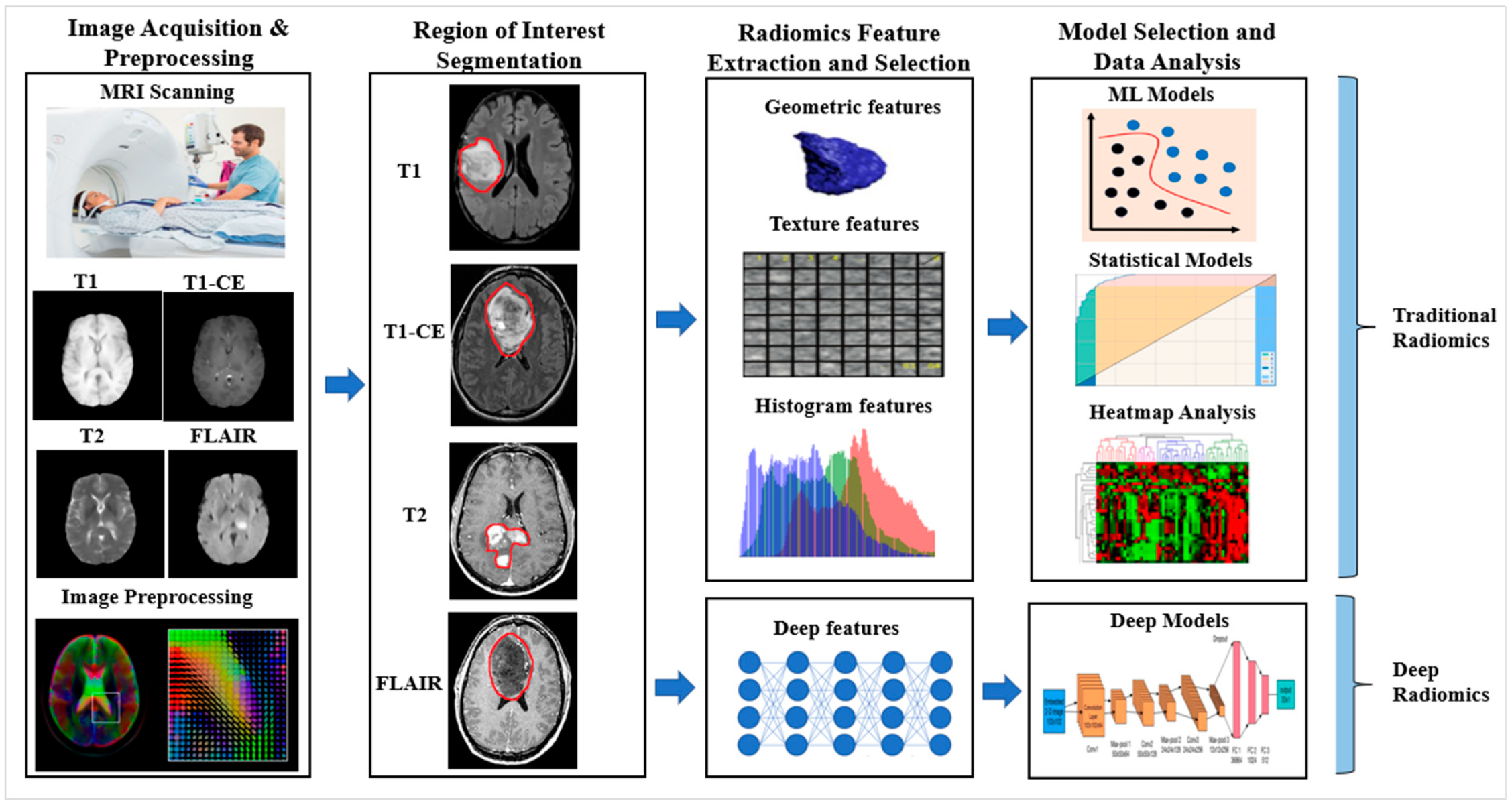
5. A Radiomics Approach to Tumor Characterization
5.1. Radiomics of Brain Tumor Characterization Using AI Framework
5.1.1. Traditional Radiomics
5.1.2. Deep Radiomics
6. AI Modelling Buffered with Genetics for BTC: A Radiogenomics Approach
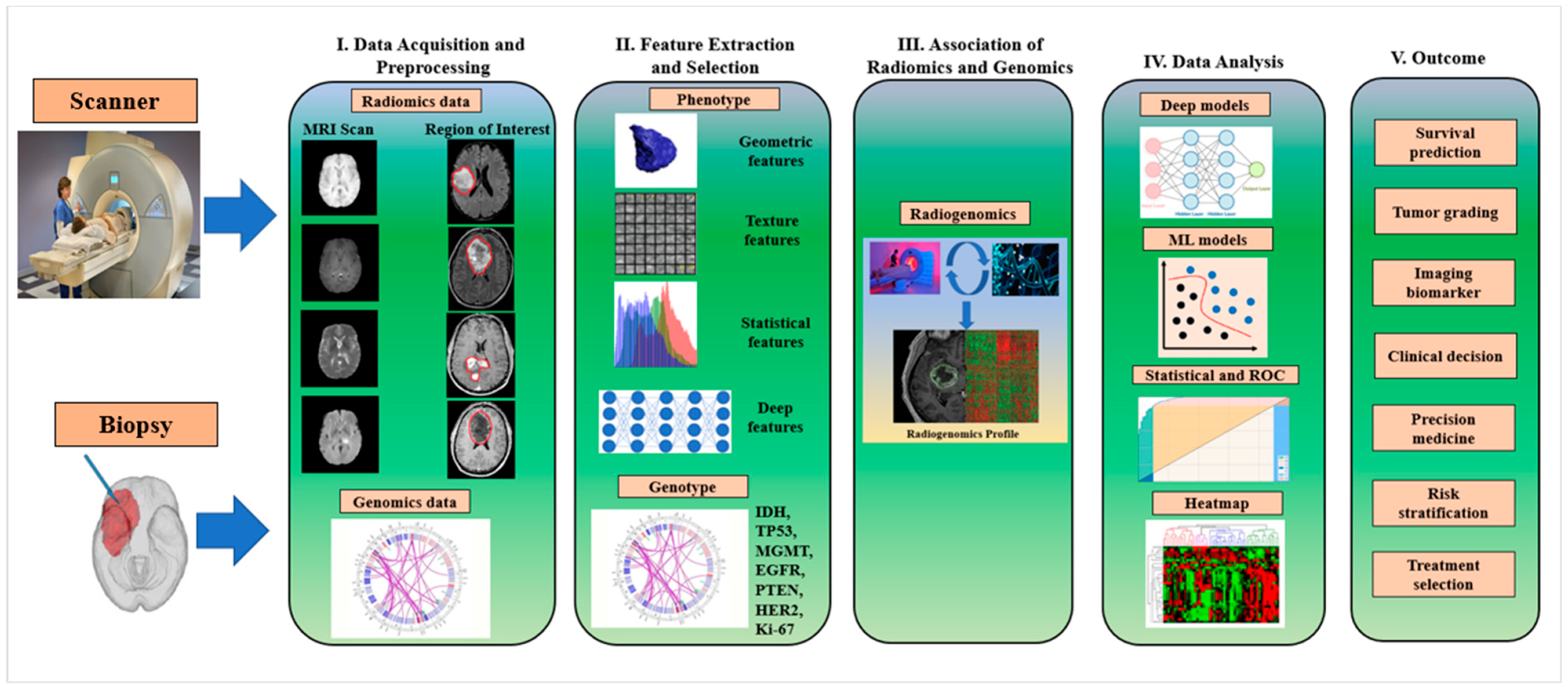
6.1. Radiogenomics Pipeline for Brain Tumor Classification Using AI Model
6.2. Challenges of Radiogenomics
7. Critical Discussion
7.1. Principal Findings
7.2. Benchmarking
7.3. Risk-of-Bias Analysis of Radiogenomics Studies
7.4. Advanced Features of Artificial Intelligence
7.5. Strength, Weakness, and Extension
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
| 1. Gliomas, glioneuronal tumors, and neuronal tumors |
| 1.1 Adult-type diffuse gliomas |
| 1.1.1 Astrocytoma, IDH-mutant 1.1.2 Glioblastoma, IDH-wildtype 1.1.3 Oligodendroglioma, IDH-mutant, and 1p/19q-co-deleted |
| 1.2 Pediatric-type diffuse low-grade gliomas |
| 1.2.1 Angiocentric glioma 1.2.2 Diffuse astrocytoma, MYB- or MYBL1-altered 1.2.3 Diffuse low-grade glioma, MAPK pathway-altered 1.2.4 Polymorphous low-grade neuroepithelial tumor of the young |
| 1.3 Pediatric-type diffuse high-grade gliomas |
| 1.3.1 Infant-type hemispheric glioma 1.3.2 Diffuse midline glioma, H3 K27-altered 1.3.3 Diffuse pediatric-type high-grade glioma, H3-wildtype and IDH-wildtype 1.3.4 Diffuse hemispheric glioma, H3 G34-mutant |
| 1.4 Circumscribed astrocytic gliomas |
| 1.4.1 Chordoid glioma 1.4.2 Pilocytic astrocytoma 1.4.3 Pleomorphic xanthoastrocytoma 1.4.4 High-grade astrocytoma with piloid features 1.4.5 Astroblastoma, MN1-altered 1.4.6 Subependymal giant cell astrocytoma |
| 1.5 Glioneuronal and neuronal tumors |
| 1.5.1 Ganglioglioma 1.5.2 Gangliocytoma 1.5.3 Central neurocytoma 1.5.4 Dysembryoplastic neuroepithelial tumor 1.5.5 Desmoplastic infantile ganglioglioma/desmoplastic infantile astrocytoma 1.5.6 Papillary glioneuronal tumor 1.5.7 Diffuse glioneuronal tumor with oligodendroglioma-like features and nuclear clusters 1.5.8 Myxoid glioneuronal tumor 1.5.9 Rosette-forming glioneuronal tumor 1.5.10 Multi-nodular and vacuolating neuronal tumor 1.5.11 Diffuse leptomeningeal glioneuronal tumor 1.5.12 Dysplastic cerebellar gangliocytoma (Lhermitte–-Duclos disease) 1.5.13 Cerebellar liponeurocytoma 1.5.14 Extraventricular neurocytoma |
| 1.6 Ependymal tumors |
| 1.6.1 Posterior fossa ependymoma 1.6.2 Posterior fossa ependymoma, group PFA 1.6.3 Posterior fossa ependymoma, group PFB 1.6.4 Supratentorial ependymoma 1.6.5 Supratentorial ependymoma, ZFTA fusion-positive 1.6.6 Supratentorial ependymoma, YAP1 fusion-positive 1.6.7 Spinal ependymoma, MYCN-amplified 1.6.8 Spinal ependymoma 1.6.9 Subependymoma 1.6.10 Myxopapillary ependymoma |
| 2. Choroid plexus tumors |
| 2.1 Choroid plexus papilloma 2.2 Choroid plexus carcinoma 2.3 Atypical choroid plexus papilloma |
| 3. Embryonal tumors |
| 3.1 Medulloblastoma |
| 3.1.1 Medulloblastomas, molecularly defined |
| 3.1.1.1 Medulloblastoma, WNT-activated 3.1.1.2 Medulloblastoma, SHH-activated and TP53-mutant 3.1.1.3 Medulloblastoma, SHH-activated and TP53-wildtype 3.1.1.4 Medulloblastoma, non-WNT/non-SHH |
| 3.1.2 Medulloblastomas, histologically defined |
| 3.2 Other CNS embryonal tumors |
| 3.2.1 Cribriform neuroepithelial tumor 3.2.2 Atypical teratoid/rhabdoid tumor 3.2.3 Embryonal tumor with multi-layered rosettes 3.2.4 CNS tumor with BCOR internal tandem duplication 3.2.5 CNS neuroblastoma, FOXR2-activated 3.2.6 CNS embryonal tumor |
| 4. Pineal tumors |
| 4.1 Pineocytoma 4.2 Pineoblastoma 4.3 Pineal parenchymal tumor of intermediate differentiation 4.4 Desmoplastic myxoid tumor of the pineal region, SMARCB1-mutant 4.5 Papillary tumor of the pineal region |
| 5. Cranial and paraspinal nerve tumors |
| 5.1 Schwannoma 5.2 Perineurioma 5.3 Neurofibroma 5.4 Malignant melanotic nerve sheath tumor 5.5 Malignant peripheral nerve sheath tumor 5.6 Hybrid nerve sheath tumor 5.7 Paraganglioma |
| 6. Meningiomas |
| 7. Mesenchymal, non-meningothelial tumors |
| 7.1 Soft tissue tumors |
| 7.1.1 Fibroblastic and myofibroblastic tumors |
| 7.1.1.1 Solitary fibrous tumor |
| 7.2 Vascular tumors |
| 7.2.1 Hemangioblastoma 7.2.2 Hemangiomas and vascular malformations |
| 7.3 Skeletal muscle tumors |
| 7.3.1 Rhabdomyosarcoma |
| 7.4 Uncertain differentiation |
| 7.4.1 Primary intracranial sarcoma, DICER1-mutant 7.4.2 Intracranial mesenchymal tumor, FET-CREB fusion-positive 7.4.3 CIC-rearranged sarcoma 7.4.4 Ewing sarcoma |
| 7.5 Chondro-osseous tumors |
| 7.5.1 Chondrogenic tumors |
| 7.5.1.1 Mesenchymal chondrosarcoma 7.5.1.2 Chondrosarcoma |
| 7.5.2 Notochordal tumors |
| 7.5.2.1 Chordoma (including poorly differentiated chordoma) |
| 8. Melanocytic tumors |
| 8.1 Diffuse meningeal melanocytic neoplasms |
| 8.1.1 Meningeal melanocytosis and meningeal melanomatosis |
| 8.2 Circumscribed meningeal melanocytic neoplasms |
| 9. Hematolymphoid tumors |
| 9.1 Lymphomas |
| 9.1.1 CNS lymphomas |
| 9.1.1.1 Lymphomatoid granulomatosis 9.1.1.2 Primary diffuse large B-cell lymphoma of the CNS 9.1.1.3 Intravascular large B-cell lymphoma 9.1.1.4 Immunodeficiency-associated CNS lymphoma |
| 9.1.2 Miscellaneous rare lymphomas in the CNS |
| 9.1.2.1 Other low-grade B-cell lymphomas of the CNS 9.1.2.2 MALT lymphoma of the dura 9.1.2.3 T-cell and NK/T-cell lymphomas 9.1.2.4 Anaplastic large cell lymphoma (ALK+/ALK−) |
| 9.2 Histiocytic tumors |
| 9.2.1 Rosai–Dorfman disease 9.2.2 Erdheim–Chester disease 9.2.3 Juvenile xanthogranuloma 9.2.4 Histiocytic sarcoma 9.2.5 Langerhans cell histiocytosis |
| 10. Germ cell tumors |
| 10.1 Germinoma 10.2 Immature teratoma 10.3 Choriocarcinoma 10.4 Mature teratoma 10.5 Embryonal carcinoma 10.6 Mixed germ cell tumor 10.7 Teratoma with somatic-type malignancy 10.8 Yolk sac tumor |
| 11. Tumors of the sellar region |
| 11.1 Pituitary blastoma 11.2 Pituitary adenoma/PitNET 11.3 Papillary craniopharyngioma 11.4 Adamantinomatous craniopharyngioma 11.5 Pituicytoma, granular cell tumor of the sellar region, and spindle cell oncocytoma |
| 12. Metastases to the CNS |
| 12.1 Metastases to the meninges 12.2 Metastases to the brain and spinal cord parenchyma |
Appendix B
Appendix B.1. Magnetic Resonance Imaging (MRI)
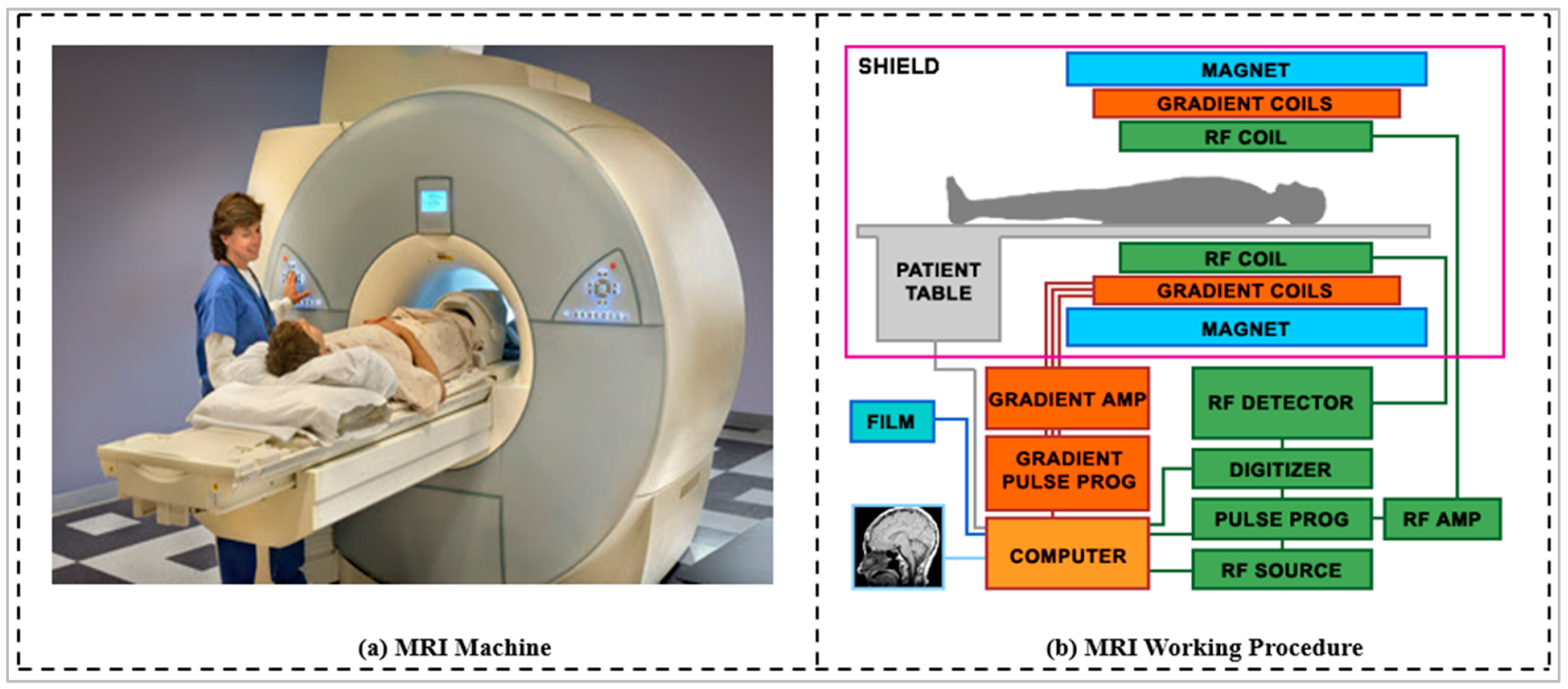
Appendix B.1.1. Image View or MRI Planes
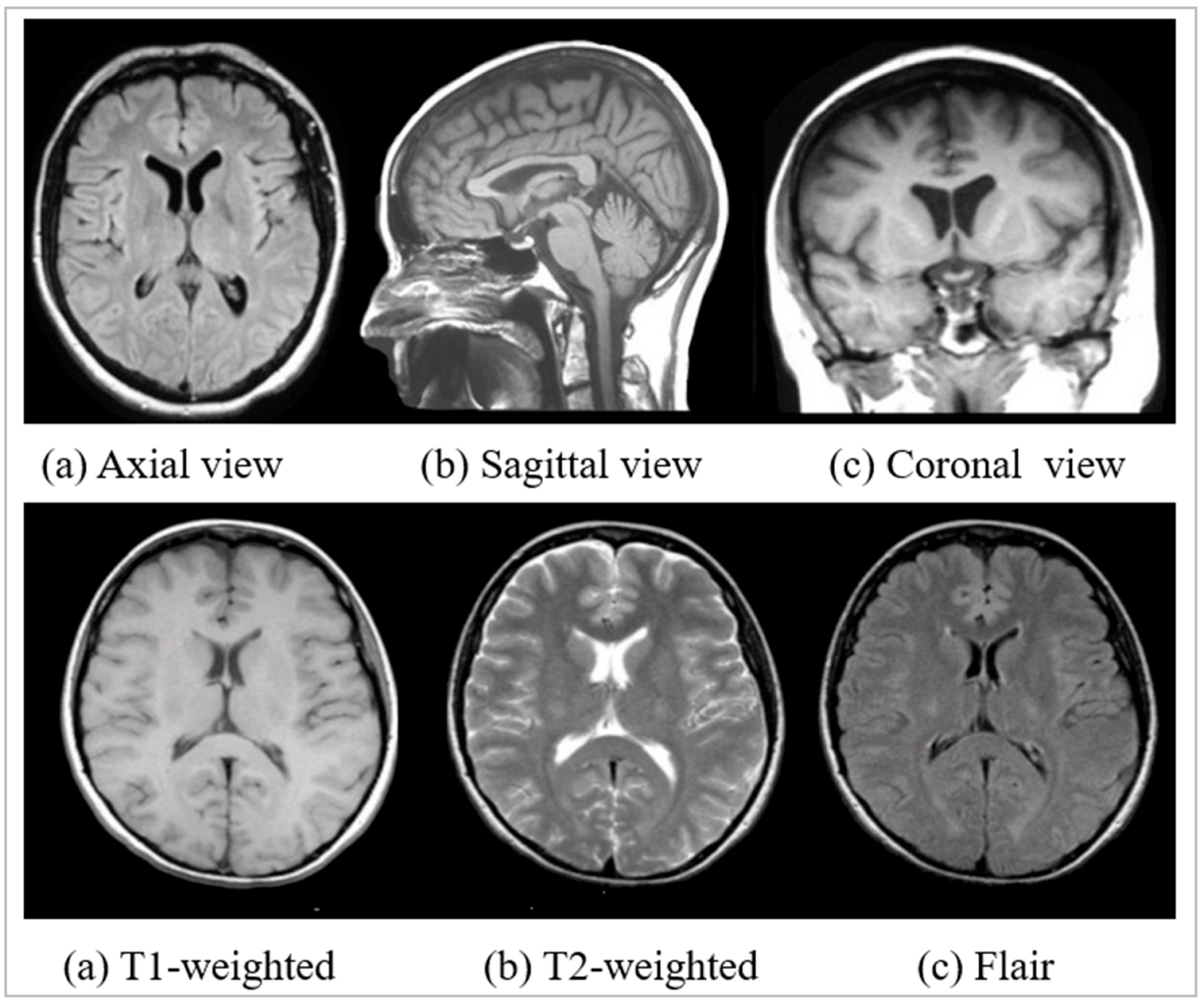
Appendix B.1.2. Image Weight or Sequence
| Brain Tissue | T1-Weighted | T2-Weighted | Flair |
|---|---|---|---|
| Cerebrospinal fluid (CSF) | Dark | Bright | Dark |
| White matter | Light | Dark Gray | Dark Gray |
| Cortex | Gray | Light Gray | Light Gray |
| Fat within the marrow | Bright | Light | Light |
| Inflammation | Dark | Bright | Bright |
| Bone | Dark | Dark | Light |
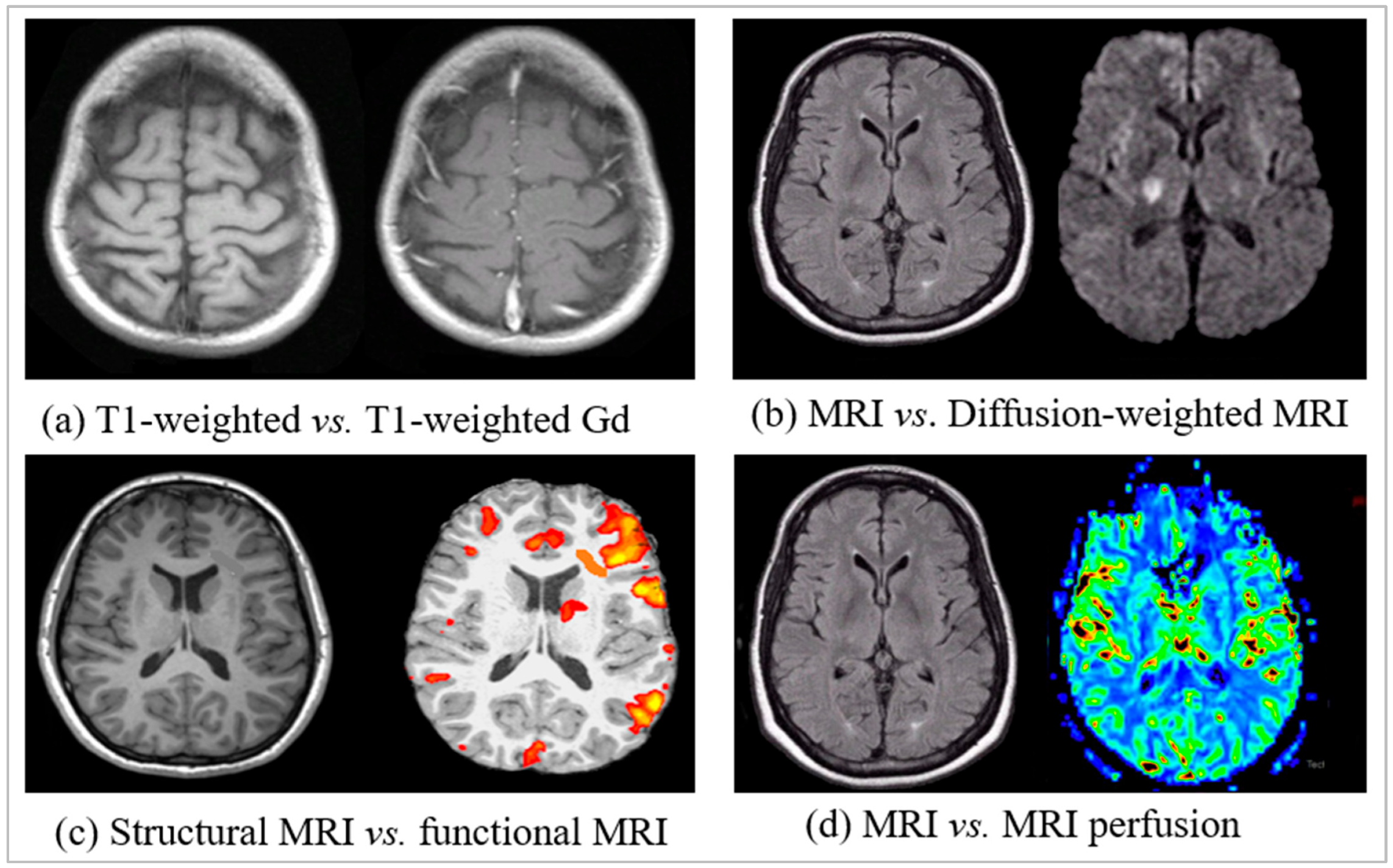
Appendix B.1.3. Functional and Perfusion, Diffusion MRI
Appendix B.1.4. Properties and Working Procedure of MRI
Appendix B.2. Other Imaging Modalities for Brain Image
Appendix B.2.1. Computed Tomography (CT)

Appendix B.2.2. Positron Emission Tomography (PET)
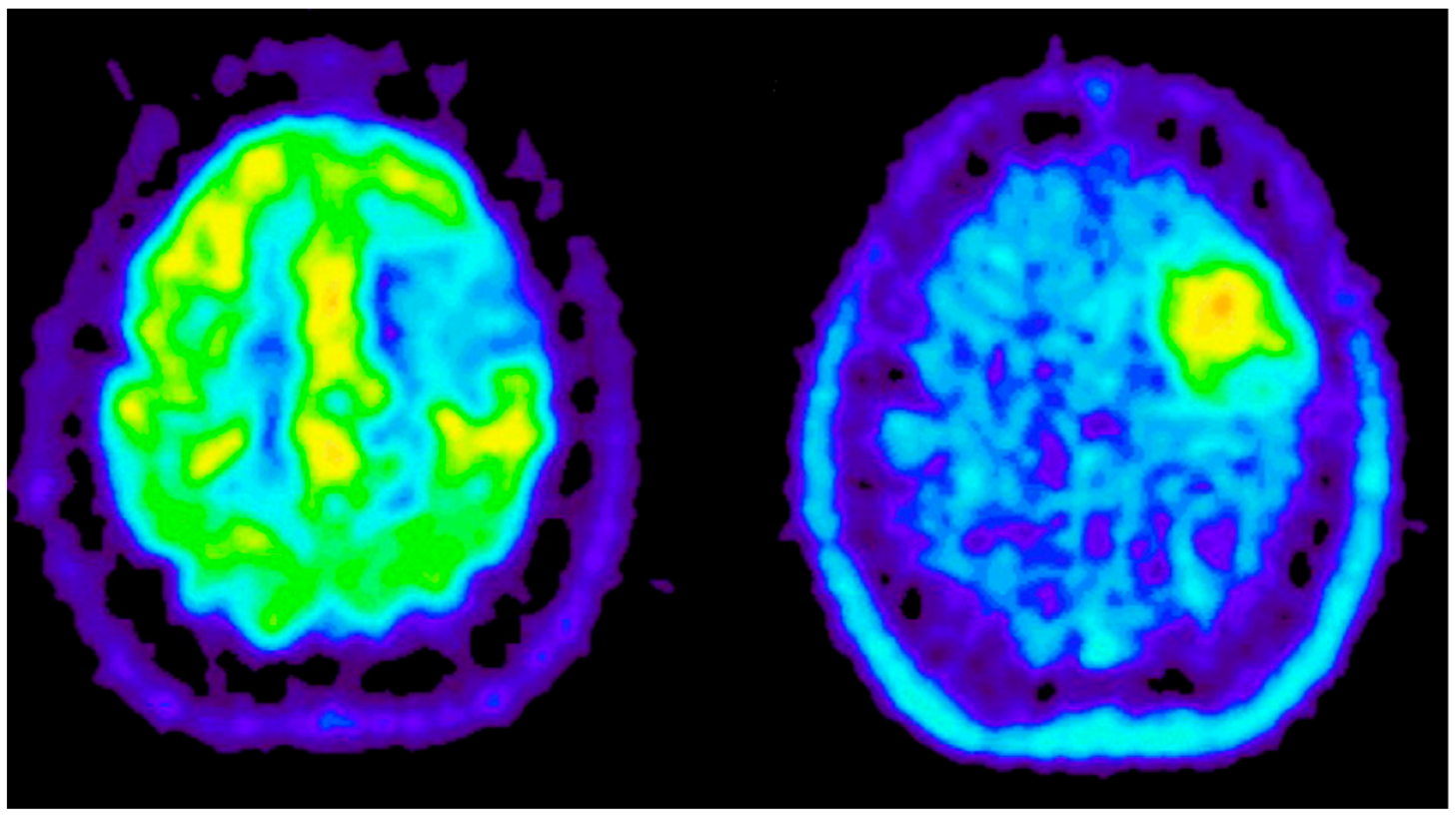
Appendix B.2.3. Single-Photon Emission Computed Tomography (SPECT)
References
- Das, S.; Bose, S.; Nayak, G.K.; Satapathy, S.C.; Saxena, S. Brain tumor segmentation and overall survival period prediction in glioblastoma multiforme using radiomic features. Concurr. Comput. Pr. Exp. 2021, 34, e6501. [Google Scholar] [CrossRef]
- Brain Tumor: Statistics. Available online: https://www.cancer.net/cancer-types/brain-tumor/statistics (accessed on 20 May 2022).
- Khazaei, Z.; Goodarzi, E.; Borhaninejad, V.; Iranmanesh, F.; Mirshekarpour, H.; Mirzaei, B.; Naemi, H.; Bechashk, S.M.; Darvishi, I.; Sarabi, R.E.; et al. The association between incidence and mortality of brain cancer and human development index (HDI): An ecological study. BMC Public Health 2020, 20, 1–7. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. The World Health Report 2001: Mental Health: New Understanding, New Hope. 2001. Available online: https://apps.who.int/iris/handle/10665/42390 (accessed on 9 December 2021).
- Jena, B.; Nayak, G.K.; Saxena, S. An empirical study of different machine learning techniques for brain tumor classification and subsequent segmentation using hybrid texture feature. Mach. Vis. Appl. 2021, 33, 1–16. [Google Scholar] [CrossRef]
- Morawski, M.; Kirilina, E.; Scherf, N.; Jäger, C.; Reimann, K.; Trampel, R.; Gavriilidis, F.; Geyer, S.; Biedermann, B.; Arendt, T.; et al. Developing 3D microscopy with CLARITY on human brain tissue: Towards a tool for informing and validating MRI-based histology. NeuroImage 2017, 182, 417–428. [Google Scholar] [CrossRef] [PubMed]
- Broocks, G.; Flottmann, F.; Ernst, M.; Faizy, T.D.; Minnerup, J.; Siemonsen, S.; Fiehler, J.; Kemmling, A. Computed Tomography–Based Imaging of Voxel-Wise Lesion Water Uptake in Ischemic Brain. Investig. Radiol. 2018, 53, 207–213. [Google Scholar] [CrossRef]
- Sanches, J.M.; Laine, A.F.; Suri, J.S. Ultrasound Imaging; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Saba, L.; Suri, J.S. Neurological Disorders and Imaging Physics, Volume 1; Application of Multiple Sclerosis; IOP Publishing Ltd.: Bristol, UK, 2019; Volume 1. [Google Scholar]
- El-Baz, A.; Suri, J.S. Application to autism spectrum disorders and Alzheimer’s. In Neurological Disorders and Imaging Physics; IOP Publishing Ltd.: Bristol, UK, 2019; Volume 3. [Google Scholar]
- El-Baz, A.; Suri, J.S. Engineering and clinical perspectives of multiple sclerosis. In Neurological Disorders and Imaging Physics; IOP Publishing Ltd.: Bristol, UK, 2019; Volume 2. [Google Scholar]
- Weiskopf, N.; Edwards, L.J.; Helms, G.; Mohammadi, S.; Kirilina, E. Quantitative magnetic resonance imaging of brain anatomy and in vivo histology. Nat. Rev. Phys. 2021, 3, 570–588. [Google Scholar] [CrossRef]
- Shehata, M.; Khalifa, F.; Soliman, A.; Shaker, S.; Shalaby, A.; El-Baz, M.; Mahmoud, A.; Dwyer, A.C.; El-Ghar, M.A.; Ghazal, M.; et al. Early Classification of Renal Rejection Types: A Deep Learning Approach. In Machine Learning in Medicine; CRC Press: Boca Raton, FL, USA, 2021; pp. 257–280. [Google Scholar] [CrossRef]
- Tandel, G.S.; Balestrieri, A.; Jujaray, T.; Khanna, N.N.; Saba, L.; Suri, J.S. Multiclass magnetic resonance imaging brain tumor classification using artificial intelligence paradigm. Comput. Biol. Med. 2020, 122, 103804. [Google Scholar] [CrossRef]
- Kuppili, V.; Biswas, M.; Sreekumar, A.; Suri, H.S.; Saba, L.; Edla, D.R.; Marinhoe, R.T.; Sanches, J.; Suri, J.S. Extreme Learning Machine Framework for Risk Stratification of Fatty Liver Disease Using Ultrasound Tissue Characterization. J. Med Syst. 2017, 41, 152. [Google Scholar] [CrossRef]
- Acharya, U.R.; Faust, O.; Sree, S.V.; Molinari, F.; Suri, J.S. ThyroScreen system: High resolution ultrasound thyroid image characterization into benign and malignant classes using novel combination of texture and discrete wavelet transform. Comput. Methods Programs Biomed. 2011, 107, 233–241. [Google Scholar] [CrossRef]
- Acharya, U.R.; Sree, S.V.; Krishnan, M.M.R.; Molinari, F.; Garberoglio, R.; Suri, J.S. Non-invasive automated 3D thyroid lesion classification in ultrasound: A class of ThyroScan™ systems. Ultrasonics 2012, 52, 508–520. [Google Scholar] [CrossRef]
- Acharya, U.R.; Sree, S.V.; Swapna, G.; Gupta, S.; Molinari, F.; Garberoglio, R.; Witkowska, A.; Suri, J.S. Effect of complex wavelet transform filter on thyroid tumor classification in three-dimensional ultrasound. Proc. Inst. Mech. Eng. Part H J. Eng. Med. 2013, 227, 284–292. [Google Scholar] [CrossRef] [PubMed]
- Suri, J.S.; Rangayyan, R.M. Recent Advances in Breast Imaging, Mammography, and Computer-Aided Diagnosis of Breast Cancer. 2006. Available online: https://spie.org/Publications/Book/651880 (accessed on 9 December 2021).
- Huang, S.-F.; Ruey-Feng, C.; Moon, W.K.; Lee, Y.-H.; dar-Ren, C.; Suri, J.S. Analysis of Tumor Vascularity Using Three-Dimensional Power Doppler Ultrasound Images. IEEE Trans. Med Imaging 2008, 27, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Acharya, U.R.; Mookiah, M.R.K.; Sree, S.V.; Yanti, R.; Martis, R.J.; Saba, L.; Molinari, F.; Guerriero, S.; Suri, J.S. Evolutionary Algorithm-Based Classifier Parameter Tuning for Automatic Ovarian Cancer Tissue Characterization and Classification. Ultraschall der Med. -Eur. J. Ultrasound 2012, 35, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Acharya, U.R.; Sree, S.V.; Kulshreshtha, S.; Molinari, F.; Koh, J.E.W.; Saba, L.; Suri, J.S. GyneScan: An Improved Online Paradigm for Screening of Ovarian Cancer via Tissue Characterization. Technol. Cancer Res. Treat. 2014, 13, 529–539. [Google Scholar] [CrossRef]
- Shrivastava, V.; Londhe, N.D.; Sonawane, R.; Suri, J.S. Reliable and accurate psoriasis disease classification in dermatology images using comprehensive feature space in machine learning paradigm. Expert Syst. Appl. 2015, 42, 6184–6195. [Google Scholar] [CrossRef]
- Shrivastava, V.; Londhe, N.D.; Sonawane, R.S.; Suri, J.S. Computer-aided diagnosis of psoriasis skin images with HOS, texture and color features: A first comparative study of its kind. Comput. Methods Programs Biomed. 2016, 126, 98–109. [Google Scholar] [CrossRef]
- Maniruzzaman; Kumar, N.; Abedin, M.; Islam, S.; Suri, H.S.; El-Baz, A.S.; Suri, J.S. Comparative approaches for classification of diabetes mellitus data: Machine learning paradigm. Comput. Methods Programs Biomed. 2017, 152, 23–34. [Google Scholar] [CrossRef]
- Maniruzzaman; Rahman, J.; Hasan, A.M.; Suri, H.S.; Abedin, M.; El-Baz, A.; Suri, J.S. Accurate Diabetes Risk Stratification Using Machine Learning: Role of Missing Value and Outliers. J. Med Syst. 2018, 42, 92. [Google Scholar] [CrossRef]
- Sharma, P.; Choudhary, K.; Gupta, K.; Chawla, R.; Gupta, D.; Sharma, A. Artificial plant optimization algorithm to detect heart rate & presence of heart disease using machine learning. Artif. Intell. Med. 2019, 102, 101752. [Google Scholar] [CrossRef]
- Acharya, U.R.; Faust, O.; S, V.S.; Alvin, A.; Krishnamurthi, G.; Seabra, J.; Sanches, J.; Suri, J.S. Understanding symptomatology of atherosclerotic plaque by image-based tissue characterization. Comput. Methods Programs Biomed. 2013, 110, 66–75. [Google Scholar] [CrossRef]
- Araki, T.; Ikeda, N.; Dey, N.; Chakraborty, S.; Saba, L.; Kumar, D.; Godia, E.C.; Jiang, X.; Gupta, A.; Radeva, P.; et al. A comparative approach of four different image registration techniques for quantitative assessment of coronary artery calcium lesions using intravascular ultrasound. Comput. Methods Programs Biomed. 2015, 118, 158–172. [Google Scholar] [CrossRef]
- Acharya, U.R.; Sree, S.V.; Krishnan, M.M.R.; Krishnananda, N.; Ranjan, S.; Umesh, P.; Suri, J.S. Automated classification of patients with coronary artery disease using grayscale features from left ventricle echocardiographic images. Comput. Methods Programs Biomed. 2013, 112, 624–632. [Google Scholar] [CrossRef] [PubMed]
- Jena, B.; Nayak, G.K.; Saxena, S. Convolutional neural network and its pretrained models for image classification and object detection: A survey. Concurr. Comput. Pr. Exp. 2021, 34, e6767. [Google Scholar] [CrossRef]
- Suri, J.S.; Biswas, M.; Kuppili, V.; Saba, L.; Edla, D.R.; Suri, H.S.; Cuadrado-Godia, E.; Laird, J.R.; Marinhoe, R.T.; Sanches, J.M.; et al. State-of-the-art review on deep learning in medical imaging. Front. Biosci. 2019, 24, 392–426. [Google Scholar] [CrossRef]
- Tandel, G.S.; Biswas, M.; Kakde, O.G.; Tiwari, A.; Suri, H.S.; Turk, M.; Laird, J.R.; Asare, C.K.; Ankrah, A.A.; Khanna, N.N.; et al. A Review on a Deep Learning Perspective in Brain Cancer Classification. Cancers 2019, 11, 111. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.K.; Sharma, N.; Saba, L.; Paraskevas, K.I.; Kalra, M.K.; Johri, A.; Laird, J.R.; Nicolaides, A.N.; Suri, J.S. Unseen Artificial Intelligence—Deep Learning Paradigm for Segmentation of Low Atherosclerotic Plaque in Carotid Ultrasound: A Multicenter Cardiovascular Study. Diagnostics 2021, 11, 2257. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.K.; Sharma, N.; Saba, L.; Paraskevas, K.I.; Kalra, M.K.; Johri, A.; Nicolaides, A.N.; Suri, J.S. Automated deep learning-based paradigm for high-risk plaque detection in B-mode common carotid ultrasound scans: An asymptomatic Japanese cohort study. Int. Angiol. 2022, 41, 9–23. [Google Scholar] [CrossRef]
- Suri, J.S.; Agarwal, S.; Elavarthi, P.; Pathak, R.; Ketireddy, V.; Columbu, M.; Saba, L.; Gupta, S.K.; Faa, G.; Singh, I.M.; et al. Inter-Variability Study of COVLIAS 1.0: Hybrid Deep Learning Models for COVID-19 Lung Segmentation in Computed Tomography. Diagnostics 2021, 11, 2025. [Google Scholar] [CrossRef]
- Suri, J.; Agarwal, S.; Pathak, R.; Ketireddy, V.; Columbu, M.; Saba, L.; Gupta, S.; Faa, G.; Singh, I.; Turk, M.; et al. COVLIAS 1.0: Lung Segmentation in COVID-19 Computed Tomography Scans Using Hybrid Deep Learning Artificial Intelligence Models. Diagnostics 2021, 11, 1405. [Google Scholar] [CrossRef]
- Skandha, S.S.; Nicolaides, A.; Gupta, S.K.; Koppula, V.K.; Saba, L.; Johri, A.M.; Kalra, M.S.; Suri, J.S. A hybrid deep learning paradigm for carotid plaque tissue characterization and its validation in multicenter cohorts using a supercomputer framework. Comput. Biol. Med. 2021, 141, 105131. [Google Scholar] [CrossRef]
- Saba, L.; Sanagala, S.S.; Gupta, S.K.; Koppula, V.K.; Johri, A.M.; Khanna, N.N.; Mavrogeni, S.; Laird, J.R.; Pareek, G.; Miner, M.; et al. Multimodality carotid plaque tissue characterization and classification in the artificial intelligence paradigm: A narrative review for stroke application. Ann. Transl. Med. 2021, 9, 1206. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.J.; Yang, Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 2009, 22, 1345–1359. [Google Scholar] [CrossRef]
- Sanagala, S.S.; Nicolaides, A.; Gupta, S.K.; Koppula, V.K.; Saba, L.; Agarwal, S.; Johri, A.M.; Kalra, M.S.; Suri, J.S. Ten Fast Transfer Learning Models for Carotid Ultrasound Plaque Tissue Characterization in Augmentation Framework Embedded with Heatmaps for Stroke Risk Stratification. Diagnostics 2021, 11, 2109. [Google Scholar] [CrossRef] [PubMed]
- Saba, L.; Sanagala, S.S.; Gupta, S.K.; Koppula, V.K.; Johri, A.M.; Sharma, A.M.; Kolluri, R.; Bhatt, D.L.; Nicolaides, A.; Suri, J.S. Ultrasound-based internal carotid artery plaque characterization using deep learning paradigm on a supercomputer: A cardiovascular disease/stroke risk assessment system. Int. J. Cardiovasc. Imaging 2021, 37, 1511–1528. [Google Scholar] [CrossRef]
- Jena, B.; Saxena, S.; Nayak, G.K.; Saba, L.; Sharma, N.; Suri, J.S. Artificial intelligence-based hybrid deep learning models for image classification: The first narrative review. Comput. Biol. Med. 2021, 137, 104803. [Google Scholar] [CrossRef]
- Saba, L.; Agarwal, M.; Patrick, A.; Puvvula, A.; Gupta, S.K.; Carriero, A.; Laird, J.R.; Kitas, G.D.; Johri, A.M.; Balestrieri, A.; et al. Six artificial intelligence paradigms for tissue characterisation and classification of non-COVID-19 pneumonia against COVID-19 pneumonia in computed tomography lungs. Int. J. Comput. Assist. Radiol. Surg. 2021, 16, 423–434. [Google Scholar] [CrossRef]
- Dong, M.; Lu, X.; Ma, Y.; Guo, Y.; Ma, Y.; Wang, K. An Efficient Approach for Automated Mass Segmentation and Classification in Mammograms. J. Digit. Imaging 2015, 28, 613–625. [Google Scholar] [CrossRef]
- Weiss, W.A. Genetics of brain tumors. Curr. Opin. Pediatrics 2000, 12, 543–548. [Google Scholar] [CrossRef]
- Soto, A.M.; Sonnenschein, C. Environmental causes of cancer: Endocrine disruptors as carcinogens. Nat. Rev. Endocrinol. 2010, 6, 363–370. [Google Scholar] [CrossRef]
- Kazerooni, A.F.; Bagley, S.J.; Akbari, H.; Saxena, S.; Bagheri, S.; Guo, J.; Chawla, S.; Nabavizadeh, A.; Mohan, S.; Bakas, S.; et al. Applications of Radiomics and Radiogenomics in High-Grade Gliomas in the Era of Precision Medicine. Cancers 2021, 13, 5921. [Google Scholar] [CrossRef]
- Habib, A.; Jovanovich, N.; Hoppe, M.; Ak, M.; Mamindla, P.; Colen, R.R.; Zinn, P. MRI-Based Radiomics and Radiogenomics in the Management of Low-Grade Gliomas: Evaluating the Evidence for a Paradigm Shift. J. Clin. Med. 2021, 10, 1411. [Google Scholar] [CrossRef] [PubMed]
- Kocak, B.; Durmaz, E.S.; Ates, E.; Ulusan, M.B. Radiogenomics in Clear Cell Renal Cell Carcinoma: Machine Learning–Based High-Dimensional Quantitative CT Texture Analysis in Predicting PBRM1 Mutation Status. Am. J. Roentgenol. 2019, 212, W55–W63. [Google Scholar] [CrossRef] [PubMed]
- Trivizakis, E.; Papadakis, G.Z.; Souglakos, I.; Papanikolaou, N.; Koumakis, L.; Spandidos, D.A.; Tsatsakis, A.; Karantanas, A.H.; Marias, K. Artificial intelligence radiogenomics for advancing precision and effectiveness in oncologic care (Review). Int. J. Oncol. 2020, 57, 43–53. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Akkus, Z.; Ali, I.; Sedlar, J.; Kline, T.L.; Agrawal, J.P.; Parney, I.F.; Giannini, C.; Erickson, B.J. Predicting 1p19q chromosomal deletion of low-grade gliomas from MR images using deep learning. arXiv 2016, arXiv:06939. [Google Scholar]
- Banerjee, I.; Crawley, A.; Bhethanabotla, M.; Daldrup-Link, H.E.; Rubin, D.L. Transfer learning on fused multiparametric MR images for classifying histopathological subtypes of rhabdomyosarcoma. Comput. Med. Imaging Graph. 2017, 65, 167–175. [Google Scholar] [CrossRef]
- Iwatate, Y.; Hoshino, I.; Yokota, H.; Ishige, F.; Itami, M.; Mori, Y.; Chiba, S.; Arimitsu, H.; Yanagibashi, H.; Nagase, H.; et al. Radiogenomics for predicting p53 status, PD-L1 expression, and prognosis with machine learning in pancreatic cancer. Br. J. Cancer 2020, 123, 1253–1261. [Google Scholar] [CrossRef]
- Park, E.K.; Lee, K.-S.; Seo, B.K.; Cho, K.R.; Woo, O.H.; Son, G.S.; Lee, H.Y.; Chang, Y.W. Machine Learning Approaches to Radiogenomics of Breast Cancer using Low-Dose Perfusion Computed Tomography: Predicting Prognostic Biomarkers and Molecular Subtypes. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Saha, A.; Harowicz, M.R.; Grimm, L.; Kim, C.E.; Ghate, S.V.; Walsh, R.; Mazurowski, M.A. A machine learning approach to radiogenomics of breast cancer: A study of 922 subjects and 529 DCE-MRI features. Br. J. Cancer 2018, 119, 508–516. [Google Scholar] [CrossRef]
- Yu, D.; Zhou, M.; Yang, F.; Dong, D.; Gevaert, O.; Liu, Z.; Shi, J.; Tian, J. Convolutional neural networks for predicting molecular profiles of non-small cell lung cancer. In Proceedings of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), Melbourne, VIC, Australia, 18–21 April 2017; pp. 569–572. [Google Scholar] [CrossRef]
- Zhou, Z.; Chen, L.; Sher, D.; Zhang, Q.; Shah, J.; Pham, N.-L.; Jiang, S.; Wang, J. Predicting Lymph Node Metastasis in Head and Neck Cancer by Combining Many-objective Radiomics and 3-dimensioal Convolutional Neural Network through Evidential Reasoning. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018. [Google Scholar] [CrossRef]
- Chang, K.; Bai, H.X.; Zhou, H.; Su, C.; Bi, W.L.; Agbodza, E.; Kavouridis, V.K.; Senders, J.T.; Boaro, A.; Beers, A.; et al. Residual Convolutional Neural Network for the Determination ofIDHStatus in Low- and High-Grade Gliomas from MR Imaging. Clin. Cancer Res. 2018, 24, 1073–1081. [Google Scholar] [CrossRef]
- Chang, P.; Grinband, J.; Weinberg, B.D.; Bardis, M.; Khy, M.; Cadena, G.; Su, M.-Y.; Cha, S.; Filippi, C.G.; Bota, D.; et al. Deep-Learning Convolutional Neural Networks Accurately Classify Genetic Mutations in Gliomas. Am. J. Neuroradiol. 2018, 39, 1201–1207. [Google Scholar] [CrossRef] [PubMed]
- Ha, R.; Mutasa, S.; Karcich, J.; Gupta, N.; Van Sant, E.P.; Nemer, J.; Sun, M.; Chang, P.; Liu, M.Z.; Jambawalikar, S. Predicting Breast Cancer Molecular Subtype with MRI Dataset Utilizing Convolutional Neural Network Algorithm. J. Digit. Imaging 2019, 32, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, R.; Takahashi, M.; Miyake, M.; Kinoshita, M.; Takahashi, S.; Ichimura, K.; Hamamoto, R.; Narita, Y.; Sese, J. Assessing Versatile Machine Learning Models for Glioma Radiogenomic Studies across Hospitals. Cancers 2021, 13, 3611. [Google Scholar] [CrossRef] [PubMed]
- Kickingereder, P.; Bonekamp, D.; Nowosielski, M.; Kratz, A.; Sill, M.; Burth, S.; Wick, A.; Eidel, O.; Schlemmer, H.-P.; Radbruch, A.; et al. Radiogenomics of Glioblastoma: Machine Learning–based Classification of Molecular Characteristics by Using Multiparametric and Multiregional MR Imaging Features. Radiology 2016, 281, 907–918. [Google Scholar] [CrossRef]
- Korfiatis, P.; Kline, T.L.; Lachance, D.H.; Parney, I.; Buckner, J.C.; Erickson, B.J. Residual Deep Convolutional Neural Network Predicts MGMT Methylation Status. J. Digit. Imaging 2017, 30, 622–628. [Google Scholar] [CrossRef]
- Li, Z.; Wang, Y.; Yu, J.; Guo, Y.; Cao, W. Deep Learning based Radiomics (DLR) and its usage in noninvasive IDH1 prediction for low grade glioma. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Liang, S.; Zhang, R.; Liang, D.; Song, T.; Ai, T.; Xia, C.; Xia, L.; Wang, Y. Multimodal 3D DenseNet for IDH Genotype Prediction in Gliomas. Genes 2018, 9, 382. [Google Scholar] [CrossRef]
- Smedley, N.F.; Hsu, W. Using deep neural networks for radiogenomic analysis. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 1529–1533. [Google Scholar]
- Wang, S.; Shi, J.; Ye, Z.; Dong, D.; Yu, D.; Zhou, M.; Liu, Y.; Gevaert, O.; Wang, K.; Zhu, Y.; et al. Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur. Respir. J. 2019, 53, 1800986. [Google Scholar] [CrossRef]
- Yoon, H.-J.; Ramanathan, A.; Alamudun, F.; Tourassi, G. Deep radiogenomics for predicting clinical phenotypes in invasive breast cancer. In Proceedings of the 14th International Workshop on Breast Imaging (IWBI 2018), Atlanta, GA, USA, 8–11 July 2018; Volume 10718. [Google Scholar] [CrossRef]
- Zhu, Z.; Albadawy, E.; Saha, A.; Zhang, J.; Harowicz, M.R.; Mazurowski, M.A. Deep learning for identifying radiogenomic associations in breast cancer. Comput. Biol. Med. 2019, 109, 85–90. [Google Scholar] [CrossRef]
- Andreassen, C.N.; Alsner, J.; Overgaard, J. Does variability in normal tissue reactions after radiotherapy have a genetic basis—where and how to look for it? Radiother. Oncol. 2002, 64, 131–140. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008, 455, 1061–1068. [Google Scholar] [CrossRef] [PubMed]
- Kalpathy-Cramer, J.; Freymann, J.B.; Kirby, J.; Kinahan, P.E.; Prior, F.W. Quantitative Imaging Network: Data Sharing and Competitive AlgorithmValidation Leveraging The Cancer Imaging Archive. Transl. Oncol. 2014, 7, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, K.; Pant, M. Hybrid particle swarm optimization-genetic algorithm trained multi-layer perceptron for classification of human glioma from molecular brain neoplasia data. Cogn. Syst. Res. 2019, 58, 173–194. [Google Scholar] [CrossRef]
- Madhavan, S.; Zenklusen, J.-C.; Kotliarov, Y.; Sahni, H.; Fine, H.A.; Buetow, K. Rembrandt: Helping Personalized Medicine Become a Reality through Integrative Translational Research. Mol. Cancer Res. 2009, 7, 157–167. [Google Scholar] [CrossRef]
- Gusev, Y.; Bhuvaneshwar, K.; Song, L.; Zenklusen, J.-C.; Fine, H.; Madhavan, S. The REMBRANDT study, a large collection of genomic data from brain cancer patients. Sci. Data 2018, 5, 180158. [Google Scholar] [CrossRef]
- Gevaert, O.; Mitchell, L.A.; Achrol, A.S.; Xu, J.; Echegaray, S.; Steinberg, G.K.; Cheshier, S.H.; Napel, S.; Zaharchuk, G.; Plevritis, S.K. Glioblastoma Multiforme: Exploratory Radiogenomic Analysis by Using Quantitative Image Features. Radiology 2014, 273, 168–174. [Google Scholar] [CrossRef]
- Wiest, R.; Reyes, M.; Meier, R.; Gutman, D.A.; Bauer, S.; Alexander, B.; Velazquez, E.R.; Dunn, W.D.; Aerts, H.J.W.L. Fully automatic GBM segmentation in the TCGA-GBM dataset: Prognosis and correlation with VASARI features. Sci. Rep. 2015, 5, 16822. [Google Scholar] [CrossRef]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Saxena, S.; Kazerooni, A.F.; Toorens, E.; Bakas, S.; Akbari, H.; Sako, C.; Mamourian, E.; Koumenis, C.; Mohan, S.; Bagley, S.J.; et al. NIMG-73. CAPTURING GLIOBLASTOMA HETEROGENEITY USING IMAGING AND DEEP LEARNING: APPLICATION TO MGMT PROMOTER METHYLATION. Neuro-Oncology 2021, 23, vi146. [Google Scholar] [CrossRef]
- Becnel, L.B.; Pereira, S.; Drummond, J.A.; Gingras, M.-C.; Covington, K.R.; Kovar, C.L.; Doddapaneni, H.V.; Hu, J.; Muzny, N.; McGuire, A.L.; et al. An open access pilot freely sharing cancer genomic data from participants in Texas. Sci. Data 2016, 3, 160010. [Google Scholar] [CrossRef] [PubMed]
- Milius, D.; Dove, E.S.; Chalmers, D.; Dyke, S.O.M.; Kato, K.; Nicolás, P.; Ouellette, B.F.; Ozenberger, B.; Rodriguez, L.L.; Zeps, N.; et al. The International Cancer Genome Consortium’s evolving data-protection policies. Nat. Biotechnol. 2014, 32, 519–523. [Google Scholar] [CrossRef] [PubMed]
- Jäkel, S.; Dimou, L. Glial Cells and Their Function in the Adult Brain: A Journey through the History of Their Ablation. Front. Cell. Neurosci. 2017, 11, 24. [Google Scholar] [CrossRef]
- Jessen, K.R. Glial cells. Int. J. Biochem. Cell Biol. 2004, 36, 1861–1867. [Google Scholar] [CrossRef]
- Hanani, M.; Spray, D.C. Emerging importance of satellite glia in nervous system function and dysfunction. Nat. Rev. Neurosci. 2020, 21, 485–498. [Google Scholar] [CrossRef]
- Glia Cell Pictotial Representation. Available online: https://courses.lumenlearning.com/wm-biology2/chapter/glial-cells/ (accessed on 14 July 2022).
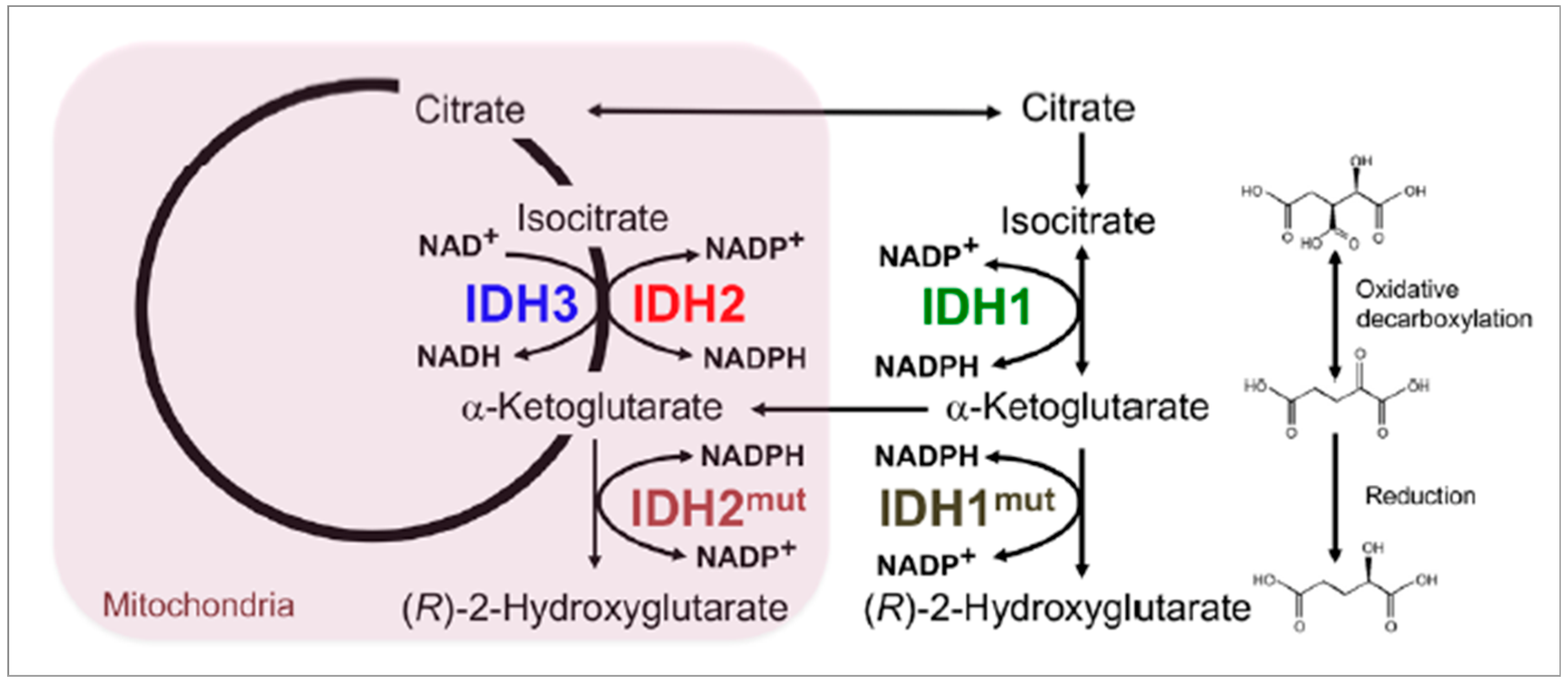
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 Mutations in Gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Mabray, M.C.; Barajas, R.F.; Cha, S. Modern Brain Tumor Imaging. Brain Tumor Res. Treat. 2015, 3, 8–23. [Google Scholar] [CrossRef]
- Suh, C.H.; Kim, H.S.; Jung, S.C.; Choi, C.G.; Kim, S.J. Imaging prediction of isocitrate dehydrogenase (IDH) mutation in patients with glioma: A systemic review and meta-analysis. Eur. Radiol. 2018, 29, 745–758. [Google Scholar] [CrossRef]
- Mack, S.C.; Hubert, C.G.; Miller, T.E.; Taylor, M.; Rich, J.N. An epigenetic gateway to brain tumor cell identity. Nat. Neurosci. 2015, 19, 10–19. [Google Scholar] [CrossRef]
- Tommasini-Ghelfi, S.; Murnan, K.; Kouri, F.M.; Mahajan, A.S.; May, J.L.; Stegh, A.H. Cancer-associated mutation and beyond: The emerging biology of isocitrate dehydrogenases in human disease. Sci. Adv. 2019, 5, eaaw4543. [Google Scholar] [CrossRef]
- Hu, N.; Richards, R.; Jensen, R. Role of chromosomal 1p/19q co-deletion on the prognosis of oligodendrogliomas: A systematic review and meta-analysis. Interdiscip. Neurosurg. 2016, 5, 58–63. [Google Scholar] [CrossRef]
- Han, Y.; Xie, Z.; Zang, Y.; Zhang, S.; Gu, D.; Zhou, M.; Gevaert, O.; Wei, J.; Li, C.; Chen, H.; et al. Non-invasive genotype prediction of chromosome 1p/19q co-deletion by development and validation of an MRI-based radiomics signature in lower-grade gliomas. J. Neuro-Oncology 2018, 140, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.; Salunke, P. Molecular markers of glioma: An update on recent progress and perspectives. J. Cancer Res. Clin. Oncol. 2012, 138, 1971–1981. [Google Scholar] [CrossRef]
- Cabrini, G.; Fabbri, E.; Lo Nigro, C.; Dechecchi, M.C.; Gambari, R. Regulation of expression of O6-methylguanine-DNA methyltransferase and the treatment of glioblastoma (Review). Int. J. Oncol. 2015, 47, 417–428. [Google Scholar] [CrossRef] [PubMed]
- Silber, J.R.; Bobola, M.S.; Blank, A.; Chamberlain, M.C. O6-Methylguanine-DNA methyltransferase in glioma therapy: Promise and problems. Biochim. Biophys. Acta 2012, 1826, 71–82. [Google Scholar] [CrossRef] [PubMed]
- González-González, R.; López-Verdín, S.; Lavalle-Carrasco, J.; Molina-Frechero, N.; Isiordia-Espinoza, M.; Carreón-Burciaga, R.G.; Bologna-Molina, R. Current concepts in ameloblastoma-targeted therapies in B-raf proto-oncogene serine/threonine kinase V600E mutation: Systematic review. World J. Clin. Oncol. 2020, 11, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Maraka, S.; Janku, F. BRAF alterations in primary brain tumors. Discov. Med. 2018, 26, 51–60. [Google Scholar]
- Werner, J.-M.; Brunn, A.; Fink, G.R.; Galldiks, N. Magnetic Resonance Imaging Reveals a Pronounced Treatment Response of a Isocitrate Dehydrogenase− and B-Raf Proto-Oncogene−Wildtype Epithelioid Glioblastoma. World Neurosurg. 2019, 127, 213–215. [Google Scholar] [CrossRef]
- Zhang, Y.; Dube, C.; Gibert, M.; Cruickshanks, N.; Wang, B.; Coughlan, M.; Yang, Y.; Setiady, I.; Deveau, C.; Saoud, K.; et al. The p53 Pathway in Glioblastoma. Cancers 2018, 10, 297. [Google Scholar] [CrossRef]
- Goldhoff, P.; Clarke, J.; Smirnov, I.; Berger, M.S.; Prados, M.D.; James, C.D.; Perry, A.; Phillips, J.J. Clinical Stratification of Glioblastoma Based on Alterations in Retinoblastoma Tumor Suppressor Protein (RB1) and Association With the Proneural Subtype. J. Neuropathol. Exp. Neurol. 2012, 71, 83–89. [Google Scholar] [CrossRef]
- Xu, H.; Zong, H.; Ma, C.; Ming, X.; Shang, M.; Li, K.; He, X.; Du, H.; Cao, L. Epidermal growth factor receptor in glioblastoma. Oncol. Lett. 2017, 14, 512–516. [Google Scholar] [CrossRef] [PubMed]
- Karsy, M.; Neil, J.A.; Guan, J.; Mahan, M.A.; Colman, H.; Jensen, R.L. A practical review of prognostic correlations of molecular biomarkers in glioblastoma. Neurosurg. Focus 2015, 38, E4. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.R.; Kuriyama, N.; Kuriyama, H.; Israel, M.A. Molecular genetics of brain tumors. Arch. Neurol. 1999, 56, 439–441. [Google Scholar] [CrossRef]
- Wechsler-Reya, R.; Scott, M.P. The Developmental Biology of Brain Tumors. Annu. Rev. Neurosci. 2001, 24, 385–428. [Google Scholar] [CrossRef]
- Yao, Y.; Mack, S.C.; Taylor, M.D. Molecular genetics of ependymoma. Chin. J. Cancer 2011, 30, 669–681. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vlaardingerbroek, M.T.; Boer, J.A. Magnetic Resonance Imaging: Theory and Practice; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- MRI Basics. Available online: https://case.edu/med/neurology/NR/MRI%20Basics.htm (accessed on 14 July 2022).
- Edelman, R.R. The History of MR Imaging as Seen through the Pages of Radiology. Radiology 2014, 273, S181–S200. [Google Scholar] [CrossRef] [PubMed]
- Corrias, G.; Micheletti, G.; Barberini, L.; Suri, J.S.; Saba, L. Texture analysis imaging “what a clinical radiologist needs to know”. Eur. J. Radiol. 2021, 146, 110055. [Google Scholar] [CrossRef]
- Wu, G.; Jochems, A.; Refaee, T.; Ibrahim, A.; Yan, C.; Sanduleanu, S.; Woodruff, H.C.; Lambin, P. Structural and functional radiomics for lung cancer. Eur. J. Pediatr. 2021, 48, 3961–3974. [Google Scholar] [CrossRef]
- Rizzo, S.; Botta, F.; Raimondi, S.; Origgi, D.; Fanciullo, C.; Morganti, A.G.; Bellomi, M. Radiomics: The facts and the challenges of image analysis. Eur. Radiol. Exp. 2018, 2, 36. [Google Scholar] [CrossRef]
- Murgia, A.; Balestrieri, A.; Crivelli, P.; Suri, J.S.; Conti, M.; Cademartiri, F.; Saba, L. Cardiac computed tomography radiomics: An emerging tool for the non-invasive assessment of coronary atherosclerosis. Cardiovasc. Diagn. Ther. 2020, 10, 2005–2017. [Google Scholar] [CrossRef]
- Srivastava, S.K.; Singh, S.K.; Suri, J.S. Effect of incremental feature enrichment on healthcare text classification system: A machine learning paradigm. Comput. Methods Programs Biomed. 2019, 172, 35–51. [Google Scholar] [CrossRef] [PubMed]
- Torigian, E.A.; Huang, S.S.; Houseni, M.; Alavi, A. Functional imaging of cancer with emphasis on molecular techniques. CA Cancer J. Clin. 2007, 57, 206–224. [Google Scholar] [CrossRef] [PubMed]
- Cau, R.; Bassareo, P.; Suri, J.S.; Saba, L. Reply to “Structural And/Or Functional Underpinnings of Magnetic Resonance Imaging Bi-Atrial Strain Impairment in Patients With Takotsubo Syndrome”. Can. Assoc. Radiol. J. 2022, 73, 599. [Google Scholar] [CrossRef] [PubMed]
- Porcu, M.; Cocco, L.; Saloner, D.; Suri, J.S.; Montisci, R.; Carriero, A.; Saba, L. Extracranial Carotid Artery Stenosis: The Effects on Brain and Cognition with a Focus on Resting-State Functional Connectivity. J. Neuroimaging 2020, 30, 736–745. [Google Scholar] [CrossRef]
- Wu, D.H.; Chen, Z.; North, J.C.; Biswas, M.; Vo, J.; Suri, J.S. Machine learning paradigm for dynamic contrast-enhanced MRI evaluation of expanding bladder. Front. Biosci. 2020, 25, 1746–1764. [Google Scholar] [CrossRef]
- Banchhor, S.K.; Londhe, N.D.; Araki, T.; Saba, L.; Radeva, P.; Laird, J.R.; Suri, J.S. Wall-based measurement features provides an improved IVUS coronary artery risk assessment when fused with plaque texture-based features during machine learning paradigm. Comput. Biol. Med. 2017, 91, 198–212. [Google Scholar] [CrossRef]
- Than, J.C.; Saba, L.; Noor, N.M.; Rijal, O.M.; Kassim, R.M.; Yunus, A.; Suri, H.S.; Porcu, M.; Suri, J.S. Lung disease stratification using amalgamation of Riesz and Gabor transforms in machine learning framework. Comput. Biol. Med. 2017, 89, 197–211. [Google Scholar] [CrossRef]
- Araki, T.; Jain, P.K.; Suri, H.S.; Londhe, N.D.; Ikeda, N.; El-Baz, A.; Shrivastava, V.K.; Saba, L.; Nicolaides, A.; Shafique, S.; et al. Stroke Risk Stratification and its Validation using Ultrasonic Echolucent Carotid Wall Plaque Morphology: A Machine Learning Paradigm. Comput. Biol. Med. 2017, 80, 77–96. [Google Scholar] [CrossRef] [PubMed]
- Negreros-Osuna, A.A.; Parakh, A.; Corcoran, R.B.; Pourvaziri, A.; Kambadakone, A.; Ryan, D.P.; Sahani, D.V. Radiomics Texture Features in Advanced Colorectal Cancer: Correlation with BRAF Mutation and 5-year Overall Survival. Radiol. Imaging Cancer 2020, 2, e190084. [Google Scholar] [CrossRef] [PubMed]
- Alzubaidi, L.; Duan, Y.; Al-Dujaili, A.; Ibraheem, I.K.; Alkenani, A.H.; Santamaría, J.; Fadhel, M.A.; Al-Shamma, O.; Zhang, J. Deepening into the suitability of using pre-trained models of ImageNet against a lightweight convolutional neural network in medical imaging: An experimental study. PeerJ Comput. Sci. 2021, 7, e715. [Google Scholar] [CrossRef]
- Sudeep, P.; Palanisamy, P.; Rajan, J.; Baradaran, H.; Saba, L.; Gupta, A.; Suri, J.S. Speckle reduction in medical ultrasound images using an unbiased non-local means method. Biomed. Signal Process. Control 2016, 28, 1–8. [Google Scholar] [CrossRef]
- Fei, B.; Zhang, S.; Savado, O.; Suri, J.; Lewin, J.S.; Wilson, D.L. Three-dimensional automatic volume registration of carotid MR images. In Proceedings of the 25th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (IEEE Cat. No.03CH37439), Cancun, Mexico, 17–21 September 2003. [Google Scholar] [CrossRef]
- Guo, Y.; Sivaramakrishna, R.; Lu, C.-C.; Suri, J.S.; Laxminarayan, S. Breast image registration techniques: A survey. Med Biol. Eng. Comput. 2006, 44, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Saxena, S.; Jena, B.; Gupta, N.; Das, S.; Sarmah, D.; Bhattacharya, P.; Nath, T.; Paul, S.; Fouda, M.M.; Kalra, M.; et al. Role of Artificial Intelligence in Radiogenomics for Cancers in the Era of Precision Medicine. Cancers 2022, 14, 2860. [Google Scholar] [CrossRef] [PubMed]
- Kazerooni, A.F.; Saxena, S.; Toorens, E.; Tu, D.; Bashyam, V.; Akbari, H.; Mamourian, E.; Sako, C.; Koumenis, C.; Verginadis, I.; et al. Clinical measures, radiomics, and genomics offer synergistic value in AI-based prediction of overall survival in patients with glioblastoma. Sci. Rep. 2022, 12, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, E.; Villanueva-Meyer, J.E.; Cha, S. A fully automated artificial intelligence method for non-invasive, imaging-based identification of genetic alterations in glioblastomas. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Kumar, P.K.; Darshan, P.; Kumar, S.; Ravindra, R.; Rajan, J.; Saba, L.; Suri, J.S. Magnetic resonance image denoising using nonlocal maximum likelihood paradigm in DCT-framework. Int. J. Imaging Syst. Technol. 2015, 25, 256–264. [Google Scholar] [CrossRef]
- Skandha, S.S.; Gupta, S.K.; Saba, L.; Koppula, V.K.; Johri, A.M.; Khanna, N.N.; Mavrogeni, S.; Laird, J.R.; Pareek, G.; Miner, M.; et al. 3-D optimized classification and characterization artificial intelligence paradigm for cardiovascular/stroke risk stratification using carotid ultrasound-based delineated plaque: Atheromatic™ 2.0. Comput. Biol. Med. 2020, 125, 103958. [Google Scholar] [CrossRef]
- Agarwal, M.; Saba, L.; Gupta, S.K.; Johri, A.M.; Khanna, N.N.; Mavrogeni, S.; Laird, J.R.; Pareek, G.; Miner, M.; Sfikakis, P.P.; et al. Wilson disease tissue classification and characterization using seven artificial intelligence models embedded with 3D optimization paradigm on a weak training brain magnetic resonance imaging datasets: A supercomputer application. Med Biol. Eng. Comput. 2021, 59, 511–533. [Google Scholar] [CrossRef]
- Kazerooni, A.F.; Bakas, S.; Rad, H.S.; Davatzikos, C. Imaging signatures of glioblastoma molecular characteristics: A radiogenomics review. J. Magn. Reson. Imaging 2019, 52, 54–69. [Google Scholar] [CrossRef]
- Alberch, P. From genes to phenotype: Dynamical systems and evolvability. Genetica 1991, 84, 5–11. [Google Scholar] [CrossRef]
- Ahnert, S.E. Structural properties of genotype–phenotype maps. J. R. Soc. Interface 2017, 14, 20170275. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Rodríguez, J.; Peel, L.; Stella, M.; Wagner, A.; Payne, J.L. The architecture of an empirical genotype-phenotype map. Evolution 2018, 72, 1242–1260. [Google Scholar] [CrossRef] [PubMed]
- Gullo, R.L.; Daimiel, I.; Morris, E.A.; Pinker, K. Combining molecular and imaging metrics in cancer: Radiogenomics. Insights Into Imaging 2020, 11, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Jena, B.; Nayak, G.K.; Paul, S.; Saxena, S. An Exhaustive Analytical Study of U-Net Architecture on Two Diverse Biomedical Imaging Datasets of Electron Microscopy Drosophila ssTEM and Brain MRI BraTS-2021 for Segmentation. SN Comput. Sci. 2022, 3, 1–13. [Google Scholar] [CrossRef]
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Jena, B.; Thakar, P.; Nayak, V.; Nayak, G.K.; Saxena, S. Malaria Parasites Detection Using Deep Neural Network. In Deep Learning Applications in Medical Imaging; IGI Global: Hershey, PA, USA, 2021; pp. 209–222. [Google Scholar] [CrossRef]
- Jena, B.; Nayak, G.K.; Saxena, S. Comprehensive Review of Abdominal Image Segmentation using Soft and Hard Computing Approaches. In Proceedings of the 2020 International Conference on Computer Science, Engineering and Applications (ICCSEA), Gunupur, India, 13–14 March 2020; pp. 1–5. [Google Scholar] [CrossRef]
- Jena, B.; Nayak, G.K.; Saxena, S. High-Performance Computing and Its Requirements in Deep Learning. In High-Performance Medical Image Processing; Apple Academic Press: Palm Bay, FL, USA, 2022; pp. 255–288. [Google Scholar] [CrossRef]
- Jena, B.; Nayak, G.K.; Saxena, S. Maximum Payload for Digital Image Steganography Obtained by Mixed Edge Detection Mechanism. In Proceedings of the 2019 International Conference on Information Technology (ICIT), Bhubaneswar, India, 19–21 December 2019; pp. 206–210. [Google Scholar] [CrossRef]
- Bodalal, Z.; Trebeschi, S.; Nguyen-Kim, T.D.L.; Schats, W.; Beets-Tan, R. Radiogenomics: Bridging imaging and genomics. Abdom. Radiol. 2019, 44, 1960–1984. [Google Scholar] [CrossRef] [PubMed]
- Shui, L.; Ren, H.; Yang, X.; Li, J.; Chen, Z.; Yi, C.; Zhu, H.; Shui, P. Era of radiogenomics in precision medicine: An emerging approach for prediction of the diagnosis, treatment and prognosis of tumors. Front. Oncol. 2020, 10, 3195. [Google Scholar]
- Singh, G.; Manjila, S.; Sakla, N.; True, A.; Wardeh, A.H.; Beig, N.; Vaysberg, A.; Matthews, J.; Prasanna, P.; Spektor, V. Radiomics and radiogenomics in gliomas: A contemporary update. Br. J. Cancer 2021, 125, 641–657. [Google Scholar] [CrossRef]
- Agarwal, M.; Agarwal, S.; Saba, L.; Chabert, G.L.; Gupta, S.; Carriero, A.; Pasche, A.; Danna, P.; Mehmedovic, A.; Faa, G.; et al. Eight pruning deep learning models for low storage and high-speed COVID-19 computed tomography lung segmentation and heatmap-based lesion localization: A multicenter study using COVLIAS 2.0. Comput. Biol. Med. 2022, 146, 105571. [Google Scholar] [CrossRef]
- Suri, J.S.; Agarwal, S.; Chabert, G.L.; Carriero, A.; Paschè, A.; Danna, P.S.C.; Saba, L.; Mehmedović, A.; Faa, G.; Singh, I.M.; et al. COVLIAS 2.0-cXAI: Cloud-Based Explainable Deep Learning System for COVID-19 Lesion Localization in Computed Tomography Scans. Diagnostics 2022, 12, 1482. [Google Scholar] [CrossRef]
- Hoffman, R.R.; Mueller, S.T.; Klein, G.; Litman, J. Metrics for explainable AI: Challenges and prospects. arXiv 2018, arXiv:04608. [Google Scholar]
- Suri, J.S.; Agarwal, S.; Carriero, A.; Paschè, A.; Danna, P.S.C.; Columbu, M.; Saba, L.; Viskovic, K.; Mehmedović, A.; Agarwal, S.; et al. COVLIAS 1.0 vs. MedSeg: Artificial Intelligence-Based Comparative Study for Automated COVID-19 Computed Tomography Lung Segmentation in Italian and Croatian Cohorts. Diagnostics 2021, 11, 2367. [Google Scholar] [CrossRef] [PubMed]
- Suri, J.S.; Agarwal, S.; Gupta, S.K.; Puvvula, A.; Viskovic, K.; Suri, N.; Alizad, A.; El-Baz, A.; Saba, L.; Fatemi, M.; et al. Systematic Review of Artificial Intelligence in Acute Respiratory Distress Syndrome for COVID-19 Lung Patients: A Biomedical Imaging Perspective. IEEE J. Biomed. Heal. Inform. 2021, 25, 4128–4139. [Google Scholar] [CrossRef] [PubMed]
- Banchhor, S.K.; Londhe, N.D.; Araki, T.; Saba, L.; Radeva, P.; Khanna, N.N.; Suri, J.S. Calcium detection, its quantification, and grayscale morphology-based risk stratification using machine learning in multimodality big data coronary and carotid scans: A review. Comput. Biol. Med. 2018, 101, 184–198. [Google Scholar] [CrossRef] [PubMed]
- Holzinger, A.; Langs, G.; Denk, H.; Zatloukal, K.; Müller, H. Causability and explainability of artificial intelligence in medicine. WIREs Data Min. Knowl. Discov. 2019, 9, e1312. [Google Scholar] [CrossRef] [PubMed]
- Castro, D.C.; Walker, I.; Glocker, B. Causality matters in medical imaging. Nat. Commun. 2020, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Chou, Y.L.; Moreira, C.; Bruza, P.; Ouyang, C.; Jorge, J. Counterfactuals and causability in explainable artificial intelligence: Theory, algorithms, and applications. Inf. Fusion 2022, 81, 59–83. [Google Scholar] [CrossRef]
- Saba, L.; Banchhor, S.K.; Londhe, N.D.; Araki, T.; Laird, J.R.; Gupta, A.; Nicolaides, A.; Suri, J.S. Web-based accurate measurements of carotid lumen diameter and stenosis severity: An ultrasound-based clinical tool for stroke risk assessment during multicenter clinical trials. Comput. Biol. Med. 2017, 91, 306–317. [Google Scholar] [CrossRef]
- El-Baz, A.; Suri, J.S. Big Data in Multimodal Medical Imaging; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar]
- Mendes, A.C.; Ribeiro, A.S.; Oros-Peusquens, A.M.; Langen, K.J.; Shah, J.; Ferreira, H.A. Brain connectivity study of brain tumor patients using MR-PET data: Preliminary results. EJNMMI Phys. 2015, 2, 1–2. [Google Scholar] [CrossRef][Green Version]
- Porcu, M.; Operamolla, A.; Scapin, E.; Garofalo, P.; Destro, F.; Caneglias, A.; Suri, J.S.; Falini, A.; DeFazio, G.; Marrosu, F.; et al. Effects of White Matter Hyperintensities on Brain Connectivity and Hippocampal Volume in Healthy Subjects According to Their Localization. Brain Connect. 2020, 10, 436–447. [Google Scholar] [CrossRef]
- Saba, L.; Sanfilippo, R.; Sannia, S.; Anzidei, M.; Montisci, R.; Mallarini, G.; Suri, J.S. Association Between Carotid Artery Plaque Volume, Composition, and Ulceration: A Retrospective Assessment With MDCT. Am. J. Roentgenol. 2012, 199, 151–156. [Google Scholar] [CrossRef]
- Suri, J.S.; Puvvula, A.; Biswas, M.; Majhail, M.; Saba, L.; Faa, G.; Singh, I.M.; Oberleitner, R.; Turk, M.; Chadha, P.S.; et al. COVID-19 pathways for brain and heart injury in comorbidity patients: A role of medical imaging and artificial intelligence-based COVID severity classification: A review. Comput. Biol. Med. 2020, 124, 103960. [Google Scholar] [CrossRef] [PubMed]
- Holdsworth, S.J.; Bammer, R. Magnetic resonance imaging techniques: fMRI, DWI, and PWI. In Seminars in Neurology; Thieme Medical Publishers: New York, NY, USA, 2008; Volume 28, pp. 395–406. [Google Scholar]
- Öz, G.; Alger, J.R.; Barker, P.B.; Bartha, R.; Bizzi, A.; Boesch, C.; Bolan, P.J.; Brindle, K.M.; Cudalbu, C.; Dinçer, A.; et al. Clinical Proton MR Spectroscopy in Central Nervous System Disorders. Radiology 2014, 270, 658–679. [Google Scholar] [CrossRef] [PubMed]
- Sourbron, S. Technical aspects of MR perfusion. Eur. J. Radiol. 2010, 76, 304–313. [Google Scholar] [CrossRef]
- Jeurissen, B.; Descoteaux, M.; Mori, S.; Leemans, A. Diffusion MRI fiber tractography of the brain. NMR Biomed. 2017, 32, e3785. [Google Scholar] [CrossRef]
- Singleton, M.J. Functional Magnetic Resonance Imaging. Yale J. Biol. Med. 2009, 82, 233. [Google Scholar]
- Buzug, T.M. Computed tomography. In Springer Handbook of Medical Technology; Springer: Berlin/Heidelberg, Germany, 2011; pp. 311–342. [Google Scholar]
- Bailey, D.L.; Maisey, M.N.; Townsend, D.W.; Valk, P.E. Positron Emission Tomography; Springer: Berlin/Heidelberg, Germany, 2005. [Google Scholar]
- Holly, T.A. Single Photon-Emission Computed Tomography; Springer: Berlin/Heidelberg, Germany, 2010. [Google Scholar]
- Catafau, A.M. Brain SPECT in clinical practice. Part I: Perfusion. J. Nucl. Med. 2001, 42, 259–271. [Google Scholar] [PubMed]

| Grading | Some Selected Types of CNS and Brain Tumor |
|---|---|
| Grade 1 | Meningioma, solitary fibrous tumor, diffuse astrocytoma (MYB- or MYBL1-altered), polymorphous low-grade neuroepithelial tumor of the young, multi-nodular and vacuolating neuronal tumor. |
| Grade 2 | Meningioma, solitary fibrous tumor, oligodendroglioma, astrocytoma (IDH-mutant), IDH-mutant, and 1p/19q-co-deleted, myxopapillary ependymoma, pleomorphic xanthoastrocytoma, supratentorial ependymoma, posterior fossa ependymoma. |
| Grade 3 | Meningioma, solitary fibrous tumor, oligodendroglioma, Astrocytoma (IDH-mutant), IDH-mutant, and 1p/19q-co-deleted, pleomorphic xanthoastrocytoma, supratentorial ependymoma posterior fossa ependymoma. |
| Grade 4 | Glioblastoma (IDH-wildtype), astrocytoma (IDH-mutant), diffuse hemispheric glioma (H3 G34-mutant). |
| SN. | Brain and CNS Tumor Types | Key Genes and Protein Alterations for Tumor |
|---|---|---|
| 1 | Astrocytoma Grade I: Pilocytic Astrocytoma | BRAF, NF1, KIAA1549-BRAF |
| 2 | Astrocytoma Grade II: Low-grade Astrocytoma | EGFR1, BRAF |
| 3 | Astrocytoma Grade III: Anaplastic Astrocytoma | IDH1/2, TP53, ATRX, CDKN2A/B |
| 4 | Astrocytoma Grade IV: Glioblastoma (GBM) | IDH1/2, TERT, chromosomes 7/10, EGFR |
| 5 | Oligodendroglioma | IDH1/2, TERT promoter, 1p/19q, NOTCH1, FUBP1, CIC |
| 6 | Angiocentric glioma | MYB |
| 7 | Diffuse astrocytoma | MYB, MYBL1 |
| 8 | Medulloblastoma | TP53, CTNNB1, PTCH1, APC, SUFU, GLI2, SMO, MYC, MYCN, PRDM6, KDM6A. |
| 9 | Meningiomas | NF2, TRAF7, AKT1, PIK3CA; SMO, SMARCE1, KLF4, BAP1 in subtypes; H3K27me3; TERT, CDKN2A/B in CNS WHO grade 3 |
| 10 | Retinoblastoma | Retinoblastoma (Rb) protein |
| 11 | Ependymomas | ZFTA, YAP1, RELA, MAML2, H3 K27me3, NF1, NF2, EZHIP, MYCN, KMT2D, RELA, FANCE, and EP300 |
| 12 | Primitive neuroectodermal tumors | Isochrome (17q) |
| 13 | Astroblastoma | MN1 |
| 14 | Chordoid glioma | PRKCA |
| 15 | Ganglion cell tumors | BRAF |
| 16 | Polymorphous low-grade neuroepithelial tumor | BRAF, FGFR family |
| 17 | Diffuse midline glioma, H3 K27-altered | TP53, H3 K27, PDGFRA, EGFR, ACVR1, EZHIP |
| 18 | Diffuse hemispheric glioma, H3 G34-mutant | TP53, H3 G34, ATRX |
| 19 | Diffuse pediatric-type high-grade glioma | IDH-wildtype, H3-wildtype, MYCN, PDGFRA, EGFR |
| 20 | Infant-type hemispheric glioma | NTRK family, ROS, ALK, MET |
| 21 | High-grade astrocytoma with piloid features | ATRX, BRAF, CDKN2A/B (methylome), NF1 |
| 22 | Pleomorphic xanthoastrocytoma | CDKN2A/B, BRAF |
| 23 | Subependymal giant cell astrocytoma | TSC1, TSC2 |
| 24 | Solitary fibrous tumor | NAB2-STAT6 |
| 25 | Meningeal melanocytic tumors | NRAS (diffuse), GNA11, GNAQ, CYSLTR2, PLCB4 |
| 26 | Atypical teratoid/rhabdoid tumor | SMARCA4, SMARCB1 |
| 27 | Embryonal tumor with multi-layered rosettes | C19MC, DICER1 |
| 28 | Glioneuronal tumor | NF1, PDFGRA, PRKCA, FGFR1, PIK3CA, KIAA1549-BRAF fusion, 1p, Chromosome 14 |
| 29 | Dysplastic cerebellar gangliocytoma | PTEN |
| 30 | Extraventricular neurocytoma | IDH-wildtype, FGFR (FGFR1-TACC1 fusion) |
| 31 | Multi-nodular and vacuolating neuronal tumor | MAPK pathway |
| 32 | Dysembryoplastic neuroepithelial tumor | FGFR1 |
| 33 | CNS neuroblastoma | FOXR2, BCOR |
| 34 | Desmoplastic myxoid tumor of the pineal region | SMARCB1 |
| Factor | MRI | CT | X-Ray | Ultrasound |
|---|---|---|---|---|
| Duration | 30–45 min | 3–7 min | 2–3 min | 5–10 min |
| Cost | High | Moderate | Low | Low |
| Soft tissue | Excellent detail | Poor detail | Poor detail | Poor detail |
| Bone | Poor detail | Excellent detail | Excellent detail | Poor detail |
| Dimension | 3 | 3 | 2 | 2 |
| Radiation | No | 10 mSv | 0.15 mSv | No |
| Author, Year and Reference | Image Modality | Radiomics Feature | Genomics Feature | AI Model Used | Result |
|---|---|---|---|---|---|
| Akkus et al. [53] (2016) | MRI: T1-CE, T2 | Deep radiomics | 1p19q deletion of LGG | DL (CNN) | Acc.: 87.7 |
| Kickingereder et al. [64] (2016) | MRI: T1, T1-CE, FLAIR, DWI, DSWCEI, PSWI | Hand-crafted | EGFR, PTEN, PDGFRA, MDM4, CDK4 CDKN2A, NF1, and RB1 | ML | Acc.: 63 AUC: 69 |
| Chang et al. [60] (2017) | MRI: T1, T1-CE, T2, FLAIR | Deep radiomics | IDH1 prediction for LGG | DL (ResNet) | Acc.: 89.1 AUC: 95 |
| Li et al. [66] (2017) | MRI: T1, T2 | Deep radiomics | IDH1 prediction for LGG | DL (CNN) | Acc.: 92.4 AUC: 95 |
| Liang et al. [67] (2017) | MRI: T1, T1-CE, T2, FLAIR | Deep radiomics | IDH1 prediction for Glioma | DL (DenseNet) | Acc.: 91.4 AUC: 94.8 |
| Korfiatis et al. [65] (2017) | MRI: T2 | Deep radiomics | MGMT status | DL (ResNet50) | Acc.: 94.9 |
| Chang et al. [61] (2018) | MRI: T1, FLAIR | Deep radiomics | IDH1, 1p/19q co-deletion, MGMT | DL (ResNet) | Acc.: 94 AUC: 91 |
| Smedley et al. [68] (2018) | MRI: T1-CE, T2, FLAIR | Deep radiomics | Tumor morphology | DL (AE) | MAE: 0.0114 |
| Calabrese et al. [131] (2020) | MRI: T1, T1-CE, T2, FLAIR, SWI, DWI, ASLPI, HARDI | Deep radiomics | ATRX, IDH, 7/10aneuploidy, CDKN2, EGFR, TERT, PTEN, TP53, MGMT | TL (CNN+ RF) | AUC: 97 |
| Kawaguchi et al. [63] (2021) | MRI: T1, T1-CE, T2, FLAIR | Hand-crafted | IDH, MGMT, TERT, 1p19q | ML | AUC: 90 |
| Author, Year and Reference | Radiomics | Radio- Genomics | AI Framework | Anatomical Cancer Discussed | Statistics and Risk-of-Bias (RoB) Analysis |
|---|---|---|---|---|---|
| Rizzo et al. (2018) [114] | ✓ | 🗶 | 🗶 | Generalized | 🗶 |
| Kazerooni et al. (2019) [135] | 🗶 | ✓ | 🗶 | Brain (Glioblastoma) | 🗶 |
| Bodalal et al. (2019) [146] | 🗶 | ✓ | ✓ | All Cancer | 🗶 |
| Trivizakis et al. (2020) [51] | 🗶 | ✓ | ✓ | All Cancer | 🗶 |
| Gullo et al. (2020) [139] | 🗶 | ✓ | 🗶 | All Cancer | Statistical analysis only |
| Shui et al. (2021) [147] | 🗶 | ✓ | ✓ | All Cancer | 🗶 |
| Singh et al. (2021) [148] | ✓ | ✓ | 🗶 | Brain (Glioma) | 🗶 |
| Wu et al. (2021) [113] | ✓ | 🗶 | ✓ | Lung | 🗶 |
| Habib et al. (2021) [49] | ✓ | ✓ | 🗶 | Brain | 🗶 |
| Jena et al. (2022) (Proposed) | ✓ | ✓ | ✓ | Brain | ✓ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jena, B.; Saxena, S.; Nayak, G.K.; Balestrieri, A.; Gupta, N.; Khanna, N.N.; Laird, J.R.; Kalra, M.K.; Fouda, M.M.; Saba, L.; et al. Brain Tumor Characterization Using Radiogenomics in Artificial Intelligence Framework. Cancers 2022, 14, 4052. https://doi.org/10.3390/cancers14164052
Jena B, Saxena S, Nayak GK, Balestrieri A, Gupta N, Khanna NN, Laird JR, Kalra MK, Fouda MM, Saba L, et al. Brain Tumor Characterization Using Radiogenomics in Artificial Intelligence Framework. Cancers. 2022; 14(16):4052. https://doi.org/10.3390/cancers14164052
Chicago/Turabian StyleJena, Biswajit, Sanjay Saxena, Gopal Krishna Nayak, Antonella Balestrieri, Neha Gupta, Narinder N. Khanna, John R. Laird, Manudeep K. Kalra, Mostafa M. Fouda, Luca Saba, and et al. 2022. "Brain Tumor Characterization Using Radiogenomics in Artificial Intelligence Framework" Cancers 14, no. 16: 4052. https://doi.org/10.3390/cancers14164052
APA StyleJena, B., Saxena, S., Nayak, G. K., Balestrieri, A., Gupta, N., Khanna, N. N., Laird, J. R., Kalra, M. K., Fouda, M. M., Saba, L., & Suri, J. S. (2022). Brain Tumor Characterization Using Radiogenomics in Artificial Intelligence Framework. Cancers, 14(16), 4052. https://doi.org/10.3390/cancers14164052










