Loss of MMP-27 Predicts Mandibular Bone Invasion in Oral Squamous Cell Carcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. mRNA Expression Analysis Using the cBioPortal Database
2.2. Patients
2.3. Assessment of Tumor Invasion
2.4. Immunohistochemical Sample Preparation and Staining Protocol
2.5. IHC Staining Assessment
2.6. Statistical Analysis
3. Results
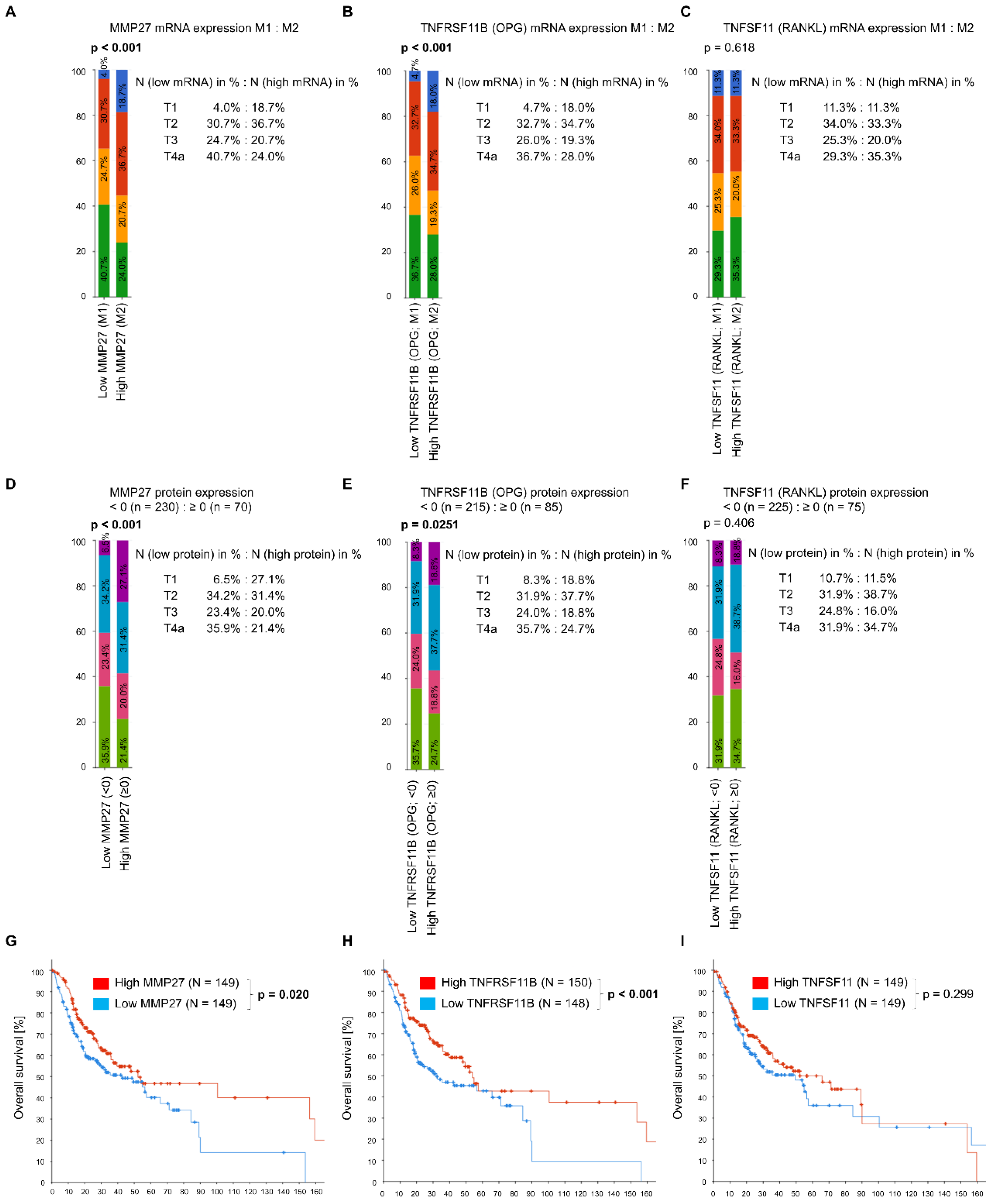
3.1. In Silico Data Analysis via the cBioPortal Database
3.2. Immunohistochemical Expression of MMP-27, RANKL and OPG in OSCC Tissue and Comparison to Clinico-Pathological Parameters
Patient Characteristics
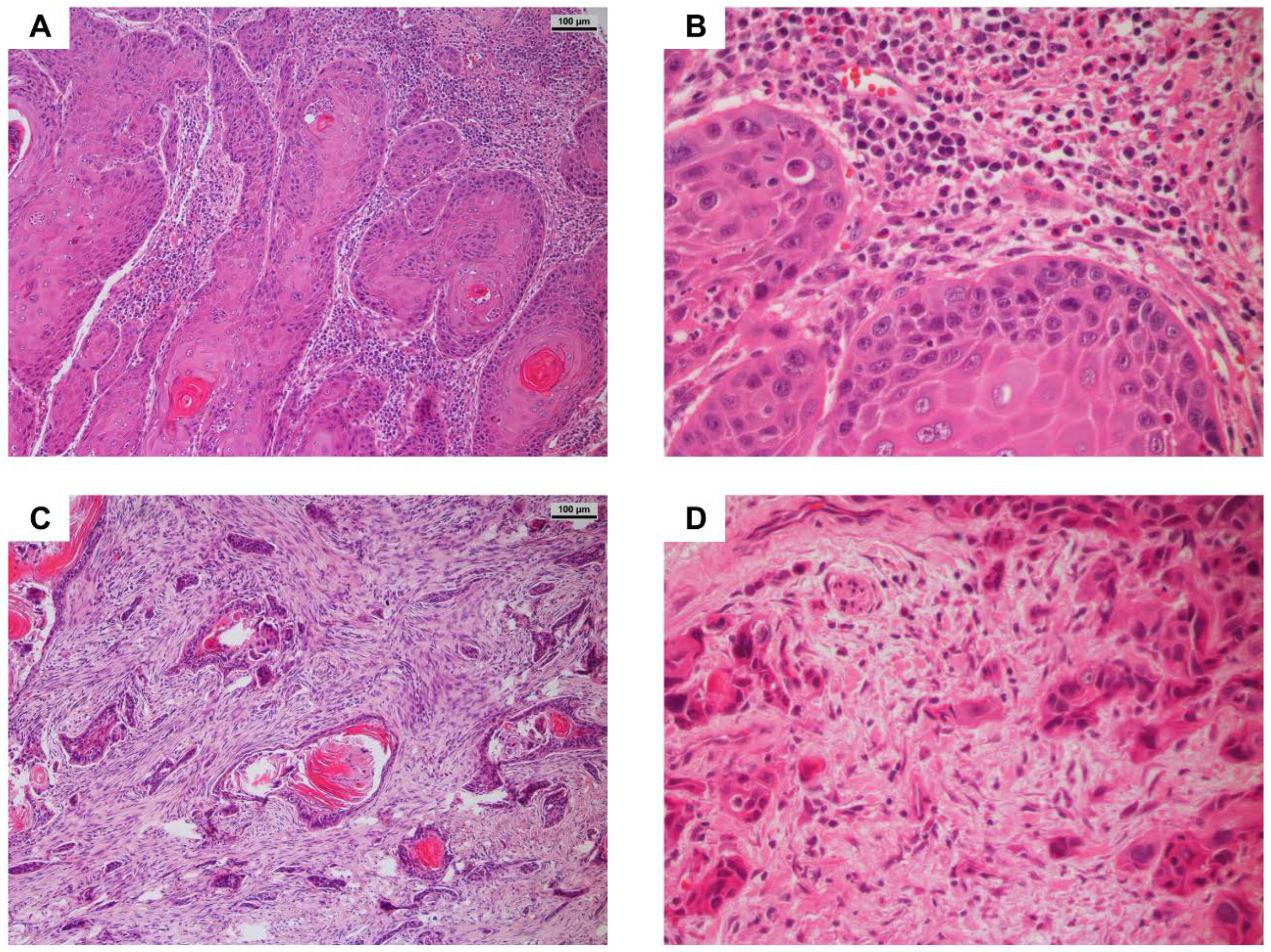
3.3. Worst Pattern of Invasion Score and Relationship to Patient Data
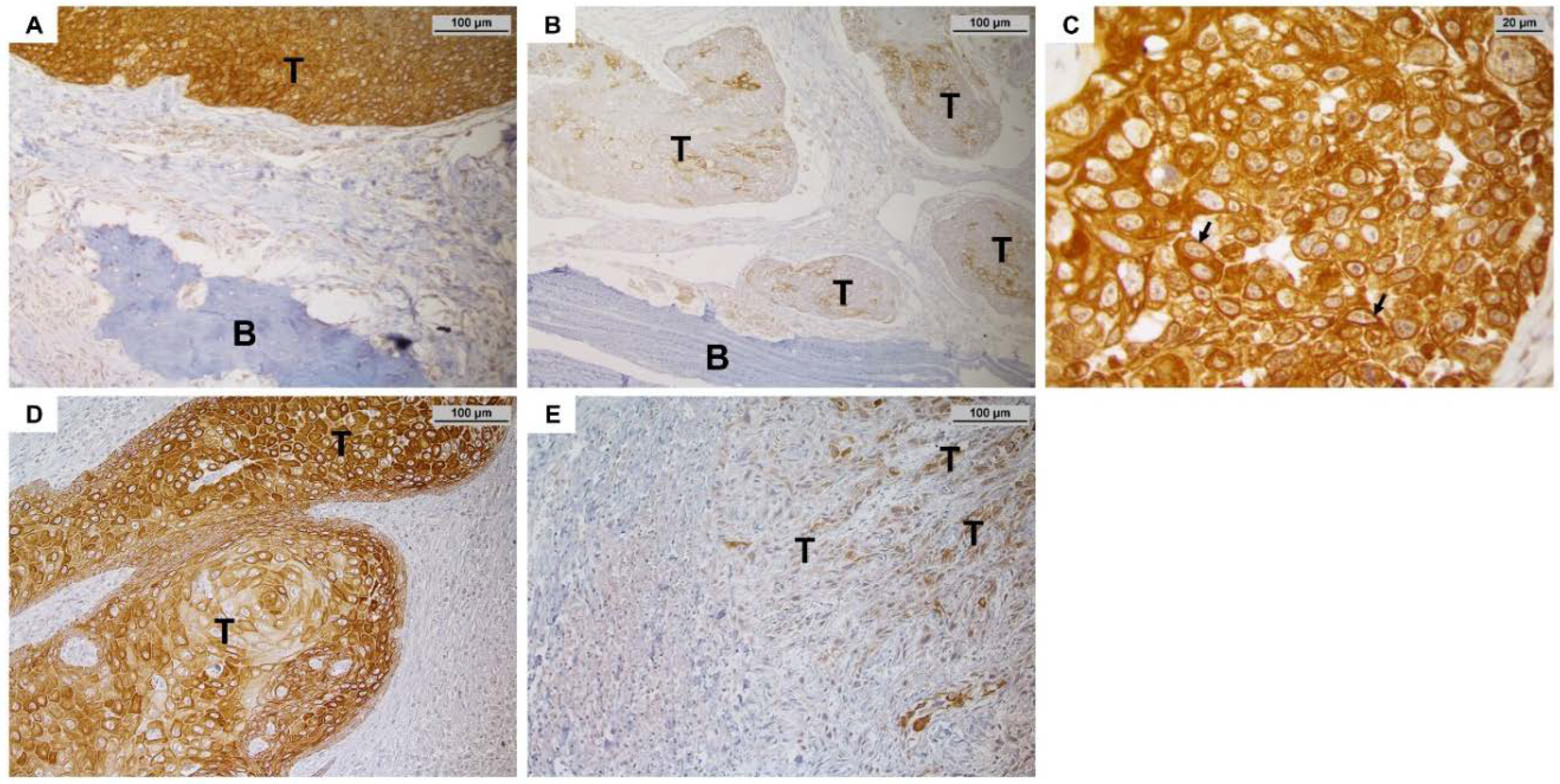
3.4. Immunohistochemical Staining of MMP-27, RANKL and OPG and Association with Clinico-Pathological Parameters, Particularly Bone Invasion
3.4.1. MMP-27 Expression
3.4.2. RANKL and OPG Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. World Cancer Report 2020; Wild, C.P., Weiderpass, E., Stewart, B.W., Eds.; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chow, L.Q.M. Head and Neck Cancer. N. Engl. J. Med. 2020, 382, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Spoerl, S.; Gerken, M.; Fischer, R.; Mamilos, A.; Spoerl, S.; Wolf, S.; Pohl, F.; Klingelhöffer, C.; Ettl, T.; Reichert, T.E.; et al. Lymphatic and vascular invasion in oral squamous cell carcinoma: Implications for recurrence and survival in a population-based cohort study. Oral Oncol. 2020, 111, 105009. [Google Scholar] [CrossRef]
- Brana, I.; Siu, L.L. Locally advanced head and neck squamous cell cancer: Treatment choice based on risk factors and optimizing drug prescription. Ann. Oncol. 2012, 23, x178–x185. [Google Scholar] [CrossRef]
- Chen, Y.-L.; Kuo, S.-W.; Fang, K.-H.; Hao, S.-P. Prognostic impact of marginal mandibulectomy in the presence of superficial bone invasion and the nononcologic outcome. Head Neck 2011, 33, 708–713. [Google Scholar] [CrossRef] [PubMed]
- Shaw, R.J.; Brown, J.S.; Woolgar, J.A.; Lowe, D.; Rogers, S.N.; Vaughan, E.D. The influence of the pattern of mandibular invasion on recurrence and survival in oral squamous cell carcinoma. Head Neck 2004, 26, 861–869. [Google Scholar] [CrossRef]
- O’Brien, C.J.; Adams, J.R.; McNeil, E.B.; Taylor, P.; Laniewski, P.; Clifford, A.; Parker, G.D. Influence of bone invasion and extent of mandibular resection on local control of cancers of the oral cavity and oropharynx. Int. J. Oral Maxillofac. Surg. 2003, 32, 492–497. [Google Scholar] [CrossRef]
- Michalek, J.; Brychtova, S.; Pink, R.; Dvorak, Z. Prognostic and predictive markers for perineural and bone invasion of oral squamous cell carcinoma. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech. Repub. 2019, 163, 302–308. [Google Scholar] [CrossRef] [Green Version]
- Jimi, E.; Furuta, H.; Matsuo, K.; Tominaga, K.; Takahashi, T.; Nakanishi, O. The cellular and molecular mechanisms of bone invasion by oral squamous cell carcinoma. Oral Dis. 2011, 17, 462–468. [Google Scholar] [CrossRef]
- Anderson, D.M.; Maraskovsky, E.; Billingsley, W.L.; Dougall, W.C.; Tometsko, M.E.; Roux, E.R.; Teepe, M.C.; DuBose, R.F.; Cosman, D.; Galibert, L. A homologue of the TNF receptor and its ligand enhance T-cell growth and dendritic-cell function. Nature 1997, 390, 175–179. [Google Scholar] [CrossRef]
- Simonet, W.S.; Lacey, D.L.; Dunstan, C.R.; Kelley, M.; Chang, M.S.; Lüthy, R.; Nguyen, H.Q.; Wooden, S.; Bennett, L.; Boone, T.; et al. Osteoprotegerin: A novel secreted protein involved in the regulation of bone density. Cell 1997, 89, 309–319. [Google Scholar] [CrossRef] [Green Version]
- Asagiri, M.; Takayanagi, H. The molecular understanding of osteoclast differentiation. Bone 2007, 40, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Leibbrandt, A.; Penninger, J.M. RANK/RANKL: Regulators of immune responses and bone physiology. Ann. N. Y. Acad. Sci. 2008, 1143, 123–150. [Google Scholar] [CrossRef]
- Cui, N.; Hu, M.; Khalil, R.A. Biochemical and Biological Attributes of Matrix Metalloproteinases. Prog. Mol. Biol. Transl. Sci. 2017, 147, 1–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, P.-F.; Pan, L.; Luo, Z.-Y.; Zhao, H.-J.; Cai, S.-X. Interrelationship of circulating matrix metalloproteinase-9, TNF-α, and OPG/RANK/RANKL systems in COPD patients with osteoporosis. COPD 2013, 10, 650–656. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.Y.; Yoshida, H.; Sarosi, I.; Tan, H.L.; Timms, E.; Capparelli, C.; Morony, S.; Oliveira-dos-Santos, A.J.; Van, G.; Itie, A.; et al. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature 1999, 397, 315–323. [Google Scholar] [CrossRef]
- Jones, D.H.; Nakashima, T.; Sanchez, O.H.; Kozieradzki, I.; Komarova, S.V.; Sarosi, I.; Morony, S.; Rubin, E.; Sarao, R.; Hojilla, C.V.; et al. Regulation of cancer cell migration and bone metastasis by RANKL. Nature 2006, 440, 692–696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, B.; Kim, H.; Jung, S.; Moon, A.; Noh, D.-Y.; Lee, Z.H.; Kim, H.J.; Kim, H.-H. A CTGF-RUNX2-RANKL Axis in Breast and Prostate Cancer Cells Promotes Tumor Progression in Bone. J. Bone Miner. Res. 2020, 35, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-M.; Kuo, C.-H.; Lai, T.-Y.; Lin, Y.-M.; Su, C.-C.; Hsu, H.-H.; Tsai, F.-J.; Tsai, C.-H.; Huang, C.-Y.; Tang, C.-H. RANKL increases migration of human lung cancer cells through intercellular adhesion molecule-1 up-regulation. J. Cell. Biochem. 2011, 112, 933–941. [Google Scholar] [CrossRef]
- Paiva, K.B.S.; Granjeiro, J.M. Matrix Metalloproteinases in Bone Resorption, Remodeling, and Repair. Prog. Mol. Biol. Transl. Sci. 2017, 148, 203–303. [Google Scholar] [CrossRef]
- Tada, T.; Jimi, E.; Okamoto, M.; Ozeki, S.; Okabe, K. Oral squamous cell carcinoma cells induce osteoclast differentiation by suppression of osteoprotegerin expression in osteoblasts. Int. J. Cancer 2005, 116, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, A.K.; Singh, M.; Bharti, A.C.; Asotra, K.; Sundaram, S.; Mehrotra, R. Genetic polymorphisms of matrix metalloproteinases and their inhibitors in potentially malignant and malignant lesions of the head and neck. J. Biomed. Sci. 2010, 17, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, B.P.; Shah, P.M.; Rawal, U.M.; Desai, A.A.; Shah, S.V.; Rawal, R.M.; Patel, P.S. Activation of MMP-2 and MMP-9 in patients with oral squamous cell carcinoma. J. Surg. Oncol. 2005, 90, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Kurkinen, M. Cloning and characterization of a novel matrix metalloproteinase (MMP), CMMP, from chicken embryo fibroblasts. CMMP, Xenopus XMMP, and human MMP19 have a conserved unique cysteine in the catalytic domain. J. Biol. Chem. 1998, 273, 17893–17900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cominelli, A.; Halbout, M.; N’Kuli, F.; Lemoine, P.; Courtoy, P.J.; Marbaix, E.; Tyteca, D.; Henriet, P. A unique C-terminal domain allows retention of matrix metalloproteinase-27 in the endoplasmic reticulum. Traffic 2014, 15, 401–417. [Google Scholar] [CrossRef]
- Hegedüs, L.; Cho, H.; Xie, X.; Eliceiri, G.L. Additional MDA-MB-231 breast cancer cell matrix metalloproteinases promote invasiveness. J. Cell. Physiol. 2008, 216, 480–485. [Google Scholar] [CrossRef]
- Palavalli, L.H.; Prickett, T.D.; Wunderlich, J.R.; Wei, X.; Burrell, A.S.; Porter-Gill, P.; Davis, S.; Wang, C.; Cronin, J.C.; Agrawal, N.S.; et al. Analysis of the matrix metalloproteinase family reveals that MMP8 is often mutated in melanoma. Nat. Genet. 2009, 41, 518–520. [Google Scholar] [CrossRef] [Green Version]
- Murugan, A.K.; Yang, C.; Xing, M. Mutational analysis of the GNA11, MMP27, FGD1, TRRAP and GRM3 genes in thyroid cancer. Oncol. Lett. 2013, 6, 437–441. [Google Scholar] [CrossRef] [Green Version]
- AJCC Cancer Staging Manual, 8th ed.; Amin, M.B.; Edge, S.B.; Greene, F.L. (Eds.) Springer: New York, NY, USA, 2017. [Google Scholar]
- Brandwein-Gensler, M.; Smith, R.V.; Wang, B.; Penner, C.; Theilken, A.; Broughel, D.; Schiff, B.; Owen, R.P.; Smith, J.; Sarta, C.; et al. Validation of the histologic risk model in a new cohort of patients with head and neck squamous cell carcinoma. Am. J. Surg. Pathol. 2010, 34, 676–688. [Google Scholar] [CrossRef]
- Li, Y.; Bai, S.; Carroll, W.; Dayan, D.; Dort, J.C.; Heller, K.; Jour, G.; Lau, H.; Penner, C.; Prystowsky, M.; et al. Validation of the risk model: High-Risk classification and tumor pattern of invasion predict outcome for patients with low-stage oral cavity squamous cell carcinoma. Head Neck Pathol. 2013, 7, 211–223. [Google Scholar] [CrossRef] [Green Version]
- Kononen, J.; Bubendorf, L.; Kallioniemi, A.; Bärlund, M.; Schraml, P.; Leighton, S.; Torhorst, J.; Mihatsch, M.J.; Sauter, G.; Kallioniemi, O.P. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat. Med. 1998, 4, 844–847. [Google Scholar] [CrossRef] [PubMed]
- Budwit-Novotny, D.A.; McCarty, K.S.; Cox, E.B.; Soper, J.T.; Mutch, D.G.; Creasman, W.T.; Flowers, J.L.; McCarty, K.S., Jr. Immunohistochemical analyses of estrogen receptor in endometrial adenocarcinoma using a monoclonal antibody. Cancer Res. 1986, 46, 5419–5425. [Google Scholar] [PubMed]
- Quattrini, I.; Pollino, S.; Pazzaglia, L.; Conti, A.; Novello, C.; Ferrari, C.; Pignotti, E.; Picci, P.; Benassi, M.S. Prognostic role of nuclear factor/IB and bone remodeling proteins in metastatic giant cell tumor of bone: A retrospective study. J. Orthop. Res. 2015, 33, 1205–1211. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.S.; Lowe, D.; Kalavrezos, N.; D’Souza, J.; Magennis, P.; Woolgar, J. Patterns of invasion and routes of tumor entry into the mandible by oral squamous cell carcinoma. Head Neck 2002, 24, 370–383. [Google Scholar] [CrossRef]
- Kessenbrock, K.; Plaks, V.; Werb, Z. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell 2010, 141, 52–67. [Google Scholar] [CrossRef] [Green Version]
- Gobin, E.; Bagwell, K.; Wagner, J.; Mysona, D.; Sandirasegarane, S.; Smith, N.; Bai, S.; Sharma, A.; Schleifer, R.; She, J.-X. A pan-cancer perspective of matrix metalloproteases (MMP) gene expression profile and their diagnostic/prognostic potential. BMC Cancer 2019, 19, 581. [Google Scholar] [CrossRef] [Green Version]
- Patel, B.P.; Shah, S.V.; Shukla, S.N.; Shah, P.M.; Patel, P.S. Clinical significance of MMP-2 and MMP-9 in patients with oral cancer. Head Neck 2007, 29, 564–572. [Google Scholar] [CrossRef]
- Ahmed Haji Omar, A.; Haglund, C.; Virolainen, S.; Häyry, V.; Atula, T.; Kontio, R.; Salo, T.; Sorsa, T.; Hagström, J. MMP-7, MMP-8, and MMP-9 in oral and cutaneous squamous cell carcinomas. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2015, 119, 459–467. [Google Scholar] [CrossRef]
- Yen, C.-Y.; Liang, S.-S.; Han, L.-Y.; Chou, H.-L.; Chou, C.-K.; Lin, S.-R.; Chiu, C.-C. Cardiotoxin III inhibits proliferation and migration of oral cancer cells through MAPK and MMP signaling. Sci. World J. 2013, 2013, 650946. [Google Scholar] [CrossRef] [Green Version]
- Quan, J.; Zhou, C.; Johnson, N.W.; Francis, G.; Dahlstrom, J.E.; Gao, J. Molecular pathways involved in crosstalk between cancer cells, osteoblasts and osteoclasts in the invasion of bone by oral squamous cell carcinoma. Pathology 2012, 44, 221–227. [Google Scholar] [CrossRef] [Green Version]
- Quan, J.; Elhousiny, M.; Johnson, N.W.; Gao, J. Transforming growth factor-β1 treatment of oral cancer induces epithelial-mesenchymal transition and promotes bone invasion via enhanced activity of osteoclasts. Clin. Exp. Metastasis 2013, 30, 659–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Björklund, M.; Koivunen, E. Gelatinase-mediated migration and invasion of cancer cells. Biochim. Biophys. Acta 2005, 1755, 37–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Kasberg, W.C.; Celo, A.; Liang, Z.; Quispe, K.; Stack, M.S. Post-translational modification of the membrane type 1 matrix metalloproteinase (MT1-MMP) cytoplasmic tail impacts ovarian cancer multicellular aggregate dynamics. J. Biol. Chem. 2017, 292, 13111–13121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Köhrmann, A.; Kammerer, U.; Kapp, M.; Dietl, J.; Anacker, J. Expression of matrix metalloproteinases (MMPs) in primary human breast cancer and breast cancer cell lines: New findings and review of the literature. BMC Cancer 2009, 9, 188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, T.; Chen, P.; Chen, J.; Deng, Y.; Huang, C. Landscape Analysis of Matrix Metalloproteinases Unveils Key Prognostic Markers for Patients With Breast Cancer. Front. Genet. 2021, 12, 809600. [Google Scholar] [CrossRef] [PubMed]
- Cui, N.; Nomura, T.; Takano, N.; Wang, E.; Zhang, W.; Onda, T.; Shibahara, T. Osteoclast-related cytokines from biopsy specimens predict mandibular invasion by oral squamous cell carcinoma. Exp. Ther. Med. 2010, 1, 755–760. [Google Scholar] [CrossRef] [Green Version]
- Sato, K.; Lee, J.-W.; Sakamoto, K.; Iimura, T.; Kayamori, K.; Yasuda, H.; Shindoh, M.; Ito, M.; Omura, K.; Yamaguchi, A. RANKL synthesized by both stromal cells and cancer cells plays a crucial role in osteoclastic bone resorption induced by oral cancer. Am. J. Pathol. 2013, 182, 1890–1899. [Google Scholar] [CrossRef]
- Rabelo, G.D.; Coutinho-Camillo, C.; Kowalski, L.P.; Portero-Muzy, N.; Roux, J.-P.; Chavassieux, P.; Alves, F.A. Evaluation of cortical mandibular bone in patients with oral squamous cell carcinoma. Clin. Oral Investig. 2018, 22, 783–790. [Google Scholar] [CrossRef]
- Yamada, T.; Tsuda, M.; Takahashi, T.; Totsuka, Y.; Shindoh, M.; Ohba, Y. RANKL expression specifically observed in vivo promotes epithelial mesenchymal transition and tumor progression. Am. J. Pathol. 2011, 178, 2845–2856. [Google Scholar] [CrossRef] [Green Version]
- Russmueller, G.; Moser, D.; Würger, T.; Wrba, F.; Christopoulos, P.; Kostakis, G.; Seemann, R.; Stadler, V.; Wimmer, G.; Kornek, G.; et al. Upregulation of osteoprotegerin expression correlates with bone invasion and predicts poor clinical outcome in oral cancer. Oral Oncol. 2015, 51, 247–253. [Google Scholar] [CrossRef]
- Takada, H.; Ibaragi, S.; Eguchi, T.; Okui, T.; Obata, K.; Masui, M.; Morisawa, A.; Takabatake, K.; Kawai, H.; Yoshioka, N.; et al. Semaphorin 4D promotes bone invasion in head and neck squamous cell carcinoma. Int. J. Oncol. 2017, 51, 625–632. [Google Scholar] [CrossRef] [Green Version]
- Montero, P.H.; Patel, S.G. Cancer of the oral cavity. Surg. Oncol. Clin. N. Am. 2015, 24, 491–508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dolens, E.d.S.; Dourado, M.R.; Almangush, A.; Salo, T.A.; Gurgel Rocha, C.A.; da Silva, S.D.; Brennan, P.A.; Coletta, R.D. The Impact of Histopathological Features on the Prognosis of Oral Squamous Cell Carcinoma: A Comprehensive Review and Meta-Analysis. Front. Oncol. 2021, 11, 784924. [Google Scholar] [CrossRef] [PubMed]
- Listl, S.; Jansen, L.; Stenzinger, A.; Freier, K.; Emrich, K.; Holleczek, B.; Katalinic, A.; Gondos, A.; Brenner, H. Survival of patients with oral cavity cancer in Germany. PLoS One 2013, 8, e53415. [Google Scholar] [CrossRef] [PubMed]
- da Silva, S.D.; Hier, M.; Mlynarek, A.; Kowalski, L.P.; Alaoui-Jamali, M.A. Recurrent oral cancer: Current and emerging therapeutic approaches. Front. Pharmacol. 2012, 3, 149. [Google Scholar] [CrossRef] [Green Version]

| Parameter | MMP-27 (N = 119) | RANKL (N = 119) | OPG (N = 85) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | Low | High | p-Value | Negative | Positive | p-Value | Low | High | p-Value | |
| Age | ||||||||||
| (Mean: 62.2 years; 38–89 years) | ||||||||||
| >62 years | 51 (42.9%) | 23 (45.1%) | 28 (54.9%) | 33 (64.7%) | 18 (35.3%) | 19 (46.3%) | 22 (53.7%) | |||
| ≤62 years | 68 (57.1%) | 34 (50.0%) | 34 (50.0%) | 0.711 | 40 (58.8%) | 28 (42.2%) | 0.571 | 32 (72.7%) | 12 (27.3%) | 0.016 |
| Sex | ||||||||||
| male | 95 (79.8%) | 44 (46.3%) | 51 (53.7%) | 58 (61.0%) | 37 (39.0%) | 42 (62.7%) | 25 (37.3%) | |||
| female | 24 (20.2%) | 13 (54.2%) | 11 (53.0%) | 0.504 | 15 (62.5%) | 9 (37.5%) | 1.000 | 9 (50.0%) | 9 (50.0%) | 0.418 |
| Anatomic Site | ||||||||||
| Gingiva | 53 (44.5%) | 26 (49.1%) | 27 (50.9%) | 30 (56.6%) | 23 (43.4%) | 24 (57.1%) | 18 (42.9%) | |||
| Floor of mouth | 66 (55.5%) | 31 (47.0%) | 35 (53.0%) | 0.855 | 43 (66.2%) | 23 (33.8%) | 0.352 | 27 (62.8%) | 16 (37.2%) | 0.661 |
| Tobacco use | ||||||||||
| Yes | 95 (73.9%) | 46 (48.4%) | 49 (51.6%) | 58 (61.0%) | 37 (39.0%) | 44 (66.7%) | 22 (33.3%) | |||
| No | 24 (20.2%) | 11 (45.8%) | 13 (54.2%) | 1.000 | 15 (62.5%) | 9 (37.5%) | 1.000 | 7 (36.8%) | 12 (63.2%) | 0.032 |
| Alcohol use | ||||||||||
| Yes | 88 (73.9%) | 43 (48.9%) | 45 (51.1%) | 54 (61.4%) | 34 (38.6%) | 38 (63.3%) | 22 (36.7%) | |||
| No | 31 (26.1%) | 14 (45.2%) | 17 (54.8%) | 0.835 | 19 (61.3%) | 12 (38.7%) | 0.655 | 13 (52.0%) | 12 (48.0%) | 0.467 |
| T-stage | ||||||||||
| pT2 + pT3 | 55 (46.6%) | 20 (36.4%) | 35 (63.6%) | 36 (65.5%) | 19 (34.5%) | 17 (48.6%) | 18 (51.4%) | |||
| pT4a (mandibular bone invasion) | 64 (53.8%) | 37 (57.8%) | 27 (42.2%) | 0.027 | 37 (57.8%) | 27 (42.2%) | 0.452 | 34 (68.0%) | 16 (32.0%) | 0.115 |
| N-stage | ||||||||||
| N0 | 57 (47.9%) | 27 (47.4%) | 30 (52.6%) | 30 (52.6%) | 27 (47.4%) | 23 (54.8%) | 19 (45.2%) | |||
| N+ | 62 (52.1%) | 30 (48.4%) | 32 (51.6%) | 1.000 | 43 (69.4%) | 19 (30.6%) | 0.089 | 28 (65.1%) | 15 (34.9%) | 0.380 |
| UICC-stage | ||||||||||
| II + III | 37 (28.8%) | 18 (48.6%) | 19 (51.4%) | 21 (56.8%) | 16 (43.2%) | 14 (53.8%) | 12 (46.2%) | |||
| IV | 82 (72.2%) | 39 (47.6%) | 43 (52.4%) | 1.000 | 52 (63.4%) | 30 (36.6%) | 0.544 | 37 (62.7%) | 22 (37.3%) | 0.478 |
| Grade | ||||||||||
| G1 + G2 | 101 (84.9%) | 44 (43.6%) | 57 (56.4%) | 64 (63.4%) | 37 (36.6%) | 41 (56.9%) | 31 (43.1%) | |||
| G3 | 18 (15.1%) | 13 (72.2%) | 5 (27.8%) | 0.039 | 9 (50.0%) | 9 (50.0%) | 0.304 | 10 (76.9%) | 3 (23.1%) | 0.277 |
| Tumor Recurrence | ||||||||||
| Yes | 36 (30.3%) | 14 (38.9%) | 22 (61.1%) | 19 (52.8%) | 17 (47.2%) | 12 (57.1%) | 9 (42.9%) | |||
| No | 83 (69.7%) | 43 (51.8%) | 40 (48.2%) | 0.233 | 53 (63.9%) | 30 (36.1%) | 0.309 | 39 (60.9%) | 25 (29.1%) | 0.801 |
| WPOI grade | ||||||||||
| Non-aggressive | 61 (51.3%) | 29 (47.5%) | 32 (52.5%) | 30 (49.2%) | 31 (50.8%) | 33 (70.2%) | 14 (29.8%) | |||
| Aggressive | 58 (48.7%) | 28 (48.3%) | 30 (51.7%) | 1.000 | 33 (56.9%) | 25 (43.1%) | 0.464 | 18 (47.4%) | 20 (52.6%) | 0.045 |
| HL | β | SE | Wald | p | Exp (β) (95% CI) | ||
|---|---|---|---|---|---|---|---|
| pT2 + pT3 vs. pT4a | 0.221 | High MMP27 | −0.828 | 0.392 | 4.459 | 0.035 | 0.437 (0.203–0.942) |
| RANKL | −0.182 | 0.415 | 0.192 | 0.662 | 0.834 (0.370–1.881) | ||
| High OPG | 0.349 | 0.427 | 0.670 | 0.413 | 1.418 (0.614–3.274) | ||
| WPOI Grade 4-5 | −0.146 | 0.402 | 0.132 | 0.717 | 0.864 (0.393–1.900) | ||
| Grade | −0.307 | 0.566 | 0.295 | 0.587 | 0.735 (0.242–2.231) |
| MMP-27 | RANKL | OPG | ||
|---|---|---|---|---|
| MMP-27 | Spearman’s ρ | 1.000 | 0.239 | 0.480 |
| p-value | - | 0.009 | <0.001 | |
| N | 119 | 119 | 85 | |
| RANKL | Spearman’s ρ | 0.239 | 1.000 | 0.381 |
| p-value | 0.009 | - | <0.001 | |
| N | 119 | 119 | 85 | |
| OPG | Spearman’s ρ | 0.480 | 0.381 | 1.000 |
| p-value | <0.001 | <0.001 | - | |
| N | 85 | 85 | 85 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eichberger, J.; Weber, F.; Spanier, G.; Gerken, M.; Schreml, S.; Schulz, D.; Fiedler, M.; Ludwig, N.; Bauer, R.J.; Reichert, T.E.; et al. Loss of MMP-27 Predicts Mandibular Bone Invasion in Oral Squamous Cell Carcinoma. Cancers 2022, 14, 4044. https://doi.org/10.3390/cancers14164044
Eichberger J, Weber F, Spanier G, Gerken M, Schreml S, Schulz D, Fiedler M, Ludwig N, Bauer RJ, Reichert TE, et al. Loss of MMP-27 Predicts Mandibular Bone Invasion in Oral Squamous Cell Carcinoma. Cancers. 2022; 14(16):4044. https://doi.org/10.3390/cancers14164044
Chicago/Turabian StyleEichberger, Jonas, Florian Weber, Gerrit Spanier, Michael Gerken, Stephan Schreml, Daniela Schulz, Mathias Fiedler, Nils Ludwig, Richard Josef Bauer, Torsten Eugen Reichert, and et al. 2022. "Loss of MMP-27 Predicts Mandibular Bone Invasion in Oral Squamous Cell Carcinoma" Cancers 14, no. 16: 4044. https://doi.org/10.3390/cancers14164044
APA StyleEichberger, J., Weber, F., Spanier, G., Gerken, M., Schreml, S., Schulz, D., Fiedler, M., Ludwig, N., Bauer, R. J., Reichert, T. E., & Ettl, T. (2022). Loss of MMP-27 Predicts Mandibular Bone Invasion in Oral Squamous Cell Carcinoma. Cancers, 14(16), 4044. https://doi.org/10.3390/cancers14164044









