Tumour Derived Extracellular Vesicles: Challenging Target to Blunt Tumour Immune Evasion
Abstract
:Simple Summary
Abstract
1. Introduction
2. TEV and CTL Reprogramming
3. TEV Characterisation
3.1. Tetraspanins
3.2. Toll-Like Receptors (TLRs)
3.3. Heat Shock Proteins (HSP)
3.4. Integrins (ITGA/ITGB)
3.5. TEV as Immunosuppressive Shuttles
3.6. Programmed Death Receptor 1 (PD-1) and Its Ligand (PD-L1)
3.7. CTLA-4/B7/CD152
3.8. T-Cell Immunoglobulin Domain and Mucin Domain 3 (TIM-3)
3.9. Cluster of Differentiation 73 (CD73)
3.10. Fas/FasL
3.11. TRAIL
3.12. Protein Corona and TEV Mechanism of Action
3.13. TGFβ1
3.14. Interleukin 10 (IL-10)
| Molecules Enriched in TEV | Cellular Effectors | Immune-Related Mechanism | Refs. |
|---|---|---|---|
| Tetraspanins (CD9, CD63, CD81) | LFA-1 | EV uptake by leukocytes | [60,61,62] |
| MHC I, MHC II | Regulation of TEV immunogenicity | [55,65] | |
| ADAM10, ADAM17 | ADAM sheddase activity | [63] | |
| HSPs | TLRs | NF-κβ activation and expression of genes involved in immune suppression in tumour cells | [8,78] |
| MDSCs-mediated production of immunosuppressive factor and induction of TAM/M2 polarisation | [85,88] | ||
| Integrins (ITGA/ITGB), CD47 | Extracellular matrix proteins (fibronectin, collagen) | Boost of fibroblast invasive phenotype, tumour spreading and immune cell recruitment | [99,100,101,102,103,104,105,106,107,108,109,110,111,112] |
| PD-L1 | PD-1 | Suppression of CTL functions and induction CD4+ Treg activity | [118,122] |
| CD80/CD86 | CTLA4 | [123,124] | |
| TIM-3 | Galectin-9 | Induction of TAM/M2 differentiation | [134,135] |
| CD73 | AMP | Suppression of T-cell clonal expansion and production of immunosuppressive cytokines by M2/TAM | [140,141] |
| FasL | Fas | Induction of T-cell apoptosis | [149,150] |
| TRAIL | TRAIL-RI/RII | [150] | |
| TGFβ | TGFβ-R1/2 | Increased expression of immunosuppressive factors, suppression of CTL functions and induction of CD4+ Treg activity | [174,175,176] |
| IL-10 | IL-10R | Suppression of CTL function, induction of CD4+ Treg activity and APC reprogramming towards tolerance | [178,179,180,181,182,183] |
3.15. TEV Non-Coding RNA Content
4. TEV and Drug Resistance
5. TEV Released by TME Cells
5.1. TEV Derived from Tumour-Educated Platelets
5.2. TEV from TEC
5.3. EV Derived from DC
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Whiteside, T.L. Exosomes and tumor-mediated immune suppression. J. Clin. Invest. 2016, 126, 1216–1223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoshino, A.; Kim, H.S.; Bojmar, L.; Gyan, K.E.; Cioffi, M.; Hernandez, J.; Zambirinis, C.P.; Rodrigues, G.; Molina, H.; Heissel, S.; et al. Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell 2020, 182, 1044–1061.e18. [Google Scholar] [CrossRef] [PubMed]
- Slingluff, C.L., Jr.; Cox, A.L.; Stover, J.M., Jr.; Moore, M.M.; Hunt, D.F.; Engelhard, V.H. Cytotoxic T-Lymphocyte Response to Autologous Human Squamous Cell Cancer of the Lung: Epitope Reconstitution with Peptides Extracted from HLA-Aw681. Cancer Res. 1994, 54, 2731–2737. [Google Scholar]
- Wolfers, J.; Lozier, A.; Raposo, G.; Regnault, A.; Théry, C.; Masurier, C.; Flament, C.; Pouzieux, S.; Faure, F.; Tursz, T.; et al. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat. Med. 2001, 7, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Echchakir, H.; Vergnon, I.; Dorothée, G.; Grunenwald, D.; Chouaib, S.; Mami-Chouaib, F. Evidence for in situ expansion of diverse antitumor-specific cytotoxic T lymphocyte clones in a human large cell carcinoma of the lung. Int. Immunol. 2000, 12, 537–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kambayashi, T.; Laufer, T.M. Atypical MHC class II-expressing antigen-presenting cells: Can anything replace a dendritic cell? Nat. Rev. Immunol. 2014, 14, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015, 17, 816–826. [Google Scholar] [CrossRef] [PubMed]
- Andersen, M.H.; Schrama, D.; Thor Straten, P.; Becker, J.C. Cytotoxic T cells. J. Invest. Dermatol. 2006, 126, 32–41. [Google Scholar] [CrossRef] [Green Version]
- Coulie, P.G.; Van den Eynde, B.J.; van der Bruggen, P.; Boon, T. Tumour antigens recognized by T lymphocytes, at the core of cancer immunotherapy. Nat. Rev. Cancer 2014, 14, 135–146. [Google Scholar] [CrossRef]
- Arina, A.; Tirapu, I.; Alfaro, C.; Rodríguez-Calvillo, M.; Mazzolini, G.; Inogés, S.; López, A.; Feijoo, E.; Bendandi, M.; Melero, I. Clinical implications of antigen transfer mechanisms from malignant to dendritic cells: Exploiting cross-priming. Exp. Hematol. 2002, 30, 1355–1364. [Google Scholar] [CrossRef]
- Wilson, N.S.; El-Sukkari, D.; Villadangos, J.A. Dendritic cells constitutively present self antigens in their immature state in vivo and regulate antigen presentation by controlling the rates of MHC class II synthesis and endocytosis. Blood 2004, 103, 2187–2195. [Google Scholar] [CrossRef] [PubMed]
- Worbs, T.; Hammerschmidt, S.I.; Förster, R. Dendritic cell migration in health and disease. Nat. Rev. Immunol. 2017, 17, 30–48. [Google Scholar] [CrossRef] [PubMed]
- Carreno, B.M.; Magrini, V.; Becker-Hapak, M.; Kaabinejadian, S.; Hundal, J.; Petti, A.A.; Ly, A.; Lie, W.-R.; Hildebrand, W.H.; Mardis, E.R.; et al. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science 2015, 348, 803–808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albert, M.L.; Sauter, B.; Bhardwaj, N. Dendritic cells acquire antigen from apoptotic cells and induce class I-restricted CTLs. Nature 1998, 392, 86–89. [Google Scholar] [CrossRef] [PubMed]
- Mallegol, J.; Van Niel, G.; Lebreton, C.; Lepelletier, Y.; Candalh, C.; Dugave, C.; Heath, J.; Raposo, G.; Cerf-Bensussan, N.; Heyman, M. T84-Intestinal Epithelial Exosomes Bear MHC Class II/Peptide Complexes Potentiating Antigen Presentation by Dendritic Cells. Gastroenterology 2007, 132, 1866–1876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Appleman, L.J.; Boussiotis, V.A. T cell anergy and costimulation. Immunol. Rev. 2003, 192, 161–180. [Google Scholar] [CrossRef] [PubMed]
- Förster, R.; Schubel, A.; Breitfeld, D.; Kremmer, E.; Renner-Müller, I.; Wolf, E.; Lipp, M. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell 1999, 99, 23–33. [Google Scholar] [CrossRef] [Green Version]
- Ohl, L.; Mohaupt, M.; Czeloth, N.; Hintzen, G.; Kiafard, Z.; Zwirner, J.; Blankenstein, T.; Henning, G.; Förster, R. CCR7 governs skin dendritic cell migration under inflammatory and steady-state conditions. Immunity 2004, 21, 279–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steinman, R.M. The control of immunity and tolerance by dendritic cell. Pathol. Biol. 2003, 51, 59–60. [Google Scholar] [CrossRef]
- Steinman, R.M.; Hawiger, D.; Liu, K.; Bonifaz, L.; Bonnyay, D.; Mahnke, K.; Iyoda, T.; Ravetch, J.; Dhodapkar, M.; Inaba, K.; et al. Dendritic cell function in vivo during the steady state: A role in peripheral tolerance. Ann. N. Y. Acad. Sci. 2003, 987, 15–25. [Google Scholar] [CrossRef]
- Shiokawa, A.; Kotaki, R.; Takano, T.; Nakajima-Adachi, H.; Hachimura, S. Mesenteric lymph node CD11b- CD103+ PD-L1High dendritic cells highly induce regulatory T cells. Immunology 2017, 152, 52–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, K.; Iyoda, T.; Saternus, M.; Kimura, Y.; Inaba, K.; Steinman, R.M. Immune Tolerance After Delivery of Dying Cells to Dendritic Cells In Situ. J. Exp. Med. 2002, 196, 1091–1097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stuart, L.M.; Lucas, M.; Simpson, C.; Lamb, J.; Savill, J.; Lacy-Hulbert, A. Inhibitory effects of apoptotic cell ingestion upon endotoxin-driven myeloid dendritic cell maturation. J. Immunol. Baltim. Md. 1950 2002, 168, 1627–1635. [Google Scholar] [CrossRef] [PubMed]
- Wallet, M.A.; Sen, P.; Flores, R.; Wang, Y.; Yi, Z.; Huang, Y.; Mathews, C.E.; Earp, H.S.; Matsushima, G.; Wang, B.; et al. MerTK is required for apoptotic cell-induced T cell tolerance. J. Exp. Med. 2008, 205, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Patente, T.A.; Pinho, M.P.; Oliveira, A.A.; Evangelista, G.C.M.; Bergami-Santos, P.C.; Barbuto, J.A.M. Human Dendritic Cells: Their Heterogeneity and Clinical Application Potential in Cancer Immunotherapy. Front. Immunol. 2018, 9, 3176. [Google Scholar] [CrossRef]
- Verweij, F.J.; Revenu, C.; Arras, G.; Dingli, F.; Loew, D.; Pegtel, D.M.; Follain, G.; Allio, G.; Goetz, J.G.; Zimmermann, P.; et al. Live Tracking of Inter-organ Communication by Endogenous Exosomes In Vivo. Dev. Cell 2019, 48, 573–589.e4. [Google Scholar] [CrossRef] [Green Version]
- Hyenne, V.; Ghoroghi, S.; Collot, M.; Bons, J.; Follain, G.; Harlepp, S.; Mary, B.; Bauer, J.; Mercier, L.; Busnelli, I.; et al. Studying the Fate of Tumor Extracellular Vesicles at High Spatiotemporal Resolution Using the Zebrafish Embryo. Dev. Cell 2019, 48, 554–572.e7. [Google Scholar] [CrossRef] [Green Version]
- Scott, A.; Sueiro Ballesteros, L.; Bradshaw, M.; Tsuji, C.; Power, A.; Lorriman, J.; Love, J.; Paul, D.; Herman, A.; Emanueli, C.; et al. In Vivo Characterization of Endogenous Cardiovascular Extracellular Vesicles in Larval and Adult Zebrafish. Arterioscler. Thromb. Vasc. Biol. 2021, 41, 2454–2468. [Google Scholar] [CrossRef]
- Grange, C.; Tapparo, M.; Tritta, S.; Deregibus, M.C.; Battaglia, A.; Gontero, P.; Frea, B.; Camussi, G. Role of HLA-G and extracellular vesicles in renal cancer stem cell-induced inhibition of dendritic cell differentiation. BMC Cancer 2015, 15, 1009. [Google Scholar] [CrossRef] [Green Version]
- Maus, R.L.G.; Jakub, J.W.; Nevala, W.K.; Christensen, T.A.; Noble-Orcutt, K.; Sachs, Z.; Hieken, T.J.; Markovic, S.N. Human Melanoma-Derived Extracellular Vesicles Regulate Dendritic Cell Maturation. Front. Immunol. 2017, 8, 358. Available online: https://www.frontiersin.org/article/10.3389/fimmu.2017.00358 (accessed on 27 May 2022). [CrossRef] [Green Version]
- Kurts, C.; Kosaka, H.; Carbone, F.R.; Miller, J.F.A.P.; Heath, W.R. Class I–restricted Cross-Presentation of Exogenous Self-Antigens Leads to Deletion of Autoreactive CD8+ T Cells. J. Exp. Med. 1997, 186, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Manicassamy, S.; Pulendran, B. Dendritic cell control of tolerogenic responses. Immunol. Rev. 2011, 241, 206–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saito, M.; Nagasawa, M.; Takada, H.; Hara, T.; Tsuchiya, S.; Agematsu, K.; Yamada, M.; Kawamura, N.; Ariga, T.; Tsuge, I.; et al. Defective IL-10 signaling in hyper-IgE syndrome results in impaired generation of tolerogenic dendritic cells and induced regulatory T cells. J. Exp. Med. 2011, 208, 235–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sela, U.; Park, C.G.; Park, A.; Olds, P.; Wang, S.; Steinman, R.M.; Fischetti, V.A. Dendritic Cells Induce a Subpopulation of IL-12Rβ2-Expressing Treg that Specifically Consumes IL-12 to Control Th1 Responses. PLoS ONE 2016, 11, e0146412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adler, A.J.; Marsh, D.W.; Yochum, G.S.; Guzzo, J.L.; Nigam, A.; Nelson, W.G.; Pardoll, D.M. CD4+ T cell tolerance to parenchymal self-antigens requires presentation by bone marrow-derived antigen-presenting cells. J. Exp. Med. 1998, 187, 1555–1564. [Google Scholar] [CrossRef] [Green Version]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef] [Green Version]
- Bousso, P.; Robey, E.A. Dynamic Behavior of T Cells and Thymocytes in Lymphoid Organs as Revealed by Two-Photon Microscopy. Immunity 2004, 21, 349–355. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.J.; Hejazi, A.S.; Wei, S.H.; Cahalan, M.D.; Parker, I. T cell repertoire scanning is promoted by dynamic dendritic cell behavior and random T cell motility in the lymph node. Proc. Natl. Acad. Sci. USA 2004, 101, 998–1003. [Google Scholar] [CrossRef] [Green Version]
- Lopatina, T.; Favaro, E.; Danilova, L.; Fertig, E.J.; Favorov, A.V.; Kagohara, L.T.; Martone, T.; Bussolati, B.; Romagnoli, R.; Albera, R.; et al. Extracellular Vesicles Released by Tumor Endothelial Cells Spread Immunosuppressive and Transforming Signals Through Various Recipient Cells. Front. Cell Dev. Biol. 2020, 8, 698. [Google Scholar] [CrossRef]
- Buschow, S.I.; Nolte-‘t Hoen, E.N.; Van Niel, G.; Pols, M.S.; Ten Broeke, T.; Lauwen, M.; Ossendorp, F.; Melief, C.J.; Raposo, G.; Wubbolts, R.; et al. MHC II in dendritic cells is targeted to lysosomes or T cell-induced exosomes via distinct multivesicular body pathways. Traffic Cph. Den. 2009, 10, 1528–1542. [Google Scholar] [CrossRef]
- Wang, X.; Shen, H.; He, Q.; Tian, W.; Xia, A.; Lu, X.-J. Exosomes derived from exhausted CD8+ T cells impaired the anticancer function of normal CD8+ T cells. J. Med. Genet. 2019, 56, 29–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okoye, I.S.; Coomes, S.M.; Pelly, V.S.; Czieso, S.; Papayannopoulos, V.; Tolmachova, T.; Seabra, M.C.; Wilson, M.S. MicroRNA-Containing T-Regulatory-Cell-Derived Exosomes Suppress Pathogenic T Helper 1 Cells. Immunity 2014, 41, 89–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smyth, L.A.; Ratnasothy, K.; Tsang, J.Y.; Boardman, D.; Warley, A.; Lechler, R.; Lombardi, G. CD73 expression on extracellular vesicles derived from CD4+CD25+Foxp3+ T cells contributes to their regulatory function. Eur. J. Immunol. 2013, 43, 2430–2440. [Google Scholar] [CrossRef] [PubMed]
- Watkins, S.K.; Zhu, Z.; Riboldi, E.; Shafer-Weaver, K.A.; Stagliano, K.E.; Sklavos, M.M.; Ambs, S.; Yagita, H.; Hurwitz, A.A. FOXO3 programs tumor-associated DCs to become tolerogenic in human and murine prostate cancer. J. Clin. Invest. 2011, 121, 1361–1372. [Google Scholar] [CrossRef] [Green Version]
- Flores-Toro, J.A.; Luo, D.; Gopinath, A.; Sarkisian, M.R.; Campbell, J.J.; Charo, I.F.; Singh, R.; Schall, T.J.; Datta, M.; Jain, R.K.; et al. CCR2 inhibition reduces tumor myeloid cells and unmasks a checkpoint inhibitor effect to slow progression of resistant murine gliomas. Proc. Natl. Acad. Sci. USA 2020, 117, 1129–1138. [Google Scholar] [CrossRef]
- Kuan, E.L.; Ziegler, S.F. A tumor–myeloid cell axis, mediated via the cytokines IL-1α and TSLP, promotes the progression of breast cancer. Nat. Immunol. 2018, 19, 366–374. [Google Scholar] [CrossRef]
- Kaplan, R.N.; Riba, R.D.; Zacharoulis, S.; Bramley, A.H.; Vincent, L.; Costa, C.; MacDonald, D.D.; Jin, D.K.; Shido, K.; Kerns, S.A.; et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature 2005, 438, 820–827. [Google Scholar] [CrossRef]
- Umakoshi, M.; Takahashi, S.; Itoh, G.; Kuriyama, S.; Sasaki, Y.; Yanagihara, K.; Yashiro, M.; Maeda, D.; Goto, A.; Tanaka, M. Macrophage-mediated transfer of cancer-derived components to stromal cells contributes to establishment of a pro-tumor microenvironment. Oncogene 2019, 38, 2162–2176. [Google Scholar] [CrossRef]
- Schiller, M.; Bekeredjian-Ding, I.; Heyder, P.; Blank, N.; Ho, A.D.; Lorenz, H.-M. Autoantigens are translocated into small apoptotic bodies during early stages of apoptosis. Cell Death Differ. 2008, 15, 183–191. [Google Scholar] [CrossRef]
- A-Gonzalez, N.; Bensinger, S.J.; Hong, C.; Beceiro, S.; Bradley, M.N.; Zelcer, N.; Deniz, J.; Ramirez, C.; Díaz, M.; Gallardo, G.; et al. Apoptotic cells promote their own clearance and immune tolerance through activation of the nuclear receptor LXR. Immunity 2009, 31, 245–258. [Google Scholar] [CrossRef] [Green Version]
- Miles, K.; Simpson, J.; Brown, S.; Cowan, G.; Gray, D.; Gray, M. Immune Tolerance to Apoptotic Self Is Mediated Primarily by Regulatory B1a Cells. Front. Immunol. 2018, 8, 1952. Available online: https://www.frontiersin.org/article/10.3389/fimmu.2017.01952 (accessed on 23 May 2022). [CrossRef] [PubMed] [Green Version]
- Al-Nedawi, K.; Meehan, B.; Micallef, J.; Lhotak, V.; May, L.; Guha, A.; Rak, J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 2008, 10, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Di Vizio, D.; Morello, M.; Dudley, A.C.; Schow, P.W.; Adam, R.M.; Morley, S.; Mulholland, D.; Rotinen, M.; Hager, M.H.; Insabato, L.; et al. Large oncosomes in human prostate cancer tissues and in the circulation of mice with metastatic disease. Am. J. Pathol. 2012, 181, 1573–1584. [Google Scholar] [CrossRef] [PubMed]
- Vagner, T.; Spinelli, C.; Minciacchi, V.R.; Balaj, L.; Zandian, M.; Conley, A.; Zijlstra, A.; Freeman, M.R.; Demichelis, F.; De, S.; et al. Large extracellular vesicles carry most of the tumour DNA circulating in prostate cancer patient plasma. J. Extracell Vesicles 2018, 7, 1505403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Escola, J.M.; Kleijmeer, M.J.; Stoorvogel, W.; Griffith, J.M.; Yoshie, O.; Geuze, H.J. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 1998, 273, 20121–20127. [Google Scholar] [CrossRef] [Green Version]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [Green Version]
- Mellman, I. Dendritic cells: Master regulators of the immune response. Cancer Immunol. Res. 2013, 1, 145–149. [Google Scholar] [CrossRef] [Green Version]
- Garg, A.D.; Nowis, D.; Golab, J.; Vandenabeele, P.; Krysko, D.V.; Agostinis, P. Immunogenic cell death, DAMPs and anticancer therapeutics: An emerging amalgamation. Biochim. Biophys. Acta 2010, 1805, 53–71. [Google Scholar] [CrossRef]
- Yáñez-Mó, M.; Barreiro, O.; Gordon-Alonso, M.; Sala-Valdés, M.; Sánchez-Madrid, F. Tetraspanin-enriched microdomains: A functional unit in cell plasma membranes. Trends Cell Biol. 2009, 19, 434–446. [Google Scholar] [CrossRef]
- Hwang, I.; Shen, X.; Sprent, J. Direct stimulation of naive T cells by membrane vesicles from antigen-presenting cells: Distinct roles for CD54 and B7 molecules. Proc. Natl. Acad. Sci. USA 2003, 100, 6670–6675. [Google Scholar] [CrossRef] [Green Version]
- Nolte-’t Hoen, E.N.M.; Buschow, S.I.; Anderton, S.M.; Stoorvogel, W.; Wauben, M.H.M. Activated T cells recruit exosomes secreted by dendritic cells via LFA-1. Blood 2009, 113, 1977–1981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segura, E.; Nicco, C.; Lombard, B.; Véron, P.; Raposo, G.; Batteux, F.; Amigorena, S.; Théry, C. ICAM-1 on exosomes from mature dendritic cells is critical for efficient naive T-cell priming. Blood 2005, 106, 216–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dornier, E.; Coumailleau, F.; Ottavi, J.F.; Moretti, J.; Boucheix, C.; Mauduit, P.; Schweisguth, F.; Rubinstein, E. TspanC8 tetraspanins regulate ADAM10/Kuzbanian trafficking and promote Notch activation in flies and mammals. J. Cell Biol. 2012, 199, 481–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Söderberg, A.; Barral, A.M.; Söderström, M.; Sander, B.; Rosén, A. Redox-signaling transmitted in trans to neighboring cells by melanoma-derived TNF-containing exosomes. Free Radic. Biol. Med. 2007, 43, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Perugini, V.R.; Zamai, M.; Gonzalez-Granado, J.M.; Barreiro, O.; Tejera, E.; Yáñez-Mó, M.; Caiolfa, V.R.; Sanchez-Madrid, F. CD81 controls sustained T cell activation signaling and defines the maturation stages of cognate immunological synapses. Mol. Cell Biol. 2013, 33, 3644–3658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chow, J.; Franz, K.M.; Kagan, J.C. PRRs are watching you: Localization of innate sensing and signaling regulators. Virology 2015, 479–480, 104–109. [Google Scholar] [CrossRef] [Green Version]
- Kalra, H.; Drummen, G.P.C.; Mathivanan, S. Focus on Extracellular Vesicles: Introducing the Next Small Big Thing. Int. J. Mol. Sci. 2016, 17, 170. [Google Scholar] [CrossRef] [Green Version]
- Hoebe, K.; Du, X.; Georgel, P.; Janssen, E.; Tabeta, K.; Kim, S.O.; Goode, J.; Lin, P.; Mann, N.; Mudd, S.; et al. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature 2003, 424, 743–748. [Google Scholar] [CrossRef]
- Murad, S. Toll-like receptor 4 in inflammation and angiogenesis: A double-edged sword. Front. Immunol. 2014, 5, 313. [Google Scholar] [CrossRef] [Green Version]
- Toll-like Receptors Activation, Signaling, and Targeting, An Overview | Bulletin of the National Research Centre | Full Text. Available online: https://bnrc.springeropen.com/articles/10.1186/s42269-019-0227-2 (accessed on 1 June 2022).
- Du, M.; Butchi, N.B.; Woods, T.; Peterson, K.E. Poly-thymidine oligonucleotides mediate activation of murine glial cells primarily through TLR7, not TLR8. PLoS ONE 2011, 6, e22454. [Google Scholar] [CrossRef]
- Ishii, K.J.; Akira, S. TLR ignores methylated RNA? Immunity 2005, 23, 111–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paschon, V.; Takada, S.H.; Ikebara, J.M.; Sousa, E.; Raeisossadati, R.; Ulrich, H.; Kihara, A.H. Interplay Between Exosomes, microRNAs and Toll-Like Receptors in Brain Disorders. Mol. Neurobiol. 2016, 53, 2016–2028. [Google Scholar] [CrossRef]
- Schaefer, T.M.; Desouza, K.; Fahey, J.V.; Beagley, K.W.; Wira, C.R. Toll-like receptor (TLR) expression and TLR-mediated cytokine/chemokine production by human uterine epithelial cells. Immunology 2004, 112, 428–436. [Google Scholar] [CrossRef]
- Fitzner, N.; Clauberg, S.; Essmann, F.; Liebmann, J.; Kolb-Bachofen, V. Human Skin Endothelial Cells Can Express All 10 TLR Genes and Respond to Respective Ligands. Clin. Vaccine Immunol. 2008, 15, 138–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, B.; Zhao, J.; Unkeless, J.C.; Feng, Z.H.; Xiong, H. TLR signaling by tumor and immune cells: A double-edged sword. Oncogene 2008, 27, 218–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeda, K.; Akira, S. TLR signaling pathways. Semin. Immunol. 2004, 16, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Zhao, J.; Li, H.; He, K.L.; Chen, Y.; Mayer, L.; Unkeless, J.C.; Xiong, H. Toll-like receptors on tumor cells facilitate evasion of immune surveillance. Cancer Res. 2005, 65, 5009–5014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elmallah, M.I.Y.; Cordonnier, M.; Vautrot, V.; Chanteloup, G.; Garrido, C.; Gobbo, J. Membrane-anchored heat-shock protein 70 (Hsp70) in cancer. Cancer Lett. 2020, 469, 134–141. [Google Scholar] [CrossRef]
- Grunberg, N.; Pevsner-Fischer, M.; Goshen-Lago, T.; Diment, J.; Stein, Y.; Lavon, H.; Mayer, S.; Levi-Galibov, O.; Friedman, G.; Ofir-Birin, Y.; et al. Cancer-Associated Fibroblasts Promote Aggressive Gastric Cancer Phenotypes via Heat Shock Factor 1–Mediated Secretion of Extracellular Vesicles. Cancer Res. 2021, 81, 1639–1653. [Google Scholar] [CrossRef]
- Hance, M.W.; Dole, K.; Gopal, U.; Bohonowych, J.E.; Jezierska-Drutel, A.; Neumann, C.A.; Liu, H.; Garraway, I.P.; Isaacs, J.S. Secreted Hsp90 is a novel regulator of the epithelial to mesenchymal transition (EMT) in prostate cancer. J. Biol. Chem. 2012, 287, 37732–37744. [Google Scholar] [CrossRef] [Green Version]
- Nolan, K.D.; Franco, O.E.; Hance, M.W.; Hayward, S.W.; Isaacs, J.S. Tumor-secreted Hsp90 subverts polycomb function to drive prostate tumor growth and invasion. J. Biol. Chem. 2015, 290, 8271–8282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asada, Y.; Tsuruta, M.; Okabayashi, K.; Shigeta, K.; Ishida, T.; Shimada, T.; Suzumura, H.; Koishikawa, K.; Akimoto, S.; Hasegawa, H.; et al. Inhibition of Heat-shock Protein 27 Reduces 5-Fluorouracil-acquired Resistance in Human Colon Cancer Cells. Anticancer Res. 2021, 41, 1283–1290. [Google Scholar] [CrossRef] [PubMed]
- Taha, E.A.; Ono, K.; Eguchi, T. Roles of Extracellular HSPs as Biomarkers in Immune Surveillance and Immune Evasion. Int. J. Mol. Sci. 2019, 20, 4588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gobbo, J.; Marcion, G.; Cordonnier, M.; Dias, A.M.M.; Pernet, N.; Hammann, A.; Richaud, S.; Mjahed, H.; Isambert, N.; Clausse, V.; et al. Restoring Anticancer Immune Response by Targeting Tumor-Derived Exosomes With a HSP70 Peptide Aptamer. J. Natl. Cancer Inst. 2016, 108, djv330. [Google Scholar] [CrossRef] [PubMed]
- Chanteloup, G.; Cordonnier, M.; Isambert, N.; Bertaut, A.; Hervieu, A.; Hennequin, A.; Luu, M.; Zanetta, S.; Coudert, B.; Bengrine, L.; et al. Monitoring HSP70 exosomes in cancer patients’ follow up: A clinical prospective pilot study. J. Extracell Vesicles 2020, 9, 1766192. [Google Scholar] [CrossRef]
- Ono, K.; Eguchi, T.; Sogawa, C.; Calderwood, S.K.; Futagawa, J.; Kasai, T.; Seno, M.; Okamoto, K.; Sasaki, A.; Kozaki, K.I. HSP-enriched properties of extracellular vesicles involve survival of metastatic oral cancer cells. J. Cell Biochem. 2018, 119, 7350–7362. [Google Scholar] [CrossRef] [Green Version]
- Chalmin, F.; Ladoire, S.; Mignot, G.; Vincent, J.; Bruchard, M.; Remy-Martin, J.P.; Boireau, W.; Rouleau, A.; Simon, B.; Lanneau, D.; et al. Membrane-associated Hsp72 from tumor-derived exosomes mediates STAT3-dependent immunosuppressive function of mouse and human myeloid-derived suppressor cells. J. Clin. Invest. 2010, 120, 457–471. [Google Scholar] [CrossRef]
- Shen, Y.; Guo, D.; Weng, L.; Wang, S.; Ma, Z.; Yang, Y.; Wang, P.; Wang, J.; Cai, Z. Tumor-derived exosomes educate dendritic cells to promote tumor metastasis via HSP72/HSP105-TLR2/TLR4 pathway. Oncoimmunology 2017, 6, e1362527. [Google Scholar] [CrossRef]
- Diao, J.; Yang, X.; Song, X.; Chen, S.; He, Y.; Wang, Q.; Chen, G.; Luo, C.; Wu, X.; Zhang, Y. Exosomal Hsp70 mediates immunosuppressive activity of the myeloid-derived suppressor cells via phosphorylation of Stat3. Med. Oncol. 2015, 32, 35. [Google Scholar] [CrossRef]
- Ramteke, A.; Ting, H.; Agarwal, C.; Mateen, S.; Somasagara, R.; Hussain, A.; Graner, M.; Frederick, B.; Agarwal, R.; Deep, G. Exosomes secreted under hypoxia enhance invasiveness and stemness of prostate cancer cells by targeting adherens junction molecules. Mol. Carcinog. 2015, 54, 554–565. [Google Scholar] [CrossRef] [Green Version]
- Fleming, V.; Hu, X.; Weller, C.; Weber, R.; Groth, C.; Riester, Z.; Hüser, L.; Sun, Q.; Nagibin, V.; Kirschning, C.; et al. Melanoma Extracellular Vesicles Generate Immunosuppressive Myeloid Cells by Upregulating PD-L1 via TLR4 Signaling. Cancer Res. 2019, 79, 4715–4728. [Google Scholar] [CrossRef] [PubMed]
- Gastpar, R.; Gehrmann, M.; Bausero, M.A.; Asea, A.; Gross, C.; Schroeder, J.A.; Multhoff, G. Heat Shock Protein 70 Surface-Positive Tumor Exosomes Stimulate Migratory and Cytolytic Activity of Natural Killer Cells. Cancer Res. 2005, 65, 5238–5247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, L.-H.; Wan, Y.-L.; Lin, Y.; Zhang, W.; Yang, M.; Li, G.-L.; Lin, H.-M.; Shang, C.-Z.; Chen, Y.-J.; Min, J. Anticancer Drugs Cause Release of Exosomes with Heat Shock Proteins from Human Hepatocellular Carcinoma Cells That Elicit Effective Natural Killer Cell Antitumor Responses in Vitro. J. Biol. Chem. 2012, 287, 15874–15885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, J.; Lee, Y.-S.; Kim, S.-H.; Ko, J.-K.; Kim, C.-W. MHC independent anti-tumor immune responses induced by Hsp70-enriched exosomes generate tumor regression in murine models. Cancer Lett. 2009, 275, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Chen, Y.; Wang, S.; Yu, L.; Shen, Y.; Zhong, H.; Yang, Y. Exosomes from heat-stressed tumour cells inhibit tumour growth by converting regulatory T cells to Th17 cells via IL-6. Immunology 2018, 154, 132–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komarova, E.Y.; Suezov, R.V.; Nikotina, A.D.; Aksenov, N.D.; Garaeva, L.A.; Shtam, T.A.; Zhakhov, A.V.; Martynova, M.G.; Bystrova, O.A.; Istomina, M.S.; et al. Hsp70-containing extracellular vesicles are capable of activating of adaptive immunity in models of mouse melanoma and colon carcinoma. Sci. Rep. 2021, 11, 21314. [Google Scholar] [CrossRef]
- Zininga, T.; Ramatsui, L.; Shonhai, A. Heat Shock Proteins as Immunomodulants. Molecules 2018, 23, 2846. [Google Scholar] [CrossRef] [Green Version]
- Chanda, D.; Otoupalova, E.; Hough, K.P.; Locy, M.L.; Bernard, K.; Deshane, J.S.; Sanderson, R.D.; Mobley, J.A.; Thannickal, V.J. Fibronectin on the Surface of Extracellular Vesicles Mediates Fibroblast Invasion. Am. J. Respir. Cell Mol. Biol. 2019, 60, 279–288. [Google Scholar] [CrossRef]
- Hoshino, A.; Costa-Silva, B.; Shen, T.-L.; Rodrigues, G.; Hashimoto, A.; Mark, M.T.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [Green Version]
- Sung, B.H.; Ketova, T.; Hoshino, D.; Zijlstra, A.; Weaver, A.M. Directional cell movement through tissues is controlled by exosome secretion. Nat. Commun. 2015, 6, 7164. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Fedele, C.; Lu, H.; Nevalainen, M.T.; Keen, J.H.; Languino, L.R. Exosome-mediated Transfer of αvβ3 Integrin from Tumorigenic to Nontumorigenic Cells Promotes a Migratory Phenotype. Mol. Cancer Res. MCR 2016, 14, 1136–1146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeRita, R.M.; Sayeed, A.; Garcia, V.; Krishn, S.R.; Shields, C.D.; Sarker, S.; Friedman, A.; McCue, P.; Molugu, S.K.; Rodeck, U.; et al. Tumor-Derived Extracellular Vesicles Require β1 Integrins to Promote Anchorage-Independent Growth. iScience 2019, 14, 199–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Purushothaman, A.; Bandari, S.K.; Liu, J.; Mobley, J.A.; Brown, E.E.; Sanderson, R.D. Fibronectin on the Surface of Myeloma Cell-derived Exosomes Mediates Exosome-Cell Interactions. J. Biol. Chem. 2016, 291, 1652–1663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Q.; Furuta, K.; Lucien, F.; Sanchez, L.H.G.; Hirsova, P.; Krishnan, A.; Kabashima, A.; Pavelko, K.D.; Madden, B.; Alhuwaish, H.; et al. Integrin β1-enriched extracellular vesicles mediate monocyte adhesion and promote liver inflammation in murine NASH. J. Hepatol. 2019, 71, 1193–1205. [Google Scholar] [CrossRef] [PubMed]
- Grigoryeva, E.S.; Savelieva, O.E.; Popova, N.O.; Cherdyntseva, N.V.; Perelmuter, V.M. Do tumor exosome integrins alone determine organotropic metastasis? Mol. Biol. Rep. 2020, 47, 8145–8157. [Google Scholar] [CrossRef]
- Altei, W.F.; Pachane, B.C.; dos Santos, P.K.; Ribeiro, L.N.M.; Sung, B.H.; Weaver, A.M.; Selistre-De-Araújo, H.S. Inhibition of αvβ3 integrin impairs adhesion and uptake of tumor-derived small extracellular vesicles. Cell Commun. Signal. 2020, 18, 158. [Google Scholar] [CrossRef]
- Soe, Z.Y.; Park, E.J.; Shimaoka, M. Integrin Regulation in Immunological and Cancerous Cells and Exosomes. Int. J. Mol. Sci. 2021, 22, 2193. [Google Scholar] [CrossRef]
- Willingham, S.B.; Volkmer, J.-P.; Gentles, A.J.; Sahoo, D.; Dalerba, P.; Mitra, S.S.; Wang, J.; Contreras-Trujillo, H.; Martin, R.; Cohen, J.D.; et al. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc. Natl. Acad. Sci. USA 2012, 109, 6662–6667. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Pu, Y.; Cron, K.R.; Deng, L.; Kline, J.; Frazier, W.A.; Xu, H.; Peng, H.; Fu, Y.-X.; Xu, M.M. CD47 blockade triggers T cell–mediated destruction of immunogenic tumors. Nat. Med. 2015, 21, 1209–1215. [Google Scholar] [CrossRef] [Green Version]
- Chao, M.P.; Weissman, I.L.; Majeti, R. The CD47–SIRPα pathway in cancer immune evasion and potential therapeutic implications. Curr. Opin. Immunol. 2012, 24, 225–232. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, A.; Sawada, K.; Kobayashi, M.; Yamamoto, M.; Yagi, T.; Kinose, Y.; Kodama, M.; Hashimoto, K.; Kimura, T. Exosomal CD47 Plays an Essential Role in Immune Evasion in Ovarian Cancer. Mol. Cancer Res. 2021, 19, 1583–1595. [Google Scholar] [CrossRef] [PubMed]
- Gaikwad, S.; Agrawal, M.Y.; Kaushik, I.; Ramachandran, S.; Srivastava, S.K. Immune checkpoint proteins: Signaling mechanisms and molecular interactions in cancer immunotherapy. Semin. Cancer Biol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Ricklefs, F.L.; Alayo, Q.; Krenzlin, H.; Mahmoud, A.B.; Speranza, M.C.; Nakashima, H.; Hayes, J.L.; Lee, K.; Balaj, L.; Passaro, C.; et al. Immune evasion mediated by PD-L1 on glioblastoma-derived extracellular vesicles. Sci. Adv. 2018, 4, eaar2766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poggio, M.; Hu, T.; Pai, C.-C.; Chu, B.; Belair, C.D.; Chang, A.; Montabana, E.; Lang, U.E.; Fu, Q.; Fong, L.; et al. Suppression of Exosomal PD-L1 Induces Systemic Anti-tumor Immunity and Memory. Cell 2019, 177, 414–427.e13. [Google Scholar] [CrossRef] [Green Version]
- Sun, Z.; Fourcade, J.; Pagliano, O.; Chauvin, J.-M.; Sander, C.; Kirkwood, J.M.; Zarour, H.M. IL10 and PD-1 Cooperate to Limit the Activity of Tumor-Specific CD8+ T Cells. Cancer Res. 2015, 75, 1635–1644. [Google Scholar] [CrossRef] [Green Version]
- Butte, M.J.; Keir, M.E.; Phamduy, T.B.; Freeman, G.J.; Sharpe, A.H. PD-L1 interacts specifically with B7-1 to inhibit T cell proliferation. Immunity 2007, 27, 111–122. [Google Scholar] [CrossRef] [Green Version]
- Kamada, T.; Togashi, Y.; Tay, C.; Ha, D.; Sasaki, A.; Nakamura, Y.; Sato, E.; Fukuoka, S.; Tada, Y.; Tanaka, A.; et al. PD-1+ regulatory T cells amplified by PD-1 blockade promote hyperprogression of cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 9999–10008. [Google Scholar] [CrossRef] [Green Version]
- Theodoraki, M.-N.; Yerneni, S.S.; Hoffmann, T.K.; Gooding, W.E.; Whiteside, T.L. Clinical Significance of PD-L1+ Exosomes in Plasma of Head and Neck Cancer Patients. Clin. Cancer Res. 2018, 24, 896–905. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.H.; Kim, H.; Choi, Y.J.; Kim, S.Y.; Lee, J.-E.; Sung, K.J.; Sung, Y.H.; Pack, C.-G.; Jung, M.-K.; Han, B.; et al. Exosomal PD-L1 promotes tumor growth through immune escape in non-small cell lung cancer. Exp. Mol. Med. 2019, 51, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Han, X. Anti-PD-1/PD-L1 therapy of human cancer: Past, present, and future. J. Clin. Invest. 2015, 125, 3384–3391. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Huang, A.C.; Zhang, W.; Zhang, G.; Wu, M.; Xu, W.; Yu, Z.; Yang, J.; Wang, B.; Sun, H.; et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 2018, 560, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Topalian, S.L.; Taube, J.M.; Anders, R.A.; Pardoll, D.M. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat. Rev. Cancer 2016, 16, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Brunner, M.C.; Chambers, C.A.; Chan, F.K.; Hanke, J.; Winoto, A.; Allison, J.P. CTLA-4-Mediated inhibition of early events of T cell proliferation. J. Immunol. Baltim. Md. 1950 1999, 162, 5813–5820. [Google Scholar]
- Carreno, B.M.; Bennett, F.; Chau, T.A.; Ling, V.; Luxenberg, D.; Jussif, J.; Baroja, M.L.; Madrenas, J. CTLA-4 (CD152) can inhibit T cell activation by two different mechanisms depending on its level of cell surface expression. J. Immunol. Baltim. Md. 1950 2000, 165, 1352–1356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnes, M.J.; Griseri, T.; Johnson, A.M.F.; Young, W.; Powrie, F.; Izcue, A. CTLA-4 promotes Foxp3 induction and regulatory T cell accumulation in the intestinal lamina propria. Mucosal Immunol. 2013, 6, 324–334. [Google Scholar] [CrossRef] [Green Version]
- Theodoraki, M.-N.; Yerneni, S.; Gooding, W.E.; Ohr, J.; Clump, D.A.; Bauman, J.E.; Ferris, R.L.; Whiteside, T.L. Circulating exosomes measure responses to therapy in head and neck cancer patients treated with cetuximab, ipilimumab, and IMRT. Oncoimmunology 2019, 8, 1593805. [Google Scholar] [CrossRef]
- Scarpa, M.; Marchiori, C.; Scarpa, M.; Castagliuolo, I. CD80 expression is upregulated by TP53 activation in human cancer epithelial cells. Oncoimmunology 2021, 10, 1907912. [Google Scholar] [CrossRef]
- Li, Y.; Bai, W.; Zhang, L. The Overexpression of CD80 and ISG15 Are Associated with the Progression and Metastasis of Breast Cancer by a Meta-Analysis Integrating Three Microarray Datasets. Pathol. Oncol. Res. POR 2020, 26, 443–452. [Google Scholar] [CrossRef]
- Azambuja, J.H.; Ludwig, N.; Yerneni, S.; Rao, A.; Braganhol, E.; Whiteside, T.L. Molecular profiles and immunomodulatory activities of glioblastoma-derived exosomes. Neuro-Oncol. Adv. 2020, 2, vdaa056. [Google Scholar] [CrossRef]
- Wang, Y.; Li, P.; Mao, S.; Mo, Z.; Cao, Z.; Luo, J.; Zhou, M.; Liu, X.; Zhang, S.; Yu, L. Exosome CTLA-4 Regulates PTEN/CD44 Signal Pathway in Spleen Deficiency Internal Environment to Promote Invasion and Metastasis of Hepatocellular Carcinoma. Front. Pharmacol. 2021, 12, 757194. [Google Scholar] [CrossRef]
- Hastings, W.D.; Anderson, D.E.; Kassam, N.; Koguchi, K.; Greenfield, E.A.; Kent, S.C.; Zheng, X.X.; Strom, T.B.; Hafler, D.A.; Kuchroo, V.K. TIM-3 is Expressed on Activated Human CD4+ T Cells and Regulates Th1 and Th17 Cytokines. Eur. J. Immunol. 2009, 39, 2492–2501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, C.; Anderson, A.C.; Kuchroo, V.K. TIM-3 and Its Regulatory Role in Immune Responses. In Negative Co-Receptors and Ligands; Ahmed, R., Honjo, T., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 1–15. [Google Scholar]
- Cheng, Z.; Wang, L.; Wu, C.; Huang, L.; Ruan, Y.; Xue, W. Tumor-derived Exosomes Induced M2 Macrophage Polarization and Promoted the Metastasis of Osteosarcoma Cells Through Tim-3. Arch. Med. Res. 2021, 52, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Qiu, X.; Li, X.; Fan, H.; Zhang, F.; Lv, T.; Song, Y. Expression profiles and clinical value of plasma exosomal Tim-3 and Galectin-9 in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2018, 498, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Allard, B.; Cousineau, I.; Spring, K.; Stagg, J. Measurement of CD73 enzymatic activity using luminescence-based and colorimetric assays. Methods Enzymol. 2019, 629, 269–289. [Google Scholar] [PubMed]
- Kordaß, T.; Osen, W.; Eichmüller, S.B. Controlling the Immune Suppressor: Transcription Factors and MicroRNAs Regulating CD73/NT5E. Front. Immunol. 2018, 9, 813. [Google Scholar] [CrossRef] [Green Version]
- Nakav, S.; Chaimovitz, C.; Sufaro, Y.; Lewis, E.C.; Shaked, G.; Czeiger, D.; Zlotnik, M.; Douvdevani, A. Anti-Inflammatory Preconditioning by Agonists of Adenosine A1 Receptor. PLoS ONE 2008, 3, e2107. [Google Scholar] [CrossRef] [Green Version]
- Yu, M.; Guo, G.; Huang, L.; Deng, L.; Chang, C.-S.; Achyut, B.R.; Canning, M.; Xu, N.; Arbab, A.S.; Bollag, R.J.; et al. CD73 on cancer-associated fibroblasts enhanced by the A2B-mediated feedforward circuit enforces an immune checkpoint. Nat. Commun. 2020, 11, 515. [Google Scholar] [CrossRef]
- Wang, M.; Jia, J.; Cui, Y.; Peng, Y.; Jiang, Y. CD73-positive extracellular vesicles promote glioblastoma immunosuppression by inhibiting T-cell clonal expansion. Cell Death Dis. 2021, 12, 1065. [Google Scholar] [CrossRef]
- Lu, T.; Zhang, Z.; Zhang, J.; Pan, X.; Zhu, X.; Wang, X.; Li, Z.; Ruan, M.; Li, H.; Chen, W.; et al. CD73 in small extracellular vesicles derived from HNSCC defines tumour-associated immunosuppression mediated by macrophages in the microenvironment. J. Extracell Vesicles 2022, 11, e12218. [Google Scholar] [CrossRef]
- Strasser, A.; Jost, P.J.; Nagata, S. The many roles of FAS receptor signaling in the immune system. Immunity 2009, 30, 180. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, M.; Lügering, N.; Pauels, H.G.; Schulze-Osthoff, K.; Domschke, W.; Kucharzik, T. IL-10 induces apoptosis in human monocytes involving the CD95 receptor/ligand pathway. Eur. J. Immunol. 2000, 30, 1769–1777. [Google Scholar] [CrossRef]
- Du, X.; Bao, G.; He, X.; Zhao, H.; Yu, F.; Qiao, Q.; Lu, J.; Ma, Q. Expression and biological significance of c-FLIP in human hepatocellular carcinomas. J. Exp. Clin. Cancer Res. 2009, 28, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luebke, T.; Schwarz, L.; Beer, Y.Y.; Schumann, S.; Misterek, M.; Sander, F.E.; Plaza-Sirvent, C.; Schmitz, I. c-FLIP and CD95 signaling are essential for survival of renal cell carcinoma. Cell Death Dis. 2019, 10, 384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plaza-Sirvent, C.; Schuster, M.; Neumann, Y.; Heise, U.; Pils, M.C.; Schulze-Osthoff, K.; Schmitz, I. c-FLIP Expression in Foxp3-Expressing Cells Is Essential for Survival of Regulatory T Cells and Prevention of Autoimmunity. Cell Rep. 2017, 18, 12–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Z.; Yang, F.; Yu, L.; Yu, Z.; Jiang, L.; Wang, Q.; Yang, Y.; Wang, L.; Cao, X.; Wang, J. Activated T Cell Exosomes Promote Tumor Invasion via Fas Signaling Pathway. J. Immunol. 2012, 188, 5954–5961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albanese, J.; Meterissian, S.; Kontogiannea, M.; Dubreuil, C.; Hand, A.; Sorba, S.; Dainiak, N. Biologically active Fas antigen and its cognate ligand are expressed on plasma membrane-derived extracellular vesicles. Blood 1998, 91, 3862–3874. [Google Scholar] [CrossRef]
- Andreola, G.; Rivoltini, L.; Castelli, C.; Huber, V.; Perego, P.; Deho, P.; Squarcina, P.; Accornero, P.; Lozupone, F.; Lugini, L.; et al. Induction of lymphocyte apoptosis by tumor cell secretion of FasL-bearing microvesicles. J. Exp. Med. 2002, 195, 1303–1316. [Google Scholar] [CrossRef]
- Huber, V.; Fais, S.; Iero, M.; Lugini, L.; Canese, P.; Squarcina, P.; Zaccheddu, A.; Colone, M.; Arancia, G.; Gentile, M.; et al. Human Colorectal Cancer Cells Induce T-Cell Death Through Release of Proapoptotic Microvesicles: Role in Immune Escape. Gastroenterology 2005, 128, 1796–1804. [Google Scholar] [CrossRef]
- Kim, J.W.; Wieckowski, E.; Taylor, D.D.; Reichert, T.E.; Watkins, S.; Whiteside, T.L. Fas ligand-positive membranous vesicles isolated from sera of patients with oral cancer induce apoptosis of activated T lymphocytes. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2005, 11, 1010–1020. [Google Scholar] [CrossRef]
- Cormier, Z. Small-molecule drug drives cancer cells to suicide. Nature 2013. [Google Scholar] [CrossRef]
- Takeda, K.; Smyth, M.J.; Cretney, E.; Hayakawa, Y.; Yamaguchi, N.; Yagita, H.; Okumura, K. Involvement of Tumor Necrosis Factor-Related Apoptosis-Inducing Ligand in NK Cell-Mediated and IFN-γ-Dependent Suppression of Subcutaneous Tumor Growth. Cell Immunol. 2001, 214, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Kischkel, F.C.; Lawrence, D.A.; Chuntharapai, A.; Schow, P.; Kim, K.J.; Ashkenazi, A. Apo2L/TRAIL-Dependent Recruitment of Endogenous FADD and Caspase-8 to Death Receptors 4 and 5. Immunity 2000, 12, 611–620. [Google Scholar] [CrossRef] [Green Version]
- Baker, S.J.; Reddy, E.P. Modulation of life and death by the TNF receptor superfamily. Oncogene 1998, 17, 3261–3270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cullen, S.P.; Martin, S.J. Fas and TRAIL ‘death receptors’ as initiators of inflammation: Implications for cancer. Semin. Cell Dev. Biol. 2015, 39, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Beyer, K.; Baukloh, A.-K.; Stoyanova, A.; Kamphues, C.; Sattler, A.; Kotsch, K. Interactions of Tumor Necrosis Factor–Related Apoptosis-Inducing Ligand (TRAIL) with the Immune System: Implications for Inflammation and Cancer. Cancers 2019, 11, 1161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Jin, T.-G.; Yang, H.; DeWolf, W.C.; Khosravi-Far, R.; Olumi, A.F. Persistent c-FLIP(L) Expression Is Necessary and Sufficient to Maintain Resistance to Tumor Necrosis Factor-Related Apoptosis-Inducing Ligand–Mediated Apoptosis in Prostate Cancer. Cancer Res. 2004, 64, 7086–7091. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Yang, X.; Xu, T.; Kong, Q.; Zhang, Y.; Shen, Y.; Wei, Y.; Wang, G.; Chang, K.-J. Overcoming resistance to TRAIL-induced apoptosis in solid tumor cells by simultaneously targeting death receptors, c-FLIP and IAPs. Int. J. Oncol. 2016, 49, 153–163. [Google Scholar] [CrossRef] [Green Version]
- Ke, C.; Hou, H.; Li, J.; Su, K.; Huang, C.; Lin, Y.; Lu, Z.; Du, Z.; Tan, W.; Yuan, Z. Extracellular Vesicle Delivery of TRAIL Eradicates Resistant Tumor Growth in Combination with CDK Inhibition by Dinaciclib. Cancers 2020, 12, 1157. [Google Scholar] [CrossRef]
- Qiu, Y.; Sun, J.; Qiu, J.; Chen, G.; Wang, X.; Mu, Y.; Li, K.; Wang, W. Antitumor Activity of Cabazitaxel and MSC-TRAIL Derived Extracellular Vesicles in Drug-Resistant Oral Squamous Cell Carcinoma. Cancer Manag. Res. 2020, 12, 10809–10820. [Google Scholar] [CrossRef]
- Shamili, F.H.; Bayegi, H.R.; Salmasi, Z.; Sadri, K.; Mahmoudi, M.; Kalantari, M.; Ramezani, M.; Abnous, K. Exosomes derived from TRAIL-engineered mesenchymal stem cells with effective anti-tumor activity in a mouse melanoma model. Int. J. Pharm. 2018, 549, 218–229. [Google Scholar] [CrossRef]
- Rivoltini, L.; Chiodoni, C.; Squarcina, P.; Tortoreto, M.; Villa, A.; Vergani, B.; Bürdek, M.; Botti, L.; Arioli, I.; Cova, A.; et al. TNF-Related Apoptosis-Inducing Ligand (TRAIL)–Armed Exosomes Deliver Proapoptotic Signals to Tumor Site. Clin. Cancer Res. 2016, 22, 3499–3512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ludwig, S.; Floros, T.; Theodoraki, M.-N.; Hong, C.-S.; Jackson, E.K.; Lang, S.; Whiteside, T.L. Suppression of Lymphocyte Functions by Plasma Exosomes Correlates with Disease Activity in Patients with Head and Neck Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017, 23, 4843–4854. [Google Scholar] [CrossRef] [Green Version]
- Setroikromo, R.; Zhang, B.; Reis, C.R.; Mistry, R.H.; Quax, W.J. Death Receptor 5 Displayed on Extracellular Vesicles Decreases TRAIL Sensitivity of Colon Cancer Cells. Front. Cell Dev. Biol. 2020, 8, 318. Available online: https://www.frontiersin.org/article/10.3389/fcell.2020.00318 (accessed on 20 June 2022). [CrossRef]
- Tóth, E.Á.; Turiák, L.; Visnovitz, T.; Cserép, C.; Mázló, A.; Sódar, B.W.; Försönits, A.I.; Petővári, G.; Sebestyén, A.; Komlósi, Z.; et al. Formation of a protein corona on the surface of extracellular vesicles in blood plasma. J. Extracell Vesicles 2021, 10, e12140. [Google Scholar] [CrossRef]
- Palviainen, M.; Saraswat, M.; Varga, Z.; Kitka, D.; Neuvonen, M.; Puhka, M.; Joenväärä, S.; Renkonen, R.; Nieuwland, R.; Takatalo, M.; et al. Extracellular vesicles from human plasma and serum are carriers of extravesicular cargo—Implications for biomarker discovery. PLoS ONE 2020, 15, e0236439. [Google Scholar] [CrossRef]
- Wolf, M.; Poupardin, R.W.; Ebner-Peking, P.; Andrade, A.C.; Blöchl, C.; Obermayer, A.; Gomes, F.G.; Vari, B.; Maeding, N.; Eminger, E.; et al. A functional corona around extracellular vesicles enhances angiogenesis, skin regeneration and immunomodulation. J. Extracell Vesicles 2022, 11, e12207. [Google Scholar] [CrossRef]
- Webber, J.; Steadman, R.; Mason, M.D.; Tabi, Z.; Clayton, A. Cancer Exosomes Trigger Fibroblast to Myofibroblast Differentiation. Cancer Res. 2010, 70, 9621–9630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yen, E.-Y.; Miaw, S.-C.; Yu, J.-S.; Lai, I.-R. Exosomal TGF-β1 is correlated with lymphatic metastasis of gastric cancers. Am. J. Cancer Res. 2017, 7, 2199–2208. [Google Scholar] [PubMed]
- Ringuette Goulet, C.; Bernard, G.; Tremblay, S.; Chabaud, S.; Bolduc, S.; Pouliot, F. Exosomes Induce Fibroblast Differentiation into Cancer-Associated Fibroblasts through TGFβ Signaling. Mol. Cancer Res. 2018, 16, 1196–1204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clayton, A.; Mitchell, J.P.; Court, J.; Mason, M.D.; Tabi, Z. Human Tumor-Derived Exosomes Selectively Impair Lymphocyte Responses to Interleukin-2. Cancer Res. 2007, 67, 7458–7466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rong, L.; Li, R.; Li, S.; Luo, R. Immunosuppression of breast cancer cells mediated by transforming growth factor-β in exosomes from cancer cells. Oncol. Lett. 2016, 11, 500–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, N.; Kuranaga, Y.; Kumazaki, M.; Shinohara, H.; Taniguchi, K.; Akao, Y. Colorectal cancer cell-derived extracellular vesicles induce phenotypic alteration of T cells into tumor-growth supporting cells with transforming growth factor-β1-mediated suppression. Oncotarget 2016, 7, 27033–27043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, D.A.; Massagué, J. TGF-beta directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell 2005, 8, 369–380. [Google Scholar] [CrossRef] [Green Version]
- Maynard, C.L.; E Harrington, L.; Janowski, K.M.; Oliver, J.R.; Zindl, C.L.; Rudensky, A.Y.; Weaver, C. Regulatory T cells expressing interleukin 10 develop from Foxp3+ and Foxp3- precursor cells in the absence of interleukin 10. Nat. Immunol. 2007, 8, 931–941. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.Y.; Flavell, R.A. Identifying Foxp3-expressing suppressor T cells with a bicistronic reporter. Proc. Natl. Acad. Sci. USA 2005, 102, 5126–5131. [Google Scholar] [CrossRef] [Green Version]
- Moore, K.W.; de Waal Malefyt, R.; Coffman, R.L.; O’Garra, A. Interleukin-10 and the interleukin-10 receptor. Annu. Rev. Immunol. 2001, 19, 683–765. [Google Scholar] [CrossRef] [PubMed]
- Rakoff-Nahoum, S.; Hao, L.; Medzhitov, R. Role of toll-like receptors in spontaneous commensal-dependent colitis. Immunity 2006, 25, 319–329. [Google Scholar] [CrossRef] [Green Version]
- Li, M.O.; Flavell, R.A. Contextual regulation of inflammation: A duet by transforming growth factor-beta and interleukin-10. Immunity 2008, 28, 468–476. [Google Scholar] [CrossRef] [Green Version]
- Levings, M.K.; Bacchetta, R.; Schulz, U.; Roncarolo, M.G. The role of IL-10 and TGF-beta in the differentiation and effector function of T regulatory cells. Int. Arch. Allergy Immunol. 2002, 129, 263–276. [Google Scholar] [CrossRef]
- Chaudhry, A.; Samstein, R.M.; Treuting, P.; Liang, Y.; Pils, M.C.; Heinrich, J.-M.; Jack, R.S.; Wunderlich, F.T.; Brüning, J.C.; Müller, W.; et al. Interleukin-10 signaling in regulatory T cells is required for suppression of Th17 cell-mediated inflammation. Immunity 2011, 34, 566–578. [Google Scholar] [CrossRef] [Green Version]
- Shu, S.; Matsuzaki, J.; Want, M.Y.; Conway, A.; Benjamin-Davalos, S.; Allen, C.L.; Koroleva, M.; Battaglia, S.; Odunsi, A.; Minderman, H.; et al. An Immunosuppressive Effect of Melanoma-derived Exosomes on NY-ESO-1 Antigen-specific Human CD8+ T Cells is Dependent on IL-10 and Independent of BRAFV600E Mutation in Melanoma Cell Lines. Immunol. Invest. 2020, 49, 744–757. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Cao, B.; Liang, X.; Li, L.; Hao, Y.; Meng, W.; He, C.; Wang, L.; Li, L. Small extracellular vesicles containing miR-192/215 mediate hypoxia-induced cancer-associated fibroblast development in head and neck squamous cell carcinoma. Cancer Lett. 2021, 506, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liu, J.; Zhang, Q.; Liu, B.; Cheng, Y.; Zhang, Y.; Sun, Y.; Ge, H.; Liu, Y. Exosome-mediated transfer of miR-93-5p from cancer-associated fibroblasts confer radioresistance in colorectal cancer cells by downregulating FOXA1 and upregulating TGFB3. J. Exp. Clin. Cancer Res. 2020, 39, 65. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Ren, H.; Dai, B.; Li, J.; Shang, L.; Huang, J.; Shi, X. Hepatocellular carcinoma-derived exosomal miRNA-21 contributes to tumor progression by converting hepatocyte stellate cells to cancer-associated fibroblasts. J. Exp. Clin. Cancer Res. 2018, 37, 324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Yuan, T.; Tschannen, M.; Sun, Z.; Jacob, H.; Du, M.; Liang, M.; Dittmar, R.L.; Liu, Y.; Liang, M.; et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genom. 2013, 14, 319. [Google Scholar] [CrossRef] [Green Version]
- van Balkom, B.W.M.; Eisele, A.S.; Pegtel, D.M.; Bervoets, S.; Verhaar, M.C. Quantitative and qualitative analysis of small RNAs in human endothelial cells and exosomes provides insights into localized RNA processing, degradation and sorting. J. Extracell Vesicles 2015, 4, 26760. [Google Scholar] [CrossRef]
- Melo, S.A.; Sugimoto, H.; O’Connell, J.T.; Kato, N.; Villanueva, A.; Vidal, A.; Qiu, L.; Vitkin, E.; Perelman, L.T.; Melo, C.A.; et al. Cancer Exosomes Perform Cell-Independent MicroRNA Biogenesis and Promote Tumorigenesis. Cancer Cell 2014, 26, 707–721. [Google Scholar] [CrossRef] [Green Version]
- Skog, J.; Würdinger, T.; Van Rijn, S.; Meijer, D.H.; Gainche, L.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Pegtel, D.M.; Cosmopoulos, K.; Thorley-Lawson, D.A.; van Eijndhoven, M.A.J.; Hopmans, E.S.; Lindenberg, J.L.; de Gruijl, T.D.; Würdinger, T.; Middeldorp, J.M. Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. USA 2010, 107, 6328–6333. [Google Scholar] [CrossRef] [Green Version]
- Deregibus, M.C.; Cantaluppi, V.; Calogero, R.; Lo Iacono, M.; Tetta, C.; Biancone, L.; Bruno, S.; Bussolati, B.; Camussi, G. Endothelial progenitor cell derived microvesicles activate an angiogenic program in endothelial cells by a horizontal transfer of mRNA. Blood 2007, 110, 2440–2448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buschmann, D.; Kirchner, B.; Hermann, S.; Märte, M.; Wurmser, C.; Brandes, F.; Kotschote, S.; Bonin, M.; Steinlein, O.K.; Pfaffl, M.W.; et al. Evaluation of serum extracellular vesicle isolation methods for profiling miRNAs by next-generation sequencing. J. Extracell Vesicles 2018, 7, 1481321. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.; Riwanto, M.; Besler, C.; Knau, A.; Fichtlscherer, S.; Röxe, T.; Zeiher, A.M.; Landmesser, U.; Dimmeler, S. Characterization of levels and cellular transfer of circulating lipoprotein-bound microRNAs. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 1392–1400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, M.; Wang, H.; Yao, X.; Zhang, D.; Xie, Y.; Cui, R.; Zhang, X. Circulating MicroRNAs in Cancer: Potential and Challenge. Front. Genet. 2019, 10, 626. [Google Scholar] [CrossRef] [Green Version]
- Mutant p53 Cancers Reprogram Macrophages to Tumor Supporting Macrophages via Exosomal miR-1246 | Nature Communications. Available online: https://www.nature.com/articles/s41467-018-03224-w (accessed on 6 June 2022).
- Chen, X.; Ying, X.; Wang, X.; Wu, X.; Zhu, Q.; Wang, X. Exosomes derived from hypoxic epithelial ovarian cancer deliver microRNA-940 to induce macrophage M2 polarization. Oncol. Rep. 2017, 38, 522–528. [Google Scholar] [CrossRef] [Green Version]
- Ying, X.; Wu, Q.; Wu, X.; Zhu, Q.; Wang, X.; Jiang, L.; Chen, X.; Wang, X. Epithelial ovarian cancer-secreted exosomal miR-222-3p induces polarization of tumor-associated macrophages. Oncotarget 2016, 7, 43076–43087. [Google Scholar] [CrossRef] [Green Version]
- Cai, J.; Qiao, B.; Gao, N.; Lin, N.; He, W. Oral squamous cell carcinoma-derived exosomes promote M2 subtype macrophage polarization mediated by exosome-enclosed miR-29a-3p. Am. J. Physiol. Cell Physiol. 2019, 316, C731–C740. [Google Scholar] [CrossRef]
- Lin, F.; Yin, H.-B.; Li, X.-Y.; Zhu, G.-M.; He, W.-Y.; Gou, X. Bladder cancer cell-secreted exosomal miR-21 activates the PI3K/AKT pathway in macrophages to promote cancer progression. Int. J. Oncol. 2019, 56, 151–164. [Google Scholar] [CrossRef] [Green Version]
- van der Vos, K.E.; Abels, E.R.; Zhang, X.; Lai, C.; Carrizosa, E.; Oakley, D.; Prabhakar, S.; Mardini, O.; Crommentuijn, M.H.; Skog, J.; et al. Directly visualized glioblastoma-derived extracellular vesicles transfer RNA to microglia/macrophages in the brain. Neuro-Oncology 2016, 18, 58–69. [Google Scholar] [CrossRef] [Green Version]
- Pyzer, A.R.; Stroopinsky, D.; Rajabi, H.; Washington, A.; Tagde, A.; Coll, M.; Fung, J.; Bryant, M.P.; Cole, L.; Palmer, K.; et al. MUC1-mediated induction of myeloid-derived suppressor cells in patients with acute myeloid leukemia. Blood 2017, 129, 1791–1801. [Google Scholar] [CrossRef] [Green Version]
- Fabbri, M.; Paone, A.; Calore, F.; Galli, R.; Gaudio, E.; Santhanam, R.; Lovat, F.; Fadda, P.; Mao, C.; Nuovo, G.J.; et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. USA 2012, 109, E2110–E2116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casadei, L.; Calore, F.; Creighton, C.J.; Guescini, M.; Batte, K.; Iwenofu, O.H.; Zewdu, A.; Braggio, D.A.; Bill, K.L.; Fadda, P.; et al. Exosome-Derived miR-25-3p and miR-92a-3p Stimulate Liposarcoma Progression. Cancer Res. 2017, 77, 3846–3856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gantier, M.; Tong, S.; Behlke, M.; Xu, D.; Phipps, S.; Foster, P.S.; Williams, B. TLR7 is involved in sequence-specific sensing of single-stranded RNAs in human macrophages. J. Immunol. Baltim. Md. 1950 2008, 180, 2117–2124. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ohto, U.; Shibata, T.; Taoka, M.; Yamauchi, Y.; Sato, R.; Shukla, N.M.; David, S.A.; Isobe, T.; Miyake, K.; et al. Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands. Cell Rep. 2018, 25, 3371–3381.e5. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Yuan, C.; Wangmo, D.; Subramanian, S. Tumor-Secreted Extracellular Vesicles Regulate T-Cell Costimulation and Can Be Manipulated To Induce Tumor-Specific T-Cell Responses. Gastroenterology 2021, 161, 560–574.e11. [Google Scholar] [CrossRef]
- Zhou, M.; Chen, J.; Zhou, L.; Chen, W.; Ding, G.; Cao, L. Pancreatic cancer derived exosomes regulate the expression of TLR4 in dendritic cells via miR-203. Cell Immunol. 2014, 292, 65–69. [Google Scholar] [CrossRef]
- Ding, G.; Zhou, L.; Qian, Y.; Fu, M.; Chen, J.; Chen, J.; Xiang, J.; Wu, Z.; Jiang, G.; Cao, L. Pancreatic cancer-derived exosomes transfer miRNAs to dendritic cells and inhibit RFXAP expression via miR-212-3p. Oncotarget 2015, 6, 29877–29888. [Google Scholar] [CrossRef] [Green Version]
- Yin, Y.; Cai, X.; Chen, X.; Liang, H.; Zhang, Y.; Li, J.; Wang, Z.; Chen, X.; Zhang, W.; Yokoyama, S.; et al. Tumor-secreted miR-214 induces regulatory T cells: A major link between immune evasion and tumor growth. Cell Res. 2014, 24, 1164–1180. [Google Scholar] [CrossRef]
- Yu, S.; Li, Y.; Liao, Z.; Wang, Z.; Wang, Z.; Li, Y.; Qian, L.; Zhao, J.; Zong, H.; Kang, B.; et al. Plasma extracellular vesicle long RNA profiling identifies a diagnostic signature for the detection of pancreatic ductal adenocarcinoma. Gut 2020, 69, 540–550. [Google Scholar] [CrossRef]
- Li, Z.; Jiang, P.; Li, J.; Peng, M.; Zhao, X.; Zhang, X.; Chen, K.; Zhang, Y.; Liu, H.; Gan, L.; et al. Tumor-derived exosomal lnc-Sox2ot promotes EMT and stemness by acting as a ceRNA in pancreatic ductal adenocarcinoma. Oncogene 2018, 37, 3822–3838. [Google Scholar] [CrossRef]
- Ghetti, M.; Vannini, I.; Storlazzi, C.T.; Martinelli, G.; Simonetti, G. Linear and circular PVT1 in hematological malignancies and immune response: Two faces of the same coin. Mol. Cancer 2020, 19, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, D.; Luo, P.; Wang, Q.; Ye, Y.; Wang, B. lncRNA PVT1 in cancer: A review and meta-analysis. Clin. Chim. Acta Int. J. Clin. Chem. 2017, 474, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Tian, X.; Wang, T.; Xia, X.; Cao, F.; Tian, J.; Xu, P.; Ma, J.; Xu, H.; Wang, S. Long noncoding RNA Pvt1 regulates the immunosuppression activity of granulocytic myeloid-derived suppressor cells in tumor-bearing mice. Mol. Cancer 2019, 18, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Khazragy, N.; Elayat, W.; Matbouly, S.; Seliman, S.; Sami, A.; Safwat, G.; Diab, A. The prognostic significance of the long non-coding RNAs ‘CCAT1, PVT1’ in t(8;21) associated Acute Myeloid Leukemia. Gene 2019, 707, 172–177. [Google Scholar] [CrossRef]
- Chen, Y.; Li, F.; Li, D.; Liu, W.; Zhang, L. Atezolizumab and blockade of LncRNA PVT1 attenuate cisplatin resistant ovarian cancer cells progression synergistically via JAK2/STAT3/PD-L1 pathway. Clin. Immunol. 2021, 227, 108728. [Google Scholar] [CrossRef] [PubMed]
- Iden, M.; Fye, S.; Li, K.; Chowdhury, T.; Ramchandran, R.; Rader, J.S. The lncRNA PVT1 Contributes to the Cervical Cancer Phenotype and Associates with Poor Patient Prognosis. PLoS ONE 2016, 11, e0156274. [Google Scholar] [CrossRef] [Green Version]
- Fan, H.; Zhu, J.; Yao, X. Long non-coding RNA PVT1 as a novel potential biomarker for predicting the prognosis of colorectal cancer. Int. J. Biol. Markers 2018, 33, 415–422. [Google Scholar] [CrossRef] [Green Version]
- Mizuno, S.; Hanamura, I.; Ota, A.; Karnan, S.; Kanasugi, J.; Nakamura, A.; Takasugi, S.; Uchino, K.; Horio, T.; Goto, M.; et al. Establishment and characterization of a novel vincristine-resistant diffuse large B-cell lymphoma cell line containing the 8q24 homogeneously staining region. FEBS Open Bio. 2018, 8, 1977–1991. [Google Scholar] [CrossRef]
- Guo, K.; Yao, J.; Yu, Q.; Li, Z.; Huang, H.; Cheng, J.; Wang, Z.; Zhu, Y. The expression pattern of long non-coding RNA PVT1 in tumor tissues and in extracellular vesicles of colorectal cancer correlates with cancer progression. Tumour Biol. J. Int. Soc Oncodevelopmental. Biol. Med. 2017, 39, 1010428317699122. [Google Scholar] [CrossRef] [Green Version]
- Lai, S.W.; Chen, M.Y.; Bamodu, O.A.; Hsieh, M.S.; Huang, T.Y.; Yeh, C.T.; Lee, W.H.; Cherng, Y.G. Exosomal lncRNA PVT1/VEGFA Axis Promotes Colon Cancer Metastasis and Stemness by Downregulation of Tumor Suppressor miR-152-3p. Oxid. Med. Cell Longev. 2021, 2021, 9959807. [Google Scholar] [CrossRef]
- Sun, C.; Wang, P.; Dong, W.; Liu, H.; Sun, J.; Zhao, L. LncRNA PVT1 promotes exosome secretion through YKT6, RAB7, and VAMP3 in pancreatic cancer. Aging 2020, 12, 10427–10440. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Chen, J.; Yang, L.; Liu, J.; Zhang, X.; Zhang, Y.; Tu, Q.; Yin, D.; Lin, D.; Wong, P.P.; et al. Extracellular vesicle-packaged HIF-1α-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat. Cell Biol. 2019, 21, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Dallavalle, S.; Dobričić, V.; Lazzarato, L.; Gazzano, E.; Machuqueiro, M.; Pajeva, I.; Tsakovska, I.; Zidar, N.; Fruttero, R. Improvement of conventional anti-cancer drugs as new tools against multidrug resistant tumors. Drug Resist. Updates 2020, 50, 100682. [Google Scholar] [CrossRef] [PubMed]
- Battista, T.; Fiorillo, A.; Chiarini, V.; Genovese, I.; Ilari, A.; Colotti, G. Roles of Sorcin in Drug Resistance in Cancer: One Protein, Many Mechanisms, for a Novel Potential Anticancer Drug Target. Cancers 2020, 12, 887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navas, T.; Kinders, R.J.; Lawrence, S.M.; Ferry-Galow, K.V.; Borgel, S.; Hollingshead, M.G.; Srivastava, A.K.; Alcoser, S.Y.; Makhlouf, H.R.; Chuaqui, R.; et al. Clinical Evolution of Epithelial-Mesenchymal Transition in Human Carcinomas. Cancer Res. 2020, 80, 304–318. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Mayea, Y.; Mir, C.; Masson, F.; Paciucci, R.; LLeonart, M.E. Insights into new mechanisms and models of cancer stem cell multidrug resistance. Semin. Cancer Biol. 2020, 60, 166–180. [Google Scholar] [CrossRef] [PubMed]
- Dean, M.; Rzhetsky, A.; Allikmets, R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 2001, 11, 1156–1166. [Google Scholar] [CrossRef]
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The Different Mechanisms of Cancer Drug Resistance: A Brief Review. Adv. Pharm. Bull. 2017, 7, 339–348. [Google Scholar] [CrossRef]
- Yoshimori, M.; Takada, H.; Imadome, K.I.; Kurata, M.; Yamamoto, K.; Koyama, T.; Shimizu, N.; Fujiwara, S.; Miura, O.; Arai, A. P-glycoprotein is expressed and causes resistance to chemotherapy in EBV-positive T-cell lymphoproliferative diseases. Cancer Med. 2015, 4, 1494–1504. [Google Scholar] [CrossRef]
- Ling, L.; Lin, Y.; Zheng, W.; Hong, S.; Tang, X.; Zhao, P.; Li, M.; Ni, J.; Li, C.; Wang, L.; et al. Circulating and tumor-infiltrating mucosal associated invariant T (MAIT) cells in colorectal cancer patients. Sci. Rep. 2016, 6, 20358. [Google Scholar] [CrossRef] [Green Version]
- Goldstein, L.J.; Galski, H.; Fojo, A.; Willingham, M.; Lai, S.-L.; Gazdar, A.; Pirker, R.; Green, A.; Crist, W.; Brodeur, G.M.; et al. Expression of a multidrug resistance gene in human cancers. J. Natl. Cancer Inst. 1989, 81, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Torreggiani, E.; Roncuzzi, L.; Perut, F.; Zini, N.; Baldini, N. Multimodal transfer of MDR by exosomes in human osteosarcoma. Int. J. Oncol. 2016, 49, 189–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, H.; Wu, H.; Liu, X.; Evans, B.R.; Medina, D.J.; Liu, C.G.; Yang, J.M. Role of MicroRNA miR-27a and miR-451 in the regulation of MDR1/P-glycoprotein expression in human cancer cells. Biochem. Pharmacol. 2008, 76, 582–588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kovalchuk, O.; Filkowski, J.; Meservy, J.; Ilnytskyy, Y.; Tryndyak, V.P.; Chekhun, V.F.; Pogribny, I.P. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol. Cancer Ther. 2008, 7, 2152–2159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghandadi, M.; Sahebkar, A. Interleukin-6, A Critical Cytokine in Cancer Multidrug Resistance. Curr. Pharm. Des. 2016, 22, 518–526. [Google Scholar] [CrossRef]
- Wang, Y.; Niu, X.L.; Qu, Y.; Wu, J.; Zhu, Y.Q.; Sun, W.J.; Li, L.Z. Autocrine production of interleukin-6 confers cisplatin and paclitaxel resistance in ovarian cancer cells. Cancer Lett. 2010, 295, 110–123. [Google Scholar] [CrossRef]
- Goulet, C.R.; Champagne, A.; Bernard, G.; Vandal, D.; Chabaud, S.; Pouliot, F.; Bolduc, S. Cancer-associated fibroblasts induce epithelial–mesenchymal transition of bladder cancer cells through paracrine IL-6 signalling. BMC Cancer 2019, 19, 137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Q.; Huang, L.; Qin, G.; Qiao, Y.; Ren, F.; Shen, C.; Wang, S.; Liu, S.; Lian, J.; Wang, D.; et al. Cancer-associated fibroblasts induce monocytic myeloid-derived suppressor cell generation via IL-6/exosomal miR-21-activated STAT3 signaling to promote cisplatin resistance in esophageal squamous cell carcinoma. Cancer Lett. 2021, 518, 35–48. [Google Scholar] [CrossRef]
- Yu, S.; Liu, C.; Su, K.; Wang, J.; Liu, Y.; Zhang, L.; Li, C.; Cong, Y.; Kimberly, R.; Grizzle, W.E.; et al. Tumor Exosomes Inhibit Differentiation of Bone Marrow Dendritic Cells. J. Immunol. 2007, 178, 6867–6875. [Google Scholar] [CrossRef] [Green Version]
- Binenbaum, Y.; Fridman, E.; Yaari, Z.; Milman, N.; Schroeder, A.; Ben David, G.; Shlomi, T.; Gil, Z. Transfer of miRNA in Macrophage-Derived Exosomes Induces Drug Resistance in Pancreatic Adenocarcinoma. Cancer Res. 2018, 78, 5287–5299. [Google Scholar] [CrossRef] [Green Version]
- Pavlyukov, M.S.; Yu, H.; Bastola, S.; Minata, M.; Shender, V.O.; Lee, Y.; Zhang, S.; Wang, J.; Komarova, S.; Wang, J.; et al. Apoptotic Cell-Derived Extracellular Vesicles Promote Malignancy of Glioblastoma Via Intercellular Transfer of Splicing Factors. Cancer Cell 2018, 34, 119–135.e10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hui, E.; Cheung, J.; Zhu, J.; Su, X.; Taylor, M.J.; Wallweber, H.A.; Sasmal, D.K.; Huang, J.; Kim, J.M.; Mellman, I.; et al. T cell costimulatory receptor CD28 is a primary target for PD-1–mediated inhibition. Science 2017, 355, 1428–1433. [Google Scholar] [CrossRef] [PubMed]
- Prat, A.; Navarro, A.; Paré, L.; Reguart, N.; Galván, P.; Pascual, T.; Martínez, A.; Nuciforo, P.; Comerma, L.; Alos, L.; et al. Immune-Related Gene Expression Profiling After PD-1 Blockade in Non–Small Cell Lung Carcinoma, Head and Neck Squamous Cell Carcinoma, and Melanoma. Cancer Res. 2017, 77, 3540–3550. [Google Scholar] [CrossRef] [Green Version]
- Aung, T.; Chapuy, B.; Vogel, D.; Wenzel, D.; Oppermann, M.; Lahmann, M.; Weinhage, T.; Menck, K.; Hupfeld, T.; Koch, R.; et al. Exosomal evasion of humoral immunotherapy in aggressive B-cell lymphoma modulated by ATP-binding cassette transporter A3. Proc. Natl. Acad. Sci. USA 2011, 108, 15336–15341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciravolo, V.; Huber, V.; Ghedini, G.C.; Venturelli, E.; Bianchi, F.; Campiglio, M.; Morelli, D.; Villa, A.; Della Mina, P.; Menard, S.; et al. Potential role of HER2-overexpressing exosomes in countering trastuzumab-based therapy. J. Cell Physiol. 2012, 227, 658–667. [Google Scholar] [CrossRef] [PubMed]
- Lorusso, G.; Rüegg, C. The tumor microenvironment and its contribution to tumor evolution toward metastasis. Histochem. Cell Biol. 2008, 130, 1091–1103. [Google Scholar] [CrossRef] [Green Version]
- Hida, K.; Maishi, N.; Annan, D.A.; Hida, Y. Contribution of Tumor Endothelial Cells in Cancer Progression. Int. J. Mol. Sci. 2018, 19, 1272. [Google Scholar] [CrossRef] [Green Version]
- Nieman, K.M.; Romero, I.L.; Van Houten, B.; Lengyel, E. Adipose tissue and adipocytes support tumorigenesis and metastasis. Biochim. Biophys. Acta BBA - Mol. Cell Biol. Lipids 2013, 1831, 1533–1541. [Google Scholar] [CrossRef] [Green Version]
- Nieman, K.M.; Kenny, H.A.; Penicka, C.V.; Ladanyi, A.; Buell-Gutbrod, R.; Zillhardt, M.R.; Romero, I.L.; Carey, M.S.; Mills, G.B.; Hotamisligil, G.S.; et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat. Med. 2011, 17, 1498–1503. [Google Scholar] [CrossRef] [Green Version]
- Luo, Y.; Zhou, H.; Krueger, J.; Kaplan, C.; Lee, S.-H.; Dolman, C.; Markowitz, D.; Wu, W.; Liu, C.; Reisfeld, R.A.; et al. Targeting tumor-associated macrophages as a novel strategy against breast cancer. J. Clin. Invest. 2006, 116, 2132–2141. [Google Scholar] [CrossRef] [Green Version]
- Ostrand-Rosenberg, S.; Sinha, P. Myeloid-Derived Suppressor Cells: Linking Inflammation and Cancer. J. Immunol. 2009, 182, 4499–4506. [Google Scholar] [CrossRef] [PubMed]
- Pyfferoen, L.; Brabants, E.; Everaert, C.; De Cabooter, N.; Heyns, K.; Deswarte, K.; Vanheerswynghels, M.; De Prijck, S.; Waegemans, G.; Dullaers, M.; et al. The transcriptome of lung tumor-infiltrating dendritic cells reveals a tumor-supporting phenotype and a microRNA signature with negative impact on clinical outcome. OncoImmunology 2017, 6, e1253655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, H.; Clauser, K.R.; Tam, W.L.; Frose, J.; Ye, X.; Eaton, E.N.; Reinhardt, F.; Donnenberg, V.S.; Bhargava, R.; Carr, S.A.; et al. A breast cancer stem cell niche supported by juxtacrine signalling from monocytes and macrophages. Nat. Cell Biol. 2014, 16, 1105–1117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raskov, H.; Orhan, A.; Christensen, J.P.; Gögenur, I. Cytotoxic CD8+ T cells in cancer and cancer immunotherapy. Br. J. Cancer 2021, 124, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Beyer, M.; Schultze, J.L. Regulatory T cells in cancer. Blood 2006, 108, 804–811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoue, S.; Leitner, W.W.; Golding, B.; Scott, D. Inhibitory Effects of B Cells on Antitumor Immunity. Cancer Res. 2006, 66, 7741–7747. [Google Scholar] [CrossRef] [Green Version]
- Neo, S.Y.; Yang, Y.; Record, J.; Ma, R.; Chen, X.; Chen, Z.; Tobin, N.; Blake, E.; Seitz, C.; Thomas, R.; et al. CD73 immune checkpoint defines regulatory NK cells within the tumor microenvironment. J. Clin. Invest. 2020, 130, 1185–1198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- GJG, S.; Wurdinger, T. Tumor-educated platelets. Blood 2019, 133, 2359–2364. [Google Scholar]
- Madeo, M.; Colbert, P.L.; Vermeer, D.W.; Lucido, C.T.; Cain, J.T.; Vichaya, E.G.; Grossberg, A.J.; Muirhead, D.; Rickel, A.P.; Hong, Z.; et al. Cancer exosomes induce tumor innervation. Nat. Commun. 2018, 9, 4284. [Google Scholar] [CrossRef] [Green Version]
- Hong, C.-S.; Funk, S.; Muller, L.; Boyiadzis, M.; Whiteside, T.L. Isolation of biologically active and morphologically intact exosomes from plasma of patients with cancer. J. Extracell Vesicles 2016, 5, 29289. [Google Scholar] [CrossRef]
- Heijnen, H.F.; Schiel, A.E.; Fijnheer, R.; Geuze, H.J.; Sixma, J.J. Activated platelets release two types of membrane vesicles: Microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood 1999, 94, 3791–3799. [Google Scholar] [CrossRef] [PubMed]
- Stone, R.L.; Nick, A.M.; McNeish, I.A.; Balkwill, F.; Han, H.D.; Bottsford-Miller, J.; Rupaimoole, R.; Armaiz-Pena, G.N.; Pecot, C.V.; Coward, J.; et al. Paraneoplastic thrombocytosis in ovarian cancer. N. Engl. J. Med. 2012, 366, 610–618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lannan, K.L.; Sahler, J.; Kim, N.; Spinelli, S.L.; Maggirwar, S.B.; Garraud, O.; Cognasse, F.; Blumberg, N.; Phipps, R.P. Breaking the Mold: Transcription Factors in the Anucleate Platelet and Platelet-Derived Microparticles. Front. Immunol. 2015, 6, 203. Available online: https://www.frontiersin.org/article/10.3389/fimmu.2015.00048 (accessed on 8 June 2022). [CrossRef] [PubMed] [Green Version]
- Denis, M.M.; Tolley, N.D.; Bunting, M.; Schwertz, H.; Jiang, H.; Lindemann, S.; Yost, C.C.; Rubner, F.J.; Albertine, K.H.; Swoboda, K.J.; et al. Escaping the Nuclear Confines: Signal-Dependent Pre-mRNA Splicing in Anucleate Platelets. Cell 2005, 122, 379–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nassa, G.; Giurato, G.; Cimmino, G.; Rizzo, F.; Ravo, M.; Salvati, A.; Nyman, T.A.; Zhu, Y.; Vesterlund, M.; Lehtiö, J.; et al. Splicing of platelet resident pre-mRNAs upon activation by physiological stimuli results in functionally relevant proteome modifications. Sci. Rep. 2018, 8, 498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nilsson, R.J.A.; Balaj, L.; Hulleman, E.; Van Rijn, S.; Pegtel, D.M.; Walraven, M.; Widmark, A.; Gerritsen, W.R.; Verheul, H.M.; Vandertop, W.P.; et al. Blood platelets contain tumor-derived RNA biomarkers. Blood 2011, 118, 3680–3683. [Google Scholar] [CrossRef]
- Calverley, D.C.; Phang, T.L.; Choudhury, Q.G.; Gao, B.; Oton, A.B.; Weyant, M.J.; Geraci, M.W. Significant downregulation of platelet gene expression in metastatic lung cancer. Clin. Transl. Sci. 2010, 3, 227–232. [Google Scholar] [CrossRef]
- Best, M.G.; Sol, N.; Kooi, I.E.; Tannous, J.; Westerman, B.A.; Rustenburg, F.; Schellen, P.; Verschueren, H.; Post, E.; Koster, J.; et al. RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. Cancer Cell 2015, 28, 666–676. [Google Scholar] [CrossRef] [Green Version]
- Best, M.G.; Sol, N.; GJG, S.; Vancura, A.; Muller, M.; Niemeijer, A.L.N.; Fejes, A.V.; Fat, L.A.T.K.; Huis, A.E.; Leurs, C.; et al. Swarm Intelligence-Enhanced Detection of Non-Small-Cell Lung Cancer Using Tumor-Educated Platelets. Cancer Cell 2017, 32, 238–252.e9. [Google Scholar] [CrossRef]
- Flaumenhaft, R. Formation and fate of platelet microparticles. Blood Cells Mol. Dis. 2006, 36, 182–187. [Google Scholar] [CrossRef]
- Varon, D.; Shai, E. Role of platelet-derived microparticles in angiogenesis and tumor progression. Discov. Med. 2009, 8, 237–241. [Google Scholar] [PubMed]
- Lazar, S.; Goldfinger, L.E. Platelets and extracellular vesicles and their cross talk with cancer. Blood 2021, 137, 3192–3200. [Google Scholar] [CrossRef] [PubMed]
- Żmigrodzka, M.; Guzera, M.; Miśkiewicz, A.; Jagielski, D.; Winnicka, A. The biology of extracellular vesicles with focus on platelet microparticles and their role in cancer development and progression. Tumour Biol. 2016, 37, 14391–14401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janowska-Wieczorek, A.; Wysoczynski, M.; Kijowski, J.; Marquez-Curtis, L.; Machalinski, B.; Ratajczak, J.; Ratajczak, M.Z. Microvesicles derived from activated platelets induce metastasis and angiogenesis in lung cancer. Int. J. Cancer 2005, 113, 752–760. [Google Scholar] [CrossRef] [PubMed]
- Labelle, M.; Begum, S.; Hynes, R.O. Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis. Cancer Cell 2011, 20, 576–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopatina, T.; Grange, C.; Cavallari, C.; Navarro-Tableros, V.; Lombardo, G.; Rosso, A.; Cedrino, M.; Pomatto, M.A.C.; Koni, M.; Veneziano, F.; et al. Targeting IL-3Rα on tumor-derived endothelial cells blunts metastatic spread of triple-negative breast cancer via extracellular vesicle reprogramming. Oncogenesis 2020, 9, 90. [Google Scholar] [CrossRef] [PubMed]
- Lombardo, G.; Gili, M.; Grange, C.; Cavallari, C.; Dentelli, P.; Togliatto, G.; Taverna, D.; Camussi, G.; Brizzi, M.F. IL-3R-alpha blockade inhibits tumor endothelial cell-derived extracellular vesicle (EV)-mediated vessel formation by targeting the β-catenin pathway. Oncogene 2018, 37, 1175–1191. [Google Scholar] [CrossRef] [Green Version]
- Lopatina, T.; Koni, M.; Grange, C.; Cedrino, M.; Femminò, S.; Lombardo, G.; Favaro, E.; Brizzi, M.F. IL-3 signalling in the tumour microenvironment shapes the immune response via tumour endothelial cell-derived extracellular vesicles. Pharmacol. Res. 2022, 179, 106206. [Google Scholar] [CrossRef]
- André, F.; Chaput, N.; Schartz, N.E.C.; Flament, C.; Aubert, N.; Bernard, J.; Lemonnier, F.; Raposo, G.; Escudier, B.; Hsu, D.-H.; et al. Exosomes as Potent Cell-Free Peptide-Based Vaccine. I. Dendritic Cell-Derived Exosomes Transfer Functional MHC Class I/Peptide Complexes to Dendritic Cells. J. Immunol. 2004, 172, 2126–2136. [Google Scholar] [CrossRef] [Green Version]
- Lindenbergh, M.F.S.; Stoorvogel, W. Antigen Presentation by Extracellular Vesicles from Professional Antigen-Presenting Cells. Annu. Rev. Immunol. 2018, 36, 435–459. [Google Scholar] [CrossRef]
- Escudier, B.; Dorval, T.; Chaput, N.; André, F.; Caby, M.P.; Novault, S.; Flament, C.; Leboulaire, C.; Borg, C.; Amigorena, S.; et al. Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes, results of thefirst phase I clinical trial. J. Transl. Med. 2005, 3, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morse, M.A.; Garst, J.; Osada, T.; Khan, S.; Hobeika, A.; Clay, T.M.; Valente, N.; Shreeniwas, R.; Sutton, M.A.; Delcayre, A.; et al. A phase I study of dexosome immunotherapy in patients with advanced non-small cell lung cancer. J. Transl. Med. 2005, 3, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segura, E.; Amigorena, S.; Théry, C. Mature dendritic cells secrete exosomes with strong ability to induce antigen-specific effector immune responses. Blood Cells Mol. Dis. 2005, 35, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Nikfarjam, S.; Rezaie, J.; Kashanchi, F.; Jafari, R. Dexosomes as a cell-free vaccine for cancer immunotherapy. J. Exp. Clin. Cancer Res. CR 2020, 39, 258. [Google Scholar] [CrossRef]
- Xiang, X.; Poliakov, A.; Liu, C.; Liu, Y.; Deng, Z.-B.; Wang, J.; Cheng, Z.; Shah, S.V.; Wang, G.-J.; Zhang, L.; et al. Induction of myeloid-derived suppressor cells by tumor exosomes. Int. J. Cancer 2009, 124, 2621–2633. [Google Scholar] [CrossRef] [Green Version]
- Valenti, R.; Huber, V.; Filipazzi, P.; Pilla, L.; Sovena, G.; Villa, A.; Corbelli, A.; Fais, S.; Parmiani, G.; Rivoltini, L. Human Tumor-Released Microvesicles Promote the Differentiation of Myeloid Cells with Transforming Growth Factor-β–Mediated Suppressive Activity on T Lymphocytes. Cancer Res. 2006, 66, 9290–9298. [Google Scholar] [CrossRef] [Green Version]
- de Paula Silva, E.; Marti, L.C.; Andreghetto, F.M.; de Sales, R.O.; Hoberman, M.; dos Santos Dias, B.; Diniz, L.F.A.; Dos Santos, A.M.; Moyses, R.A.; Curioni, O.A.; et al. Extracellular vesicles cargo from head and neck cancer cell lines disrupt dendritic cells function and match plasma microRNAs. Sci. Rep. 2021, 11, 18534. [Google Scholar] [CrossRef]
- Zitvogel, L.; Regnault, A.; Lozier, A.; Wolfers, J.; Flament, C.; Tenza, D.; Ricciardi-Castagnoli, P.; Raposo, G.; Amigorena, S. Eradication of established murine tumors using a novel cell-free vaccine: Dendritic cell-derived exosomes. Nat. Med. 1998, 4, 594–600. [Google Scholar] [CrossRef]
- Fu, C.; Peng, P.; Loschko, J.; Feng, L.; Pham, P.; Cui, W.; Lee, K.P.; Krug, A.B.; Jiang, A. Plasmacytoid dendritic cells cross-prime naive CD8 T cells by transferring antigen to conventional dendritic cells through exosomes. Proc. Natl. Acad. Sci. USA 2020, 117, 23730–23741. [Google Scholar] [CrossRef]
- Viaud, S.; Ploix, S.; Lapierre, V.; Théry, C.; Commere, P.-H.; Tramalloni, D.; Gorrichon, K.; Virault-Rocroy, P.; Tursz, T.; Lantz, O.; et al. Updated Technology to Produce Highly Immunogenic Dendritic Cell-derived Exosomes of Clinical Grade: A Critical Role of Interferon-γ. J. Immunother. 2011, 34, 65–75. [Google Scholar] [CrossRef]
- Pan, B.T.; Johnstone, R.M. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: Selective externalization of the receptor. Cell 1983, 33, 967–978. [Google Scholar] [CrossRef]
- Barz, D.; Goppelt, M.; Szamel, M.; Schirrmacher, V.; Resch, K. Characterization of cellular and extracellular plasma membrane vesicles from a non-metastasizing lymphoma (Eb) and its metastasizing variant (ESb). Biochim. Biophys. Acta 1985, 814, 77–84. [Google Scholar] [CrossRef]
- Schirrmacher, V.; Barz, D. Characterization of cellular and extracellular plasma membrane vesicles from a low metastatic lymphoma (Eb) and its high metastatic variant (ESb): Inhibitory capacity in cell-cell interaction systems. Biochim. Biophys. Acta 1986, 860, 236–242. [Google Scholar] [CrossRef]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef]
- Andre, F.; Schartz, N.E.; Movassagh, M.; Flament, C.; Pautier, P.; Morice, P.; Pomel, C.; Lhomme, C.; Escudier, B.; Le Chevalier, T.; et al. Malignant effusions and immunogenic tumour-derived exosomes. Lancet 2002, 360, 295–305. [Google Scholar] [CrossRef]
- Chaput, N.; Schartz, N.E.C.; Andre, F.; Taïeb, J.; Novault, S.; Bonnaventure, P.; Aubert, N.; Bernard, J.; Lemonnier, F.; Merad, M.; et al. Exosomes as potent cell-free peptide-based vaccine. II. Exosomes in CpG adjuvants efficiently prime naive Tc1 lymphocytes leading to tumor rejection. J. Immunol. Baltim. Md. 1950 2004, 172, 2137–2146. [Google Scholar] [CrossRef] [Green Version]
- Altieri, S.L.; Khan, A.N.H.; Tomasi, T.B. Exosomes from plasmacytoma cells as a tumor vaccine. J. Immunother. Hagerstown Md. 1997 2004, 27, 282–288. [Google Scholar] [CrossRef]
- Peinado, H.; Alečković, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; García-Santos, G.; Ghajar, C.M.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef] [Green Version]
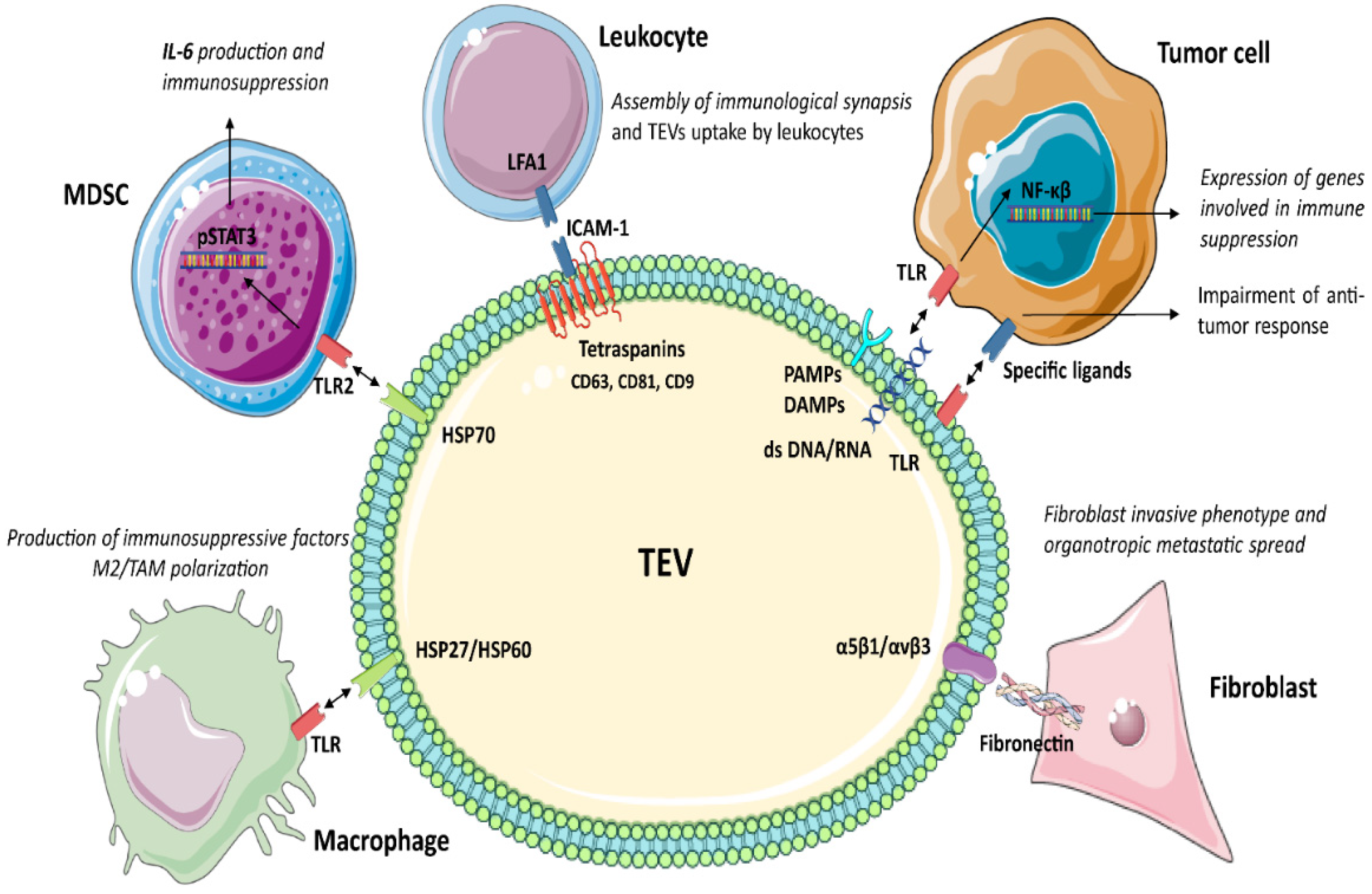
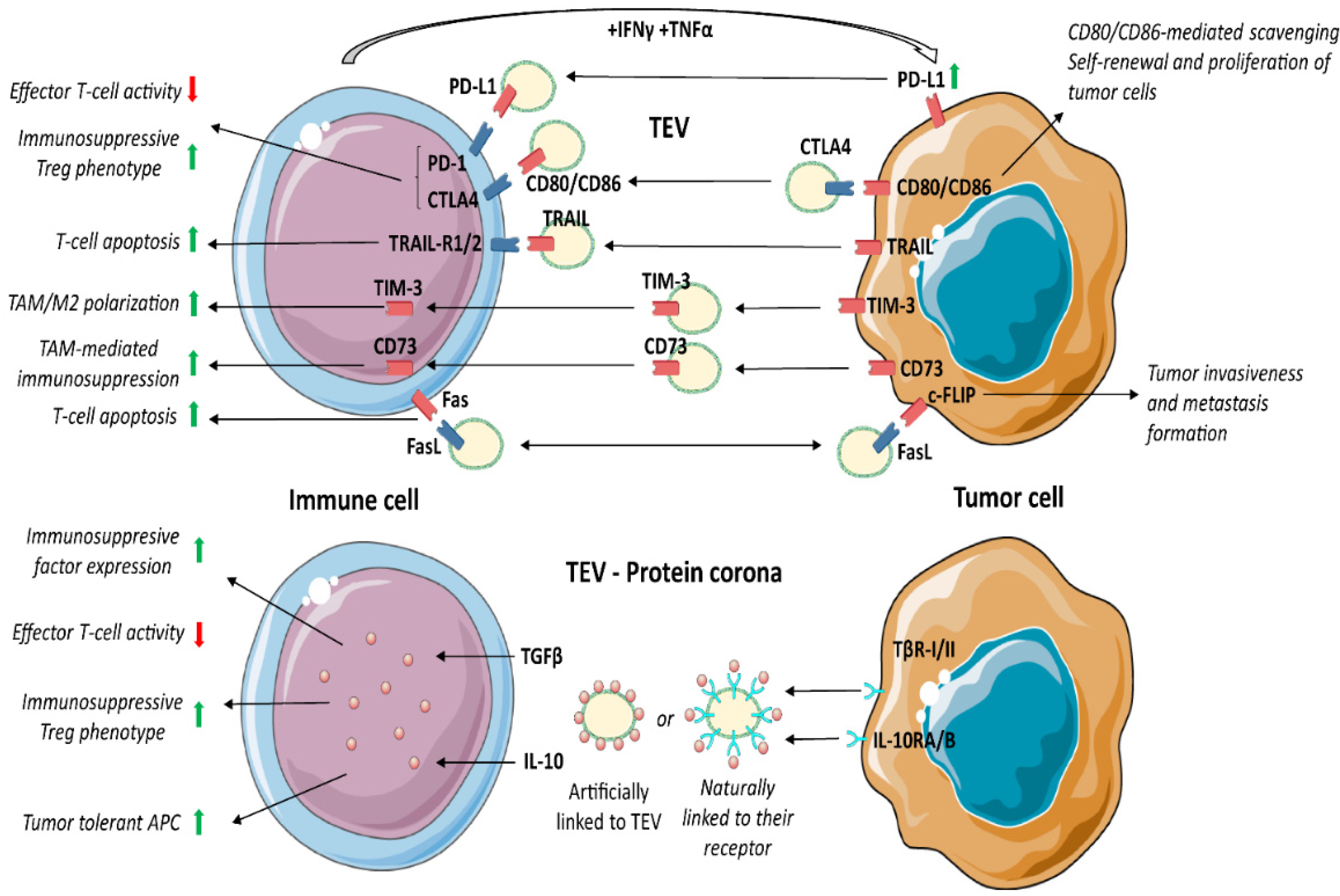
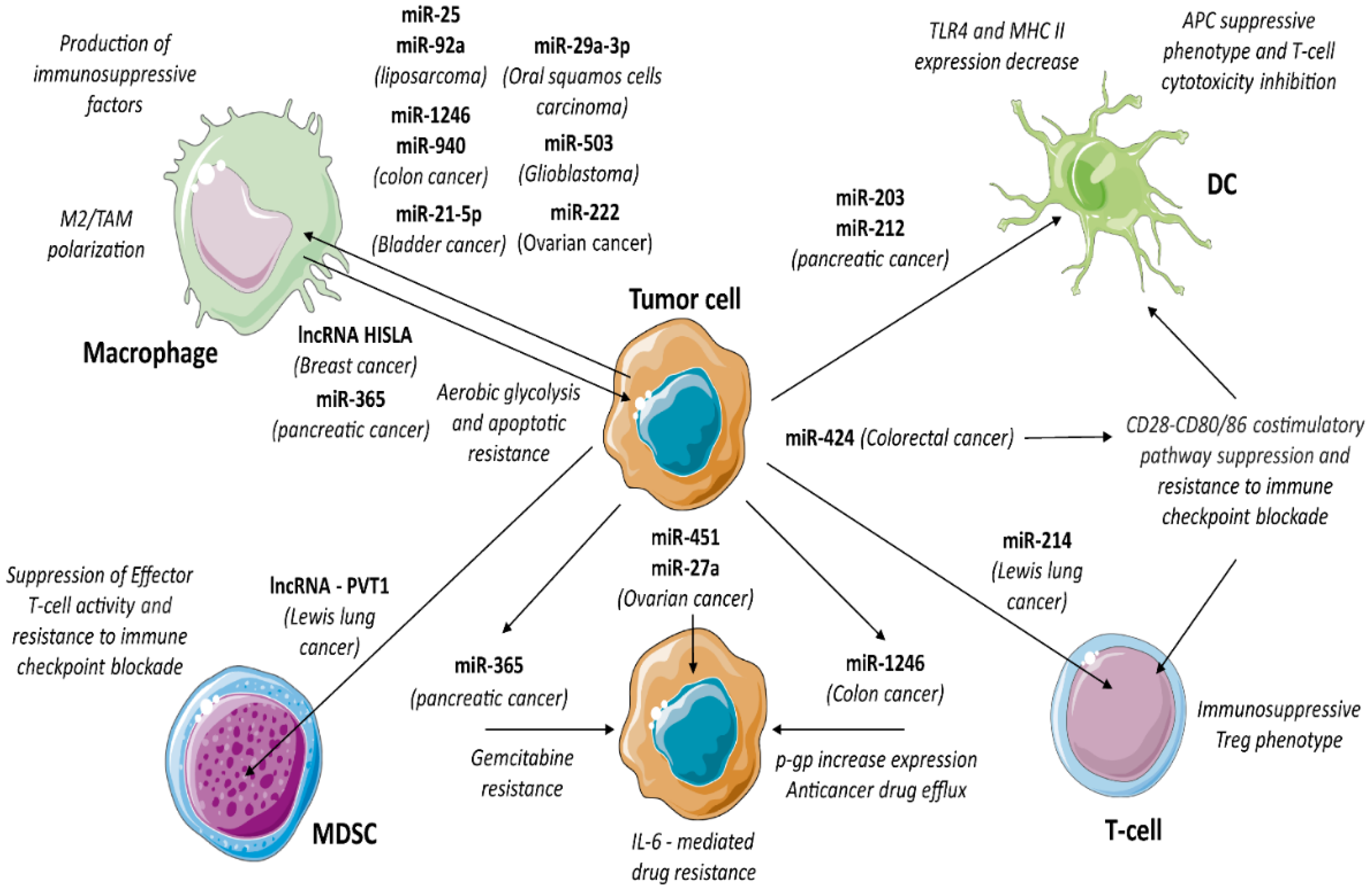
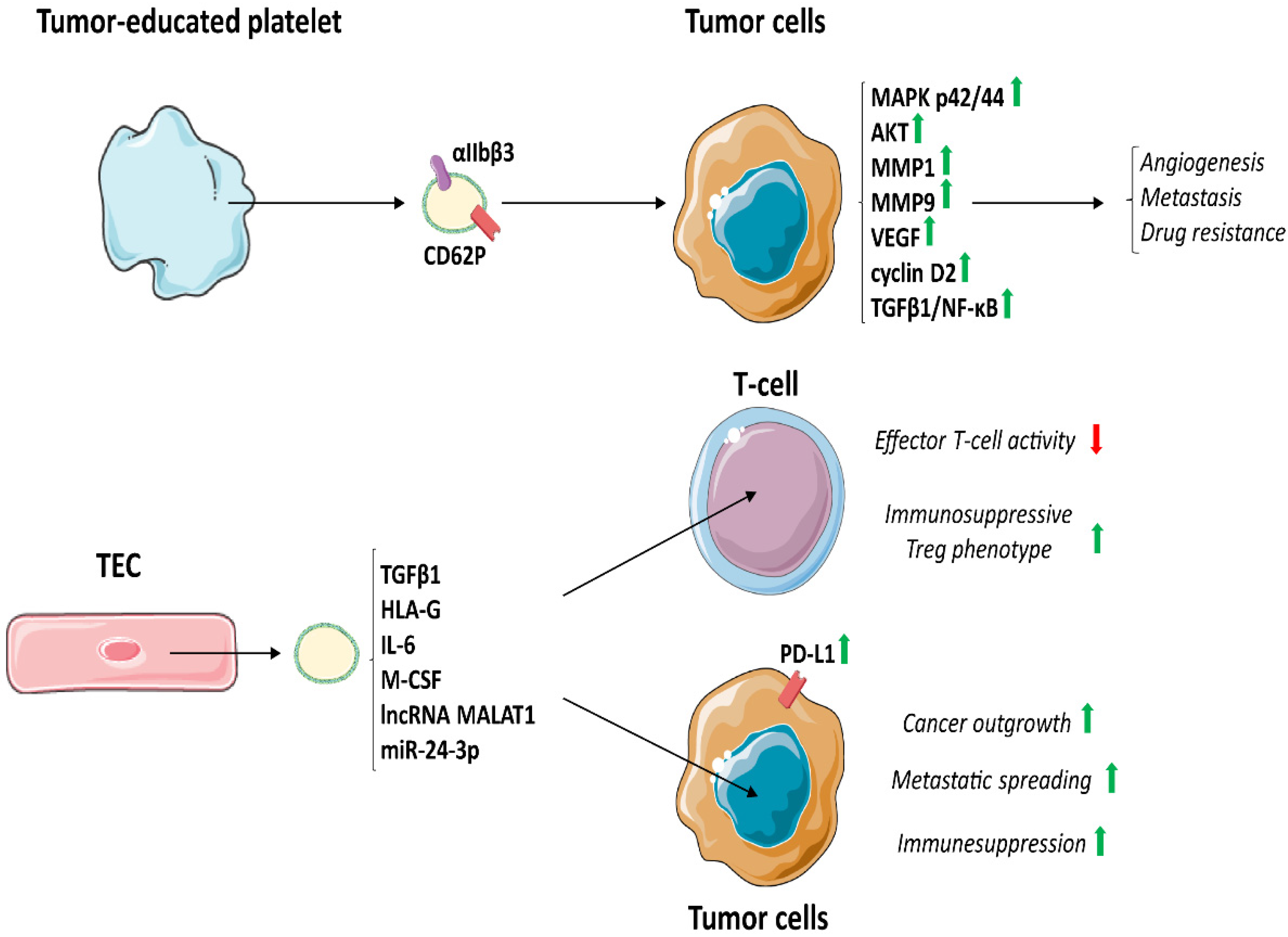
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lopatina, T.; Sarcinella, A.; Brizzi, M.F. Tumour Derived Extracellular Vesicles: Challenging Target to Blunt Tumour Immune Evasion. Cancers 2022, 14, 4020. https://doi.org/10.3390/cancers14164020
Lopatina T, Sarcinella A, Brizzi MF. Tumour Derived Extracellular Vesicles: Challenging Target to Blunt Tumour Immune Evasion. Cancers. 2022; 14(16):4020. https://doi.org/10.3390/cancers14164020
Chicago/Turabian StyleLopatina, Tatiana, Alessandro Sarcinella, and Maria Felice Brizzi. 2022. "Tumour Derived Extracellular Vesicles: Challenging Target to Blunt Tumour Immune Evasion" Cancers 14, no. 16: 4020. https://doi.org/10.3390/cancers14164020
APA StyleLopatina, T., Sarcinella, A., & Brizzi, M. F. (2022). Tumour Derived Extracellular Vesicles: Challenging Target to Blunt Tumour Immune Evasion. Cancers, 14(16), 4020. https://doi.org/10.3390/cancers14164020






