Volatilomics: An Emerging and Promising Avenue for the Detection of Potential Prostate Cancer Biomarkers
Abstract
:Simple Summary
Abstract
1. Introduction
2. Prostate Cancer Biomarkers
3. Contribution of the OMICs Science
3.1. Volatilomics—An Emerging Yet Challenging Approach
Challenges of Volatilomics
3.2. Volatilome of Prostate Cancer—A Promising Approach for Biomarkers’ Detection
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Rawla, P. Epidemiology of Prostate Cancer. World J. Oncol. 2019, 10, 63–89. [Google Scholar] [CrossRef] [PubMed]
- Beltran, H.; Demichelis, F. Intrapatient heterogeneity in prostate cancer. Nat. Rev. Urol. 2015, 12, 430–431. [Google Scholar] [CrossRef]
- Dudka, I.; Thysell, E.; Lundquist, K.; Antti, H.; Iglesias-Gato, D.; Flores-Morales, A.; Bergh, A.; Wikström, P.; Gröbner, G. Comprehensive metabolomics analysis of prostate cancer tissue in relation to tumor aggressiveness and TMPRSS2-ERG fusion status. BMC Cancer 2020, 20, 437. [Google Scholar] [CrossRef] [PubMed]
- Ostrander, E.A.; Johannesson, B. Prostate cancer susceptibility loci: Finding the genes. Adv. Exp. Med. Biol. 2008, 617, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Kgatle, M.M.; Kalla, A.A.; Islam, M.M.; Sathekge, M.; Moorad, R. Prostate Cancer: Epigenetic Alterations, Risk Factors, and Therapy. Prostate Cancer 2016, 2016, 1–11. [Google Scholar] [CrossRef]
- Salciccia, S.; Capriotti, A.L.; Lagana, A.; Fais, S.; Logozzi, M.; De Berardinis, E.; Busetto, G.M.; Di Pierro, G.B.; Ricciuti, G.P.; Del Giudice, F.; et al. Biomarkers in Prostate Cancer Diagnosis: From Current Knowledge to the Role of Metabolomics and Exosomes. Int. J. Mol. Sci. 2021, 22, 4367. [Google Scholar] [CrossRef]
- Culp, M.B.; Soerjomataram, I.; Efstathiou, J.A.; Bray, F.; Jemal, A. Recent Global Patterns in Prostate Cancer Incidence and Mortality Rates. Eur. Urol. 2020, 77, 38–52. [Google Scholar] [CrossRef]
- Lee, S.; Ku, J.Y.; Kang, B.J.; Kim, K.H.; Ha, H.K.; Kim, S. A Unique Urinary Metabolic Feature for the Determination of Bladder Cancer, Prostate Cancer, and Renal Cell Carcinoma. Metabolites 2021, 11, 591. [Google Scholar] [CrossRef]
- Lima, A.R.; Pinto, J.; Amaro, F.; Bastos, M.L.; Carvalho, M.; Guedes de Pinho, P. Advances and Perspectives in Prostate Cancer Biomarker Discovery in the Last 5 Years through Tissue and Urine Metabolomics. Metabolites 2021, 11, 181. [Google Scholar] [CrossRef]
- Louie, K.S.; Seigneurin, A.; Cathcart, P.; Sasieni, P. Do prostate cancer risk models improve the predictive accuracy of PSA screening? A meta-analysis. Ann. Oncol. 2015, 26, 848–864. [Google Scholar] [CrossRef] [PubMed]
- Das, C.J.; Razik, A.; Sharma, S.; Verma, S. Prostate biopsy: When and how to perform. Clin. Radiol. 2019, 74, 853–864. [Google Scholar] [CrossRef]
- Spur, E.M.; Decelle, E.A.; Cheng, L.L. Metabolomic imaging of prostate cancer with magnetic resonance spectroscopy and mass spectrometry. Eur. J. Nucl. Med. Mol. Imaging 2013, 40 (Suppl. S1), S60–S71. [Google Scholar] [CrossRef] [PubMed]
- Morrison, G.J.; Goldkorn, A. Development and Application of Liquid Biopsies in Metastatic Prostate Cancer. Curr. Oncol. Rep. 2018, 20, 35. [Google Scholar] [CrossRef]
- Kretschmer, A.; Tilki, D. Biomarkers in prostate cancer–Current clinical utility and future perspectives. Crit. Rev. Oncol. Hematol. 2017, 120, 180–193. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.-H.; Deng, J.-L.; Wang, G.; Zhu, Y.-S. Long non-coding RNAs in prostate cancer: Functional roles and clinical implications. Cancer Lett. 2019, 464, 37–55. [Google Scholar] [CrossRef]
- Matuszczak, M.; Schalken, J.A.; Salagierski, M. Prostate Cancer Liquid Biopsy Biomarkers’ Clinical Utility in Diagnosis and Prognosis. Cancers 2021, 13, 3373. [Google Scholar] [CrossRef]
- Oto, J.; Fernández-Pardo, Á.; Royo, M.; Hervás, D.; Martos, L.; Vera-Donoso, C.D.; Martínez, M.; Heeb, M.J.; España, F.; Medina, P.; et al. A predictive model for prostate cancer incorporating PSA molecular forms and age. Sci. Rep. 2020, 10, 2463. [Google Scholar] [CrossRef]
- Auprich, M.; Bjartell, A.; Chun, F.K.; de la Taille, A.; Freedland, S.J.; Haese, A.; Schalken, J.; Stenzl, A.; Tombal, B.; van der Poel, H. Contemporary role of prostate cancer antigen 3 in the management of prostate cancer. Eur. Urol. 2011, 60, 1045–1054. [Google Scholar] [CrossRef]
- Parekh, D.J.; Punnen, S.; Sjoberg, D.D.; Asroff, S.W.; Bailen, J.L.; Cochran, J.S.; Concepcion, R.; David, R.D.; Deck, K.B.; Dumbadze, I.; et al. A multi-institutional prospective trial in the USA confirms that the 4Kscore accurately identifies men with high-grade prostate cancer. Eur. Urol. 2015, 68, 464–470. [Google Scholar] [CrossRef]
- Nordström, T.; Vickers, A.; Assel, M.; Lilja, H.; Grönberg, H.; Eklund, M. Comparison Between the Four-kallikrein Panel and Prostate Health Index for Predicting Prostate Cancer. Eur. Urol. 2015, 68, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Zappala, S.M.; Scardino, P.T.; Okrongly, D.; Linder, V.; Dong, Y. Clinical performance of the 4Kscore Test to predict high-grade prostate cancer at biopsy: A meta-analysis of us and European clinical validation study results. Rev. Urol. 2017, 19, 149–155. [Google Scholar] [CrossRef]
- White, J.; Shenoy, B.V.; Tutrone, R.F.; Karsh, L.I.; Saltzstein, D.R.; Harmon, W.J.; Broyles, D.L.; Roddy, T.E.; Lofaro, L.R.; Paoli, C.J.; et al. Clinical utility of the Prostate Health Index (phi) for biopsy decision management in a large group urology practice setting. Prostate Cancer Prostatic Dis. 2018, 21, 78–84. [Google Scholar] [CrossRef]
- Saidi, S.; Al Riyami, N.; Marhoon, M.; Saraf, M.; Busaidi, S.; Mula-Abed, W.-A.; Bayoumi, R. Validity of Prostate Health Index and Percentage of [-2] Pro-Prostate-Specific Antigen as Novel Biomarkers in the Diagnosis of Prostate Cancer: Omani Tertiary Hospitals Experience. Oman Med. J. 2017, 32, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, S.V.M.; García-Perdomo, H.A. Diagnostic accuracy of prostate cancer antigen 3 (PCA3) prior to first prostate biopsy: A systematic review and meta-analysis. Can. Urol. Assoc. J. 2020, 14, E214–E219. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, A.; Mahon, J.; Boland, A.; Beale, S.; Dwan, K.; Fleeman, N.; Hockenhull, J.; Dundar, Y. The clinical effectiveness and cost-effectiveness of the PROGENSA® prostate cancer antigen 3 assay and the Prostate Health Index in the diagnosis of prostate cancer: A systematic review and economic evaluation. Health Technol. Assess. 2015, 19, 1–191. [Google Scholar] [CrossRef]
- Van Neste, L.; Hendriks, R.J.; Dijkstra, S.; Trooskens, G.; Cornel, E.B.; Jannink, S.A.; de Jong, H.; Hessels, D.; Smit, F.P.; Melchers, W.J.; et al. Detection of High-grade Prostate Cancer Using a Urinary Molecular Biomarker-Based Risk Score. Eur. Urol. 2016, 70, 740–748. [Google Scholar] [CrossRef]
- Tomlins, S.A.; Day, J.R.; Lonigro, R.J.; Hovelson, D.H.; Siddiqui, J.; Kunju, L.P.; Dunn, R.L.; Meyer, S.; Hodge, P.; Groskopf, J.; et al. Urine TMPRSS2:ERG Plus PCA3 for Individualized Prostate Cancer Risk Assessment. Eur. Urol. 2016, 70, 45–53. [Google Scholar] [CrossRef]
- BridgeSpan. Gene-Based Tests for Screening, Detection, and/or Management of Prostate Cancer. In Genetic Testing, Policy No. 17; BridgeSpan: Boston, MA, USA, 2020. [Google Scholar]
- McKiernan, J.; Donovan, M.J.; O’Neill, V.; Bentinck, S.; Noerholm, M.; Belzer, S.; Skog, J.; Kattan, M.W.; Partin, A.; Andriole, G.; et al. A Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer at Initial Biopsy. JAMA Oncol. 2016, 2, 882–889. [Google Scholar] [CrossRef]
- Ross, A.E.; Feng, F.Y.; Ghadessi, M.; Echo, N.; Crisan, A.; Buerki, C.; Sundi, D.; Mitra, A.P.; Vergara, I.A.; Thompson, D.J.; et al. A genomic classifier predicting metastatic disease progression in men with biochemical recurrence after prostatectomy. Prostate Cancer Prostatic Dis. 2014, 17, 64–69. [Google Scholar] [CrossRef]
- Narayan, V.M.; Konety, B.R.; Warlick, C. Novel biomarkers for prostate cancer: An evidence-based review for use in clinical practice. Int. J. Urol. 2017, 24, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Chistiakov, D.A.; Myasoedova, V.A.; Grechko, A.V.; Melnichenko, A.A.; Orekhov, A.N. New biomarkers for diagnosis and prognosis of localized prostate cancer. Semin. Cancer Biol. 2018, 52, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Porzycki, P.; Ciszkowicz, E. Modern biomarkers in prostate cancer diagnosis. Cent. Eur. J. Urol. 2020, 73, 300–306. [Google Scholar] [CrossRef]
- Crawford, E.D.; Ventii, K.; Shore, N.D. New biomarkers in prostate cancer. Oncology 2014, 28, 135–142. [Google Scholar] [PubMed]
- Clinton, T.N.; Bagrodia, A.; Lotan, Y.; Margulis, V.; Raj, G.V.; Woldu, S.L. Tissue-based biomarkers in prostate cancer. Expert Rev. Precis. Med. Drug Dev. 2017, 2, 249–260. [Google Scholar] [CrossRef]
- Monteiro, M.; Carvalho, M.; Bastos, M.; Guedes de Pinho, P. Biomarkers in renal cell carcinoma: A metabolomics approach. Metabolomics 2014, 10, 1210–1222. [Google Scholar] [CrossRef]
- Trock, B.J. Application of Metabolomics to Prostate Cancer. Urol. Oncol. Semin. Orig. Investig. 2011, 29, 572–581. [Google Scholar] [CrossRef]
- Lima, A.R.; Pinto, J.; Azevedo, A.I.; Barros-Silva, D.; Jerónimo, C.; Henrique, R.; de Lourdes Bastos, M.; Guedes de Pinho, P.; Carvalho, M. Identification of a biomarker panel for improvement of prostate cancer diagnosis by volatile metabolic profiling of urine. Br. J. Cancer 2019, 121, 857–868. [Google Scholar] [CrossRef]
- Rudaz, S. Identification and Data-Processing Methods in Metabolomics. In Identification and Data Processing Methods in Metabolomics; Future Science Book Series; Future Science Ltd.: London, UK, 2015; pp. 2–5. [Google Scholar]
- Lima, A.R.; Bastos, M.d.L.; Carvalho, M.; Guedes de Pinho, P. Biomarker Discovery in Human Prostate Cancer: An Update in Metabolomics Studies. Transl. Oncol. 2016, 9, 357–370. [Google Scholar] [CrossRef]
- Silva, C.; Perestrelo, R.; Silva, P.; Tomás, H.; Câmara, J.S. Breast Cancer Metabolomics: From Analytical Platforms to Multivariate Data Analysis. A Review. Metabolites 2019, 9, 102. [Google Scholar] [CrossRef]
- Janfaza, S.; Khorsand, B.; Nikkhah, M.; Zahiri, J. Digging deeper into volatile organic compounds associated with cancer. Biol. Methods Protoc. 2019, 4, bpz014. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Lee, W.-Y. Urinary metabolites for urological cancer detection: A review on the application of volatile organic compounds for cancers. Am. J. Clin. Exp. Urol. 2019, 7, 232–248. [Google Scholar] [PubMed]
- Spratlin, J.L.; Serkova, N.J.; Gail Eckhardt, S. Clinical Applications of Metabolomics in Oncology: A Review. Clin. Cancer Res. 2009, 15, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, P.; Baker, J.; Boyd, M.T.; Coyle, S.; Probert, C.; Chapman, E.A. Optimisation of Urine Sample Preparation for Headspace-Solid Phase Microextraction Gas Chromatography-Mass Spectrometry: Altering Sample pH, Sulphuric Acid Concentration and Phase Ratio. Metabolites 2020, 10, 482. [Google Scholar] [CrossRef] [PubMed]
- Živković Semren, T.; Brčić Karačonji, I.; Safer, T.; Brajenović, N.; Tariba Lovaković, B.; Patent, A. Gas chromatographic-mass spectrometric analysis of urinary volatile organic metabolites: Optimization of the HS-SPME procedure and sample storage conditions. Talanta 2018, 176, 537–543. [Google Scholar] [CrossRef]
- Lima, A.R.; Araújo, A.M.; Pinto, J.; Jerónimo, C.; Henrique, R.; Bastos, M.d.L.; Carvalho, M.; Guedes de Pinho, P. Discrimination between the human prostate normal and cancer cell exometabolome by GC-MS. Sci. Rep. 2018, 8, 5539. [Google Scholar] [CrossRef]
- Câmara, J.S.; Perestrelo, R.; Berenguer, C.V.; Andrade, C.F.P.; Gomes, T.M.; Olayanju, B.; Kabir, A.; MR Rocha, C.; Teixeira, J.A.; Pereira, J.A.M. Green Extraction Techniques as Advanced Sample Preparation Approaches in Biological, Food, and Environmental Matrices: A Review. Molecules 2022, 27, 2953. [Google Scholar] [CrossRef]
- Arthur, C.L.; Pawliszyn, J. Solid phase microextraction with thermal desorption using fused silica optical fibers. Anal. Chem. 1990, 62, 2145–2148. [Google Scholar] [CrossRef]
- Silva, C.L.; Passos, M.; Camara, J.S. Solid phase microextraction, mass spectrometry and metabolomic approaches for detection of potential urinary cancer biomarkers--a powerful strategy for breast cancer diagnosis. Talanta 2012, 89, 360–368. [Google Scholar] [CrossRef]
- Porto-Figueira, P.; Pereira, J.; Miekisch, W.; Câmara, J.S. Exploring the potential of NTME/GC-MS, in the establishment of urinary volatomic profiles. Lung cancer patients as a case study. Sci. Rep. 2018, 8, 13113. [Google Scholar] [CrossRef]
- Capelli, L.; Taverna, G.; Bellini, A.; Eusebio, L.; Buffi, N.; Lazzeri, M.; Guazzoni, G.; Bozzini, G.; Seveso, M.; Mandressi, A.; et al. Application and Uses of Electronic Noses for Clinical Diagnosis on Urine Samples: A Review. Sensors 2016, 16, 1708. [Google Scholar] [CrossRef] [PubMed]
- Filianoti, A.; Costantini, M.; Bove, A.M.; Anceschi, U.; Brassetti, A.; Ferriero, M.; Mastroianni, R.; Misuraca, L.; Tuderti, G.; Ciliberto, G.; et al. Volatilome Analysis in Prostate Cancer by Electronic Nose: A Pilot Monocentric Study. Cancers 2022, 14, 2927. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, H.; Daulton, E.; Bannaga, A.S.; Arasaradnam, R.P.; Covington, J.A. Urinary Volatiles and Chemical Characterisation for the Non-Invasive Detection of Prostate and Bladder Cancers. Biosensors 2021, 11, 437. [Google Scholar] [CrossRef] [PubMed]
- Bax, C.; Taverna, G.; Eusebio, L.; Sironi, S.; Grizzi, F.; Guazzoni, G.; Capelli, L. Innovative Diagnostic Methods for Early Prostate Cancer Detection through Urine Analysis: A Review. Cancers 2018, 10, 123. [Google Scholar] [CrossRef] [PubMed]
- Opitz, P.; Herbarth, O. The volatilome–investigation of volatile organic metabolites (VOM) as potential tumor markers in patients with head and neck squamous cell carcinoma (HNSCC). J. Otolaryngol.-Head Neck Surg. 2018, 47, 42. [Google Scholar] [CrossRef] [PubMed]
- Taware, R.; Taunk, K.; Pereira, J.A.M.; Dhakne, R.; Kannan, N.; Soneji, D.; Câmara, J.S.; Nagarajaram, H.A.; Rapole, S. Investigation of urinary volatomic alterations in head and neck cancer: A non-invasive approach towards diagnosis and prognosis. Metabolomics 2017, 13, 111. [Google Scholar] [CrossRef]
- Silva, C.L.; Passos, M.; Camara, J.S. Investigation of urinary volatile organic metabolites as potential cancer biomarkers by solid-phase microextraction in combination with gas chromatography-mass spectrometry. Br. J. Cancer 2011, 105, 1894–1904. [Google Scholar] [CrossRef]
- Wang, D.; Wang, C.; Pi, X.; Guo, L.; Wang, Y.; Li, M.; Feng, Y.; Lin, Z.; Hou, W.; Li, E. Urinary volatile organic compounds as potential biomarkers for renal cell carcinoma. Biomed. Rep. 2016, 5, 68–72. [Google Scholar] [CrossRef]
- Daulton, E.; Wicaksono, A.N.; Tiele, A.; Kocher, H.M.; Debernardi, S.; Crnogorac-Jurcevic, T.; Covington, J.A. Volatile organic compounds (VOCs) for the non-invasive detection of pancreatic cancer from urine. Talanta 2021, 221, 121604. [Google Scholar] [CrossRef]
- Rudnicka, J.; Kowalkowski, T.; Buszewski, B. Searching for selected VOCs in human breath samples as potential markers of lung cancer. Lung Cancer 2019, 135, 123–129. [Google Scholar] [CrossRef]
- Kischkel, S.; Miekisch, W.; Fuchs, P.; Schubert, J.K. Breath analysis during one-lung ventilation in cancer patients. Eur. Respir. J. 2012, 40, 706–713. [Google Scholar] [CrossRef] [PubMed]
- Markar, S.R.; Brodie, B.; Chin, S.T.; Romano, A.; Spalding, D.; Hanna, G.B. Profile of exhaled-breath volatile organic compounds to diagnose pancreatic cancer. Br. J. Surg. 2018, 105, 1493–1500. [Google Scholar] [CrossRef] [PubMed]
- Altomare, D.F.; Di Lena, M.; Porcelli, F.; Trizio, L.; Travaglio, E.; Tutino, M.; Dragonieri, S.; Memeo, V.; de Gennaro, G. Exhaled volatile organic compounds identify patients with colorectal cancer. Br. J. Surg. 2013, 100, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.J.; Seo, H.S.; Kim, J.H.; Song, K.Y.; Park, C.H.; Lee, H.H. Advanced Diagnostic Technology of Volatile Organic Compounds Real Time analysis Analysis From Exhaled Breath of Gastric Cancer Patients Using Proton-Transfer-Reaction Time-of-Flight Mass Spectrometry. Front. Oncol. 2021, 11, 560591. [Google Scholar] [CrossRef]
- Taware, R.; Taunk, K.; Pereira, J.A.M.; Shirolkar, A.; Soneji, D.; Câmara, J.S.; Nagarajaram, H.A.; Rapole, S. Volatilomic insight of head and neck cancer via the effects observed on saliva metabolites. Sci. Rep. 2018, 8, 17725. [Google Scholar] [CrossRef]
- Bel’skaya, L.V.; Sarf, E.A.; Shalygin, S.P.; Postnova, T.V.; Kosenok, V.K. Identification of salivary volatile organic compounds as potential markers of the stomach and colorectal cancer: A pilot study. J. Oral Biosci. 2020, 62, 212–221. [Google Scholar] [CrossRef]
- Cavaco, C.; Pereira, J.A.M.; Taunk, K.; Taware, R.; Rapole, S.; Nagarajaram, H.; Camara, J.S. Screening of salivary volatiles for putative breast cancer discrimination: An exploratory study involving geographically distant populations. Anal. Bioanal. Chem. 2018, 410, 4459–4468. [Google Scholar] [CrossRef]
- Cavaco, C.; Perestrelo, R.; Silva, C.; Aveiro, F.; Pereira, J.; Câmara, J.S. Establishment of the Saliva Volatomic Profile as an Exploratory and Non-invasive Strategy to Find Potential Breast Cancer Biomarkers. Int. Labmate. 2014, 39, 4–5. [Google Scholar]
- Monedeiro, F.; Monedeiro-Milanowski, M.; Zmysłowski, H.; De Martinis, B.S.; Buszewski, B. Evaluation of salivary VOC profile composition directed towards oral cancer and oral lesion assessment. Clin. Oral Investig. 2021, 25, 4415–4430. [Google Scholar] [CrossRef]
- Shirasu, M.; Touhara, K. The scent of disease: Volatile organic compounds of the human body related to disease and disorder. J. Biochem. 2011, 150, 257–266. [Google Scholar] [CrossRef]
- Pereira, J.; Porto-Figueira, P.; Cavaco, C.; Taunk, K.; Rapole, S.; Dhakne, R.; Nagarajaram, H.; Camara, J.S. Breath analysis as a potential and non-invasive frontier in disease diagnosis: An overview. Metabolites 2015, 5, 3–55. [Google Scholar] [CrossRef] [PubMed]
- Tienpont, B.; David, F.; Bicchi, C.; Sandra, P. High capacity headspace sorptive extraction. J. Microcolumn Sep. 2000, 12, 577–584. [Google Scholar] [CrossRef]
- Pugliese, G.; Trefz, P.; Brock, B.; Schubert, J.K.; Miekisch, W. Extending PTR-based breath analysis to real-time monitoring of reactive volatile organic compounds. Analyst 2019, 144, 7359–7367. [Google Scholar] [CrossRef] [PubMed]
- Streckfus, C.F.; Brown, R.E.; Bull, J.M. Proteomics, morpho-proteomics phosphoproteomics, saliva and breast cancer: An emerging approach to guide the delivery of individualised thermal therapy, thermochemotherapy and monitor therapy response. Int. J. Hyperth. 2010, 26, 649–661. [Google Scholar] [CrossRef] [PubMed]
- Amann, A.; Costello, B.d.L.; Miekisch, W.; Schubert, J.; Buszewski, B.; Pleil, J.; Ratcliffe, N.; Risby, T. The human volatilome: Volatile organic compounds (VOCs) in exhaled breath, skin emanations, urine, feces and saliva. J. Breath Res. 2014, 8, 034001. [Google Scholar] [CrossRef]
- Malathi, N.; Mythili, S.; Vasanthi, H.R. Salivary Diagnostics: A Brief Review. ISRN Dent. 2014, 2014, 1–8. [Google Scholar] [CrossRef]
- Pereira, J.A.M.; Taware, R.; Porto-Figueira, P.; Rapole, S.; Câmara, J.S. The Salivary Volatome in Breast Cancer; Elsevier: Amsterdam, The Netherlands, 2020; pp. 301–307. [Google Scholar]
- Lima, A.R.; Pinto, J.; Carvalho-Maia, C.; Jerónimo, C.; Henrique, R.; Bastos, M.d.L.; Carvalho, M.; Guedes de Pinho, P. A Panel of Urinary Volatile Biomarkers for Differential Diagnosis of Prostate Cancer from Other Urological Cancers. Cancers 2020, 12, 2017. [Google Scholar] [CrossRef]
- da Costa, B.R.B.; De Martinis, B.S. Analysis of urinary VOCs using mass spectrometric methods to diagnose cancer: A review. Clin. Mass Spectrom 2020, 18, 27–37. [Google Scholar] [CrossRef]
- Dai, C.; Heemers, H.; Sharifi, N. Androgen Signaling in Prostate Cancer. Cold Spring Harb. Perspect. Med. 2017, 7, a030452. [Google Scholar] [CrossRef]
- Koppenol, W.H.; Bounds, P.L.; Dang, C.V. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat. Rev. Cancer 2011, 11, 325–337. [Google Scholar] [CrossRef]
- Lima, A.R.; Pinto, J.; Barros-Silva, D.; Jerónimo, C.; Henrique, R.; Bastos, M.d.L.; Carvalho, M.; Guedes Pinho, P. New findings on urinary prostate cancer metabolome through combined GC–MS and 1H NMR analytical platforms. Metabolomics 2020, 16, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.C.; Sakaki, T.; Yamamoto, K.; Kittaka, A. The Roles of Cytochrome P450 Enzymes in Prostate Cancer Development and Treatment. Anticancer. Res. 2012, 32, 291. [Google Scholar] [PubMed]
- Maksymchuk, O.V.; Kashuba, V.I. Altered expression of cytochrome P450 enzymes involved in metabolism of androgens and vitamin D in the prostate as a risk factor for prostate cancer. Pharmacol. Rep. 2020, 72, 1161–1172. [Google Scholar] [CrossRef]
- Khalid, T.; Aggio, R.; White, P.; De Lacy Costello, B.; Persad, R.; Al-Kateb, H.; Jones, P.; Probert, C.S.; Ratcliffe, N. Urinary Volatile Organic Compounds for the Detection of Prostate Cancer. PLoS ONE 2015, 10, e0143283. [Google Scholar] [CrossRef]
- Struck-Lewicka, W.; Kordalewska, M.; Bujak, R.; Yumba Mpanga, A.; Markuszewski, M.; Jacyna, J.; Matuszewski, M.; Kaliszan, R.; Markuszewski, M.J. Urine metabolic fingerprinting using LC-MS and GC-MS reveals metabolite changes in prostate cancer: A pilot study. J. Pharm. Biomed. Anal. 2015, 111, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Su, X.; Annabi, M.H.; Schreiter, B.R.; Prince, T.; Ackerman, A.; Morgas, S.; Mata, V.; Williams, H.; Lee, W.-Y. Application of Urinary Volatile Organic Compounds (VOCs) for the Diagnosis of Prostate Cancer. Clin. Genitourin. Cancer 2019, 17, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Pacheco, A.; Salinero-Bachiller, M.; Iribar, M.C.; López-Luque, A.; Miján-Ortiz, J.L.; Peinado, J.M. Furan and p-xylene as candidate biomarkers for prostate cancer. Urol. Oncol. Semin. Orig. Investig. 2018, 36, 243.e221–243.e227. [Google Scholar] [CrossRef]
- Peng, G.; Hakim, M.; Broza, Y.Y.; Billan, S.; Abdah-Bortnyak, R.; Kuten, A.; Tisch, U.; Haick, H. Detection of lung, breast, colorectal, and prostate cancers from exhaled breath using a single array of nanosensors. Br. J. Cancer 2010, 103, 542–551. [Google Scholar] [CrossRef]
- Taverna, G.; Grizzi, F.; Tidu, L.; Bax, C.; Zanoni, M.; Vota, P.; Lotesoriere, B.J.; Prudenza, S.; Magagnin, L.; Langfelder, G.; et al. Accuracy of a new electronic nose for prostate cancer diagnosis in urine samples. Int. J. Urol. 2022, 29, 890–896. [Google Scholar] [CrossRef]
- Capelli, L.; Bax, C.; Grizzi, F.; Taverna, G. Optimization of training and measurement protocol for eNose analysis of urine headspace aimed at prostate cancer diagnosis. Sci. Rep. 2021, 11, 20898. [Google Scholar] [CrossRef]
- Bannaga, A.S.; Kvasnik, F.; Persaud, K.; Arasaradnam, R.P. Differentiating cancer types using a urine test for volatile organic compounds. J. Breath Res. 2020, 15, 017102. [Google Scholar] [CrossRef] [PubMed]
- Waltman, C.G.; Marcelissen, T.A.T.; van Roermund, J.G.H. Exhaled-breath Testing for Prostate Cancer Based on Volatile Organic Compound Profiling Using an Electronic Nose Device (Aeonose™): A Preliminary Report. Eur. Urol. Focus 2020, 6, 1220–1225. [Google Scholar] [CrossRef] [PubMed]
- Van Poppel, H.; Hogenhout, R.; Albers, P.; van den Bergh, R.C.N.; Barentsz, J.O.; Roobol, M.J. A European Model for an Organised Risk-stratified Early Detection Programme for Prostate Cancer. Eur. Urol. Oncol. 2021, 4, 731–739. [Google Scholar] [CrossRef] [PubMed]
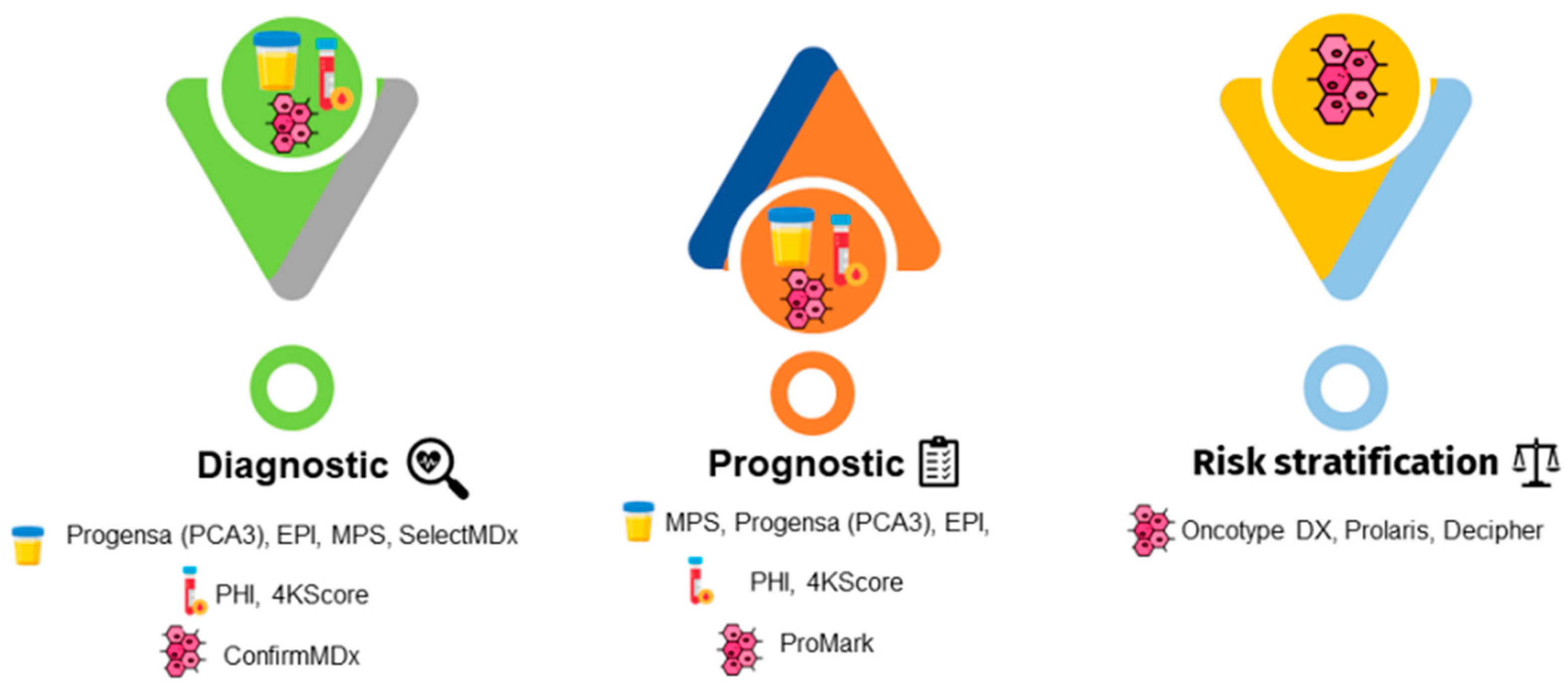


| Biomarker Test | Molecular Markers | Potential Clinical Utility | Characteristics | Availability |
|---|---|---|---|---|
| Serum biomarkers | ||||
| PSA | PSA | Treatment monitoring | Sensitivity: 60% [18] Specificity: 79% [18] AUC: 0.55 [19] | |
| 4KScore | Total PSA, free PSA, intact PSA, hK2 | Unnecessary biopsy reduction of 43% [20] Risk prediction of PCa metastases Previous negative biopsy | Sensitivity: 75% [21] Specificity: 63% [21] AUC: 0.71 [22] | CLIA-certified |
| PHI | Total PSA, free PSA, p2PSA isoform | Unnecessary biopsy reduction of 40% [23] Prediction of high-grade PCa Active supervision monitoring | Sensitivity: 82% [24] Specificity: 80% [24] AUC: 0.71 [21] | FDA-approved |
| Urinary biomarkers | ||||
| Progensa (PCA3) | Long non-coding RNAs (ratio of PCA3 mRNA:PSA mRNA) | Unnecessary biopsy reduction of 23–38% [25] PCa detection, staging, and prognosis Previous negative biopsy | Sensitivity: 69% [26] Specificity: 65% [26] AUC: 0.73 [26] | FDA-approved |
| SelectMDx | HOXC6 and DLX1 mRNA | Unnecessary biopsy reduction of 53% [27] Prediction of high-grade PCa | Sensitivity: 91% [27] Specificity: 36% [27] AUC: 0.71–0.81 [27] | CLIA-certified |
| MPS | PCA3 and TMPRSS2-ERG mRNA | Unnecessary biopsy reduction of 35–47% [28] Predict the risk of PCa and high-grade PCa | Sensitivity: 93% [29] Specificity: 33% [29] AUC: 0.69 [28] | CLIA-certified |
| EPI | Exosomal RNA (SPDEF, PCA3, ERG) | Unnecessary biopsy reduction of 27% [30] Improved identification of high-grade PCa | Sensitivity: 92% [30] Specificity: 34% [30] AUC: 0.70 [30] | CLIA-certified |
| Tissue biomarkers | ||||
| ConfirmMDx | DNA hypermethylation GSTP1, APC, and RASSF1 | Prediction of true negative prostate biopsies | Sensitivity: 68% [24] Specificity: 64% [24] AUC: 0.74 [24] | Not FDA-approved yet |
| OncotypeDX | mRNA expression (17 genes) | Monitoring of tumor aggressiveness | AUC: 0.73 [26] | Not FDA-approved yet |
| Prolaris | mRNA expression (31 genes) | Monitoring of tumor aggressiveness | AUC: 0.78 [26] | FDA-approved |
| Decipher | mRNA expression (22 genes) | Treatment monitoring | Sensitivity: 73% [31] Specificity: 74% [31] AUC: 0.79 [31] | CLIA-certified |
| ProMark | Protein biomarker test (8 proteins) | Monitoring of tumor aggressiveness | Sensitivity: 90% [27] Specificity: 85% [27] AUC: 0.72 [27] | CLIA-certified |
| Cancer Type | Analytical Approach | Biomarker’s Candidates | Prediction Model | Validation Characteristics | Reference |
|---|---|---|---|---|---|
| Urine | |||||
| Head and neck | HS-SPME/GC–MS | m-cresol, benzene, nonanal, acetone | PLS-DA | NA | [57] |
| Head and neck | HS-SPME/GC–MS | 2,6-dimethyl-7-octen-2-ol, 1-butanol, p-xylene, 4-methyl-2-heptanone | PLS-DA, ROC | NA | [58] |
| Leukaemia, colorectal, lymphoma | dHS-SPME/GC–qMS | 16 VOMs were found statistically significant | PCA | NA | [59] |
| Breast | dHS-SPME/GC–qMS | Heptanal, dimethyl disulfide and 2-methyl-3-phenyl-2-propenal | PCA | NA | [51] |
| Renal cell carcinoma | HS-SPME/GC–MS | 11 VOMs | PCA, PLSDA | NA | [60] |
| Pancreatic | TD-GC–TOF-MS GC–IMS | 2,6-dimethyl-octane, nonanal, 4-ethyl-1,2-dimethyl-benzene, 2-pentanone | Repeated 10-Fold CV | NA | [61] |
| Exhaled breath | |||||
| Lung | HS-SPME/GC–MS | Acetone, methyl acetate, isoprene, methyl vinyl ketone, cyclohexane, 2-methylheptane, cyclohexanone | DFA, ANN | Sensitivity: 80% Specificity: 91.23% AUC: NA | [62] |
| Lung | HS-SPME/GC–MS | Caprolactam and propanoic acid | PCA, OPLS-DA, PLSDA | NA | [63] |
| Pancreatic | TD-GC–MS | Formaldehyde, acetone, acetoin, undecane, isopropyl alcohol, pentane, n-hexane, 1-butanol, 1-(methylthio)-propane, benzaldehyde, tetradecane, amylene hydrate | ROC | Sensitivity: 81% Specificity: 58% AUC: 0.736 | [64] |
| Colorectal | TD-GC–MS | 15 specific VOMs | PNN | Sensitivity: 86% Specificity: 83% AUC: 0.852 | [65] |
| Gastric | PTR-TOF-MS | Propanal, aceticamide, isoprene, 1,3-propanediol | ROC | Sensitivity: 61% Specificity: 94% AUC: 0.842 | [66] |
| Saliva | |||||
| Head and neck | HS-SPME/GC–MS | 1,4-dichlorobenzene, 1,2-decanediol, 2,5-bis1,1-dimethylethylphenol, E-3-decen-2-ol | ROC, OPLS-DA | NA | [67] |
| Colorectal/stomach | GC–FID | Acetaldehyde, acetone, 2-propanol, ethanol, methanol | ROC | Sensitivity: 95.7% Specificity: 90.9% AUC: 0.857/0.839 | [68] |
| Breast | HS-SPME/GC–MS | 3-methyl-pentanoic acid, 4-methyl-pentanoic acid, phenol, p-tert-butyl-phenol (Portuguese samples) and acetic, propanoic, benzoic acids, 1,2-decanediol, 2-decanone, decanal (Indian samples) | PLS-DA, OPLS-DA | NA | [69] |
| Breast | dHS-SPME/GC–qMS | Phenol, 2-ethyl-1-hexanol | PCA | NA | [70] |
| Oral | HS-SPME/GC–MS | 1-octen-3-ol, hexanoic acid, E-2-octenal, heptanoic acid, octanoic acid, E-2-nonenal, nonanoic acid, 2,4-decadienal, 9-undecenoic acid | PCA | Sensitivity: 100% Specificity: 100% AUC: 1 | [71] |
| Sample Groups | Analytical Approach | Biomarker’s Candidates | Prediction Model | Validation Characteristics | Reference |
|---|---|---|---|---|---|
| Urine | |||||
| PCa: 59 HC: 43 | HS-SPME/GC–MS | 2,6-dimethyl-7-octen-2-ol, pentanal, 3-octanone, 2-octanone | Repeated 10-Fold CV, Repeated Double CV | NA | [87] |
| PCa: 58 HC: 60 | HS-SPME/GC–MS | hexanal, 2,5-dimethylbenzaldehyde, 4-methylhexan-3-one, dihydroedulan IA, methylglyoxal, 3-phenylpropionaldehyde | PLS-DA, ROC | Sensitivity: 89% Specificity: 83% AUC: 0.904 | [39] |
| PCa: 20 BC: 20 RC: 20 HC: 20 | HS-SPME/GC–MS | methylglyoxal, hexanal, 3-phenylpropionaldehyde, 4-methylhexan-3-one, 2,5-dimethylbenzaldehyde, dihydroedulan IA, ethylbenzene, heptan-2-one, heptan-3-one, 4-(2-methylpropoxy)butan-2-one, methyl benzoate, 3-methyl-benzaldehyde | PLS-DA | Sensitivity: 76% Specificity: 97% AUC: 0.90 | [80] |
| PCa: 32 HC: 32 | GC–MS | VOMs involved in amino acids, purine, glucose, urea, Krebs cycle biochemical pathways | PCA, PLS-DA | NA | [88] |
| PCa: 108 HC: 75 | SBSE/TD-GC–MS | 11 VOMs | ROC | Sensitivity: 87% Specificity: 77% AUC: 0.86 | [89] |
| PCa: 29 BPH: 21 | HS-SPME/GC–MS | furan, p-xylene | - | NA | [90] |
| BC: 15 PCa: 55 HC: 36 | GC–TOF-MS and GC–IMS | 35 VOMs | ROC, Repeated 10-Fold CV | GC–IMS method Sensitivity: 87% Specificity: 92% AUC: 0.95 GC–TOF-MS method Sensitivity: 78% Specificity: 88% AUC: 0.94 | [55] |
| PCa: 88 HC: 86 | Urine headspace conditioning, followed by e-nose analysis | The study tested the ability of urinary volatilome profiling to distinguish patients with PCa from HC | ROC | Sensitivity: 85.2% Specificity: 79.1% AUC: 0.82 | [92] |
| PCa: 133 HC: 139 | Urine headspace conditioning, followed by e-nose analysis (Cyranose C320) | The study tested the ability of urinary volatilome profiling to distinguish patients with PCa from HC | PCA, ROC | Sensitivity: 82.7% Specificity: 88.5% AUC: 0.90 | [54] |
| PCa: 132 HC: 60 | Urine headspace conditioning, followed by e-nose analysis (Cyranose C320) | The study tested the ability of urinary volatilome profiling to distinguish patients with PCa from HC | PCA | Sensitivity: 82% Specificity: 87% AUC: NA | [93] |
| HCC: 31 PCa: 62 BC: 29 HC: 18 | SPME, followed by analysis with polymer tabs sensor | The study tested the ability of urinary volatilome profiling to distinguish patients with PCa from HC | PCA, ROC | PCa detection Sensitivity: 70% Specificity: NA AUC: 0.70 | [94] |
| Exhaled breath | |||||
| LC: 30 CC: 26 BTC: 22 PCa: 18 HC: 22 | HS-SPME/GC–MS | 6 VOMs for LC, 6 VOMs for CC, 5 VOMs for BTC, 4 VOMs for PCa | PCA | NA | [91] |
| PCa: 32 HC: 53 | E-nose analysis (Cyranose C320) | The study tested the ability of exhaled breath volatilome profiling to distinguish patients with PCa from HC | ANN | Sensitivity: 84% Specificity: 70 AUC: 0.79 | [95] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berenguer, C.V.; Pereira, F.; Pereira, J.A.M.; Câmara, J.S. Volatilomics: An Emerging and Promising Avenue for the Detection of Potential Prostate Cancer Biomarkers. Cancers 2022, 14, 3982. https://doi.org/10.3390/cancers14163982
Berenguer CV, Pereira F, Pereira JAM, Câmara JS. Volatilomics: An Emerging and Promising Avenue for the Detection of Potential Prostate Cancer Biomarkers. Cancers. 2022; 14(16):3982. https://doi.org/10.3390/cancers14163982
Chicago/Turabian StyleBerenguer, Cristina V., Ferdinando Pereira, Jorge A. M. Pereira, and José S. Câmara. 2022. "Volatilomics: An Emerging and Promising Avenue for the Detection of Potential Prostate Cancer Biomarkers" Cancers 14, no. 16: 3982. https://doi.org/10.3390/cancers14163982
APA StyleBerenguer, C. V., Pereira, F., Pereira, J. A. M., & Câmara, J. S. (2022). Volatilomics: An Emerging and Promising Avenue for the Detection of Potential Prostate Cancer Biomarkers. Cancers, 14(16), 3982. https://doi.org/10.3390/cancers14163982







