Simple Summary
MicroRNAs (miRNAs) have been under the spotlight for the last three decades. These non-coding RNAs seem to be dynamic regulators of mRNA stability and translation, in addition to interfering with transcription. Circulating miRNAs play a critical role in cell-to-cell interplay; therefore, they can serve as disease biomarkers. Meta-analysis of published data revealed that the CC genotype of rs4938723 in pri-miR-34b/c and the TT genotype of rs543412 in miR-100 confer protection against acute lymphoblastic leukemia (ALL) in children. Reanalysis of small RNA-seq data with novel tools identified significantly overexpressed members of the miR-128, miR-181, miR-130 and miR-17 families and significantly lower expression of miR-30, miR-24-2 and miR143~145 clusters, miR-574 and miR-618 in pediatric T-ALL cases compared with controls. Inconsistencies in methodology and study designs in most published material preclude reproducibility, and further cohort studies need to be conducted in order to empower novel tools, such as ALLSorts and RNAseqCNV.
Abstract
MicroRNAs (miRNAs) have been implicated in childhood acute lymphoblastic leukemia (ALL) pathogenesis. We performed a systematic review and meta-analysis of miRNA single-nucleotide polymorphisms (SNPs) in childhood ALL compared with healthy children, which revealed (i) that the CC genotype of rs4938723 in pri-miR-34b/c and the TT genotype of rs543412 in miR-100 confer protection against ALL occurrence in children; (ii) no significant association between rs2910164 genotypes in miR-146a and childhood ALL; and (iii) SNPs in DROSHA, miR-449b, miR-938, miR-3117 and miR-3689d-2 genes seem to be associated with susceptibility to B-ALL in childhood. A review of published literature on differential expression of miRNAs in children with ALL compared with controls revealed a significant upregulation of the miR-128 family, miR-130b, miR-155, miR-181 family, miR-210, miR-222, miR-363 and miR-708, along with significant downregulation of miR-143 and miR-148a, seem to have a definite role in childhood ALL development. MicroRNA signatures among childhood ALL subtypes, along with differential miRNA expression patterns between B-ALL and T-ALL cases, were scrutinized. With respect to T-ALL pediatric cases, we reanalyzed RNA-seq datasets with a robust and sensitive pipeline and confirmed the significant differential expression of hsa-miR-16-5p, hsa-miR-19b-3p, hsa-miR-92a-2-5p, hsa-miR-128-3p (ranked first), hsa-miR-130b-3p and -5p, hsa-miR-181a-5p, -2-3p and -3p, hsa-miR-181b-5p and -3p, hsa-miR-145-5p and hsa-miR-574-3p, as described in the literature, along with novel identified miRNAs.
1. Introduction
Acute lymphoblastic leukemia (ALL) remains the predominant cancer type of childhood; despite the advancements in ALL treatment and genetic characterization, there is a need for reliable biomarkers to further improve survival beyond 93% [1,2,3,4,5]. Conventional diagnostics of ALL are based on suggestive findings in peripheral blood confirmed by bone marrow aspiration and biopsy, flow cytometry, cytogenetics and molecular studies. Chemotherapy and radiation therapy are the cornerstones of childhood ALL treatment, whereas targeted therapies have been recently added to the clinicians’ armamentaria with favorable outcomes [5,6]. The discovery of the first microRNA (miRNA) in 1993 was succeeded by numerous studies on the role of miRNAs in disease. MicroRNAs are regulatory small non-coding RNAs, and their mature form is an average of 22 nucleotides in length. According to the latest miRBase release (http://mirbase.org/, accessed on 16 August 2022), 1917 human miRNAs have been documented to date, most of which have distinct biological features and gene targets. miRNAs were recently identified as key regulators of lymphoid differentiation, playing a major role in leukemia biology. Upregulated miRNAs and downregulated miRNAs seem to act as oncogenes and tumor suppressor genes, respectively, whereas miRNA signatures can be utilized to recognize ALL patients and discrimination ALL subtypes. Figure 1 displays the canonical pathway of miRNA biogenesis and describes their mechanism of action [7,8,9]. We conducted a review of the literature, in addition to a meta-analysis of miRNA single-nucleotide polymorphisms (SNPs) and a reanalysis of small RNA-seq datasets, to investigate and highlight the potential role of miRNAs as biomarkers of childhood ALL.
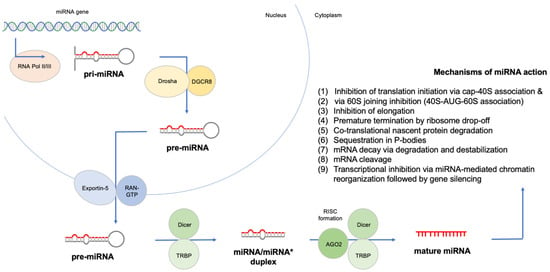
Figure 1.
Canonical biogenesis and mechanism of action of miRNAs.
2. Materials and Methods
We have searched PubMed, Scopus and Cochrane Library for “[(microRNA or miRNA) AND (childhood) AND (lymphoblastic leukemia)]” until March 2022 using the corresponding MeSH terms. Abstracts from both International Society of Paediatric Oncology (SIOP) and American Society of Hematology (ASH) meetings were also scanned for relevant content. An additional search was conducted for every miR of interest through HMDD v3.2: the Human microRNA Disease Database (http://www.cuilab.cn/hmdd, accessed on 16 August 2022) and miRbase. References of extracted studies were thoroughly scanned for relevant literature.
We further reanalyzed data from the only available small RNA-seq study that included bone marrow (BM) samples from 34 children with T-ALL at diagnosis, along with 5 BM samples from healthy donors [10,11]. Samples were processed using the small RNA-seq pipeline implemented in the bcbio-nextgen project [12]. Raw reads were examined for quality issues using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/, accessed on 16 August 2022) to ensure that library generation and sequencing were suitable for further analysis. Adapter sequences were detected using DNApi [13]. Adapter sequences and other contaminant sequences, such as polyA tails and low-quality sequences with PHRED quality scores <5, were trimmed from reads using Atropos [14]. Trimmed reads were aligned to Homo sapiens GRCh38 miRBase v21 with SeqBuster [8,15]. The aligned reads were used with SeqCluster to characterize the whole small RNA transcriptome and classify reads into rRNA, miRNA, repeats, genes, tRNAs and others with UCSC annotation [16,17]. Alignments were checked for evenness of coverage, rRNA content, genomic context of alignments (for example, alignments in known transcripts and introns), complexity and other quality checks using a combination of FastQC and MultiQC custom codes inside the bcbio-nextgen pipeline [18]. Data were loaded into R using the bcbioSmallRna R package and isomiRs’ Bioconductor package to obtain normalized expression values. Small RNA-seq differential expression analysis was performed using DESeq2 [19].
Review Manager (RevMan) version 5.4.1 (The Cochrane Collaboration, London, UK) was used to validate and investigate miRNA gene SNPs with respect to susceptibility to childhood ALL that had published results in more than one study.
3. Results
A total of 73 studies were identified based on the inclusion criteria (only pediatric population, only human samples and only case–control studies involving miRNAs): 62 publications concerned differential expression of miRNAs (19 with only one type of ALL and 43 regarding ALL cases of all types; Table 1), whereas 14 studies were focused on predisposition to ALL based on SNPs in known miRNAs (Table 2). Four studies had mixed adult and pediatric population samples, but their results will only be discussed and not be further analyzed [20,21,22,23]. The heterogeneity between the above studies is remarkable, and previous reviews of the field have failed to be accurate [24].

Table 1.
Childhood ALL diagnosis and miRNAs.
3.1. miRNA Polymorphisms and Childhood ALL
As mentioned above, polymorphisms in several miRNA genes have been implicated in childhood ALL pathogenesis (Table 2). The aim of this systematic review is to investigate the role of microRNA SNPs in childhood ALL occurrence, adhering to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines (PROSPERO registration: 353499). With respect to SNPs, contradictory results have been published for miR-146a, miR-196a-2, miR-499 and miR-612 genes. A previous meta-analysis reported increased risk for childhood ALL in CC carriers of miR-146a rs2910164 in Asian populations, although significant association has only been found when using the additive model (CC vs. GG: OR, 1.598; 95% CI: 1.003 to 2.545; p = 0.049) and no other model [87]. We investigated all six studies included in the latter systematic review (one study reported increased risk for childhood ALL in carriers of the CC genotype of miR-146a rs2910164, one study found increased risk for CC and CG genotypes and one study supported the protective role of GG genotype, whereas four studies revealed non-significant associations), plus one study from Brazil that found no significant association of miR-146a rs2910164 genotypes with susceptibility to childhood ALL [88].
We performed a meta-analysis including the seven studies mentioned above regarding data for rs2910164 genotypes in miR-146a and found no significant associations for any of the models. The additive model that had been previously utilized concluded that children carrying the CC genotype of miR-146a rs2910164 had a 50% increased chance of being diagnosed with ALL compared with GG carriers (OR, 1.52; 95% CI: 0.93 to 2.49; I2 = 75%), but this association was not significant (p = 0.09). SNP rs57095329 A>G in miR146A has been previously associated with reduced binding of the transcription factor V-Ets oncogene homolog 1 (Ets-1) and decreased miR-146a expression, whereas rs2910164 G>C seems to reduce pre-miR-146a nuclear-processing efficiency, thus reducing the expression of mature miR-146a and resulting in less efficient inhibition of the target genes (by affecting binding specificity), including tumor necrosis factor (TNF), TNF receptor-associated factor 6 (TRAF6) and interleukin 1 receptor-associated kinase 1 (IRAK1) [47,89]. On the other hand, miR-146a overexpression (i.e., with G allele) can inhibit IRAK1 and TRAF6 expression, impair the activity of their downstream molecule, NF-ĸB, and suppress several NF-ĸB-targeted inflammatory genes, such as IL6, CXCL8 alias IL-8, IL1B and TNF [90]. With respect to other repeatedly studied miRNA SNPs and predisposition to childhood ALL, only the CC genotype of rs4938723 in pri-miR-34b/c has been linked significantly with protection against the disease. Forest plots of studies including results for SNPs in miRNA genes in childhood ALL cases and controls are shown in Figure 2. In summary, our systematic review and meta-analysis showed that SNPs in pri-miR-34b/c and miR-100 genes have been linked with protection again childhood ALL, whereas SNPs rs3805500 in DROSHA, rs3746444 in miR-449b, rs2505901 in miR-938, rs12402181in miR-3117 and rs62571442 in miR-3689d-2 genes have been associated with susceptibility to B-ALL (Table 2).
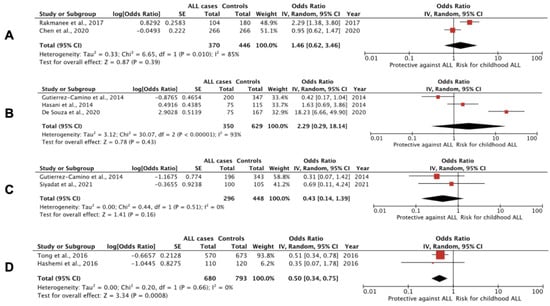
Figure 2.
Forest plots of studies, including results for SNPs in miRNA genes in childhood ALL cases and controls. (A) T>C polymorphism of rs11614913 in the miR-196a-2 gene: CC vs. CT+CC; (B) A>G polymorphism of rs37464444 in miR-499 gene -only B-ALL cases in de Souza et al, 2021, and Gutierrez-Camino et al., 2014 [88,91]: GG vs. GA+AA; (C) G>A polymorphism of rs12803915 in the miR-612 gene: AA vs. AG+GG; (D) T>C polymorphism of rs4938723 in the pri-miR-34b/c gene: CC vs. CT+TT.

Table 2.
Predisposition to childhood ALL and miRNA SNPs.
Table 2.
Predisposition to childhood ALL and miRNA SNPs.
| Study | Cohort Characteristics | Methods | Outcomes |
|---|---|---|---|
| Siyadat et al., 2021 [92] | 100 childhood B-ALL patients and 105 controls; Iran | RT-PCR, HRM and Sanger sequencing; BM or whole blood | No significant relationship was confirmed between rs12803915 variants of miR-612 and susceptibility to B-ALL. |
| Pei et al., 2020 [90] | 266 childhood ALL patients and 266 controls; Taiwan | PCR-RFLP; whole blood | The G allele of miR-146a polymorphism rs2910164 seems to be a protective biomarker for childhood ALL (OR, 0.66; 95% CI: 0.52 to 0.85; p = 0.0011), whereas patients with the GG genotype were associated with decreased susceptibility to ALL (OR, 0.4; 95% CI: 0.23 to 0.67; p = 0.0004) compared with healthy controls. Conspicuously, children bearing CG and GG genotypes were found to have earlier onset compared to those with CC genotype (p = 0.025 and p = 0.0001, respectively). The G>C polymorphism leads to reduced amounts of mature miR-146a, whereas the C allele has reduced capacity to inhibit target genes. |
| C.C. Chen et al., 2020 [93] | 266 childhood ALL patients and 266 controls; Taiwan | PCR-RFLP; whole blood | The T>C polymorphism of rs11614913 in miR-196a-2 does not seem to be associated with susceptibility to childhood ALL. |
| De Souza et al., 2020 [88] | 100 childhood B-ALL patients and 180 adult controls; Brazil | qRT-PCR; whole blood | The mutant homozygote (AA) genotype of DROSHA rs3805500 was associated with a threefold increase in the risk of developing ALL (recessive model: GG + GA vs. AA; OR, 2.913; 95% CI: 1.415 to 5.998; p = 0.004). The mutant homozygote (GG) genotype of the miR-499 gene rs3746444 was associated with a 17.8-fold increase in the risk of developing ALL (recessive model: AA + AG vs. GG; 95% CI: 5.55 to 57.016; p < 0.001). In contrast, the wild homozygous (CC) genotype of the miR-938 gene rs2505901 seems to confer a protective effect against developing ALL (dominant model: CC vs. CT + TT; OR, 0.359; 95% CI: 0.160 to 0.805; p = 0.013). No significant associations for AGO1 rs636832, MIR219A1 rs213210 and rs107822, MIR146A rs2910164, MIR330 rs12894467 and MIR608 rs4919510. |
| Jemimah Devanandan et al., 2019 [47] | 71 childhood ALL patients and 74 controls; India | qRT-PCR; whole blood | No statistically significant association was found between miR-146a gene SNPs rs2910164 G>C and rs57095329 A>G and ALL risk. |
| Xue et al., 2019 [42] | 831 childhood ALL patients and 1079 controls; validation: 88 childhood ALL cases and 99 controls; China | qRT-PCR; whole blood; plasma from validation cohort | Subjects carrying the mutant homozygous TT genotype of miR-100 rs543412 had a statistically significantly decreased risk of childhood ALL (OR, 0.73; 95% CI: 0.55 to 0.97; p = 0.029). No association was been found for miR-146a rs2910164 and miR-210 rs7395206 polymorphisms and ALL. |
| Gutierrez-Camino et al., 2018 [94] | 217 children with B-ALL and 330 controls from Spain and 75 children with B-ALL and 96 controls from Slovenia | GoldenGate genotyping assay and OpenArray genotyping; whole blood or BM | The AA genotype of rs12402181 in miR-3117-3p was associated with B-ALL risk (OR, 1.44; 95% CI: 1.01 to 2.08; p = 0.047; GG vs. AG vs. AA) in the Spanish cohort. The same effect was observed in the Slovenian cohort (OR, 2.01; 95% CI: 1.02 to 3.95; p = 0.041). When both populations were analyzed together, they displayed a more significant trend (OR, 1.53; 95% CI: 1.12 to 2.09; p = 0.006). With respect to allele frequency analysis, minor allele A showed a 1.51-fold increased risk of B-ALL in total (95% CI: 1.11 to 2.05; p = 0.007). The CT/CC genotype in rs62571442 of miR-3689d-2 was associated with a significantly increased risk of developing B-ALL (OR, 1.48; 95% CI: 1.02 to 2.15; p = 0.039) in the Spanish cohort, whereas the risk was even greater in the Slovenian cohort (OR, 3.57; 95% CI: 1.57 to 8.12; p = 0.001). Analyzed together, both cohorts confirmed the latter association (OR, 1.31; 95% CI: 1.06 to 1.60; p = 0.011). Allele frequency analysis in both populations showed that minor allele C is associated with a 1.31-fold increased risk of B-ALL (95% CI: 1.06 to 1.6; p = 0.012). |
| Liu et al., 2018 [95] | 200 childhood ALL patients and 100 controls; China | RFLP; whole blood | The miR-146a rs2910164 CC or CG genotype significantly increased the risk of ALL (frequency of GG, GC and CC genotypes in the patient group and in the control group was 16%, 44.5%, 39.5%, and 29%, 41%, 30%, respectively). The expression of GC/CC genotypes were significantly higher in patients than in controls (GG genotype as reference; for the GC genotype: OR, 1.967; 95% CI: 1.054 to 3.672; p = 0.037; and for the CC genotype: OR, 2.386; 95%CI: 1.239 to 4.595; p = 0.012). |
| Rakmanee et al., 2017 [96] | 104 childhood ALL patients and 180 controls; Thailand | PCR-RFLP; whole blood | Variant CC of rs11614913 in miR-196a-2 (OR, 4.321; 95% CI: 2.091 to 8.930; p < 0.001), TC heterozygote (OR, 2.248; 95% CI: 1.103 to 4.579; p = 0.024) and CC+TC genotypes (OR, 2.921; 95% CI: 1.504 to 5.673; p = 0.001) were associated with childhood ALL susceptibility compared with the TT wild type. CC homozygotes were associated with significantly increased miR-196a-2 expression. |
| Chansing et al., 2016 [97] | 100 childhood ALL patients and 200 controls; Thailand | PCR-RFLP; whole blood | There was no association between miR-146a rs2910164 G>C polymorphism and susceptibility to childhood ALL. |
| Hashemi et al., 2016 [98] | 110 childhood ALL patients and 120 controls; Iran | PCR-RFLP; whole blood | A study of polymorphism rs4938723 in pri-miR-34b/c in ALL and healthy children revealed decreased risk of ALL in heterozygous (OR, 0.48; 95% CI: 0.28 to 0.84; p = 0.012; TC vs. TT), and overdominant (OR, 0.51; 95% CI: 0.3 to 0.89; p = 0.02; TC vs. TT+CC) inheritance models. The C allele significantly decreased the risk of childhood ALL compared to the T allele (OR, 0.52; 95% CI: 0.33 to 0.83; p = 0.006). |
| Tong et al., 2016 [99] | 570 childhood ALL patients at diagnosis and 673 controls; China | RT-PCR; whole blood | The CC genotype in rs4938723 of pri-miR-34b/c was associated with significantly reduced ALL risk (CC vs. TT: OR, 0.51; 95% CI: 0.33 to 0.8; p = 0.003, and CC vs. TT+TC: OR, 0.49; 95% CI: 0.32 to 0.75; p = 0.002). |
| Gutierrez-Camino et al., 2014 [91] | 213 childhood precursor B-ALL patients and 387 adult controls; Spain | OpenArray; remission BM or whole blood | The A allele of rs12803915 in premature mir-612 was found to be protective (GG vs. GA vs. AA; OR, 0.61; 95% CI: 0.42 to 0.88; p = 0.007) against ALL risk. The G allele of rs3746444 in the seed region of mature miR-499-3p was also found to be protective (AA vs. AG vs. GG; OR, 0.67; 95% CI: 0.49 to 0.91; p = 0.009). An association with rs10061133 in mir-449b was also identified (AA vs. GA+GG; OR, 0.52; 95% CI: 0.31 to 0.89; p = 0.012). Eight SNPs present in the six miRNA biogenesis pathway genes (TNRC6B, DROSHA, DGCR8, EIF2C1 or AGO1, CNOT1 and CNOT6) were also identified. |
| Hasani et al., 2014 [89] | 75 childhood ALL patients and 115 controls; Iran | T-ARMS-PCR; whole blood | The G>C variant of rs2910164 in hsa-miR-146a was found to significantly increase risk of ALL (CC vs. GG; OR, 4.24; 95% CI: 1.52 to 11.87; p = 0.006, and GC vs. GG; OR, 3.55; 95% CI: 1.41 to 8.93; p = 0.007; C vs. G; OR 1.73; 95% CI: 1.13 to 2.67; p = 0.012). No association was found between rs3746444 of miR-499 and ALL risk. |
AGO1 = argonaut RISC component 1; CNOT1/6 = carbon catabolite repression 4 protein-NOT transcription complex subunit 1/6; DGCR8 = DiGeorge syndrome critical region gene 8 Microprocessor complex subunit; HRM = high-resolution melt analysis; RFLP = restriction fragment length polymorphism; T-ARMS = tetra-primer amplification refractory mutation system; TNRC6B = trinucleotide repeat-containing adaptor 6B. MicroRNAs in bold indicate statistically significant results.
Analysis of all 43 publications with data for miRNA expression in pediatric ALL patients versus controls revealed significant differential expression for several miRNAs (Figure 3). The top list was built according to published data and emphasis was placed on studies with a large sample size (n > 100), studies with validated results (e.g., microarrays validated by qRT-PCR in the same or another cohort), studies that used next-generation sequencing methods (e.g., RNA-seq) and studies with minimal risk of bias (e.g., exclusion of studies with adult or sick controls, studies with ALL cases undergoing chemotherapy, studies with disproportionate numbers of B- and T-ALL cases, etc.).
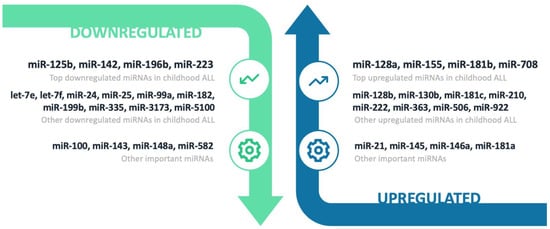
Figure 3.
Schematic representation of significantly differentially expressed miRNAs in childhood ALL diagnosis compared to controls.
3.2. Downregulated miRNAs
3.2.1. miR-125b
Downregulation of miR-125b was reported in four case–control studies (two of them with more than 100 cases), whereas one study reported otherwise [51,57,77,83]. Some pediatric BCP-ALL cases displayed upregulation of miR-125b: ETV6-RUNX1, BCR-ABL1, ERG-deregulated and t(11;14)(q24;q32) cases [29,46,100,101,102,103,104]. Although pediatric T-ALL cases are associated with lower miR-125b levels, TLX1- and TLX3-driven cases seem to have higher levels compared to other T-ALL cases, and infants with T-ALL have significantly higher miR-125b levels than the respective childhood cases [57,105,106]. miR-125b belongs to a family comprising eight members: miR-10a, miR-10b, miR-99a, miR-99b, miR-100, miR-125a, miR-125b-1 and miR-125b-2. In humans, miR-125 homologs are organized into three distinct clusters: (i) miR-125b-1, let-7a-2 and miR-100 in chromosome 11; (ii) miR-125b-2, let-7c and miR-99a in chromosome 21; and (iii) miR-125a, let-7e and miR-99b in chromosome 19; this suggests that let-7 and miR125 families have a cooperative biological function related to leukemogenesis. The diverse role of miR-125 in hematopoiesis relies on the fact that it enhances proliferation of early progenitors but also blocks their further differentiation [103]. A significant elevation of miR-125b levels after induction of treatment compared with diagnosis levels is noted, but elevated day 33/diagnosis miR-125b ratio has been linked with relapse and poor outcome [57]. Extra genetic material with respect to chromosome 21 (high hyperdiploid, Down syndrome and iAMP ALL) does not seem to contribute to increased expression levels of the miR-125b cluster, whereas its upregulation is typically featured in ETV6-RUNX1 cases, conferring survival advantage to growth inhibitory signals, independent of p53. Moreover, knockdown of miR-125b in cell lines bearing the previous fusion demonstrated increased apoptosis rates after treatment with either doxorubicin or staurosporine [101]. With respect to TLX3-positive cases, overexpression of miR-125b and miR-99a provokes the transactivation of the lncRNA gene LINC00478, which hosts the miR-125b-2 cluster and therefore leads to differentiation arrest and expansion of transformed T-cells [106]. In addition, a recent study showed that circPVT2 can promote ALL cell proliferation, migration and invasion by reducing miR-125b and enhancing the NF-κB signaling pathway [107]. Upregulation of miR-125b in childhood ALL is associated with the methylation of the promoter regions of tumor suppressor genes PPP1CA (encoding protein phosphatase 1 catalytic subunit alpha), BTG2 (gene of B-Cell translocation gene anti-proliferation factor 2) and PTEN (encoding phosphatase and tensin homolog) and their downregulation (reversed by DNA methyltransferase inhibitor decitabine) [108]. In vitro experiments support the oncogenic role of miR-125b, as it seems to repress BCL2 (and thus promote proliferation in response to CD40 ligand, i.e., CD154 in human leukemic B-cells) and to enhance proliferation and block differentiation by targeting ARID3A (AT-rich interaction domain 3A or BRIGHT, i.e., B-cell regulator of immunoglobulin heavy-chain transcription) in t(11;14)(q24;q32) B-ALL [109,110].
3.2.2. miR-142
Significant downregulation of miR-142 in children with ALL compared with controls was reported in three studies [40,54,86], whereas two other study groups reported a significant upregulation of this miRNA [77,83]. Significant upregulation of miR-142 has also been reported in T-ALL, pre-B-ALL, KMT2A-r and B-other cases compared to controls [79,83,85]. miR-142 clusters with miR-4736 in chromosome 17 and targets BCLAF1 (which encodes BCL2-associated transcription factor 1) and KMT2A-AFF1 fusion protein (formerly known as MLL-AF4) in t(4;11) leukemia [77,111]. Expression of miR-142 has been associated with glucocorticoid (GC) resistance in ALL, mainly by direct targeting of 3′-UTR of GRα mRNA (glucocorticoid receptor alpha encoded by NR3C1) [112,113]. The oncogenic role of miR-142 in T-ALL (relatively resistant to GCs) is supported by the findings that high miR-142 expression results in low levels of cAMP (cyclic adenosine monophosphate) and weak protein kinase A (PKA) activity, thus alleviating its inhibitory effect on T-cell proliferation [113]. An early report of an aggressive B-ALL case bearing a masked t(8;17) translocation indicated overactive MYC as a result of miR-142 promoter rearrangement [114]. Moreover, a murine model demonstrated that miR-142 knockdown results in altered lymphopoiesis and immunodeficiency [115].
3.2.3. miR-196b
Downregulation of miR-196b is a common finding in childhood ALL [35,54,69,76,78,82,83]. Conversely, upregulation of miR-196b is prominent in pediatric KMT2A-r cases [29,83,100,116] and in some T-ALL subgroups (such as HOXA-r, PICALM-MLLT10 or CALM-AF10, inv(7)(p15q35) and SET-NUP214 cases), especially when aberrant expression of HOXA genes is evident [20,83,116]. HOXA9 and MEIS1 homeobox oncogenes (adjacent and coexpressed with miR-196b), along with FAS tumor suppressor, are direct targets of miR-196b and drive carcinogenesis in KMT2A-r cases in a bidirectional manner [117]. As expected, miR-196b levels seem to be significantly higher in pediatric T-ALL compared with B-ALL cases, whereas miR-196b is significantly overexpressed in children with T-ALL compared with infantile T-ALL cases [76,105,116,118]. The tumor-suppressive role of miR-196b is supported by the findings that it regulates MYC and ERG and that it binds the 3′-UTR region of IGFBP3 mRNA (reversible by resveratrol in vitro) [78,119]. A ChIP (chromatin immunoprecipitation) study of 179 children with ALL from Spain reported high levels of K9H3me2 and/or low levels of K4H3me3 near to CpG islands in the miR-196b gene, indicating its downregulation. Treatment with decitabine led to the successful upregulation of its levels in vitro, suggesting a major role of methylation status in early leukemogenesis [120].
3.2.4. miR-223
Downregulation of miR-223 in childhood ALL compared with controls has been disclosed multiple times in the literature [34,46,54,69,77,84]. Only one small study has reported otherwise [40]. An inconsistent finding is that many childhood T-ALL subgroups exhibit a different profile with considerable overexpression of miR-223 compared with controls: TLX1- and TLX3-deregulated, SIL-TAL1, PICALM-MLLT10 and inv(7) cases [79]. Nevertheless, two studies involving children with T-ALL reported downregulation of miR-223 compared to healthy subjects [10,20]. In the same context, infants with T-ALL have been found to have significantly higher miR-223 levels than childhood T-ALL cases, whereas high levels of miR-223 in KMT2A-r cases are apparent [79,105]. Hyperdiploid cases display the highest and TCF3-r cases (E2A or Transcription Factor 3), the lowest miR-223 levels among BCP-ALL cases [29,100]. Notwithstanding, low expression of plasma miR-223 at admission has been associated with poor prognosis in childhood ALL [121]. This miRNA seems to inhibit cell proliferation, migration and invasion, and to promote apoptosis by targeting FOXO1 (encoding forkhead box O1 protein) [34]. Expression of miR-223 is uniformly downregulated in B-cell lymphoproliferative disorders and is stage-specific in B-cell differentiation, with higher levels in naive and memory cells compared with germinal center cells [122]. Furthermore, an early in vitro study demonstrated that miR-223 can reversibly regulate erythroid and megakaryocytic differentiation via downmodulation of LMO2 [123]. Conversely, a study involving pediatric samples reported that TAL1-mediated upregulation of miR-223 leads to promotion of a malignant phenotype in T-ALL through repression of the FBXW7 tumor suppressor (encoding F-box and WD repeat domain-containing 7 protein) [124]. In particular, elevated MYB levels in T-ALL can arise either directly through TCR-mediated MYB proto-oncogene translocations, MYB duplications and enhanced TAL1 complex binding at the MYB locus or indirectly through the TAL1/miR-223/FBXW7 regulatory axis [125]. A study including children suggested myeloid-specific features for miR-223, reporting significantly lower expression in ALL compared with AML samples [126]. Intriguingly, monitoring of miR-223 demonstrated significantly higher levels in 13 children (3 with T-ALL and 10 with BCP-ALL) after 24 h of GC monotherapy compared with their diagnosis levels [127].
3.2.5. Other Downregulated miRNAs
Significantly lower expression of 2 of the 12 members of the let-7 gene family, i.e., let-7e and let-7f-1, has been implicated in childhood ALL pathogenesis. MicroRNA let-7e is clustered with miR-99b and miR-125a, let-7f-1 is clustered with let-7a-1 and let-7d and let-7f-2 is clustered with miR-98 [59,76,83]. The let-7 family is known for its tumor-suppressor activity (by inhibiting NRAS, HMG2A and MYC protooncogenes), and their reduced levels are typically associated with cancer stemness [128,129]. In the same context, significantly lower levels of let-7b and let-7d compared with controls have been documented in children with KMT2A-r ALL and B-ALL, respectively [67,69,130]. Conversely, let-7c, originating from chromosome 21 and clustered with miR-99a, seems to be upregulated in pediatric ALL cases involving fusions in this chromosome, i.e., ETV6-RUNX1- and ERG-related cases [100,101,102]. Whereas miR-99a is generally downregulated in pediatric ALL, its upregulation in BCP-ALL cases has been identified as the strongest indicator of ETV6-RUNX1 fusion presence [29,100,101]. Restoration of underexpressed levels of miR-99a and miR-100 in ALL cells in vitro resulted in suppression of cell proliferation and increased cell apoptosis after dexamethasone treatment. Moreover, elevated expression of miR-99a and miR-100 seems to target the FKBP5 (FK506-binding protein prolyl isomerase 5) signaling pathway (in turn influence GC receptor activity) and to inhibit expression of IGF1R (insulin-like growth factor 1 receptor) and MTOR (mechanistic target of rapamycin kinase), along with their downstream MCL1 (myeloid cell leukemia sequence 1 apoptosis regulator, BCL2 family member) oncogene [75].
Downregulation of miR-24 has also been suggested as a hallmark of ALL pathogenesis in children [70,86]. Two studies on children with T-ALL reported significantly reduced expression of miR-24 compared with healthy children [10,68]. On the contrary, significantly elevated miR-24 levels in children with ALL compared with controls were reported in recent small study in Egypt [30]. The miR-24 miRNA family consists of miR-24-1 (clustered with miR-23b, miR-27b and miR-3074 in chromosome 9) and miR-24-2 (clustered with miR-23a and miR-27a in chromosome 19). Low miR-24 expression was previously associated with a diminished tumor-suppressor effect of miR-31 and PAX5 (paired box 5 protein) deletion. In the same study, high expression of miR-24 was associated with PAR1 (F2R; encoding coagulation factor II thrombin receptor) deletion [131]. Beyond BCL2, miR-24-2 was demonstrated to target a number of proteins functionally important for cell survival, such as YWHAZ (tyrosine 3-monooxygenase/tryptophan 5-mono-oxygenase activation protein zeta), TP53 (Tumor Protein P53), SMAD3 (SMAD Family Member 3), ESR1 (Estrogen Receptor 1) and CREBBP (binding protein of cAMP-response element binding protein) [132]. As demonstrated by acute leukemia cell lines and primary samples (of unknown age and leukemia type), the expression of the miR-24-2 cluster was found to be significantly decreased compared to CD34+ PBMCs from normal adult donors (86% of pre-B-ALL and 100% of T-ALL) [133]. A study on acute erythroid leukemia suggested that the miR-24-2 cluster mediates the inactivation of multiple targets in the IL6ST/JAK1/STAT3 pathway (interleukin 6 cytokine family signal transducer or GP130, Janus kinase 1 and signal transducer and activator of transcription 3, respectively) and promotes GATA1 expression, thereby facilitating apoptosis and restraining adverse proliferation. The same study pointed out that JAK1 inhibitor ruxolitinib can rescue phenotypic changes induced by low miR-24-2 [134]. Significant downregulation of miR-25 has been also reported in pediatric ALL cases compared with healthy controls [59,62]. miR-24-2 belongs to a family comprising miR-92a-1, miR-92a-2 and miR-92b, whereas it is found clustered with miR-93 and miR-106b. On the contrary, a recent study identified miR-25 as a suitable endogenous normalizer for qRT-PCR in T-ALL [135].
As discussed below, miR-182 was found to be upregulated in some T-ALL subgroups (Supplementary Table S1), although its downregulation seems to efficiently discriminate pediatric ALL cases from healthy controls [50]. miR-182 clustered with miR-96, and miR-183 seems to be involved in GC treatment response by targeting FOXO3A (forkhead box O3 protein) and its downstream target, BIM (alias BCL2L11) [112,136]. Significant overexpression of miR-182 (before treatment) was calculated in pediatric GC-sensitive ALL cases compared with GC-resistant cases [137]. In the same context, the BM miR-143/miR-182 ratio seems to be significantly decreased in childhood ALL patients at diagnosis, whereas it is increased in more than 90% of patients at the end of induction (EoI) [50]. On the contrary, GC resistance in T-ALL might be explained by the upregulation of glycolysis, oxidative phosphorylation, cholesterol synthesis, glutamic acid synthesis, fast growth rate and activation of PI3K/AKT/mTOR (phosphoinositide 3-kinases, AKT serine/threonine kinases and mTOR pathway) and the MYC-signaling pathways. Therefore, miR-182, along with miR-185, is likely to enhance GC sensitivity and GC receptor auto-upregulation by suppressing mTOR activity [137]. Altogether, restoration of miR-182 levels could be a promising therapeutic strategy for childhood ALL.
Significant downregulation of miR-199b in children with ALL compared with healthy children has been also noted by several studies [27,84,86]. This miRNA is clustered with miR-3154, which was also found to be significantly downregulated in children with T-ALL compared with controls [10]. Another significantly downregulated miRNA in childhood ALL is miR-335 [37,43,69]. Fluctuations of miR-335 levels among childhood ALL cytogenetic groups was also reported (highest in ETV6-RUNX1- and lowest in BCR-ABL1-positive cases) [138]. It has been also reported that miR-335 specifically targets 3′-UTR of RB1 (retinoblastoma transcriptional corepressor 1) and activates the p53 tumor-suppressor pathway [139]. Moreover, downregulation of miR-335 seems to decrease the sensitivity of ALL cells to prednisone (by enhancing its target MAPK1), whereas treatment with Ras-Raf-MEK-ERK pathway inhibitors can activate BIM and promote prednisolone-induced cell death [112,138].
Significant downregulation of miR-3173 and miR-5100 in children with ALL compared to healthy individuals has been documented in the literature, but data are scarce [59,61]. With respect to miR-3173, it has been shown to target the 3′-UTR of PTK2 in B-ALL cells, whereas its downregulation seems to promote cell proliferation, migration and invasion via PTK2 overexpression [61]. However, there are insufficient data to justify annotation confidence for miR-5100 [8].
As mentioned above, downregulation of miR-100 seems to be a hallmark of ALL development in children [38,75,76,83]. Paradoxically, one study reported significant upregulation of miR-100 in children with ALL compared with controls, although the study sample included a large proportion (33%) of biphenotypic ALL cases [65]. Among pediatric ALL cases, the highest levels of miR-100 have been measured in ETV6-RUNX1-positive cases, whereas miR-100 abundance is unlikely in hyperdiploid cases [29,76,100]. In addition, high miR-100 expression has been linked with low WBC levels (white blood cell levels; <50,000/mm3) at diagnosis and in vincristine- and daunorubicin-resistant cases (synergistically with miR-125b and miR-99a overexpression) [29,65,76,140].
Reduced expression of miR-143 has been associated with childhood ALL phenotype multiple times in the literature [10,28,50,84,86]. Most importantly, in vitro studies have illustrated a pathogenetic link between the notoriously poor prognosis of t(4;11) ALL and miR-143 elimination. Repression of miR-143 via methylation seems to enhance KMT2A-AFF1 oncogene expression, whereas upregulation of this miRNA could be a therapeutic target for this ALL subgroup [141]. In the same context, miR-143 overexpression in ALL cells in vitro resulted in growth inhibition and induction of apoptosis by either silencing DNMT3A (DNA methyltransferase 3 alpha) or by downregulating BCR-ABL1 expression in the respective cases [142,143]. A recent study reported significant upregulation of miR-145, which is clustered with miR-143, although this qRT-PCR study included only 13 pediatric ALL cases and 5 controls [36]. Regarding this cluster, genetic network analysis of 51 cancer types identified the miR-17~92 family as amplified and the miR-143~145 cluster as deleted, designating the latter cluster as a hallmark of tumorigenesis [144].
Significant reductions in miR-148a levels has been noted in children with ALL compared with controls [78,80,82,86]. A recent case–control study from Egypt supported the tumor-suppressive properties of miR-148a by calculating significant upregulation of miR-148a levels in treatment responders [30]. Many reports in recent years have established the role of miR-148a as a tumor suppressor through its ability to inhibit proliferation by directly suppressing DNMT1 (encoding DNA methyltransferase 1) [145]. Copy-number loss in 7p15.2 of miR-148a was linked with adult ALL, whereas this miRNA downregulation seems to be characteristic of adult KMT2A-r cases [146]. The role of miR-582 downregulation in childhood ALL occurrence is debatable. The controversy lies on the fact that case–control studies have reported both significantly high and low expression of miR-582 in ALL cases compared to controls [10,20,83,86]. The dubious finding of significant upregulation of miR-582 in a study by Schotte et al. (2009) may be explained in part by the inhomogeneous sample (KMT2A-r cases: 22%) and the small number of controls (two children with brain tumors) [83].
3.3. Upregulated miRNAs
3.3.1. miR-128 Family
This family consists of two members: miR-128-1 (formerly known as miR-128a), with its gene located in 2q21.3; and miR-128-2 (also known as miR-128b), with its gene residing in 3p22.3. Significant upregulation of both miR-128-1 and miR-128-2 has been associated with pediatric ALL cases compared with controls [10,28,46,53,68,76,83,84,86]. Two studies including both adults and children reported significant upregulation of miR-128 family members compared with normal CD19+ cells and endogenous snoRNA U6 content, respectively [126,147]. Among pediatric ALL cases, excessive upregulation of miR-128-1 is evident in t(4;11)- and TCF3-PBX1-positive cases, whereas the lowest values of miR-128-2 have been observed in IKZF1 del, PAX5 del and KMT2A-AFF1 fusion-positive cases [76,131,148,149]. In agreement with the latter findings, significantly higher levels of both miRNAs were been documented in infantile T-ALL compared with childhood T-ALL cases [105]. Expression levels of miR-128-2 seem to play a pivotal role in GC resistance. Re-expression of miR-128-2 in ALL cell lines bearing the KMT2A-AFF1 fusion resulted in sensitization to GCs in vitro, whereas target analysis has shown that miR-128-2 downregulates KMT2A, AFF1 and their fusion gene [112,127,148,150]. A13G mutation in the MIR128-2 gene seems to significantly reduce the processing of pri-miR-128-2 and therefore reduces dexamethasone-induced apoptosis, i.e., leads to GC resistance of t(4;11) ALL cells [150]. In addition, systemic GC monotherapy for 24 h resulted in significant downregulation of miR-128-2 levels in children with ALL, suggesting its critical role in GC response [127]. These oncomirs lead to leukemogenesis by affecting several pathways, mainly by inhibiting TERT (telomerase reverse transcriptase) mRNA, PHF6 (PHD finger protein 6, especially in NOTCH1-driven T-ALL models), BMI1 proto-oncogene and UBE2W (encoding ubiquitin-conjugating enzyme E2 W), an ubiquitin ligase involved in the BRCA DNA repair associated-pathway [46,149,151,152].
3.3.2. miR-155
Upregulation of miR-155 is another important hallmark of childhood ALL, and many studies have cited significantly increased expression of miR-155 in pediatric ALL cases compared with healthy controls [25,27,28,36,39,44,46,62,68,69,77,86]. Among pediatric ALL cases, the lowest expression levels for miR-155 have been recorded in ETV6-RUNX1-positive and ERG-related cases (ETS transcription factor ERG or p55) [102,153]. This miRNA promotes the development of ALL by regulating the CBL-mediated IRF4/CDK6 axis (Cbl proto-oncogene protein and Interferon regulatory factor 4/cyclin-dependent kinase 6, respectively; by inhibiting CBL, ubiquitination of IRF4 and thus inhibition of CDK6) [25]. Furthermore, miR-155 seems to promote ALL cell proliferation by targeting ZBTB18 (also known as ZNF238; encoding zinc finger and BTB domain-containing 18 protein), whereas anti-miR-155 treatment seems to prevent CD154-mediated repression (i.e., CD40 ligand) of BCL2 and to reduce proliferation in leukemic cells in vitro [27,110]. Extremely high expression of miR-155 has been associated with poor prognosis, whereas in vitro experiments have shown that anti-miR-155 administration seems to be regulatory for ALL cell viability, proliferation, cell cycle and apoptosis; this antileukemic effect has been reinforced by concomitant prednisolone treatment [44,154]. High miR-155 levels were correlated with KMT2A-r cases, although miR-155 expression is not essential for leukemia development, so treatment with antagomirs in the respective cases would probably fail [155].
3.3.3. miR-181 Family
Significant upregulation of miR-181 family members has been associated with ALL cases compared with healthy children [10,27,28,35,46,58,69,74,76,83,84,86]. This miRNA family consists of six members: miR-181a-1 clustered with miR-181b-1 in chromosome 1, miR-181a-2 clustered with miR-181b-2 in chromosome 9 and miR-181c clustered with miR-181d in chromosome 19 [8].
Significantly reduced levels of miR-181a-1, miR-181c and miR-181d in ETV6-RUNX1-positive cases compared with other pediatric BCP-ALL cases can be explained by the fact that this fusion gene and MIR181A1 regulate each other, whereas a double-negative loop involving these two genes seems to contribute to ETV6-RUNX1-driven arrest of differentiation in pre-B ALL [153]. In vitro studies in ALL cell lines have demonstrated that treatment with miR-181a induces a significant antileukemic effect against the ETV6-RUNX1 fusion gene [156]. Besides its role as an oncomir, miR-181a also seems to be involved in GC response. Anti-miR-181a treatment of ALL cells has achieved favorable results with respect to proliferation and apoptosis rates, whereas this effect was augmented by prednisolone administration [154]. Expression levels of this miRNA family are associated with pediatric KMT2A-r cases (specifically with miR-181b), whereas specimens with deregulated TLX1 seem to be the only case of significantly low miR-181d at diagnosis compared to healthy individuals [76,79]. Inhibition of miR-181a expression in ALL cells revealed an influence on CD4 and HMGB1 (high-mobility group box 1 protein) levels, indicating a possible role of miR-181a in immunogenicity [157]. Expression levels of miR-181a and miR-181b have been negatively correlated with target genes involved in immune response (IL1RN, BCL6, CCR1, PTAFR, FN1, CCL2, TLR5, TLR8, LTB4R, IL6R, and TNFSF1 or LTA encoding lymphotoxin alpha), implying their role in escaping immune surveillance [158]. The interplay between miR-181b-1 and NF-κB was recently described as an amplifying loop linking inflammation to cancer [159]. STAT3 is not only a downstream target of IL-6, but, along with miR-181b-1, miR-21, PTEN and CYLD, lysine 63 deubiquitinase is part of the positive feedback loop regulating the epigenetic switch linking inflammation to cancer [160]. In addition, miR-181a overexpression in T-ALL cells seems to downregulate EGR1 expression (early growth response 1 zinc finger transcription factor), thereby increasing cell proliferation and enhancing cell cycle progression from the G1 to S phase [161]. Bioinformatics analysis in B-ALL samples revealed that miR-181c regulates lncRNA XIST (X inactive-specific transcript) and that miR-181a-2 and miR-181b-2 are regulated by transcription factors including CDX2 (caudal-type homeobox 2) [162]. miR-181 family levels have been associated with OS (overall survival) in ALL [163]. In the same context, miR-181b and miR-708 levels seem to be significantly higher in high-risk common-ALL compared with standard- and intermediate-risk common-ALL [74].
3.3.4. miR-708
Significant upregulation of miR-708 has been documented in children with ALL compared with controls [27,28,66,74,83]. Despite the latter fact, BCR-ABL1-positive and KMT2A-r pediatric cases seem to have significantly lower miR-708 levels compared with other pediatric ALL cases [29,81,83,164]. Most importantly, lower miR-708 levels seem to efficiently discriminate pediatric T-ALL from B-ALL cases more accurately than any other miRNA [29,66,81,118,131,164,165]. Low expression of miR-708 has been also associated with PAX5 deletion, whereas high miR-708 expression has been correlated with ETV6 deletion [131]. As mentioned above, significantly higher miR-708 values have been found in children with high-risk common-ALL, whereas miR-708 seems to target the 3΄-UTR of CNTFR, NNAT and GNG12 (encoding ciliary neurotrophic factor receptor, neuronatin and G protein subunit gamma 12, respectively) [74]. Beyond identification of ALL cases, miR-708 levels can be utilized in disease risk stratification and prediction of GC response [112,131,164].
3.3.5. Other Upregulated miRNAs
Significant elevation of miR-130b levels has been documented in children with ALL compared with controls [10,28,84,86]. This miRNA gene resides in chromosome 22 and is clustered with miR-301b, whereas their miRNA family comprises two more clustered miRNAs, miR-130a and miR-301a [8]. Complementary to these reports, it is also known that pediatric ETV6-RUNX1- and BCR-ABL1-positive cases have significantly lower levels of miR-130b compared with other BCP-ALL cases [100,153]. Apparently, downregulation of miR-130a and miR-130b in BCR-ABL1 kinase presence is chaperoned by a decrease in CCN3 (cellular communication network factor 3), a negative growth regulator [166].
Upregulation of miR-222 seems to be another trait of pediatric ALL cases when compared with healthy children [36,46,69,77,85]. This miRNA is clustered with miR-221 in chromosome X [8]. Both of these miRNAs seem to target cyclin-dependent kinase inhibitors CDKN1B and CDKN1C (also known as p27 and p57, respectively), and when growth factors are scarce, they lead to quiescence bypass and induce precocious S-phase entry [167]. PTEN is a target of miR-221 and miR-222, and their levels can affect radiation sensitivity via the PTEN/AKT pathway [168]. Similarly, treatment with anti-miR-222 inhibitor has resulted an in vitro decrease in cell proliferation, migration, tube formation and induced apoptosis by upregulation of ARID1A (AT-Rich Interaction Domain 1A) and PTEN [169]. Downregulation of the ETS1 proto-oncogene is a hallmark of ETP-ALL (early T-cell precursor ALL) and is associated with the miR-221~222 cluster. Significant elevation of this cluster seems to efficiently discriminate ETP-ALL from non-ETP T-ALL cases, whereas in vitro studies have shown that miR-222 seems to regulate proliferation, cell cycle and apoptosis in leukemia cells [170]. Another upregulated miRNA in pediatric ALL is miR-210. Significantly elevated expression of miR-210 compared with healthy children has been documented in many studies [10,27,38,74,86]. Among pediatric BCP-ALL cases, the lowest values of miR-210 are found in BCR-ABL1-positive cases [29]. Low miR-210 expression has been associated with poor prognosis and resistance to treatment [171]. Interestingly, an hypoxic environment leads to excessive miR-210 upregulation; therefore, it is considered the miRNA of hypoxia. Other roles of miR-210 include (i) immune response: negative feedback regulator in lipopolysaccharide/toll-like receptor 4 (LPS/TLR4) signaling; (ii) transformation, leukemogenesis and chemotactic invasion: positive feedback loop with NF-κB, IL-6 and STAT3; (ii) DNA repair of double-stranded breaks in cases of hypoxia-induced genomic instability by targeting RAD52; and (iv) activation of the PI3K/AKT pathway by targeting SHIP1 (also known as INPP5 inositol polyphosphate-5-phosphatase D) has been correlated with MDS (myelodysplastic syndrome) development [172].
Significant elevation of miR-363 levels seems to be a unique trait of children with ALL when compared with controls [10,27,69,84,86]. This miRNA is clustered with miR-18b, miR-19b-2, miR-20b, miR-92a-2 and miR-106a in chromosome X [8]. Although studies regarding miR-363 levels in pediatric ALL are lacking, network analysis was recently used to decipher the interplay between this miRNA and NOTCH1 as a pathogenetic marker for cancer development [173]. Another miRNA originating from chromosome X, miR-506, has been associated with childhood ALL occurrence. This miRNA is clustered with miR-507, miR-508 and miR-513a-2 and belongs to the miR-506~514b family, which comprises 18 members [8]. A recent large case–control study reported significantly higher expression levels of serum miR-506 and miR-922 in children with ALL than in controls [31]. However, this finding might be explained in part by the extremely high levels of both of these miRNAs in pediatric ETV6-RUNX1-positive and KMT2A-r cases, corresponding to one-fifth of total pediatric ALL cases [31,174].
Circulating levels of miR-21 seem to be a cancer biomarker according to a relevant meta-analysis. In particular, individuals with high levels of miR-21 in plasma or serum have a 17-fold increased chance of being diagnosed with cancer [175]. Significantly elevated expression of miR-21 in children with ALL has been reported in four studies [30,36,46,60], whereas two studies have reported otherwise [56,68]. Known targets of miR-21 include SERPINB5 (also known as maspin) and PTEN, along with tumor suppressors TPM1 (tropomyosin 1) and PDCD4 (programmed cell death 4 protein) [175]. Moreover, miR-21 downregulation seems to decrease cellular viability and proliferation, whereas targeting of miR-21 results in autophagy induction and sensitivity of leukemic cells to etoposide and doxorubicin [176]. Inhibition of miR-21 has been also been associated with enhancement of daunorubicin toxicity against leukemia cells mediated by PTEN upregulation and the subsequent suppression of the PI3K/AKT pathway [177,178]. With respect to T-ALL development, two pathogenetic links have been described to date: (a) the signaling axis involving miR-21 and PDCD4 in Notch-mediated induction of T-ALL and (b) miR-21-mediated STAT3 inhibition, prompting increased proliferation and invasion and diminishing the apoptosis rate [179,180]. Recently, a study on children with B-ALL correlated upregulation of miR-21 with poor response to induction therapy, as well as shorter DFS (disease-free survival) and OS (overall survival) [60].
Two other important miRNAs in childhood ALL are miR-145 and miR-146a, both originating from chromosome 5. Significant higher levels of miR-145 in children with ALL compared with healthy individuals have been reported three times in the literature [27,28,36]. On the contrary, one study reported low levels of miR-145 in a pediatric ALL population [86]. Significant decreases in miR-145 have also been reported in children with T-ALL compared to controls [10,28]. As mentioned above, cancer development is associated with eradication of the miR-143~145 cluster [144]. A mouse model demonstrated that loss of miR-145 results in leukemia, whereas another study has positively correlated miR-145 levels with the expression of proapoptotic genes PRKAA1, PTEN, FOXO3 (encoding Forkhead Box O3 protein), BCL2L11 (aka BIM), DIDO1 (encoding death inducer-obliterator 1) and BCL2L13 [158,181]. Cell cycle, proliferation, apoptosis and invasion are among the cellular processes that miR-145, as a potent tumor suppressor, regulates, whereas p53 has been shown to induce miR-145 levels, which, in turn, inhibits MYC and MUC1 (mucin 1 cell surface-associated protein) [182,183]. The complex interplay between leukemic cells and the microenvironment (along with methodological discrepancies) may explain the contradictory results with respect to the miR-143~145 cluster in childhood ALL [184]. Published data regarding miR-146a are far more transparent [27,28,33,36,46,48,65,69,77,86]. Upregulation of miR-146a seems to be ubiquitous across childhood ALL, with the exception of the TCF3-r BCP-ALL subtype [29,100]. Two studies that have reported otherwise are disputable (very small sample size and not significant results, respectively) [40,47]. As analyzed in the corresponding section, miR-146a expression levels may vary depending upon polymorphisms [47,89]. This miRNA seems to promote cell proliferation, migration and invasion, in addition to inhibiting cell apoptosis by downregulating its target, CNTFR (ciliary neurotrophic factor receptor), whereas low CNTFR expression leads to LIF inhibition (LIF interleukin 6 family cytokine) and JAK2/STAT3 pathway activation [48]. High expression of miR-146a is most prevalent among biphenotypic ALL patients, whereas some evidence associates this miRNA with WBC at diagnosis [65]. Furthermore, miR-146a plasma levels in children with poor-prognosis ALL seem to be significantly higher than in children with favorably prognostic characteristics; this finding may be attributed to the fact that miR-146a interferes with prednisolone efficacy [121,154]. Besides its major role in T-ALL, miR-146a seems to be implicated in B-cell maturation through miR-the 146a/NFKB1/BCL11A pathway and in megakaryopoiesis regulated by the ZBTB16/miR-146a/CXCR4 axis (ZBTB16 or zinc finger and BTB domain-containing 16 protein alias PLZF of ZNF145; C-X-C motif chemokine receptor 4 alias CD184 or fusin) [185,186]. This miRNA has been also used to discriminate ALL from AML cases, but in both cases, miR-146a seems to confer poor OS [163].
3.4. miRNAs and ALL Subtypes
We investigated the literature for differentially expressed miRNAs across childhood ALL subtypes and found considerable heterogeneity among studies, even in terms of defining subgroups within ALL. A review of published data (Supplementary Table S1) revealed the involvement of several miRNAs in T-ALL development (reported by ≥2 studies), i.e., childhood T-ALL versus controls has been associated with upregulation of miR-16, miR-19b, miR-92a, miR-130b, miR-146a, miR-181a, miR-181b and miR-221 and with downregulation of miR-29a, miR-145 and miR-574.
To verify the importance of specific miRNAs in leukemogenesis, we reanalyzed the only available small RNA-seq dataset of pediatric T-ALL cases compared with controls (Supplementary Table S2) [10]: hsa-miR-16-5p (rank #57; p = 0.0012), hsa-miR-19b-3p (#90; p = 0.011), hsa-miR-92a-2-5p (#48; p < 0.001), hsa-miR-130b-3p and -5p (#14 and #127; p < 0.001 and p = 0.04, respectively), hsa-miR-181a-5p, -2-3p and -3p (#15, #18 and #29; p < 0.001), hsa-miR-181b-5p and -3p (#2 and #25; p < 0.001), hsa-miR-145-5p (#20; p < 0.001) and hsa-miR-574-3p (#9; p < 0.001). Adjusted p values for hsa-miR-221-3p and -5p, hsa-miR-29a-3p and hsa-miR-146a-5p did not reach the significance level. Interestingly, analysis of effect size estimates highlighted the importance of differentially expressed hsa-miR-574-3p, hsa-miR-618, hsa-miR-504-5p, hsa-miR-4488, hsa-miR-6500-3p, hsa-miR-153-3p, hsa-miR-548a-3p, hsa-miR-2682-5p, hsa-miR-466, hsa-miR-4492, hsa-miR-106a-3p, hsa-miR-3196, hsa-miR-135a-5p and hsa-miR-1281 (log2 fold change > 5) in children with T-ALL compared with controls. hsa-miR-128-3p ranked first among differentially expressed miRNAs (lowest adjusted p value).
Downregulation of miR-16 has been long known to be a main event in CLL (chronic lymphocytic leukemia), but its upregulation seems to play a pivotal role in regulating BCL2 (by binding the 3′-UTR region of BCL2) and in mediating prednisolone-induced apoptosis in childhood T-ALL [79,187,188,189,190]. In addition, miR-16-1 levels have been proposed as a prognostic biomarker for OS, although discrepancies do exist [187,190]. Some BCL2 inhibitors (venetoclax or ABT-199, navitoclax or ABT-263 and ABT-737) have achieved favorable outcomes in specific pediatric ALL groups, especially in ETP-ALL, hypodiploid B-ALL and BCR-ABL1-positive cases; this confirms a major role for BCL2 and the miR-16 precursor family (miR-15a, miR-15b, miR-16-1, miR-16-2, miR-195 and miR-497) in leukemogenesis and apoptosis [191,192].
The miR-17~92 cluster in chromosome 13, a polycistronic family comprising miR-17, miR-18a, miR-19a, miR-19b-1, miR-20a and miR-92a-1, is implicated in many hematological cancers. In specific, miR-19 has been proven sufficient to promote leukemogenesis in Notch1-induced T-ALL in vivo and to influence cell cycle progression by targeting cyclin-dependent kinase inhibitor CDKN1A (p21), whereas inhibition of miR-19b seems to lead Jurkat cells to increased apoptosis [193,194]. Interestingly, miR-19 targets BIM (BCL2-like 11 protein) encoded by BCL2L11, PRKAA1 (encoding protein kinase AMP-activated catalytic subunit alpha 1), PTEN (encoding phosphatase and tensin homolog) and PPP2R5E (encoding protein phosphatase 2 regulatory subunit B’epsilon), whereas miR-92 regulates FBX7 (encoding F-box protein 7), BCL2L11, IKZF1 (encoding IKAROS family zinc finger 1 protein), PTEN and NF1 (encoding Neurofibromin 1), indicating that regulators of PI3Ks are crucial for childhood T-ALL [79,193]. Moreover, a recurrent gain of 13q31.2-q31.2 in the aforementioned cluster’s region has been noted as a common finding (14%) in BM samples of untreated pediatric patients with T-ALL, resulting in increased miR-19 expression levels [194]. Oncomir properties of miR-19b and miR-92a in childhood T-ALL have been substantiated by case–control studies [20,79].
Upregulation of 130b has been implicated in T-ALL occurrence, especially in TAL-r T-ALL pediatric cases [10,28]. This miRNA has been associated with drug resistance, and it is an essential driver of KMT2A-AFF1 leukemia, integrated with the aberrant overexpression of key downstream targets, i.e., the antiapoptotic factor BCL2 and proto-oncogene MYC [195,196]. The role of miR-146a in childhood ALL is discussed above in the text.
Overexpression of clustered miR-181a and miR-181b seems to be universal for childhood ALL, except for ETV6-RUNX1-positive cases [10,28,153]. miR-181a has been implicated in T-ALL development and has also exhibited regulatory effects towards the development and differentiation of B-cells and cytotoxic T-cells by diminishing the expression of genes involved in thymocyte maturation (such as BCL2, CD69 and those of TCR) [28]. Silencing of exosomal miR-181a reverses childhood ALL cell proliferation in vitro by upregulating proliferative (PCNA-encoding proliferating cell nuclear antigen and MKI67-encoding marker of proliferation Ki-67) and pro-survival genes (MCL1 apoptosis regulator and BCL2) and by suppressing proapoptotic genes (BAD of BCL2-associated agonist of cell death and BAX of BCL2-associated X apoptosis regulator) [197]. With respect to miR-181b, its upregulation in childhood BCP-ALL specimens has been associated with methylation of the promoter regions and subsequent downregulation of tumor suppressor genes PPP1CA, BTG2 and PTEN, and these effects have been restored with decitabine treatment in vitro [108]. Among others, ALL-related targets of miR-181a and miR-181b include ERK/MAPK, mTOR and NF-κB signaling pathways, whereas the expression of this cluster seems to be abundant in the early B-cell stage and to decline progressively with maturation [198].
Significant overexpression of miR-221 was associated with T-ALL diagnosis in two case–control studies [33,73]. High levels of both miR-221 and miR-146a seem to accurately discriminate T-ALL pediatric cases from healthy controls [33]. This miRNA is embroiled in hematopoietic stem cell differentiation and DNA damage response, and it is known to downregulate CDKN1B [148]. An in vitro study reported that miR-221 may exert inhibitory effects towards bone marrow mononuclear cells and their abilities to proliferate, migrate and invade in childhood ALL cases, all by inhibiting the Wnt/β-catenin signaling pathway, as demonstrated by reduced PCNA (proliferating cell nuclear antigen), cyclin D1 and MMP9 (matrix metallopeptidase 9). In the context of previous reports, miR-221 presence has been linked with a significant increase in cells in the G0/G1 phase and an increase in apoptotic cells accompanied by increased caspase 3 levels [199]. Overexpression of miR-221 concerns solely T-ALL (and AML), whereas there are indications that it is downregulated in B-ALL (especially in ETV6-RUNX1 and KMT2A-r cases) [29,69,100,148,153,200]. In addition, children with T-ALL/CD56+ seem to have significantly higher miR-221 levels compared with T-ALL/CD56- patients (271.4-fold; CD56 or NCAM1 i.e., neural cell adhesion molecule 1 expression confers poor prognosis in T-ALL), and pediatric immature T-ALL cases have significantly higher miR-221 levels than all other T-ALL subgroups [20,73].
Low expression of miR-29a in pediatric T-ALL has been documented in two studies [71,79]. In general, miR-29a acts as a tumor suppressor and affects the methylation status of certain target genes, thus contributing to T-ALL pathogenesis [71]. This miRNA, together with miR-7 and miR-195,s seem to play a crucial role in pediatric T-ALL, invariably downregulated in most T-ALL subgroups: TLX1-, TLX3- and TAL1-deregulated, PICALM-MLLT10 and inv(7) cases [79]. Expression of miR-29a among pediatric B-ALL cases varies significantly (high levels in BCR-ABL1 cases and low levels in TCF3-r and KMT2A-r cases) [29,62,69,79,100]. Leukemia patients with TP53 abnormalities display deregulated miR-29 family expression profiles, and downregulation of miR-29c has been associated with upregulation of MCL1 and TCL1 (encoding TCL1 family AKT coactivator A) proto-oncogenes [201]. Significantly lower levels of both miR-145 and miR-574 have been reported in children with T-ALL compared to controls in two independent studies [10,28]. Low miR-574 expression seems to efficiently discriminate childhood T-ALL from B-ALL patients, whereas TLX1-deregulated cases seem to be excluded from this pattern [79,165]. Notably, significantly higher levels of miR-574 have been found in BCP-ALL, KMT2A-r and TCF3-PBX1 subtypes in comparison with other B-ALL cases [202].
Investigation of differentially expressed miRNAs in T-ALL subgroups (compared with controls, ALL or T-ALL samples) revealed a prevalent miRNA signature across pediatric T-ALL subgroups: upregulation of miR-146a, miR-182, miR-196b, miR-223, miR-376a and miR-662,and downregulation of miR-7, miR-29a, miR-99a, miR-195, miR-296 and miR-422a [20,48,79,106,116]. The aforementioned signature of T-ALL subgroups is in agreement with a literature review regarding T-ALL case–control studies in the context of miR-146a upregulation and miR-29a downregulation. However, reanalysis of the small RNA-seq dataset failed to confirm the significant differential expression of miR-146a and miR-29a. Further large cohort studies are needed to confirm our results.
Further analysis of miRNA signatures in other childhood ALL subgroups is beyond the scope of this review and is covered in Supplementary Table S1. A recent meta-analysis of differentially expressed miRNAs in B-ALL compared with controls across 25 studies (including both adult and pediatric samples) revealed no consensus miRNA signature. Eight miRNAs in the latter meta-analysis showed promising insight with respect to diagnosing B-ALL: upregulation of miR-128-1, miR-128-2, miR-142, miR-155, miR-181a, miR-181b and miR-181c and downregulation of miR-451a [198]. Our review of miRNA levels in pediatric B-ALL compared with controls revealed that miR-155 is the most frequently reported upregulated miRNA (followed by miR-146a, miR-181b, miR-222 and miR-708; two citations for miR-16, miR-21, miR-34a, miR-100, miR-128-1, miR-181a, miR-181c, miR-195, miR-210, miR-320a and miR-660), whereas the most frequently reported downregulated miRNA is miR-374a (two citations for miR-27a, miR-30c, miR-196b, miR-223 and miR-494). With respect to miR-374a, it is clustered with miR-545 at the X-chromosome inactivation center and seems to regulate cell growth and differentiation, although there are inconsistent reports in literature [203].
MicroRNAs have also been employed to discriminate pediatric T-ALL from B-ALL cases, and differential expression analysis designated miR-196b as the top upregulated miRNA (followed by miR-542) and miR-708 as the top downregulated miRNA (followed by miR-151a, miR-425 and miR-497; Supplementary Table S1).
4. Discussion
Most of case–control studies that have been carried out regarding childhood ALL have traits that limit reproducibility and increase bias. The main flaws in methodology that have been determined can be categorized in seven tiers:
[i] Sample issues: Sixteen publications in this systematic review had considerably small sample sizes (30 or less; 26%) [36,40,41,46,48,49,58,64,68,69,77,78,80,82,84,86]. Larger sample sizes are required to achieve sufficient statistical power and allow for solid conclusions to be drawn. Another problem is the small number of healthy participants in case–control studies; 76% of included studies had 30 or fewer controls (Table 1). The proportion between the number of children with ALL and controls in also of interest and may affect measured expression levels and outcomes. In seven studies, participating healthy controls accounted for 10% or less of children with ALL (2.2%, lowest ratio) [35,53,54,66,75,76,83]. In one case, controls were adults and not age-matched [69], whereas in four studies, preserved cells or cell subsets obtained by donors of unknown age and health status were used [66,78,79,82]. One qRT-PCR from the Netherlands utilized CD34+ progenitor cells from two children with brain tumor as controls, and one another study used cancer-free children with fractures as controls, although it is unclear whether these practices could interfere with measured outcomes [59,83]. It is common sense and in line with reports that associate miRNA levels with tumorigenesis and inflammation that only samples from completely healthy children (age-matched if applicable) should be obtained as controls. Another issue regarding samples that might influence miRNA expression measurements is the proportion of each ALL subtype within each sample. Figure 4 represents our current knowledge on representation of each subtype within childhood ALL [1,2,3,4,5]. For example, it is erroneous to include more KMT2A-r cases of T-ALL in the same ALL sample; instead, separate calculations should be employed. Weighted samples in terms of cytogenetics and subtype classification should be always considered, excluding cases investigating specific subtypes alone [67,72,83]. Previous research in the field has shown that population characteristics (ethnicity, race, etc.) may also play a role in childhood ALL susceptibility, especially when studying the effects of specific SNPs on the disease.
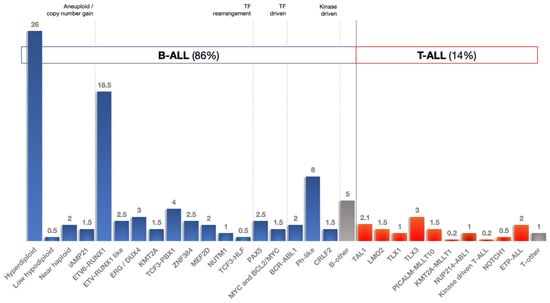
Figure 4.
Childhood ALL subtypes.
[ii] Origin of samples: Expression of miRNAs has been demonstrated to be tissue-specific. Heterogeneity of results could be explained to some extent by measurements in different specimens: BM, PBMCs, serum or plasma. Simultaneous study of miRNA expression patterns in both BM and peripheral blood has resulted in contradictory results; one study reported no significant difference, whereas two other studies reported significant fluctuations [28,46,68]. Accordingly, a qRT-PCR study on adults with ALL reported significantly lower levels of oncomiR miR-92a in fresh leukemia cells compared with CD34+ cells obtained from healthy volunteers (p = 0.0014), significantly lower expression of miR-92a in ALL subjects compared to controls (p = 0.001) and a significantly higher cell-to-plasma ratio of miR-92a in ALL cells (p < 0.0001) compared with those in peripheral blood specimens obtained from healthy individuals, indicating that leukemia cells retain miR-92a [204]. Circulating miRNAs in serum and plasma show significant stability, rendering them reliable biomarkers for discrimination of childhood ALL cases from healthy children [24].
[iii] Time point in therapy: A sample size comprising children with ALL in different stages of therapy is not appropriate and may tamper miRNA expression profiles. In terms of prognosis and drug sensitivity, study of matched samples before and after therapy is strongly encouraged. Newly diagnosed children with ALL, as well as relapsed and remitted cases, were detected together in five case–control studies included in the present review [32,41,43,46,54].
[iv] Handling of outliers: With respect to children from distinct cytogenetic groups, it is essential to keep the respective proportions within the ALL group or study subgroups individually, as stated above. Another problem is cases with high leukemic blast burden; therefore, predetermined cutoff values are recommended, and enrichment should be applied in cases with inadequate blasts [24].
[v] Methods and validation: The most commonly used method to determine expression levels of miRNAs is qRT-PCR followed by microarrays (Table 1). Nevertheless, RNA-seq and other NGS (next-generation sequencing) methods with revolutionize miRNA detection and quantification in the coming years. Substantial technical requirements, extensive amplification, time and cost are the main disadvantages of these sequencing methods, but identification of isomiRs and novel miRNAs are in the armamentarium of RNA-seq. Poor primer and probe design, along with selection of inappropriate normalization controls, is among the most common flaws in qRT-PCR. Validation of NGS results with RT-PCR is considered a gold standard [205].
[vi] Reproducibility: Data of future studies must be transparent and reproducible in subsequent studies. The data of most large-scale studies included in this review are not deposited in GEO (Gene Expression Omnibus) or similar platforms.
Various miRNA SNPs seem to be associated with predisposition to childhood ALL in the literature. The systematic review of the literature and meta-analysis performed in the present study on all relevant data revealed significant protective roles of SNPs in pri-miR-34b/c and miR-100, whereas predisposition to B-ALL was associated with SNPs in DROSHA, miR-449b, miR-938, miR-3117 and miR-3689d-2 genes (Figure 2). Variant allele rs1573613 T>C of ETV6 seems to weaken the binding of miR-34c-5p and miR-449b-5p (resulting in 17% and 33% increase protein levels relative to the T allele, respectively), whereas these interactions double the risk for childhood ALL (OR, 1.9; 95% CI: 1.16 to 3.11; p < 0.05) [206]. The expression of miR-34b/c in childhood ALL is inhibited by methylation of its promoter, which in turn impairs the restraining effects of miR-34b on cell proliferation and response to prednisone [63,120]. Moreover, miR-34b/c seems to be a tumor suppressor (part of the p53 network), and the C allele of rs4938723 in pri-miR-34b/c confers increased transcriptional activity of the miR-34b/c promoter [98]. The AA genotype of rs12402181 in miR-3117-3p affects the seed region of miR-3117-3p, whereas the CT/CC genotype in rs62571442 of miR-3689d-2 leads to changes in the respective pre-miRNA. Both of these SNPs lead to energy changes, alterations in secondary miRNA structure and aberrant activation of the MAPK/ERK pathway [94]. Although studies are lacking, miR-3117 seems to target CD274 (alias PD-L1 or programmed cell death 1 ligand 1 gene), and miR-3689d-2 targets CAPN6 (encoding calpain 6), which is implicated in the NF-κB and B-cell receptor signaling pathways [8,207]. The role of miR-100 in childhood ALL was elaborated earlier in the text.
Evaluation of published data on differential expression of miRNAs in children with ALL compared with controls generated some interesting conclusions: (i) there are substantial differences in study designs of published material, making it impossible to conduct an unbiased meta-analysis; (ii) upregulation of the miR-128 family, miR-130 family, miR-155, miR-181 family, miR-210, miR-222, miR-363 and miR-708, along with downregulation of miR-143 and miR-148a, seems to play a definite role in childhood ALL development; (iii) other important miRNAs involved in ALL pathogenesis in children include differentially expressed let-7e, let-7f, miR-24, miR-99a, miR-100, miR-125b, miR-145, miR-146a, miR-182, miR-196b, miR-199b, miR-223 and miR-335; and (iv) significant upregulation of miR-130b, miR-181a and miR-181b and downregulation of miR-145 and miR-574 were confirmed by our re-analysis as putative biomarkers of T-ALL presence in childhood.
The performed reanalysis of small RNA-seq data derived from pediatric T-ALL cases and controls revealed a top-10 signature that could be utilized for diagnostic purposes in the future: (i & ii) hsa-miR-128-3p and hsa-miR-181b-5p are major regulators of early lymphoid differentiation that prevent stem-progenitor cells from maturation and contribute to escape from immune surveillance [46,158,159]; (iii) hsa-miR-130a-3p induce modulation of cell survival programs by regulating autophagic flux and inhibition of DICER1, a major component of RNA-induced silencing complex (RISC) [208]; (iv & vi) hsa-miR-106a-5p and hsa-miR-20b-5p, clustered together, are members of the miR-17 family and major drivers of tumorigenesis in various cancers [144]; (v) hsa-miR-30a-5p, the downregulation of which results in apoptosis inhibition, promotes proliferation and stimulates migration and invasion through PI3K/AKT/mTOR and miR-30a/NOTCH1/MYC pathways [209,210]; (vii) hsa-miR-24-3p, the downregulation of which has been associated with PAX5 deletion, whereas its deregulated levels interfere with the expression of both MYC, BCL2 and HIF1A (hypoxia-inducible factor 1 subunit alpha) oncogenes and p21 and p53 tumor suppressor proteins [131,211]; (viii) hsa-miR-143-3p induced the deletion of the miR-143~145 cluster has been designated as a hallmark of tumorigenesis in various cancers [144]; (ix) has-miR-574-3p, the reduced expression of which seems to promote proliferation and inhibit apoptosis in leukemic cells by targeting the IL6/JAK/STAT3 pathway [212]; (x) hsa-miR-618, the downregulation of which has been previously reported in childhood ALL, in addition to being implicated in lymphoma pathogenesis via p53, STAT3, HDAC3 (histone deacetylase 3), CUL4A (cullin 4A) and FKBP3 [10,27,213].
Recently, ALLSorts, a method that utilizes RNA-seq data to classify pediatric B-ALL samples into 18 known subtypes and 5 meta-subtypes, was made publicly available [214]. Subtype-specific miRNA signatures bear resemblance with those previously published in the literature (Supplementary Table S1; Figure 4). RNAseqCNV is another tool that can detect large-scale CNVs (copy number variations) from RNA-seq data. This method is consistent with DNA-based techniques and more effective than conventional cytogenetic studies in determining ALL subtypes [215].
5. Conclusions
In conclusion, many miRNA case–control studies in the field of childhood ALL suffer from high risk of bias and limitations of evidence, including inconsistencies in methodology and study designs that preclude reproducibility and proper meta-analysis. Larger cohort studies and utilization of novel NGS tools will revolutionize childhood ALL diagnosis and subtype classification henceforth.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers14163976/s1, Supplementary Table S1: Upregulated and downregulated miRNAs in childhood ALL subtypes; Supplementary Table S2: Results of reanalysis of small RNA-seq data of pediatric T-ALL cases compared with controls. References [216,217,218,219,220,221,222,223] are cited in Supplementary Materials.
Author Contributions
Conceptualization, I.K. and A.T.; methodology, I.K., K.K. and A.T.; software, K.K. and A.T.; validation, I.K. and K.K.; formal analysis, I.K. and K.K.; investigation, I.K. and K.K.; resources, K.K. and A.T.; data curation, I.K. and K.K.; writing—original draft preparation, I.K. and K.K.; writing—review and editing, A.T.; visualization, I.K.; supervision, A.T.; project administration, A.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hunger, S.P.; Mullighan, C.G. Redefining ALL classification: Toward detecting high-risk ALL and implementing precision medicine. Blood 2015, 125, 3977–3987. [Google Scholar] [CrossRef] [PubMed]
- Qian, M.; Zhang, H.; Kham, S.K.Y.; Liu, S.; Jiang, C.; Zhao, X.; Lu, Y.; Goodings, C.; Lin, T.N.; Zhang, R.; et al. Whole-transcriptome sequencing identifies a distinct subtype of acute lymphoblastic leukemia with predominant genomic abnormalities of EP300 and CREBBP. Genome Res. 2017, 27, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Li, J.F.; Dai, Y.T.; Lilljebjörn, H.; Shen, S.H.; Cui, B.W.; Bai, L.; Liu, Y.F.; Qian, M.X.; Kubota, Y.; Kiyoi, H.; et al. Transcriptional landscape of B cell precursor acute lymphoblastic leukemia based on an international study of 1223 cases. Proc. Natl. Acad. Sci. USA 2018, 115, E11711–E11720. [Google Scholar] [CrossRef] [PubMed]
- Roberts, K.G. Genetics and prognosis of ALL in children vs. adults. Hematology 2018, 2018, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Malard, F.; Mohty, M. Acute lymphoblastic leukaemia. Lancet 2020, 395, 1146–1162. [Google Scholar] [CrossRef]
- Kyriakidis, I.; Vasileiou, E.; Rossig, C.; Roilides, E.; Groll, A.H.; Tragiannidis, A. Invasive Fungal Diseases in Children with Hematological Malignancies Treated with Therapies That Target Cell Surface Antigens: Monoclonal Antibodies, Immune Checkpoint Inhibitors and CAR T-Cell Therapies. J. Fungi 2021, 7, 186. [Google Scholar] [CrossRef]
- Morozova, N.; Zinovyev, A.; Nonne, N.; Pritchard, L.L.; Gorban, A.N.; Harel-Bellan, A. Kinetic signatures of microRNA modes of action. RNA 2012, 18, 1635–1655. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. MiRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Rodriguez, P.D.; Paculova, H.; Kogut, S.; Heath, J.; Schjerven, H.; Frietze, S. Non-coding rna signatures of b-cell acute lymphoblastic leukemia. Int. J. Mol. Sci. 2021, 22, 2683. [Google Scholar] [CrossRef]
- Dawidowska, M.; Jaksik, R.; Drobna, M.; Szarzyńska-Zawadzka, B.; Kosmalska, M.; Sędek, Ł.; Machowska, L.; Lalik, A.; Lejman, M.; Ussowicz, M.; et al. Comprehensive Investigation of miRNome Identifies Novel Candidate miRNA-mRNA Interactions Implicated in T-Cell Acute Lymphoblastic Leukemia. Neoplasia 2019, 21, 294–310. [Google Scholar] [CrossRef]
- Olson, N.D.; Wagner, J.; McDaniel, J.; Stephens, S.H.; Westreich, S.T.; Prasanna, A.G.; Johanson, E.; Boja, E.; Maier, E.J.; Serang, O.; et al. PrecisionFDA Truth Challenge V2: Calling variants from short and long reads in difficult-to-map regions. Cell Genom. 2022, 2, 100129. [Google Scholar] [CrossRef]
- Chapman, B.; Kirchner, R.; Pantano, L.; Naumenko, S.; De Smet, M.; Beltrame, L.; Khotiainsteva, T.; Sytchev, I.; Guimera, R.V.; Kern, J.; et al. Bcbio/Bcbio-Nextgen. Available online: https://doi.org/10.5281/ZENODO.5781867 (accessed on 16 August 2022).
- Tsuji, J.; Weng, Z. DNApi: A De Novo Adapter Prediction Algorithm for Small RNA Sequencing Data. PLoS ONE 2016, 11, e0164228. [Google Scholar] [CrossRef] [PubMed]
- Didion, J.P.; Martin, M.; Collins, F.S. Atropos: Specific, sensitive, and speedy trimming of sequencing reads. PeerJ 2017, 2017, e3720. [Google Scholar] [CrossRef]
- Pantano, L.; Estivill, X.; Martí, E. SeqBuster, a bioinformatic tool for the processing and analysis of small RNAs datasets, reveals ubiquitous miRNA modifications in human embryonic cells. Nucleic Acids Res. 2010, 38, e34. [Google Scholar] [CrossRef] [PubMed]
- Pantano, L.; Estivill, X.; Martí, E. A non-biased framework for the annotation and classification of the non-miRNA small RNA transcriptome. Bioinformatics 2011, 27, 3202–3203. [Google Scholar] [CrossRef]
- Mangan, M.E.; Williams, J.M.; Kuhn, R.M.; Lathe, W.C. The UCSC Genome Browser: What Every Molecular Biologist Should Know. Curr. Protoc. Mol. Biol. 2014, 107, 19.9.1–19.9.28. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Wallaert, A.; Van Loocke, W.; Hernandez, L.; Taghon, T.; Speleman, F.; Van Vlierberghe, P. Comprehensive miRNA expression profiling in human T-cell acute lymphoblastic leukemia by small RNA-sequencing. Sci. Rep. 2017, 7, 7901. [Google Scholar] [CrossRef]
- Tavakoli, F.; Jaseb, K.; Far, M.A.J.; Soleimani, M.; Khodadi, E.; Saki, N. Evaluation of microRNA-146a expression in acute lymphoblastic leukemia. Front. Biol. 2016, 11, 53–58. [Google Scholar] [CrossRef]
- Miao, M.H.; Ji, X.Q.; Zhang, H.; Xu, J.; Zhu, H.; Shao, X.J. miR-590 promotes cell proliferation and invasion in T-cell acute lymphoblastic leukaemia by inhibiting RB1. Oncotarget 2016, 7, 39527–39534. [Google Scholar] [CrossRef] [PubMed]
- Mi, S.; Lu, J.; Sun, M.; Li, Z.; Zhang, H.; Neilly, M.B.; Wang, Y.; Qian, Z.; Jin, J.; Zhang, Y.; et al. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 2007, 104, 19971–19976. [Google Scholar] [CrossRef] [PubMed]
- Rashed, W.M.; Hammad, A.M.; Saad, A.M.; Shohdy, K.S. MicroRNA as a diagnostic biomarker in childhood acute lymphoblastic leukemia; systematic review, meta-analysis and recommendations. Crit. Rev. Oncol. Hematol. 2019, 136, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Guan, G.; Dai, Y.; Zhao, P.; Liu, L.; Wang, Q.; Li, X. microRNA-155-5p initiates childhood acute lymphoblastic leukemia by regulating the IRF4/CDK6/CBL axis. Lab. Investig. 2021, 102, 411–421. [Google Scholar] [CrossRef]
- Zamani, A.; Fattahi Dolatabadi, N.; Houshmand, M.; Nabavizadeh, N. miR-324-3p and miR-508-5p expression levels could serve as potential diagnostic and multidrug-resistant biomarkers in childhood acute lymphoblastic leukemia. Leuk. Res. 2021, 109, 106643. [Google Scholar] [CrossRef]
- Liang, C.; Li, Y.; Wang, L.N.; Zhang, X.L.; Luo, J.S.; Peng, C.J.; Tang, W.Y.; Huang, L.B.; Tang, Y.L.; Luo, X.Q. Up-regulated miR-155 is associated with poor prognosis in childhood acute lymphoblastic leukemia and promotes cell proliferation targeting ZNF238. Hematology 2021, 26, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Duyu, M.; Durmaz, B.; Gunduz, C.; Vergin, C.; Yilmaz Karapinar, D.; Aksoylar, S.; Kavakli, K.; Cetingul, N.; Irken, G.; Yaman, Y.; et al. Prospective evaluation of whole genome microrna expression profiling in childhood acute lymphoblastic leukemia. BioMed Res. Int. 2014, 2014, 967585. [Google Scholar] [CrossRef] [PubMed]
- Schotte, D.; de Menezes, R.X.; Moqadam, F.A.; Khankahdani, L.M.; Lange-Turenhout, E.; Chen, C.; Pieters, R.; den Boer, M.L. MicroRNA characterize genetic diversity and drug resistance in pediatric acute lymphoblastic leukemia. Haematologica 2011, 96, 703–711. [Google Scholar] [CrossRef]
- El-maadawy, E.A.; Bakry, R.M.; Moussa, M.M.; El-Naby, S.H.; Talaat, R.M. Alteration in miRNAs expression in paediatric acute lymphocyticleukaemia: Insight into patients’ therapeutic response. Clin. Exp. Pharmacol. Physiol. 2021, 48, 35–43. [Google Scholar] [CrossRef]
- Zhu, Y.-Y.; Wu, R.-N.; Li, X.; Chen, X.-B. Value of serum miR-922 and miR-506 expression levels in the diagnosis and prognostic assessment of childhood acute lymphoblastic leukemia. Chin. J. Contemp. Pediatr. 2021, 23, 1021–1026. [Google Scholar] [CrossRef]
- Fayed, D.; Donia, T.; El-Shanshory, M.; Ali, E.M.M.; Mohamed, T.M. Evaluation of MicroRNA92, MicroRNA638 in Acute Lymphoblastic Leukemia of Egyptian Children. Asian Pac. J. Cancer Prev. 2021, 22, 1567–1572. [Google Scholar] [CrossRef] [PubMed]
- Li, S.W.; Li, H.; Zhang, Z.P.; Zhuo, F.; Li, Z.X. Expression and Clinical Significance of MiR-146a and MiR-221 in Childhood Acute T Lymphoblastic Leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2020, 28, 436–441. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhao, T.; Nie, L.; Zou, Y.; Zhang, Q. MicroRNA-223 decreases cell proliferation, migration, invasion, and enhances cell apoptosis in childhood acute lymphoblastic leukemia via targeting forkhead box O 1. Biosci. Rep. 2020, 40, BSR20200485. [Google Scholar] [CrossRef] [PubMed]
- Shafik, R.E.; Wahab, N.A.; El Mokhtar, M.M.; Taweel, M.A.E.; Ebeid, E. Expression of microRNA-181a and microRNA-196b in Egyptian Pediatric acute Lymphoblastic Leukemia. Asian Pac. J. Cancer Prev. 2020, 21, 3429–3434. [Google Scholar] [CrossRef]
- Akpinar, F.; Polat, A.; Balci, Y.I.; Akca, H.; Senol, H.; Tokgun, O. Microrna expression profiles and changes with treatment on childhood leukemias. UHOD—Uluslar. Hematol. Derg. 2020, 30, 72–80. [Google Scholar] [CrossRef]
- Chen, L.; Shi, Y.; Li, J.; Yang, X.; Li, R.; Zhou, X.; Zhu, L. LncRNA CDKN2B-AS1 contributes to tumorigenesis and chemoresistance in pediatric T-cell acute lymphoblastic leukemia through miR-335-3p/TRAF5 axis. Anticancer Drugs 2020. [Google Scholar] [CrossRef]
- Hassan, N.M.; Refaat, L.A.; Ismail, G.N.; Abdellateif, M.; Fadel, S.A.; AbdelAziz, R.S. Diagnostic, prognostic and predictive values of miR-100 and miR-210 in pediatric acute lymphoblastic Leukemia. Hematology 2020, 25, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.S.; El-Khazragy, N.; Elshimy, A.A.; Aboelhussein, M.M.; Saleh, S.A.; Fadel, S.; Atia, H.A.; Matbouly, S.; Tamer, N. In vitro knock-out of miR-155 suppresses leukemic and HCV virus loads in pediatric HCV-4–associated acute lymphoid leukemia: A promising target therapy. J. Cell. Biochem. 2020, 121, 2811–2817. [Google Scholar] [CrossRef]
- Al Nakeeb, R.H.; Al- Rubaye, D. The expression of different micrornas in iraqi patients with childhood acute leukemia and their association to C/EBP-Β serum level. Iraqi J. Sci. 2020, 61, 2879–2887. [Google Scholar] [CrossRef]
- Sheybani, Z.; Rahgozar, S.; Ghodousi, E.S. The hedgehog signal transducer smoothened and microRNA-326: Pathogenesis and regulation of drug resistance in pediatric B-cell acute lymphoblastic leukemia. Cancer Manag. Res. 2019, 11, 7621–7630. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Yang, X.; Hu, S.; Kang, M.; Chen, J.; Fang, Y. A genetic variant in miR-100 is a protective factor of childhood acute lymphoblastic leukemia. Cancer Med. 2019, 8, 2553–2560. [Google Scholar] [CrossRef] [PubMed]
- Pouyanrad, S.; Rahgozar, S.; Ghodousi, E.S. Dysregulation of miR-335-3p, targeted by NEAT1 and MALAT1 long non-coding RNAs, is associated with poor prognosis in childhood acute lymphoblastic leukemia. Gene 2019, 692, 35–43. [Google Scholar] [CrossRef] [PubMed]
- El-Khazragy, N.; Noshi, M.A.; Abdel-Malak, C.; Zahran, R.F.; Swellam, M. miRNA-155 and miRNA-181a as prognostic biomarkers for pediatric acute lymphoblastic leukemia. J. Cell. Biochem. 2019, 120, 6315–6321. [Google Scholar] [CrossRef] [PubMed]
- El-Khazragy, N.; Elshimy, A.A.; Hassan, S.S.; Matbouly, S.; Safwat, G.; Zannoun, M.; Riad, R.A. Dysregulation of miR-125b predicts poor response to therapy in pediatric acute lymphoblastic leukemia. J. Cell. Biochem. 2019, 120, 7428–7438. [Google Scholar] [CrossRef] [PubMed]
- Rzepiel, A.; Kutszegi, N.; Gézsi, A.; Sági, J.C.; Egyed, B.; Péter, G.; Butz, H.; Nyíro, G.; Müller, J.; Kovács, G.T.; et al. Circulating microRNAs as minimal residual disease biomarkers in childhood acute lymphoblastic leukemia. J. Transl. Med. 2019, 17, 372. [Google Scholar] [CrossRef] [PubMed]
- Jemimah Devanandan, H.; Venkatesan, V.; Scott, J.X.; Magatha, L.S.; Durairaj Paul, S.F.; Koshy, T. MicroRNA 146a Polymorphisms and Expression in Indian Children with Acute Lymphoblastic Leukemia. Lab Med. 2019, 50, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, H.; Lei, D. Microrna-146a promotes growth of acute leukemia cells by downregulating ciliary neurotrophic factor receptor and activating jak2/stat3 signaling. Yonsei Med. J. 2019, 60, 924–934. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, H.; Zhang, B.; Zhang, X. Expression and role of microRNA-663b in childhood acute lymphocytic leukemia and its mechanism. Open Med. 2019, 14, 863–871. [Google Scholar] [CrossRef]
- Piatopoulou, D.; Avgeris, M.; Drakaki, I.; Marmarinos, A.; Xagorari, M.; Baka, M.; Pourtsidis, A.; Kossiva, L.; Gourgiotis, D.; Scorilas, A. Clinical utility of miR-143/miR-182 levels in prognosis and risk stratification specificity of BFM-treated childhood acute lymphoblastic leukemia. Ann. Hematol. 2018, 97, 1169–1182. [Google Scholar] [CrossRef] [PubMed]
- Swellam, M.; Hashim, M.; Mahmoud, M.S.; Ramadan, A.; Hassan, N.M. Aberrant Expression of Some Circulating miRNAs in Childhood Acute Lymphoblastic Leukemia. Biochem. Genet. 2018, 56, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Ghodousi, E.S.; Rahgozar, S. MicroRNA-326 and microRNA-200c: Two novel biomarkers for diagnosis and prognosis of pediatric acute lymphoblastic leukemia. J. Cell. Biochem. 2018, 119, 6024–6032. [Google Scholar] [CrossRef] [PubMed]
- Shafik, R.E.; Abd El Wahab, N.; Senoun, S.A.; Ebeid, E.; El Taweel, M.A. Expression of micro-RNA 128 and let-7b in pediatric acute lymphoblastic leukemia cases. Asian Pac. J. Cancer Prev. 2018, 19, 2263–2267. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Lu, X.; Huang, P.; Gao, C.; Zhao, X.; Xing, T.; Li, G.; Bao, S.; Zheng, H. Expression of miR-652-3p and Effect on Apoptosis and Drug Sensitivity in Pediatric Acute Lymphoblastic Leukemia. BioMed Res. Int. 2018, 2018, 5724686. [Google Scholar] [CrossRef] [PubMed]
- Zang, Y.; Yu, R.; Bai, Y.; Chen, X. MicroRNA-9 suppresses cancer proliferation and cell cycle progression in acute lymphoblastic leukemia with inverse association of neuropilin-1. J. Cell. Biochem. 2018, 119, 6604–6613. [Google Scholar] [CrossRef] [PubMed]
- Asnafi, A.A.; Khodadi, E.; Golchin, N.; Alghasi, A.; Tavakolifar, Y.; Saki, N. Association between microRNA-21, microRNA-150, and micro-RNA-451 expression and clinical outcome of patients with acute lymphoblastic leukemia. Front. Biol. 2017, 12, 63–70. [Google Scholar] [CrossRef]
- Piatopoulou, D.; Avgeris, M.; Marmarinos, A.; Xagorari, M.; Baka, M.; Doganis, D.; Kossiva, L.; Scorilas, A.; Gourgiotis, D. MiR-125b predicts childhood acute lymphoblastic leukaemia poor response to BFM chemotherapy treatment. Br. J. Cancer 2017, 117, 801–812. [Google Scholar] [CrossRef]
- Nabhan, M.; Louka, M.L.; Khairy, E.; Tash, F.; Ali-Labib, R.; El-Habashy, S. MicroRNA-181a and its target Smad 7 as potential biomarkers for tracking child acute lymphoblastic leukemia. Gene 2017, 628, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.; Wu, S.; Huang, S.; Jiang, W.; Ke, Y. Association between expression of plasma miRNA and the risk of childhood acute lymphocytic leukemia. Chin. J. Epidemiol. 2017, 38, 1252–1258. [Google Scholar] [CrossRef]
- Labib, H.A.; Elantouny, N.G.; Ibrahim, N.F.; Alnagar, A.A. Upregulation of microRNA-21 is a poor prognostic marker in patients with childhood B cell acute lymphoblastic leukemia. Hematology 2017, 22, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Cao, J.; Ji, Q.; Zhang, C.; Qian, T.; Song, X.; Huang, B.; Tian, X. The downregulation of miR-3173 in B-cell acute lymphoblastic leukaemia promotes cell invasion via PTK2. Biochem. Biophys. Res. Commun. 2017, 494, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Ramani, R.; Megason, G.; Schallheim, J.; Karlson, C.; Vijayakumar, V.; Vijayakumar, S.; Hicks, C. Integrative analysis of microRNA-mediated gene signatures and pathways modulating white blood cell count in childhood acute lymphoblastic leukemia. Biomark. Insights 2017, 12, 1177271917702895. [Google Scholar] [CrossRef]
- Cao, L.; Wang, N.; Pan, J.; Hu, S.; Zhao, W.; He, H.; Wang, Y.; Gu, G.; Chai, Y. Clinical significance of microRNA-34b expression in pediatric acute leukemia. Mol. Med. Rep. 2016, 13, 2777–2784. [Google Scholar] [CrossRef] [PubMed]
- Lou, Y.; Liu, L.; Zhan, L.; Wang, X.; Fan, H. MIR-187-5p regulates cell growth and apoptosis in acute lymphoblastic Leukemia via DKK2. Oncol. Res. 2016, 24, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Swellam, M.; El-Khazragy, N. Clinical impact of circulating microRNAs as blood-based marker in childhood acute lymphoblastic leukemia. Tumor Biol. 2016, 37, 10571–10576. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, J.C.; Scrideli, C.A.; Brassesco, M.S.; Yunes, J.A.; Brandalise, S.R.; Tone, L.G. MiR-708-5p is differentially expressed in childhood acute lymphoblastic leukemia but not strongly associated to clinical features. Pediatr. Blood Cancer 2015, 62, 177–178. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Eguchi-Ishimae, M.; Yagi, C.; Iwabuki, H.; Gao, W.; Tauchi, H.; Inukai, T.; Sugita, K.; Ishii, E.; Eguchi, M. HMGA2 as a potential molecular target in KMT2A-AFF1-positive infant acute lymphoblastic leukaemia. Br. J. Haematol. 2015, 171, 818–829. [Google Scholar] [CrossRef] [PubMed]
- Nemes, K.; Csóka, M.; Nagy, N.; Márk, Á.; Váradi, Z.; Dankó, T.; Kovács, G.; Kopper, L.; Sebestyén, A. Expression of Certain Leukemia/Lymphoma Related microRNAs and its Correlation with Prognosis in Childhood Acute Lymphoblastic Leukemia. Pathol. Oncol. Res. 2015, 21, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Luna-Aguirre, C.M.; Martinez-Fierro, M.D.L.L.; Mar-Aguilar, F.; Garza-Veloz, I.; Treviño-Alvaradod, V.; Rojas-Martinez, A.; Jaime-Perez, J.C.; Malagon-Santiago, G.I.; Gutierrez-Aguirre, C.H.; Gonzalez-Llano, O.; et al. Circulating microRNA expression profile in B-cell acute lymphoblastic leukemia. Cancer Biomark. 2015, 15, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Organista-Nava, J.; Gómez-Gómez, Y.; Illades-Aguiar, B.; Del Carmen Alarcón-Romero, L.; Saavedra-Herrera, M.V.; Rivera-Ramírez, A.B.; Garzón-Barrientos, V.H.; Leyva-Vázquez, M.A. High miR-24 expression is associated with risk of relapse and poor survival in acute leukemia. Oncol. Rep. 2015, 33, 1639–1649. [Google Scholar] [CrossRef]
- Oliveira, L.H.; Schiavinato, J.L.; Fráguas, M.S.; Lucena-Araujo, A.R.; Haddad, R.; Araújo, A.G.; Dalmazzo, L.F.; Rego, E.M.; Covas, D.T.; Zago, M.A.; et al. Potential roles of microRNA-29a in the molecular pathophysiology of T-cell acute lymphoblastic leukemia. Cancer Sci. 2015, 106, 1264–1277. [Google Scholar] [CrossRef]
- Malik, D.; Kaul, D.; Chauhan, N.; Marwaha, R.K. MiR-2909-mediated regulation of KLF4: A novel molecular mechanism for differentiating between B-cell and T-cell pediatric acute lymphoblastic leukemias. Mol. Cancer 2014, 13, 175. [Google Scholar] [CrossRef] [PubMed]
- Gimenes-Teixeira, H.L.; Lucena-Araujo, A.R.; dos Santos, G.A.; Zanette, D.L.; Scheucher, P.S.; Oliveira, L.C.; Dalmazzo, L.F.; Silva-Júnior, W.A.; Falcão, R.P.; Rego, E.M. Increased expression of miR-221 is associated with shorter overall survival in T-cell acute lymphoid leukemia. Exp. Hematol. Oncol. 2013, 2, 10. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, D.; Zhuang, Y.; Shi, Q.; Wei, W.; Zhang, H.; Ju, X. li [The expression and regulatory mechanism of microRNA-708 in pediatric common B-cell acute lymphoblastic leukemia]. Zhonghua Xue Ye Xue Za Zhi 2013, 34, 138–143. [Google Scholar] [PubMed]
- Li, X.J.; Luo, X.Q.; Han, B.W.; Duan, F.T.; Wei, P.P.; Chen, Y.Q. MicroRNA-100/99a, deregulated in acute lymphoblastic leukaemia, suppress proliferation and promote apoptosis by regulating the FKBP51 and IGF1R/mTOR signalling pathways. Br. J. Cancer 2013, 109, 2189–2198. [Google Scholar] [CrossRef]
- De Oliveira, J.C.; Scrideli, C.A.; Brassesco, M.S.; Morales, A.G.; Pezuk, J.A.; de Paula Queiroz, R.; Yunes, J.A.; Brandalise, S.R.; Tone, L.G. Differential MiRNA expression in childhood acute lymphoblastic leukemia and association with clinical and biological features. Leuk. Res. 2012, 36, 293–298. [Google Scholar] [CrossRef]
- Borze, I.; Guled, M.; Musse, S.; Raunio, A.; Elonen, E.; Saarinen-Pihkala, U.; Karjalainen-Lindsberg, M.L.; Lahti, L.; Knuutila, S. MicroRNA microarrays on archive bone marrow core biopsies of leukemias-Method validation. Leuk. Res. 2011, 35, 188–195. [Google Scholar] [CrossRef]
- Bhatia, S.; Kaul, D.; Varma, N. Functional genomics of tumor suppressor miR-196b in T-cell acute lymphoblastic leukemia. Mol. Cell. Biochem. 2011, 346, 103–116. [Google Scholar] [CrossRef]
- Mavrakis, K.J.; Van Der Meulen, J.; Wolfe, A.L.; Liu, X.; Mets, E.; Taghon, T.; Khan, A.A.; Setti, M.; Rondou, P.; Vandenberghe, P.; et al. A cooperative microRNA-tumor suppressor gene network in acute T-cell lymphoblastic leukemia (T-ALL). Nat. Genet. 2011, 43, 673–678. [Google Scholar] [CrossRef]
- Stumpel, D.J.P.M.; Schotte, D.; Lange-Turenhout, E.A.M.; Schneider, P.; Seslija, L.; De Menezes, R.X.; Marquez, V.E.; Pieters, R.; Den Boer, M.L.; Stam, R.W. Hypermethylation of specific microRNA genes in MLL-rearranged infant acute lymphoblastic leukemia: Major matters at a micro scale. Leukemia 2011, 25, 429–439. [Google Scholar] [CrossRef]
- Schotte, D.; Moqadam, F.A.; Lange-Turenhout, E.A.M.; Chen, C.; Van Ijcken, W.F.J.; Pieters, R.; Den Boer, M.L. Discovery of new microRNAs by small RNAome deep sequencing in childhood acute lymphoblastic leukemia. Leukemia 2011, 25, 1389–1399. [Google Scholar] [CrossRef]
- Bhatia, S.; Kaul, D.; Varma, N. Potential tumor suppressive function of miR-196b in B-cell lineage acute lymphoblastic leukemia. Mol. Cell. Biochem. 2010, 340, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Schotte, D.; Chau, J.C.K.; Sylvester, G.; Liu, G.; Chen, C.; van der Velden, V.H.J.; Broekhuis, M.J.C.; Peters, T.C.J.M.; Pieters, R.; den Boer, M.L. Identification of new microRNA genes and aberrant microRNA profiles in childhood acute lymphoblastic leukemia. Leukemia 2009, 23, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yang, J.H.; Zheng, Y.S.; Zhang, P.; Chen, X.; Wu, J.; Xu, L.; Luo, X.Q.; Ke, Z.Y.; Zhou, H.; et al. Genome-wide analysis of small RNA and novel microRNA discovery in human acute lymphoblastic leukemia based on extensive sequencing approach. PLoS ONE 2009, 4, e6849. [Google Scholar] [CrossRef]
- Ju, X.; Li, D.; Shi, Q.; Hou, H.; Sun, N.; Shen, B. Differential microRNA expression in childhood B-cell precursor acute lymphoblastic leukemia. Pediatr. Hematol. Oncol. 2009, 26, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Luo, X.Q.; Zhang, P.; Huang, L.B.; Zheng, Y.S.; Wu, J.; Zhou, H.; Qu, L.H.; Xu, L.; Chen, Y.Q. MicroRNA patterns associated with clinical prognostic parameters and CNS relapse prediction in pediatric acute leukemia. PLoS ONE 2009, 4, e7826. [Google Scholar] [CrossRef]
- Zou, D.; Yin, J.; Ye, Z.; Zeng, Q.; Tian, C.; Wang, Y.; Chen, Q.; Chen, R. Association Between the miR-146a Rs2910164 Polymorphism and Childhood Acute Lymphoblastic Leukemia Susceptibility in an Asian Population. Front. Genet. 2020, 11, 886. [Google Scholar] [CrossRef]
- De Souza, T.P.; de Carvalho, D.C.; Wanderley, A.V.; Fernandes, S.M.; Gomes Rodrigues, J.C.; Cohen-Paes, A.; Fernandes, M.R.; Mello, F.A.R.; Pastana, L.F.; Vinagre, L.W.M.S.; et al. Influence of variants of the drosha, mir499a, and mir938 genes on susceptibility to acute lymphoblastic leukemia in an admixed population from the brazilian amazon. Am. J. Transl. Res. 2021, 12, 8216–8224. [Google Scholar]
- Hasani, S.S.; Hashemi, M.; Eskandari-Nasab, E.; Naderi, M.; Omrani, M.; Sheybani-Nasab, M. A functional polymorphism in the miR-146a gene is associated with the risk of childhood acute lymphoblastic leukemia: A preliminary report. Tumor Biol. 2014, 35, 219–225. [Google Scholar] [CrossRef]
- Pei, J.S.; Chang, W.S.; Hsu, P.C.; Chen, C.C.; Chin, Y.T.; Huang, T.L.; Hsu, Y.N.; Kuo, C.C.; Wang, Y.C.; Tsai, C.W.; et al. Significant association βetween the mIR146a genotypes and susceptibility to childhood acute lymphoblastic leukemia in Taiwan. Cancer Genom. Proteom. 2020, 17, 175–180. [Google Scholar] [CrossRef]
- Gutierrez-Camino, A.; Lopez-Lopez, E.; Martin-Guerrero, I.; Piñan, M.A.; Garcia-Miguel, P.; Sanchez-Toledo, J.; Carbone Bañeres, A.; Uriz, J.; Navajas, A.; Garcia-Orad, A. Noncoding RNA-related polymorphisms in pediatric acute lymphoblastic leukemia susceptibility. Pediatr. Res. 2014, 75, 767–773. [Google Scholar] [CrossRef]
- Siyadat, P.; Ayatollahi, H.; Barati, M.; Sheikhi, M.; Shahidi, M. High Resolution Melting Analysis for Evaluation of mir-612 (Rs12803915) Genetic Variant with Susceptibility to Pediatric Acute Lymphoblastic Leukemia. Rep. Biochem. Mol. Biol. 2021, 9, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.C.; Hsu, P.C.; Shih, L.C.; Hsu, Y.N.; Kuo, C.C.; Chao, C.Y.; Chang, W.S.; Tsai, C.W.; Bau, D.T.; Pei, J.S. MiR-196a-2 Genotypes determine the susceptibility and early onset of childhood acute lymphoblastic leukemia. Anticancer Res. 2020, 40, 4465–4469. [Google Scholar] [CrossRef]
- Gutierrez-Camino, A.; Martin-Guerrero, I.; Dolzan, V.; Jazbec, J.; Carbone-Bañeres, A.; de Andoin, N.G.; Sastre, A.; Astigarraga, I.; Navajas, A.; Garcia-Orad, A. Involvement of SNPs in miR-3117 and miR-3689d2 in childhood acute lymphoblastic leukemia risk. Oncotarget 2018, 9, 22907–22914. [Google Scholar] [CrossRef] [PubMed]
- LIU, X.; LIU, L.; CAO, Z.; GUO, B.; LI, M. Association between miR146a(rs2910164)G>C polymorphism and susceptibility to acute lymphoblastic leuke-mia in children. Chin. J. Appl. Clin. Pediatr. 2018, 24, 200–202. [Google Scholar] [CrossRef]
- Rakmanee, S.; Pakakasama, S.; Hongeng, S.; Sanguansin, S.; Thongmee, A.; Pongstaporn, W. Increased risk of Thai childhood acute lymphoblastic leukemia with the MiR196a2 T > C polymorphism. Asian Pac. J. Cancer Prev. 2017, 18, 1117–1120. [Google Scholar] [CrossRef]
- Chansing, K.; Pakakasama, S.; Hongeng, S.; Thongmee, A.; Pongstaporn, W. Lack of association between the MiR146a polymorphism and susceptibility to Thai childhood acute lymphoblastic leukemia. Asian Pac. J. Cancer Prev. 2016, 17, 2435–2438. [Google Scholar] [CrossRef]
- Hashemi, M.; Bahari, G.; Naderi, M.; Sadeghi-Bojd, S.; Taheri, M. Pri-miR-34b/c rs4938723 polymorphism is associated with the risk of childhood acute lymphoblastic leukemia. Cancer Genet. 2016, 209, 493–496. [Google Scholar] [CrossRef]
- Tong, N.; Chu, H.; Wang, M.; Xue, Y.; Du, M.; Lu, L.; Zhang, H.; Wang, F.; Fang, Y.; Li, J.; et al. Pri-miR-34b/c rs4938723 polymorphism contributes to acute lymphoblastic leukemia susceptibility in Chinese children. Leuk. Lymphoma 2016, 57, 1436–1441. [Google Scholar] [CrossRef]
- Akbari Moqadam, F.; Lange-Turenhout, E.A.M.; Van Der Veer, A.; Marchante, J.R.M.; Boer, J.M.; Pieters, R.; Den Boer, M. MicroRNA signature in BCR-ABL1-like and BCR-ABL1-positive childhood acute lymphoblastic leukemia: Similarities and dissimilarities. Leuk. Lymphoma 2014, 55, 1942–1945. [Google Scholar] [CrossRef]
- Gefen, N.; Binder, V.; Zaliova, M.; Linka, Y.; Morrow, M.; Novosel, A.; Edry, L.; Hertzberg, L.; Shomron, N.; Williams, O.; et al. Hsa-mir-125b-2 is highly expressed in childhood ETV6/RUNX1 (TEL/AML1) leukemias and confers survival advantage to growth inhibitory signals independent of p53. Leukemia 2010, 24, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Vendramini, E.; Giordan, M.; Giarin, E.; Michielotto, B.; Fazio, G.; Cazzaniga, G.; Biondi, A.; Silvestri, D.; Valsecchi, M.G.; Muckenthaler, M.U.; et al. High expression of miR-125b-2 and SNORD116 noncoding RNA clusters characterize ERG-related B cell precursor acute lymphoblastic leukemia. Oncotarget 2017, 8, 42398–42413. [Google Scholar] [CrossRef]
- Shaham, L.; Binder, V.; Gefen, N.; Borkhardt, A.; Izraeli, S. MiR-125 in normal and malignant hematopoiesis. Leukemia 2012, 26, 2011–2018. [Google Scholar] [CrossRef] [PubMed]
- Tassano, E.; Acquila, M.; Tavella, E.; Micalizzi, C.; Panarello, C.; Morerio, C. MicroRNA-125b-1 and BLID upregulation resulting from a novel IGH translocation in childhood B-cell precursor acute lymphoblastic leukemia. Genes Chromosom. Cancer 2010, 49, 682–687. [Google Scholar] [CrossRef]
- Doerrenberg, M.; Kloetgen, A.; Hezaveh, K.; Wössmann, W.; Bleckmann, K.; Stanulla, M.; Schrappe, M.; McHardy, A.C.; Borkhardt, A.; Hoell, J.I. T-cell acute lymphoblastic leukemia in infants has distinct genetic and epigenetic features compared to childhood cases. Genes Chromosom. Cancer 2017, 56, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Renou, L.; Boelle, P.Y.; Deswarte, C.; Spicuglia, S.; Benyoucef, A.; Calvo, J.; Uzan, B.; Belhocine, M.; Cieslak, A.; Landman-Parker, J.; et al. Homeobox protein TLX3 activates miR-125b expression to promote T-cell acute lymphoblastic leukemia. Blood Adv. 2017, 1, 733–747. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Hu, H.; Gu, W. Regulation Effects of Circular RNA CircPVT1 and miR-125b on NF-κB Signal Pathway in Childhood ALL. Clin. Lab. 2021, 67, 2491–2500. [Google Scholar] [CrossRef] [PubMed]
- Vafadar, A.; Mokaram, P.; Erfani, M.; Yousefi, Z.; Farhadi, A.; Elham Shirazi, T.; Tamaddon, G. The effect of decitabine on the expression and methylation of the PPP1CA, BTG2, and PTEN in association with changes in miR-125b, miR-17, and miR-181b in NALM6 cell line. J. Cell. Biochem. 2019, 120, 13156–13167. [Google Scholar] [CrossRef]
- Puissegur, M.P.; Eichner, R.; Quelen, C.; Coyaud, E.; Mari, B.; Lebrigand, K.; Broccardo, C.; Nguyen-Khac, F.; Bousquet, M.; Brousset, P. B-cell regulator of immunoglobulin heavy-chain transcription (Bright)/ARID3a is a direct target of the oncomir microRNA-125b in progenitor B-cells. Leukemia 2012, 26, 2224–2232. [Google Scholar] [CrossRef] [PubMed]
- Willimott, S.; Wagner, S.D. miR-125b and miR-155 contribute to BCL2 repression and proliferation in response to CD40 ligand (CD154) in human leukemic B-cells. J. Biol. Chem. 2012, 287, 2608–2617. [Google Scholar] [CrossRef] [PubMed]
- Dou, L.; Li, J.; Zheng, D.; Li, Y.; Gao, X.; Xu, C.; Gao, L.; Wang, L.; Yu, L. MicroRNA-142-3p inhibits cell proliferation in human acute lymphoblastic leukemia by targeting the MLL-AF4 oncogene. Mol. Biol. Rep. 2013, 40, 6811–6819. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, D.; Zhang, G.; Li, X.; Liang, Y.; Kasukurthi, M.V.; Li, S.; Borchert, G.M.; Huang, J. A semantics-oriented computational approach to investigate microRNA regulation on glucocorticoid resistance in pediatric acute lymphoblastic leukemia. BMC Med. Inform. Decis. Mak. 2018, 18, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Lv, M.; Zhang, X.; Jia, H.; Li, D.; Zhang, B.; Zhang, H.; Hong, M.; Jiang, T.; Jiang, Q.; Lu, J.; et al. An oncogenic role of miR-142-3p in human T-cell acute lymphoblastic leukemia (T-ALL) by targeting glucocorticoid receptor-α and cAMP/PKA pathways. Leukemia 2012, 26, 769–777. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Croce, C.M. MicroRNA-cancer connection: The beginning of a new tale. Cancer Res. 2006, 66, 7390–7394. [Google Scholar] [CrossRef] [PubMed]
- Kramer, N.J.; Wang, W.L.; Reyes, E.Y.; Kumar, B.; Chen, C.C.; Ramakrishna, C.; Cantin, E.M.; Vonderfecht, S.L.; Taganov, K.D.; Chau, N.; et al. Altered lymphopoiesis and immunodeficiency in miR-142 null mice. Blood 2015, 125, 3720–3730. [Google Scholar] [CrossRef] [PubMed]
- Schotte, D.; Lange-Turenhout, E.A.M.; Stumpel, D.J.P.M.; Stam, R.W.; Buijs-Gladdines, J.G.C.A.M.; Meijerink, J.P.P.; Pieters, R.; den Boer, M.L. Expression of miR-196b is not exclusively MLL-driven but is especially linked to activation of HOXA genes in pediatric acute lymphoblastic leukemia. Haematologica 2010, 95, 1675–1682. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Huang, H.; Chen, P.; He, M.; Li, Y.; Arnovitz, S.; Jiang, X.; He, C.; Hyjek, E.; Zhang, J.; et al. miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia. Nat. Commun. 2012, 3, 688. [Google Scholar] [CrossRef] [PubMed]
- Avigad, S.; Verly, I.R.; Lebel, A.; Kordi, O.; Shichrur, K.; Ohali, A.; Hameiri-Grossman, M.; Kaspers, G.J.; Cloos, J.; Fronkova, E.; et al. miR expression profiling at diagnosis predicts relapse in pediatric precursor B-cell acute lymphoblastic leukemia. Genes Chromosom. Cancer 2016, 55, 328–339. [Google Scholar] [CrossRef]
- Zhou, W.; Shunqing, W.; Yi, Y.; Zhou, R.; Mao, P. MiR-196b/miR-1290 participate in the antitumor effect of resveratrol via regulation of IGFBP3 expression in acute lymphoblastic leukemia. Oncol. Rep. 2017, 37, 1075–1083. [Google Scholar] [CrossRef]
- Roman-Gomez, J.; Agirre, X.; Jiménez-Velasco, A.; Arqueros, V.; Vilas-Zornoza, A.; Rodriguez-Otero, P.; Martin-Subero, I.; Garate, L.; Corden, L.; José-Eneriz, E.S.; et al. Epigenetic regulation of MicroRNAs in acute lymphoblastic leukemia. J. Clin. Oncol. 2009, 27, 1316–1322. [Google Scholar] [CrossRef]
- Hu, X.-Y.; Yin, J.-J.; Chao, R.; Zhu, S.-D.; Deng, W.; Pan, M. [Correlation of Plasma MiR-146a, MiR-223 Levels with the Prognosis of Childhood with Acute Lymphoblastic Leukemia]. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2021, 29, 1727–1732. [Google Scholar] [CrossRef]
- Zhou, K.; Yi, S.; Yu, Z.; Li, Z.; Wang, Y.; Zou, D.; Qi, J.; Zhao, Y.; Qiu, L. MicroRNA-223 expression is uniformly down-regulated in B cell lymphoproliferative disorders and is associated with poor survival in patients with chronic lymphocytic leukemia. Leuk. Lymphoma 2012, 53, 1155–1161. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.Y.; Wang, F.; Yu, J.; Yang, G.H.; Liu, X.L.; Zhang, J.W. MicroRNA-223 reversibly regulates erythroid and megakaryocytic differentiation of K562 cells. J. Cell. Mol. Med. 2009, 13, 4551–4559. [Google Scholar] [CrossRef] [PubMed]
- Mansour, M.R.; Sanda, T.; Lawton, L.N.; Li, X.; Kreslavsky, T.; Novina, C.D.; Brand, M.; Gutierrez, A.; Kelliher, M.A.; Jamieson, C.H.M.; et al. The TAL1 complex targets the FBXW7 tumor suppressor by activating miR-223 in human T cell acute lymphoblastic leukemia. J. Exp. Med. 2013, 210, 1545–1557. [Google Scholar] [CrossRef] [PubMed]
- Mets, E.; Van Der Meulen, J.; Van Peer, G.; Boice, M.; Mestdagh, P.; Van De Walle, I.; Lammens, T.; Goossens, S.; De Moerloose, B.; Benoit, Y.; et al. MicroRNA-193b-3p acts as a tumor suppressor by targeting the MYB oncogene in T-cell acute lymphoblastic leukemia. Leukemia 2015, 29, 798–806. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.D.; Wang, L.; Sun, C.; Fan, L.; Zhu, D.X.; Fang, C.; Wang, Y.H.; Zou, Z.J.; Zhang, S.J.; Li, J.Y.; et al. Distinctive microRNA signature is associated with the diagnosis and prognosis of acute leukemia. Med. Oncol. 2012, 29, 2323–2331. [Google Scholar] [CrossRef] [PubMed]
- Rainer, J.; Ploner, C.; Jesacher, S.; Ploner, A.; Eduardoff, M.; Mansha, M.; Wasim, M.; Panzer-Grümayer, R.; Trajanoski, Z.; Niederegger, H.; et al. Glucocorticoid-regulated microRNAs and mirtrons in acute lymphoblastic leukemia. Leukemia 2009, 23, 746–752. [Google Scholar] [CrossRef] [PubMed]
- McCarty, M.F. Metformin may antagonize Lin28 and/or Lin28B activity, thereby boosting let-7 levels and antagonizing cancer progression. Med. Hypotheses 2012, 78, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Silver, S.J.; Hagen, J.W.; Okamura, K.; Perrimon, N.; Lai, E.C. Functional screening identifies miR-315 as a potent activator of Wingless signaling. Proc. Natl. Acad. Sci. USA 2007, 104, 18151–18156. [Google Scholar] [CrossRef] [PubMed]
- Nishi, M.; Eguchi-Ishimae, M.; Wu, Z.; Gao, W.; Iwabuki, H.; Kawakami, S.; Tauchi, H.; Inukai, T.; Sugita, K.; Hamasaki, Y.; et al. Suppression of the let-7b microRNA pathway by DNA hypermethylation in infant acute lymphoblastic leukemia with MLL gene rearrangements. Leukemia 2013, 27, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Krzanowski, J.; Madzio, J.; Pastorczak, A.; Tracz, A.; Braun, M.; Tabarkiewicz, J.; Pluta, A.; Młynarski, W.; Zawlik, I. Selected miRNA levels are associated with IKZF1 microdeletions in pediatric acute lymphoblastic leukemia. Oncol. Lett. 2017, 14, 3853–3861. [Google Scholar] [CrossRef] [PubMed]
- Manvati, S.; Mangalhara, K.C.; Kalaiarasan, P.; Srivastava, N.; Bamezai, R.N.K. miR-24-2 regulates genes in survival pathway and demonstrates potential in reducing cellular viability in combination with docetaxel. Gene 2015, 567, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Scheibner, K.A.; Teaboldt, B.; Hauer, M.C.; Chen, X.; Cherukuri, S.; Guo, Y.; Kelley, S.M.; Liu, Z.; Baer, M.R.; Heimfeld, S.; et al. MiR-27a Functions as a Tumor Suppressor in Acute Leukemia by Regulating 14-3-3θ. PLoS ONE 2012, 7, e50895. [Google Scholar] [CrossRef] [PubMed]
- Su, R.; Dong, L.; Zou, D.; Zhao, H.; Ren, Y.; Li, F.; Yi, P.; Li, L.; Zhu, Y.; Ma, Y.; et al. microRNA-23a, -27a and -24 synergistically regulate JAK1/Stat3 cascade and serve as novel therapeutic targets in human acute erythroid leukemia. Oncogene 2016, 35, 6001–6014. [Google Scholar] [CrossRef]
- Drobna, M.; Szarzyńska-Zawadzka, B.; Daca-Roszak, P.; Kosmalska, M.; Jaksik, R.; Witt, M.; Dawidowska, M. Identification of Endogenous Control miRNAs for RT-qPCR in T-Cell Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2018, 19, 2858. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Ma, J.; Wu, M.; Qin, W.; Zhao, B.; Shi, Y.; Jin, Y.; Xie, Y. Aberrant microRNA-182 expression is associated with glucocorticoid resistance in lymphoblastic malignancies. Leuk. Lymphoma 2012, 53, 2465–2473. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Shen, T.; Wang, H.; Ke, Z.; Liang, Y.; Ouyang, J.; Jiang, T. MicroRNA-185-5p restores glucocorticoid sensitivity by suppressing the mammalian target of rapamycin complex (mTORC) signaling pathway to enhance glucocorticoid receptor autoregulation. Leuk. Lymphoma 2017, 58, 2657–2667. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Jiang, N.; Huang, G.; Tay, J.L.S.; Lin, B.; Bi, C.; Koh, G.S.; Li, Z.; Tan, J.; Chung, T.H.; et al. Deregulated MIR335 that targets MAPK1 is implicated in poor outcome of paediatric acute lymphoblastic leukaemia. Br. J. Haematol. 2013, 163, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Scarola, M.; Schoeftner, S.; Schneider, C.; Benetti, R. miR-335 directly targets Rb1 (pRb/p105) in a proximal connection to p53-dependent stress response. Cancer Res. 2010, 70, 6925–6933. [Google Scholar] [CrossRef]
- Akbari Moqadam, F.; Lange-Turenhout, E.A.M.; Ariës, I.M.; Pieters, R.; den Boer, M.L. MiR-125b, miR-100 and miR-99a co-regulate vincristine resistance in childhood acute lymphoblastic leukemia. Leuk. Res. 2013, 37, 1315–1321. [Google Scholar] [CrossRef]
- Dou, L.; Zheng, D.; Li, J.; Li, Y.; Gao, L.; Wang, L.; Yu, L. Methylation-mediated repression of microRNA-143 enhances MLL-AF4 oncogene expression. Oncogene 2012, 31, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.; Song, Y.; Zheng, W.; Ma, W. miRNA143 Induces K562 Cell Apoptosis Through Downregulating BCR-ABL. Med. Sci. Monit. 2016, 22, 2761–2767. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.Z.; Zhang, Y.Y.; Fu, H.Y.; Wu, D.S.; Zhou, H.R. Overexpression of microRNA-143 inhibits growth and induces apoptosis in human leukemia cells. Oncol. Rep. 2014, 31, 2035–2042. [Google Scholar] [CrossRef] [PubMed]
- Volinia, S.; Galasso, M.; Costinean, S.; Tagliavini, L.; Gamberoni, G.; Drusco, A.; Marchesini, J.; Mascellani, N.; Sana, M.E.; Jarour, R.A.; et al. Reprogramming of miRNA networks in cancer and leukemia. Genome Res. 2010, 20, 589–599. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Ma, T.; Huang, C.; Zhang, L.; Xu, T.; Hu, T.; Li, J. MicroRNA-148a: A potential therapeutic target for cancer. Gene 2014, 533, 456–457. [Google Scholar] [CrossRef] [PubMed]
- Ninomiya, S.; Tyybäkinoja, A.; Borze, I.; Räty, R.; Saarinen-Pihkala, U.M.; Usvasalo, A.; Elonen, E.; Knuutila, S. Integrated analysis of gene copy number, copy neutral LOH, and microRNA profiles in adult acute lymphoblastic leukemia. Cytogenet. Genome Res. 2012, 136, 246–255. [Google Scholar] [CrossRef]
- Zanette, D.L.; Rivadavia, F.; Molfetta, G.A.; Barbuzano, F.G.; Proto-Siqueira, R.; Falcão, R.P.; Zago, M.A.; Silva, W.A. miRNA expression profiles in chronic lymphocytic and acute lymphocytic leukemia. Braz. J. Med. Biol. Res. 2007, 40, 1435–1440. [Google Scholar] [CrossRef] [PubMed]
- Kotani, A.; Ha, D.; Hsieh, J.; Rao, P.K.; Schotte, D.; Den Boer, M.L.; Armstrong, S.A.; Lodish, H.F. miR-128b is a potent glucocorticoid sensitizer in MLL-AF4 acute lymphocytic leukemia cells and exerts cooperative effects with miR-221. Blood 2009, 114, 4169–4178. [Google Scholar] [CrossRef] [PubMed]
- Fulci, V.; Colombo, T.; Chiaretti, S.; Messina, M.; Citarella, F.; Tavolaro, S.; Guarini, A.; Foà, R.; Macino, G. Characterization of B- and T-lineage acute lymphoblastic leukemia by integrated analysis of microRNA and mRNA expression profiles. Genes Chromosom. Cancer 2009, 48, 1069–1082. [Google Scholar] [CrossRef] [PubMed]
- Kotani, A.; Ha, D.; Schotte, D.; Den Boer, M.L.; Armstrong, S.A.; Lodish, H.F. A novel mutation in the miR-128b gene reduces miRNA processing and leads to glucocorticoid resistance of MLL-AF4 acute lymphocytic leukemia cells. Cell Cycle 2010, 9, 1037–1042. [Google Scholar] [CrossRef] [PubMed]
- Mets, E.; Van Peer, G.; Van der Meulen, J.; Boice, M.; Taghon, T.; Goossens, S.; Mestdagh, P.; Benoit, Y.; De Moerloose, B.; Van Roy, N.; et al. MicroRNA-128-3p is a novel oncomiR targeting PHF6 in T-cell acute lymphoblastic leukemia. Haematologica 2014, 99, 1326–1333. [Google Scholar] [CrossRef] [PubMed]
- Guzman, H.; Sanders, K.; Idica, A.; Bochnakian, A.; Jury, D.; Daugaard, I.; Zisoulis, D.G.; Pedersen, I.M. miR-128 inhibits telomerase activity by targeting TERT mRNA. Oncotarget 2018, 9, 13244–13253. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.L.; Yen, C.T.; Pai, C.H.; Chen, H.Y.; Yu, S.L.; Lin, C.Y.; Hu, C.Y.; Jou, S.T.; Lin, D.T.; Lin, S.R.; et al. A double negative loop comprising ETV6/RUNX1 and MIR181A1 contributes to differentiation block in t(12;21)-positive acute lymphoblastic leukemia. PLoS ONE 2015, 10, e0142863. [Google Scholar] [CrossRef] [PubMed]
- Durmaz, B.; Bagca, B.G.; Cogulu, O.; Susluer, S.Y.; Alpay, A.; Aksoylar, S.; Gunduz, C. Antileukemic Effects of Anti-miR-146a, Anti-miR-155, Anti-miR-181a, and Prednisolone on Childhood Acute Lymphoblastic Leukemia. BioMed Res. Int. 2021, 2021, 3207328. [Google Scholar] [CrossRef]
- Schneider, E.; Staffas, A.; Röhner, L.; Krowiorz, K.; Heuser, M.; Döhner, K.; Bullinger, L.; Döhner, H.; Fogelstrand, L.; Rouhi, A.; et al. MicroRNA-155 is upregulated in MLL-rearranged AML but its absence does not affect leukemia development. Exp. Hematol. 2016, 44, 1166–1171. [Google Scholar] [CrossRef]
- Hickey, C.; Schwind, S.; Becker, H.; Alachkar, H.; Garzon, R.; Wu, Y.-Z.; Liu, S.; Perrotti, D.; Marcucci, G. MicroRNA-181a Targets TEL/AML1 Expression and Impairs Cell Proliferation in t(12;21) Acute Lymphocytic Leukemia (ALL) Cells. Blood 2009, 114, 766. [Google Scholar] [CrossRef]
- Dahlhaus, M.; Schult, C.; Lange, S.; Freund, M.; Junghanss, C. MicroRNA 181a influences the expression of HMGB1 and CD4 in acute Leukemias. Anticancer Res. 2013, 33, 445–452. [Google Scholar] [PubMed]
- Havelange, V.; Stauffer, N.; Heaphy, C.C.E.; Volinia, S.; Andreeff, M.; Marcucci, G.; Croce, C.M.; Garzon, R. Functional implications of microRNAs in acute myeloid leukemia by integrating microRNA and messenger RNA expression profiling. Cancer 2011, 117, 4696–4706. [Google Scholar] [CrossRef]
- Markopoulos, G.S.; Roupakia, E.; Tokamani, M.; Alabasi, G.; Sandaltzopoulos, R.; Marcu, K.B.; Kolettas, E. Roles of NF-κB Signaling in the Regulation of miRNAs Impacting on Inflammation in Cancer. Biomedicines 2018, 6, 40. [Google Scholar] [CrossRef]
- Iliopoulos, D.; Jaeger, S.A.; Hirsch, H.A.; Bulyk, M.L.; Struhl, K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol. Cell 2010, 39, 493–506. [Google Scholar] [CrossRef]
- Verduci, L.; Azzalin, G.; Gioiosa, S.; Carissimi, C.; Laudadio, I.; Fulci, V.; Macino, G. MicroRNA-181a enhances cell proliferation in acute lymphoblastic leukemia by targeting EGR1. Leuk. Res. 2015, 39, 479–485. [Google Scholar] [CrossRef]
- Zeng, M.N.; Ma, W.L.; Zheng, W.L. [Bioinformatics analysis of microRNA comprehensive regulatory network in B- cell acute lymphoblastic leukemia]. Zhonghua Xue Ye Xue Za Zhi 2016, 37, 585–590. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Z.; He, C.; Wang, D.; Yuan, X.; Chen, J.; Jin, J. MicroRNAs expression signatures are associated with lineage and survival in acute leukemias. Blood Cells Mol. Dis. 2010, 44, 191–197. [Google Scholar] [CrossRef]
- Han, B.W.; Feng, D.D.; Li, Z.G.; Luo, X.Q.; Zhang, H.; Li, X.J.; Zhang, X.J.; Zheng, L.L.; Zeng, C.W.; Lin, K.Y.; et al. A set of miRNAs that involve in the pathways of drug resistance and leukemic stem-cell differentiation is associated with the risk of relapse and glucocorticoid response in childhood ALL. Hum. Mol. Genet. 2011, 20, 4903–4915. [Google Scholar] [CrossRef] [PubMed]
- Almeida, R.S.; Costa e Silva, M.; Coutinho, L.L.; Garcia Gomes, R.; Pedrosa, F.; Massaro, J.D.; Donadi, E.A.; Lucena-Silva, N. MicroRNA expression profiles discriminate childhood T- from B-acute lymphoblastic leukemia. Hematol. Oncol. 2019, 37, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Suresh, S.; McCallum, L.; Lu, W.; Lazar, N.; Perbal, B.; Irvine, A.E. MicroRNAs 130a/b are regulated by BCR-ABL and downregulate expression of CCN3 in CML. J. Cell Commun. Signal. 2011, 5, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Medina, R.; Zaidi, S.K.; Liu, C.G.; Stein, J.L.; VanWijnen, A.J.; Croce, C.M.; Stein, G.S. MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer Res. 2008, 68, 2773–2780. [Google Scholar] [CrossRef]
- Zhang, C.; Kang, C.; Wang, P.; Cao, Y.; Lv, Z.; Yu, S.; Wang, G.; Zhang, A.; Jia, Z.; Han, L.; et al. MicroRNA-221 and -222 regulate radiation sensitivity by targeting the PTEN pathway. Int. J. Radiat. Oncol. Biol. Phys. 2011, 80, 240–248. [Google Scholar] [CrossRef]
- Wang, M.; Ge, X.; Zheng, J.; Li, D.; Liu, X.; Wang, L.; Jiang, C.; Shi, Z.; Qin, L.; Liu, J.; et al. Role and mechanism of miR-222 in arsenic-transformed cells for inducing tumor growth. Oncotarget 2016, 7, 17805–17814. [Google Scholar] [CrossRef] [PubMed]
- Coskun, E.; Neumann, M.; Schlee, C.; Liebertz, F.; Heesch, S.; Goekbuget, N.; Hoelzer, D.; Baldus, C.D. MicroRNA profiling reveals aberrant microRNA expression in adult ETP-ALL and functional studies implicate a role for miR-222 in acute leukemia. Leuk. Res. 2013, 37, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Gao, C.; Wang, K.; Cui, L.; Li, W.; Zhao, X.; Liu, F.; Wu, M.; Deng, G.; Ding, W.; et al. Effect of microRNA-210 on prognosis and response to chemotherapeutic drugs in pediatric acute lymphoblastic leukemia. Cancer Sci. 2014, 105, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Tili, E.; Michaille, J.J.; Croce, C.M. MicroRNAs play a central role in molecular dysfunctions linking inflammation with cancer. Immunol. Rev. 2013, 253, 167–184. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Yan, J.; Liu, C.; Zhou, H.; Zheng, Y. Tumor Suppressor Role of miR-363-3p in Gastric Cancer. Med. Sci. Monit. 2015, 21, 4074–4080. [Google Scholar] [CrossRef] [PubMed]
- Busche, S.; Ge, B.; Vidal, R.; Spinella, J.F.; Saillour, V.; Richer, C.; Healy, J.; Chen, S.H.; Droit, A.; Sinnett, D.; et al. Integration of high-resolution methylome and transcriptome analyses to dissect epigenomic changes in childhood acute lymphoblastic leukemia. Cancer Res. 2013, 73, 4323–4336. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Li, L.; Li, S. Circulating microRNA-21 as a biomarker for the detection of various carcinomas: An updated meta-analysis based on 36 studies. Tumour Biol. 2015, 36, 1973–1981. [Google Scholar] [CrossRef] [PubMed]
- Seca, H.; Lima, R.; Lopes-Rodrigues, V.; Guimaraes, J.; Gabriela, G.; Vasconcelos, M. Targeting miR-21 induces autophagy and chemosensitivity of leukemia cells. Curr. Drug Targets 2013, 14, 1135–1143. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.H.; Yang, L.; Liu, X.J.; Nie, Z.Y.; Luo, J.M. [Targeted suppression of miRNA-21 inhibit K562 cells growth through PTEN-PI3K/AKT signaling pathway]. Zhonghua Xue Ye Xue Za Zhi 2016, 37, 982–986. [Google Scholar] [CrossRef]
- Bai, H.; Xu, R.; Cao, Z.; Wei, D.; Wang, C. Involvement of miR-21 in resistance to daunorubicin by regulating PTEN expression in the leukaemia K562 cell line. FEBS Lett. 2011, 585, 402–408. [Google Scholar] [CrossRef]
- Shi, C.; Zhang, X.; Li, X.; Zhang, L.; Li, L.; Sun, Z.; Fu, X.; Wu, J.; Chang, Y.; Li, W.; et al. Effects of microRNA-21 on the biological functions of T-cell acute lymphoblastic lymphoma/leukemia. Oncol. Lett. 2016, 12, 4173–4180. [Google Scholar] [CrossRef]
- Aster, J.C. Dicing up T-ALL. Blood 2015, 126, 929–930. [Google Scholar] [CrossRef][Green Version]
- Starczynowski, D.T.; Morin, R.; McPherson, A.; Lam, J.; Chari, R.; Wegrzyn, J.; Kuchenbauer, F.; Hirst, M.; Tohyama, K.; Humphries, R.K.; et al. Genome-wide identification of human microRNAs located in leukemia-associated genomic alterations. Blood 2011, 117, 595–607. [Google Scholar] [CrossRef]
- Cui, S.Y.; Wang, R.; Chen, L.B. MicroRNA-145: A potent tumour suppressor that regulates multiple cellular pathways. J. Cell. Mol. Med. 2014, 18, 1913–1926. [Google Scholar] [CrossRef] [PubMed]
- Sachdeva, M.; Mo, Y.Y. miR-145-mediated suppression of cell growth, invasion and metastasis. Am. J. Transl. Res. 2010, 2, 170. [Google Scholar] [PubMed]
- Almeida, M.I.; Calin, G.A. The miR-143/miR-145 cluster and the tumor microenvironment: Unexpected roles. Genome Med. 2016, 8, 29. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Zhang, Q.; Zhang, H.M.; Liu, W.; Liu, C.J.; Li, Q.; Guo, A.Y. Transcription factor and miRNA co-regulatory network reveals shared and specific regulators in the development of B cell and T cell. Sci. Rep. 2015, 5, 15215. [Google Scholar] [CrossRef] [PubMed]
- Labbaye, C.; Spinello, I.; Quaranta, M.T.; Pelosi, E.; Pasquini, L.; Petrucci, E.; Biffoni, M.; Nuzzolo, E.R.; Billi, M.; Foà, R.; et al. A three-step pathway comprising PLZF/miR-146a/CXCR4 controls megakaryopoiesis. Nat. Cell Biol. 2008, 10, 788–801. [Google Scholar] [CrossRef]
- Tong, L.G.; Wu, W.Z.; Zhang, Y.P.; Zhou, Z.G.; Chen, Y.F.; Huang, W.J.; Xu, H.; Su, Q.Q. [Expression of miR-16 in patients with T lymphoblastic lymphoma/acute lymphoblastic leukemia]. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2014, 22, 99–103. [Google Scholar] [CrossRef]
- Azimi, A.; Hagh, M.F.; Yousefi, B.; Rahnama, M.A.; Khorrami, A.; Heydarabad, M.Z.; Najafpour, M.; Hallajzadeh, J.; Ghahremani, A. The Effect of Prednisolone on miR 15a and miR16-1 Expression Levels and Apoptosis in Acute Lymphoblastic Leukemia Cell Line: CCRF-CEM. Drug Res. 2016, 66, 432–435. [Google Scholar] [CrossRef]
- Kaddar, T.; Chien, W.W.; Bertrand, Y.; Pages, M.P.; Rouault, J.P.; Salles, G.; Ffrench, M.; Magaud, J.P. Prognostic value of miR-16 expression in childhood acute lymphoblastic leukemia relationships to normal and malignant lymphocyte proliferation. Leuk. Res. 2009, 33, 1217–1223. [Google Scholar] [CrossRef]
- Li, J.; Li, P.; Wang, J.; Xi, Y.X. [Significance of microRNA-16 and bcl-2 expression in T lymphoblastic lymphoma/leukemia and its relation with prognosis]. Zhonghua Bing Li Xue Za Zhi 2013, 42, 748–752. [Google Scholar] [CrossRef]
- McBride, A.; Houtmann, S.; Wilde, L.; Vigil, C.; Eischen, C.M.; Kasner, M.; Palmisiano, N. The role of inhibition of apoptosis in acute leukemias and myelodysplastic syndrome. Front. Oncol. 2019, 9, 192. [Google Scholar] [CrossRef]
- Xi, Y.; Li, J.; Zan, L.; Wang, J.; Wang, G.; Ning, Y. Micro-RNA-16 expression in paraffin-embedded specimen correlates with overall survival of T-lymphoblastic lymphoma/leukemia. Hum. Pathol. 2013, 44, 1011–1016. [Google Scholar] [CrossRef] [PubMed]
- Mavrakis, K.J.; Wolfe, A.L.; Oricchio, E.; Palomero, T.; De Keersmaecker, K.; McJunkin, K.; Zuber, J.; James, T.; Chang, K.; Khan, A.A.; et al. Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nat. Cell Biol. 2010, 12, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Slovak, M.L.; Mannoor, K.; Chen, C.; Hunger, S.P.; Carroll, A.J.; Schultz, R.A.; Shaffer, L.G.; Ballif, B.C.; Ning, Y. Microarray detection of multiple recurring submicroscopic chromosomal aberrations in pediatric T-cell acute lymphoblastic leukemia. Leukemia 2011, 25, 1042–1046. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mesrian Tanha, H.; Mojtabavi Naeini, M.; Rahgozar, S.; Moafi, A.; Honardoost, M.A. Integrative computational in-depth analysis of dysregulated miRNA-mRNA interactions in drug-resistant pediatric acute lymphoblastic leukemia cells: An attempt to obtain new potential gene-miRNA pathways involved in response to treatment. Tumor Biol. 2016, 37, 7861–7872. [Google Scholar] [CrossRef]
- Malouf, C.; Antunes, E.T.B.; O’Dwyer, M.; Jakobczyk, H.; Sahm, F.; Landua, S.L.; Anderson, R.A.; Soufi, A.; Halsey, C.; Ottersbach, K. miR-130b and miR-128a are essential lineage-specific codrivers of t(4;11) MLL-AF4 acute leukemia. Blood 2021, 138, 2066–2092. [Google Scholar] [CrossRef]
- Haque, S.; Vaiselbuh, S.R. Silencing of exosomal miR-181a reverses pediatric acute lymphocytic leukemia cell proliferation. Pharmaceuticals 2020, 13, 241. [Google Scholar] [CrossRef]
- Longjohn, M.N.; Squires, W.R.B.; Christian, S.L. Meta-analysis of microRNA profiling data does not reveal a consensus signature for B cell acute lymphoblastic leukemia. Gene 2022, 821, 146211. [Google Scholar] [CrossRef]
- Shi, L.H.; Tian, L.; Wang, Y.F.; Liu, J.S.; Guo, M.F.; Liu, W. Effects of MiR-221-Mediated Wnt/β- Catenin Signaling Pathway on Biological Activity of Childhood Acute Lymphoblastic Leukemia Cells. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2019, 27, 1367–1373. [Google Scholar] [CrossRef]
- De Leeuw, D.C.; Van Den Ancker, W.; Denkers, F.; De Menezes, R.X.; Westers, T.M.; Ossenkoppele, G.J.; Van De Loosdrecht, A.A.; Smit, L. MicroRNA profiling can classify acute leukemias of ambiguous lineage as either acute myeloid leukemia or acute lymphoid leukemia. Clin. Cancer Res. 2013, 19, 2187–2196. [Google Scholar] [CrossRef]
- Mraz, M.; Pospisilova, S.; Malinova, K.; Slapak, I.; Mayer, J. MicroRNAs in chronic lymphocytic leukemia pathogenesis and disease subtypes. Leuk. Lymphoma 2009, 50, 506–509. [Google Scholar] [CrossRef]
- Alsuwaidi, L.; Hachim, M.; Senok, A. Novel Markers in Pediatric Acute Lymphoid Leukemia: The Role of ADAM6 in B Cell Leukemia. Front. Cell Dev. Biol. 2021, 9, 1680. [Google Scholar] [CrossRef] [PubMed]
- Bian, H.; Zhou, Y.; Zhou, D.; Zhang, Y.; Shang, D.; Qi, J. The latest progress on miR-374 and its functional implications in physiological and pathological processes. J. Cell. Mol. Med. 2019, 23, 3063–3076. [Google Scholar] [CrossRef] [PubMed]
- Ohyashiki, J.H.; Umezu, T.; Kobayashi, C.; Hamamura, R.S.; Tanaka, M.; Kuroda, M.; Ohyashiki, K. Impact on cell to plasma ratio of miR-92a in patients with acute leukemia: In vivo assessment of cell to plasma ratio of miR-92a. BMC Res. Notes 2010, 3, 347. [Google Scholar] [CrossRef] [PubMed]
- Git, A.; Dvinge, H.; Salmon-Divon, M.; Osborne, M.; Kutter, C.; Hadfield, J.; Bertone, P.; Caldas, C. Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA 2010, 16, 991. [Google Scholar] [CrossRef] [PubMed]
- Dzikiewicz-Krawczyk, A.; Macieja, A.; Mały, E.; Januszkiewicz-Lewandowska, D.; Mosor, M.; Fichna, M.; Strauss, E.; Nowak, J. Polymorphisms in microRNA target sites modulate risk of lymphoblastic and myeloid leukemias and affect microRNA binding. J. Hematol. Oncol. 2014, 7, 43. [Google Scholar] [CrossRef]
- GeneCards—Human Genes | Gene Database | Gene Search. Available online: https://www.genecards.org/ (accessed on 23 February 2021).
- Kovaleva, V.; Mora, R.; Park, Y.J.; Plass, C.; Chiramel, A.I.; Bartenschlager, R.; Doḧner, H.; Stilgenbauer, S.; Pscherer, A.; Lichter, P.; et al. miRNA-130a targets ATG2B and DICER1 to inhibit autophagy and trigger killing of chronic lymphocytic leukemia cells. Cancer Res. 2012, 72, 1763–1772. [Google Scholar] [CrossRef]
- Jiang, L.H.; Zhang, H.D.; Tang, J.H. MiR-30a: A Novel Biomarker and Potential Therapeutic Target for Cancer. J. Oncol. 2018, 2018, 5167829. [Google Scholar] [CrossRef]
- Ortega, M.; Bhatnagar, H.; Lin, A.P.; Wang, L.; Aster, J.C.; Sill, H.; Aguiar, R.C.T. A microRNA-mediated regulatory loop modulates NOTCH and MYC oncogenic signals in B- and T-cell malignancies. Leukemia 2015, 29, 968–976. [Google Scholar] [CrossRef]
- Mukherjee, S.; Shelar, B.; Krishna, S. Versatile role of miR-24/24-1*/24-2* expression in cancer and other human diseases. Am. J. Transl. Res. 2022, 14, 20. [Google Scholar]
- Yang, H.; Zhang, J.; Li, J.; Zhao, F.; Shen, Y.; Xing, X. Overexpression of miR-574-3p suppresses proliferation and induces apoptosis of chronic myeloid leukemia cells via targeting IL6/JAK/STAT3 pathway. Exp. Ther. Med. 2018, 16, 4296–4302. [Google Scholar] [CrossRef]
- Fu, A.; Hoffman, A.E.; Liu, R.; Jacobs, D.I.; Zheng, T.; Zhu, Y. Targetome profiling and functional genetics implicate miR-618 in lymphomagenesis. Epigenetics 2014, 9, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, B.M.; Brown, L.M.; Ryland, G.; Lonsdale, A.; Kosasih, H.J.; Ludlow, L.E.; Majewski, I.J.; Blombery, P.; Ekert, P.G.; Davidson, N.; et al. ALLSorts: A RNA-Seq subtype classifier for B-Cell Acute Lymphoblastic Leukemia. Blood Adv. 2022, 6, 4093–4097. [Google Scholar] [CrossRef] [PubMed]
- Bařinka, J.; Hu, Z.; Wang, L.; Wheeler, D.A.; Rahbarinia, D.; McLeod, C.; Gu, Z.; Mullighan, C.G. RNAseqCNV: Analysis of large-scale copy number variations from RNA-seq data. Leukemia 2022, 36, 1492–1498. [Google Scholar] [CrossRef]
- Li, X.; Sanda, T.; Thomas Look, A.; Novina, C.D.; von Boehmer, H. Repression of tumor suppressor miR-451 is essential for NOTCH1-induced oncogenesis in T-ALL. J. Exp. Med. 2011, 208, 663–675. [Google Scholar] [CrossRef] [PubMed]
- Akbari Moqadam, F.; Boer, J.M.; Lange-Turenhout, E.A.M.; Pieters, R.; Den Boer, M.L. Altered expression of miR-24, miR-126 and miR-365 does not affect viability of childhood TCF3-rearranged leukemia cells. Leukemia 2014, 28, 1008–1014. [Google Scholar] [CrossRef]
- Diakos, C.; Zhong, S.; Xiao, Y.; Zhou, M.; Vasconcelos, G.M.; Krapf, G.; Yeh, R.F.; Zheng, S.; Kang, M.; Wiencke, J.K.; et al. TEL-AML1 regulation of survivin and apoptosis via miRNA-494 and miRNA-320a. Blood 2010, 116, 4885–4893. [Google Scholar] [CrossRef]
- Scherr, M.; Elder, A.; Battmer, K.; Barzan, D.; Bomken, S.; Ricke-Hoch, M.; Schröder, A.; Venturini, L.; Blair, H.J.; Vormoor, J.; et al. Differential expression of miR-17∼92 identifies BCL2 as a therapeutic target in BCR-ABL-positive B-lineage acute lymphoblastic leukemia. Leukemia 2014, 28, 554–565. [Google Scholar] [CrossRef]
- Agueli, C.; Cammarata, G.; Salemi, D.; Dagnino, L.; Nicoletti, R.; La Rosa, M.; Messana, F.; Marfia, A.; Bica, M.G.; Coniglio, M.L.; et al. 14q32/miRNA clusters loss of heterozygosity in acute lymphoblastic leukemia is associated with up-regulation of BCL11a. Am. J. Hematol. 2010, 85, 575–578. [Google Scholar] [CrossRef]
- Amankwah, E.K.; Devidas, M.; Teachey, D.T.; Rabin, K.R.; Brown, P.A. Six Candidate miRNAs Associated With Early Relapse in Pediatric B-Cell Acute Lymphoblastic Leukemia. Anticancer Res. 2020, 40, 3147–3153. [Google Scholar] [CrossRef]
- Gutierrez-Camino, A.; Richer, C.; St-Onge, P.; Lopez-Lopez, E.; Bañeres, A.C.; De Andoin, N.G.; Sastre, A.; Astigarraga, I.; Martin-Guerrero, I.; Sinnett, D.; et al. Role of rs10406069 in miR-5196 in hyperdiploid childhood acute lymphoblastic leukemia. Epigenomics 2020, 12, 1949–1955. [Google Scholar] [CrossRef]
- Piroozian, F.; Bagheri Varkiyani, H.; Koolivand, M.; Ansari, M.; Afsa, M.; AtashAbParvar, A.; MalekZadeh, K. The impact of variations in transcription of DICER and AGO2 on exacerbation of childhood B-cell lineage acute lymphoblastic leukaemia. Int. J. Exp. Pathol. 2019, 100, 184–191. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).