Elevated miR-29c-5p Expression in Nipple Aspirate Fluid Is Associated with Extremely High Mammographic Breast Density
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Nipple Aspirate Fluid Collection and Processing
2.2. RNA Isolation, Reverse Transcription, and Pre-Amplification
2.3. Discovery Phase: Taqman OpenArray Profiling Analysis of Nipple Aspirate Fluid
2.4. Validation Phase: Individual TaqMan Advanced miRNA qPCR Assays
2.5. Statistical Analysis
2.6. Target Analysis of Differentially Expressed miRNAs
3. Results
3.1. Study Subject and NAF Characteristics Per Cohort
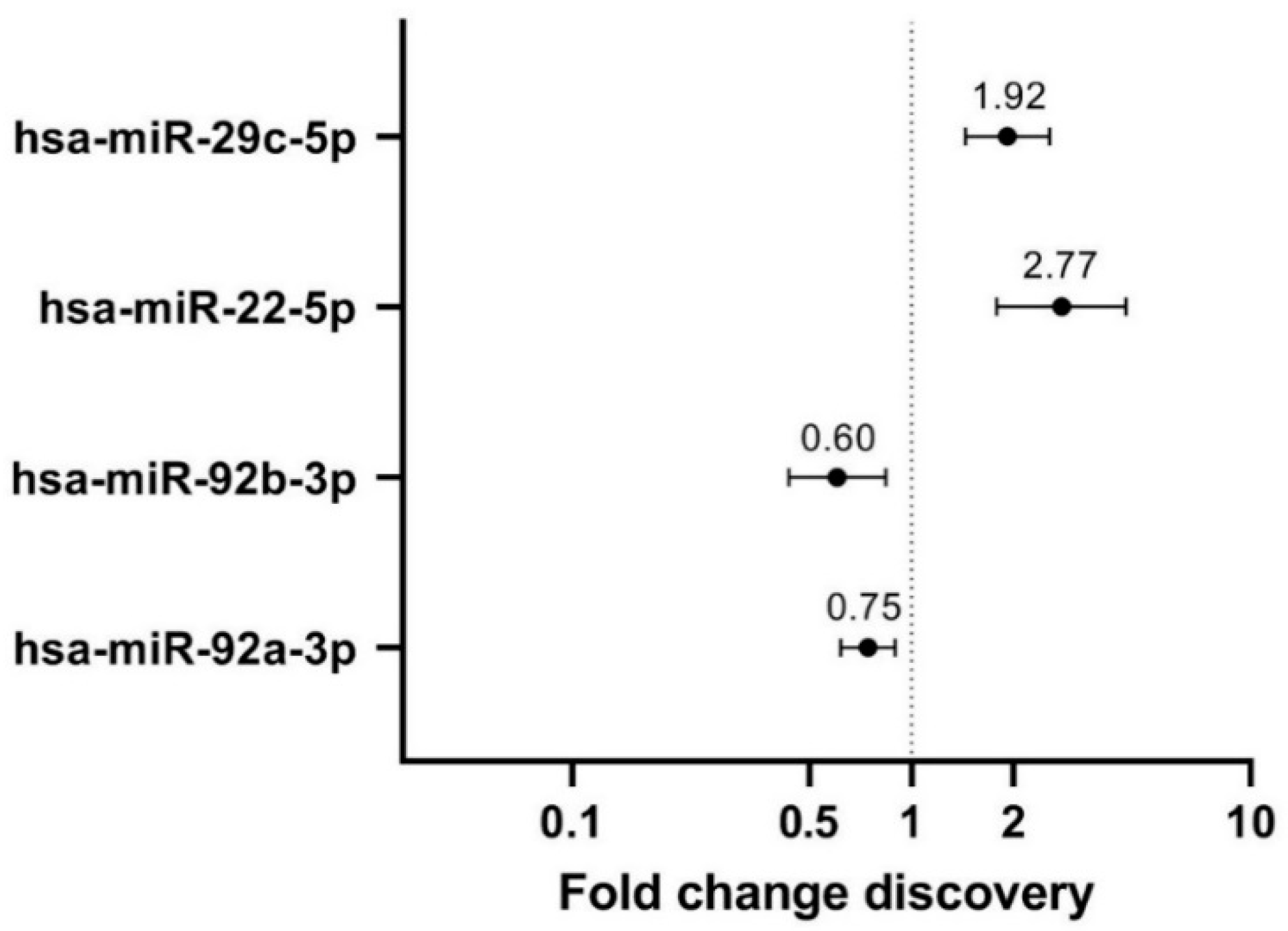
3.2. Discovery: Four Differentially Expressed miRNAs between Extremely High and Very Low MD
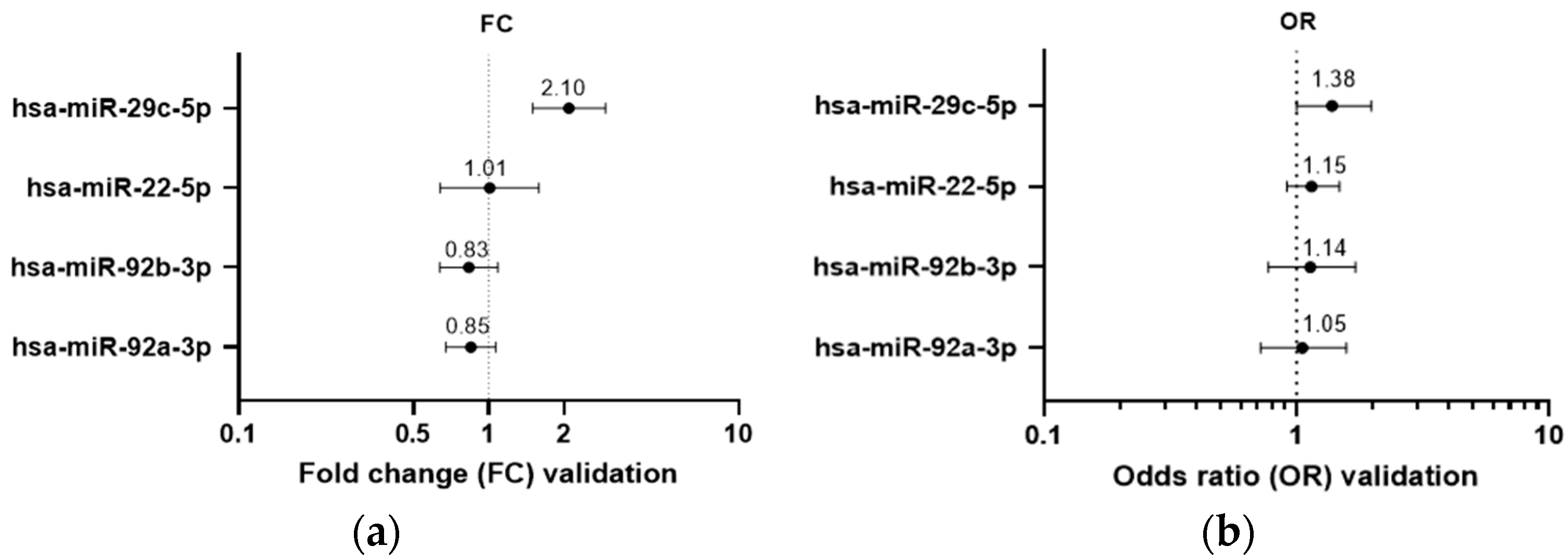
3.3. Validation: Hsa-miR-29c-5p Is Differentially Expressed between Extremely High and Very Low MD
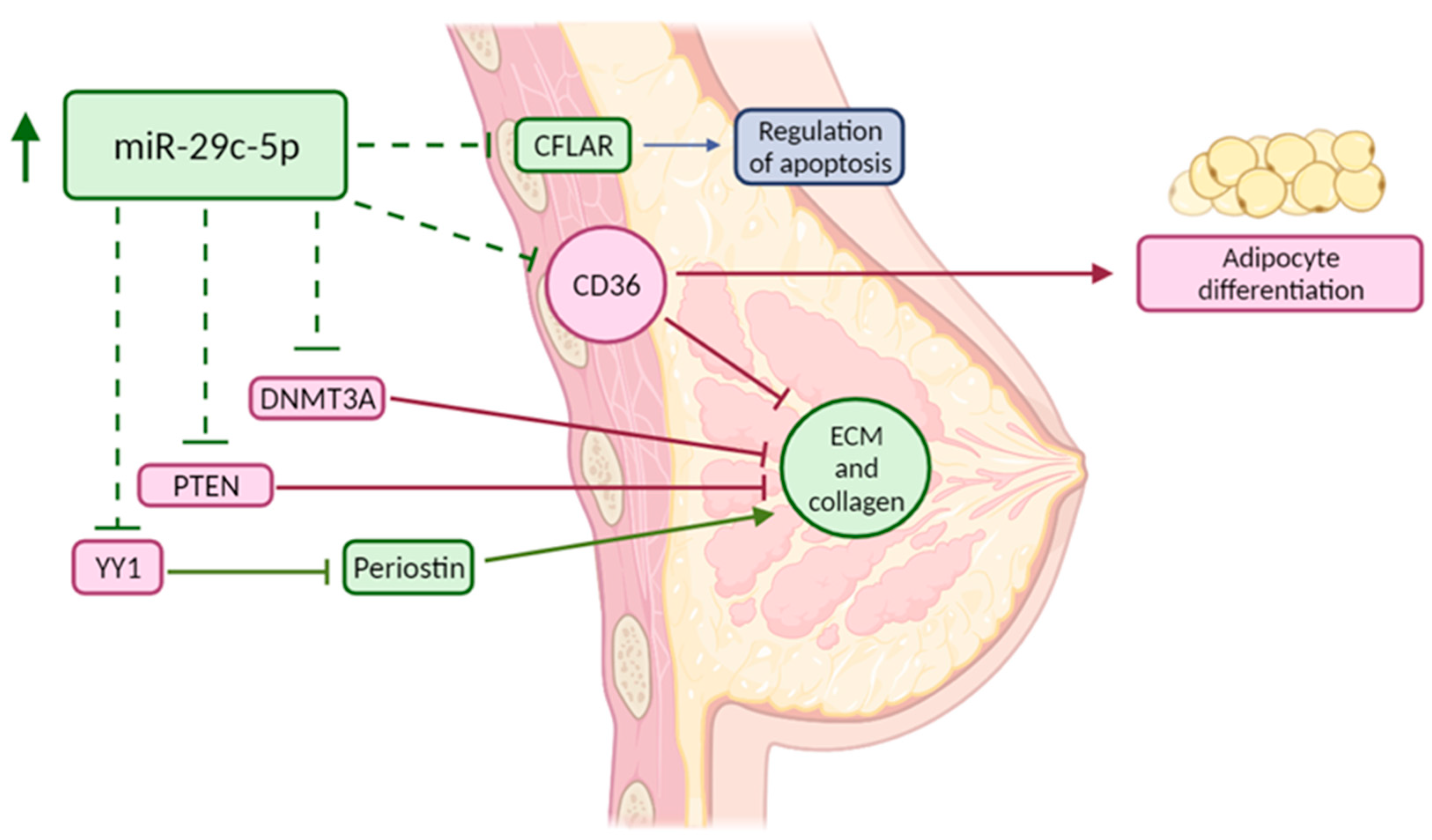
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sherratt, M.J.; McConnell, J.C.; Streuli, C.H. Raised mammographic density: Causative mechanisms and biological consequences. Breast Cancer Res. 2016, 18, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spak, D.A.; Plaxco, J.S.; Santiago, L.; Dryden, M.J.; Dogan, B.E. BI-RADS® fifth edition: A summary of changes. Diagn. Interv. Imaging 2017, 98, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Nazari, S.S.; Mukherjee, P. An overview of mammographic density and its association with breast cancer. Breast Cancer 2018, 25, 259–267. [Google Scholar] [CrossRef] [Green Version]
- Huo, C.W.; Chew, G.; Hill, P.; Huang, D.; Ingman, W.; Hodson, L.; Brown, K.A.; Magenau, A.; Allam, A.H.; McGhee, E.; et al. High mammographic density is associated with an increase in stromal collagen and immune cells within the mammary epithelium. Breast Cancer Res. 2015, 17, 79. [Google Scholar] [CrossRef] [Green Version]
- Huo, C.W.; Chew, G.L.; Britt, K.L.; Ingman, W.V.; Henderson, M.A.; Hopper, J.L.; Thompson, E.W. Mammographic density—A review on the current understanding of its association with breast cancer. Breast Cancer Res. Treat. 2014, 144, 479–502. [Google Scholar] [CrossRef] [PubMed]
- McCormack, V.A.; dos Santos Silva, I. Breast density and parenchymal patterns as markers of breast cancer risk: A meta-analysis. Cancer Epidemiol. Biomark. Prev. 2006, 15, 1159–1169. [Google Scholar] [CrossRef] [Green Version]
- Alowami, S.; Troup, S.; Al-Haddad, S.; Kirkpatrick, I.; Watson, P.H. Mammographic density is related to stroma and stromal proteoglycan expression. Breast Cancer Res. 2003, 5, R129–R135. [Google Scholar] [CrossRef]
- DeFilippis, R.A.; Chang, H.; Dumont, N.; Rabban, J.T.; Chen, Y.Y.; Fontenay, G.V.; Berman, H.K.; Gauthier, M.L.; Zhao, J.; Hu, D.; et al. CD36 repression activates a multicellular stromal program shared by high mammographic density and tumor tissues. Cancer Discov. 2012, 2, 826–839. [Google Scholar] [CrossRef] [Green Version]
- DeFilippis, R.A.; Fordyce, C.; Patten, K.; Chang, H.; Zhao, J.; Fontenay, G.V.; Kerlikowske, K.; Parvin, B.; Tlsty, T.D. Stress Signaling from Human Mammary Epithelial Cells Contributes to Phenotypes of Mammographic Density. Cancer Res. 2014, 74, 5032–5044. [Google Scholar] [CrossRef] [Green Version]
- Huo, C.W.; Hill, P.; Chew, G.; Neeson, P.J.; Halse, H.; Williams, E.D.; Henderson, M.A.; Thompson, E.W.; Britt, K.L. High mammographic density in women is associated with protumor inflammation. Breast Cancer Res. 2018, 20, 92. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blondal, T.; Nielsen, S.J.; Baker, A.; Andreasen, D.; Mouritzen, P.; Teilum, M.W.; Dahlsveen, I.K. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods 2013, 59, S1–S6. [Google Scholar] [CrossRef] [PubMed]
- Patuleia, S.I.; van Gils, C.H.; Oneto Cao, A.M.; Bakker, M.F.; van Diest, P.J.; van der Wall, E.; Moelans, C.B. The Physiological MicroRNA Landscape in Nipple Aspirate Fluid: Differences and Similarities with Breast Tissue, Breast Milk, Plasma and Serum. Int. J. Mol. Sci. 2020, 21, 8466. [Google Scholar] [CrossRef] [PubMed]
- Suijkerbuijk, K.P.; Van Der Wall, E.; Meijrink, H.; Pan, X.; Borel Rinkes, I.H.; Ausems, M.G.; Van Diest, P.J. Successful oxytocin-assisted nipple aspiration in women at increased risk for breast cancer. Fam. Cancer 2010, 9, 321–325. [Google Scholar] [CrossRef] [Green Version]
- Emaus, M.J.; Bakker, M.F.; Peeters, P.H.; Loo, C.E.; Mann, R.M.; De Jong, M.D.; Bisschops, R.H.; Veltman, J.; Duvivier, K.M.; Lobbes, M.B.; et al. MR Imaging as an Additional Screening Modality for the Detection of Breast Cancer in Women Aged 50–75 Years with Extremely Dense Breasts: The DENSE Trial Study Design. Radiology 2015, 277, 527–537. [Google Scholar] [CrossRef] [Green Version]
- Bakker, M.F.; de Lange, S.V.; Pijnappel, R.M.; Mann, R.M.; Peeters, P.H.; Monninkhof, E.M.; Emaus, M.J.; Loo, C.E.; Bisschops, R.H.; Lobbes, M.B.; et al. Supplemental MRI Screening for Women with Extremely Dense Breast Tissue. N. Engl. J. Med. 2019, 381, 2091–2102. [Google Scholar] [CrossRef]
- Moore, H.M.; Kelly, A.B.; Jewell, S.D.; McShane, L.M.; Clark, D.P.; Greenspan, R.; Hayes, D.F.; Hainaut, P.; Kim, P.; Mansfield, E.A.; et al. Biospecimen reporting for improved study quality (BRISQ). Cancer Cytopathol. 2011, 119, 92–102. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- D’haene, B.; Mestdagh, P.; Hellemans, J.; Vandesompele, J. miRNA expression profiling: From reference genes to global mean normalization. Methods Mol. Biol. 2012, 822, 261–272. [Google Scholar] [CrossRef]
- Mestdagh, P.; Van Vlierberghe, P.; De Weer, A.; Muth, D.; Westermann, F.; Speleman, F.; Vandesompele, J. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009, 10, R64. [Google Scholar] [CrossRef] [Green Version]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- McCall, M.N.; McMurray, H.R.; Land, H.; Almudevar, A. On non-detects in qPCR data. Bioinformatics 2014, 30, 2310–2316. [Google Scholar] [CrossRef] [PubMed]
- Patuleia, S.I.; van der Wall, E.; van Gils, C.H.; Bakker, M.F.; Jager, A.; Voorhorst-Ogink, M.M.; van Diest, P.J.; Moelans, C.B. The changing microRNA landscape by color and cloudiness: A cautionary tale for nipple aspirate fluid biomarker analysis. Cell. Oncol. 2021, 44, 1339–1349. [Google Scholar] [CrossRef]
- Huang, H.Y.; Lin, Y.C.D.; Cui, S.; Huang, Y.; Tang, Y.; Xu, J.; Bao, J.; Li, Y.; Wen, J.; Zuo, H.; et al. miRTarBase update 2022: An informative resource for experimentally validated miRNA–target interactions. Nucleic Acids Res. 2021, 50, D222–D230. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Sticht, C.; Torre, C.D.L.; Parveen, A.; Gretz, N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS ONE 2018, 13, e0206239. [Google Scholar] [CrossRef]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.P.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef]
- Kwon, J.J.; Factora, T.D.; Dey, S.; Kota, J. A Systematic Review of miR-29 in Cancer. Mol. Ther. Oncolytics 2019, 12, 173–194. [Google Scholar] [CrossRef] [Green Version]
- Aure, M.R.; Fleischer, T.; Bjørklund, S.; Ankill, J.; Castro-Mondragon, J.A.; Børresen-Dale, A.L.; Tost, J.; Sahlberg, K.K.; Mathelier, A.; Tekpli, X.; et al. Crosstalk between microRNA expression and DNA methylation drives the hormone-dependent phenotype of breast cancer. Genome Med. 2021, 13, 72. [Google Scholar] [CrossRef]
- Li, J.; Chen, W.; Luo, L.; Liao, L.; Deng, X.; Wang, Y. The microRNA miR-29c-5p inhibits cell proliferation and migration by targeting TMEM98 in head and neck carcinoma. Aging 2020, 13, 769–781. [Google Scholar] [CrossRef]
- Shu, Y.J.; Bao, R.F.; Jiang, L.; Wang, Z.; Wang, X.A.; Zhang, F.; Liang, H.B.; Li, H.F.; Ye, Y.Y.; Xiang, S.S.; et al. MicroRNA-29c-5p suppresses gallbladder carcinoma progression by directly targeting CPEB4 and inhibiting the MAPK pathway. Cell Death Differ. 2017, 24, 445–457. [Google Scholar] [CrossRef] [PubMed]
- Chou, J.; Lin, J.H.; Brenot, A.; Kim, J.; Provot, S.; Werb, Z. GATA3 suppresses metastasis and modulates the tumour microenvironment by regulating microRNA-29b expression. Nat. Cell Biol. 2013, 15, 201–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enerly, E.; Steinfeld, I.; Kleivi, K.; Leivonen, S.K.; Aure, M.R.; Russnes, H.G.; Rønneberg, J.A.; Johnsen, H.; Navon, R.; Rødland, E.; et al. miRNA-mRNA Integrated Analysis Reveals Roles for miRNAs in Primary Breast Tumors. PLoS ONE 2011, 6, e16915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Yi, J.; Zheng, X.; Liu, S.; Fu, W.; Ren, L.; Li, L.; Hoon, D.S.; Wang, J.; Du, G. miR-29c plays a suppressive role in breast cancer by targeting the TIMP3/STAT1/FOXO1 pathway. Clin. Epigenet. 2018, 10, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, X.; Chen, Z.; Huang, R.; Yao, Y.; Ma, G. Transforming growth factor β1 induces the expression of collagen type I by DNA methylation in cardiac fibroblasts. PLoS ONE 2013, 8, e60335. [Google Scholar] [CrossRef] [Green Version]
- Byström, S.; Eklund, M.; Hong, M.G.; Fredolini, C.; Eriksson, M.; Czene, K.; Hall, P.; Schwenk, J.M.; Gabrielson, M. Affinity proteomic profiling of plasma for proteins associated to area-based mammographic breast density. Breast Cancer Res. 2018, 20, 14. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Q.; Khoshdeli, M.; Ferguson, B.S.; Jabbari, K.; Zang, C.; Parvin, B. YY1 is a cis-regulator in the organoid models of high mammographic density. Bioinformatics 2020, 36, 1663–1667. [Google Scholar] [CrossRef]
- Zhu, H.; Wang, G.; Zhu, H.; Xu, A. ITGA5 is a prognostic biomarker and correlated with immune infiltration in gastrointestinal tumors. BMC Cancer 2021, 21, 269. [Google Scholar] [CrossRef]
- Pantano, F.; Croset, M.; Driouch, K.; Bednarz-Knoll, N.; Iuliani, M.; Ribelli, G.; Bonnelye, E.; Wikman, H.; Geraci, S.; Bonin, F.; et al. Integrin alpha5 in human breast cancer is a mediator of bone metastasis and a therapeutic target for the treatment of osteolytic lesions. Oncogene 2021, 40, 1284–1299. [Google Scholar] [CrossRef]
- Wu, J.; Feng, Z.; Wang, R.; Li, A.; Wang, H.; He, X.; Shen, Z. Integration of bioinformatics analysis and experimental validation identifies plasma exosomal miR-103b/877-5p/29c-5p as diagnostic biomarkers for early lung adenocarcinoma. Cancer Med. 2022. [Google Scholar] [CrossRef] [PubMed]
- Lin, G.B.; Zhang, C.M.; Chen, X.Y.; Wang, J.; Chen, S.; Tang, S.; Yu, T. Identification of circulating miRNAs as novel prognostic biomarkers for bladder cancer. Math. Biosci. Eng. 2019, 17, 834–844. [Google Scholar] [CrossRef] [PubMed]
- Tsegaye, M.A.; He, J.; McGeehan, K.; Murphy, I.M.; Nemera, M.; Schafer, Z.T. Oncogenic signaling inhibits c-FLIP expression and promotes cancer cell survival during ECM-detachment. bioRxiv 2021. [Google Scholar] [CrossRef]
- Jones, C.E.; Sharick, J.T.; Colbert, S.E.; Shukla, V.C.; Zent, J.M.; Ostrowski, M.C.; Ghadiali, S.N.; Sizemore, S.T.; Leight, J.L. Pten regulates collagen fibrillogenesis by fibroblasts through SPARC. PLoS ONE 2021, 16, e0245653. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.E.; Hammer, A.M.; Cho, Y.; Sizemore, G.M.; Cukierman, E.; Yee, L.D.; Ghadiali, S.N.; Ostrowski, M.C.; Leight, J.L. Stromal PTEN Regulates Extracellular Matrix Organization in the Mammary Gland. Neoplasia 2018, 21, 132–145. [Google Scholar] [CrossRef] [PubMed]
- Sarvagalla, S.; Kolapalli, S.P.; Vallabhapurapu, S. The Two Sides of YY1 in Cancer: A Friend and a Foe. Front. Oncol. 2019, 9, 1230. Available online: https://www.frontiersin.org/article/10.3389/fonc.2019.01230 (accessed on 27 May 2022). [CrossRef] [Green Version]
- Romeo, F.; Falbo, L.; Di Sanzo, M.; Misaggi, R.; Faniello, M.C.; Barni, T.; Cuda, G.; Viglietto, G.; Santoro, C.; Quaresima, B.; et al. Negative transcriptional regulation of the human periostin gene by YingYang-1 transcription factor. Gene 2011, 487, 129–134. [Google Scholar] [CrossRef]
- McConnell, J.C.; O’Connell, O.V.; Brennan, K.; Weiping, L.; Howe, M.; Joseph, L.; Knight, D.; O’Cualain, R.; Lim, Y.; Leek, A.; et al. Increased peri-ductal collagen micro-organization may contribute to raised mammographic density. Breast Cancer Res. 2016, 18, 5. [Google Scholar] [CrossRef]
- Chen, T.; Li, Z.; Tu, J.; Zhu, W.; Ge, J.; Zheng, X.; Yang, L.; Pan, X.; Yan, H.; Zhu, J. MicroRNA-29a regulates pro-inflammatory cytokine secretion and scavenger receptor expression by targeting LPL in oxLDL-stimulated dendritic cells. FEBS Lett. 2011, 585, 657–663. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.-Y.; Wang, F.-S.; Yang, Y.-L.; Huang, Y.-H. MicroRNA-29a Suppresses CD36 to Ameliorate High Fat Diet-Induced Steatohepatitis and Liver Fibrosis in Mice. Cells 2019, 8, 1298. [Google Scholar] [CrossRef] [Green Version]
- Massart, J.; Sjögren, R.J.; Lundell, L.S.; Mudry, J.M.; Franck, N.; O’Gorman, D.J.; Egan, B.; Zierath, J.R.; Krook, A. Altered miR-29 Expression in Type 2 Diabetes Influences Glucose and Lipid Metabolism in Skeletal Muscle. Diabetes 2017, 66, 1807–1818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanders, A.P.; Burris, H.H.; Just, A.C.; Motta, V.; Svensson, K.; Mercado-Garcia, A.; Pantic, I.; Schwartz, J.; Tellez-Rojo, M.M.; Wright, R.O.; et al. microRNA expression in the cervix during pregnancy is associated with length of gestation. Epigenetics 2015, 10, 221–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klinge, C.M. Estrogen Regulation of MicroRNA Expression. Curr. Genom. 2009, 10, 169–183. [Google Scholar] [CrossRef] [Green Version]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Huang, R.S.; Gamazon, E.R.; Ziliak, D.; Wen, Y.; Im, H.K.; Zhang, W.; Wing, C.; Duan, S.; Bleibel, W.K.; Cox, N.J.; et al. Population differences in microRNA expression and biological implications. RNA Biol. 2011, 8, 692–701. [Google Scholar] [CrossRef] [Green Version]
| (a) Discovery Cohort (N = 41) | ||||
| High Density | Low Density | p Value | ||
| Age | Median (range) | N = 21 55 (51–72) | N = 20 54.5 (50–60) | 0.29 |
| BMI | Median (range) | N = 21 21.7 (18.4–28.9) | N = 17 27.6 (21.6–38.8) | <0.0001 |
| Age at first live birth | Median (range) | N = 15 30 (21–34) | N = 18 29 (18–38) | 0.81 |
| Age at menarche | Median (range) | N = 21 13 (11–16) | N = 20 13 (9–18) | 0.76 |
| Parity | Nulliparous (n = 8) | 6 (29%) | 2 (10%) | 0.24 |
| Parous (n = 33) | 15 (71%) | 18 (90%) | ||
| First degree BC | Yes (n = 8) | 4 (31%) | 4 (22%) | 0.69 |
| No (n = 23) | 9 (69%) | 14 (78%) | ||
| NAF color | Clear white/yellow (n = 11) | 3 (14%) | 8 (40%) | 0.028 |
| Turbid white/yellow (n = 3) | 1 (5%) | 2 (10%) | ||
| Bloody/orange/pink (n = 13) | 11 (52%) | 2 (10%) | ||
| Green/brown (n = 14) | 6 (29%) | 8 (40%) | ||
| (b) Validation Cohort (N = 170) | ||||
| High Density | Low Density | p Value | ||
| Age | Median (range) | N = 89 56 (50–74) | N = 81 55 (50–60) | 0.05 |
| BMI | Median (range) | N = 84 21.8 (17.0–34.5) | N = 73 29.0 (23.1–49.6) | <0.0001 |
| Age at first live birth | Median (range) | N = 67 28 (19–42) | N = 72 27 (20–42) | 0.23 |
| Age at menarche | Median (range) | N = 82 14 (10–17) | N = 80 13 (9–16) | 0.001 |
| Parity | Nulliparous (n = 25) | 17 (20%) | 8 (10%) | 0.07 |
| Parous (n = 139) | 67 (80%) | 72 (90%) | ||
| First degree BC | Yes (n = 26) | 15 (25%) | 11 (15%) | 0.12 |
| No (n = 108) | 44 (75%) | 64 (85%) | ||
| NAF color | Clear white/yellow (n = 79) | 41 (47%) | 38 (47%) | 0.237 |
| Turbid white/yellow (n = 45) | 22 (25%) | 23 (28%) | ||
| Bloody/orange/pink (n = 31) | 18 (20%) | 13 (16%) | ||
| Green/brown (n = 14) | 7 (8%) | 7 (9%) | ||
| Hsa-miR-29c-5p Targets | Protein Class | Relevant GO BP and Reactome Pathways |
|---|---|---|
| CPEB4 | mRNA polyadenylation factor | regulation of translation, translational elongation, ionotropic glutamate receptor signaling pathway, response to ischemia |
| TMEM98 | Transmembrane protein | protein localization to nucleus, protein processing, negative regulator of FRAT2 mediated Wnt/ß-catenin signaling |
| CD36 * | Membrane trafficking regulatory protein | positive regulation of NF-kappaB TF activity, Toll-like receptor cascades, regulation of ERK1/2 cascade, regulation of gene expression, regulation of cell death, regulation of cell-matrix adhesion, phagocytosis, immune response, transcriptional regulation of white adipocyte differentiation, triglyceride transport, fatty acid/lipid metabolic process, lipid storage |
| CFLAR * | Protease | positive regulation of ERK1 and ERK2 cascade, positive regulation of I-kappaB kinase/NF-kappaB signaling, apoptotic signaling pathway, regulation of necroptotic process, negative regulation of ROS biosynthetic process, negative regulation of cellular response to TGF-β stimulus, wound healing, cellular response to estradiol, testosterone, hypoxia and EGF stimulus, proteolysis, regulation of ECM organization |
| DNMT3A * | DNA methyltransferase | epigenetic regulation of gene expression, chromatin organization, metabolism of proteins, SUMOylation, mitotic cell cycle, response to estradiol, positive regulation of cell death, cellular response to hypoxia/ toxic substance |
| YY1 * | Transcription factor | (regulation of) DNA repair, estrogen-dependent gene expression, nucleotide excision repair, RNA localization, regulation of transcription, regulation of cell cycle |
| PTEN * | Protein phosphatase | negative regulation of PI3-kinase and AKT signaling, PDGFR signaling pathway, p53 pathway, regulation of apoptotic signaling pathway, canonical Wnt signaling pathway, regulation of ERK1 and ERK2 cascade, protein dephosphorylation, angiogenesis, regulation of cell population proliferation, response to glucose, regulation of gene expression, negative regulation of EMT, negative regulation of cell migration, response to estradiol/hypoxia/insulin-like growth factor stimulus, negative regulation of G1/S phase transition |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vissers, T.A.C.M.; Piek, L.; Patuleia, S.I.S.; Duinmeijer, A.J.; Bakker, M.F.; van der Wall, E.; van Diest, P.J.; van Gils, C.H.; Moelans, C.B. Elevated miR-29c-5p Expression in Nipple Aspirate Fluid Is Associated with Extremely High Mammographic Breast Density. Cancers 2022, 14, 3805. https://doi.org/10.3390/cancers14153805
Vissers TACM, Piek L, Patuleia SIS, Duinmeijer AJ, Bakker MF, van der Wall E, van Diest PJ, van Gils CH, Moelans CB. Elevated miR-29c-5p Expression in Nipple Aspirate Fluid Is Associated with Extremely High Mammographic Breast Density. Cancers. 2022; 14(15):3805. https://doi.org/10.3390/cancers14153805
Chicago/Turabian StyleVissers, Tessa A. C. M., Leonie Piek, Susana I. S. Patuleia, Aafke J. Duinmeijer, Marije F. Bakker, Elsken van der Wall, Paul J. van Diest, Carla H. van Gils, and Cathy B. Moelans. 2022. "Elevated miR-29c-5p Expression in Nipple Aspirate Fluid Is Associated with Extremely High Mammographic Breast Density" Cancers 14, no. 15: 3805. https://doi.org/10.3390/cancers14153805
APA StyleVissers, T. A. C. M., Piek, L., Patuleia, S. I. S., Duinmeijer, A. J., Bakker, M. F., van der Wall, E., van Diest, P. J., van Gils, C. H., & Moelans, C. B. (2022). Elevated miR-29c-5p Expression in Nipple Aspirate Fluid Is Associated with Extremely High Mammographic Breast Density. Cancers, 14(15), 3805. https://doi.org/10.3390/cancers14153805






