Current Developments of Artificial Intelligence in Digital Pathology and Its Future Clinical Applications in Gastrointestinal Cancers
Abstract
:Simple Summary
Abstract
1. Introduction
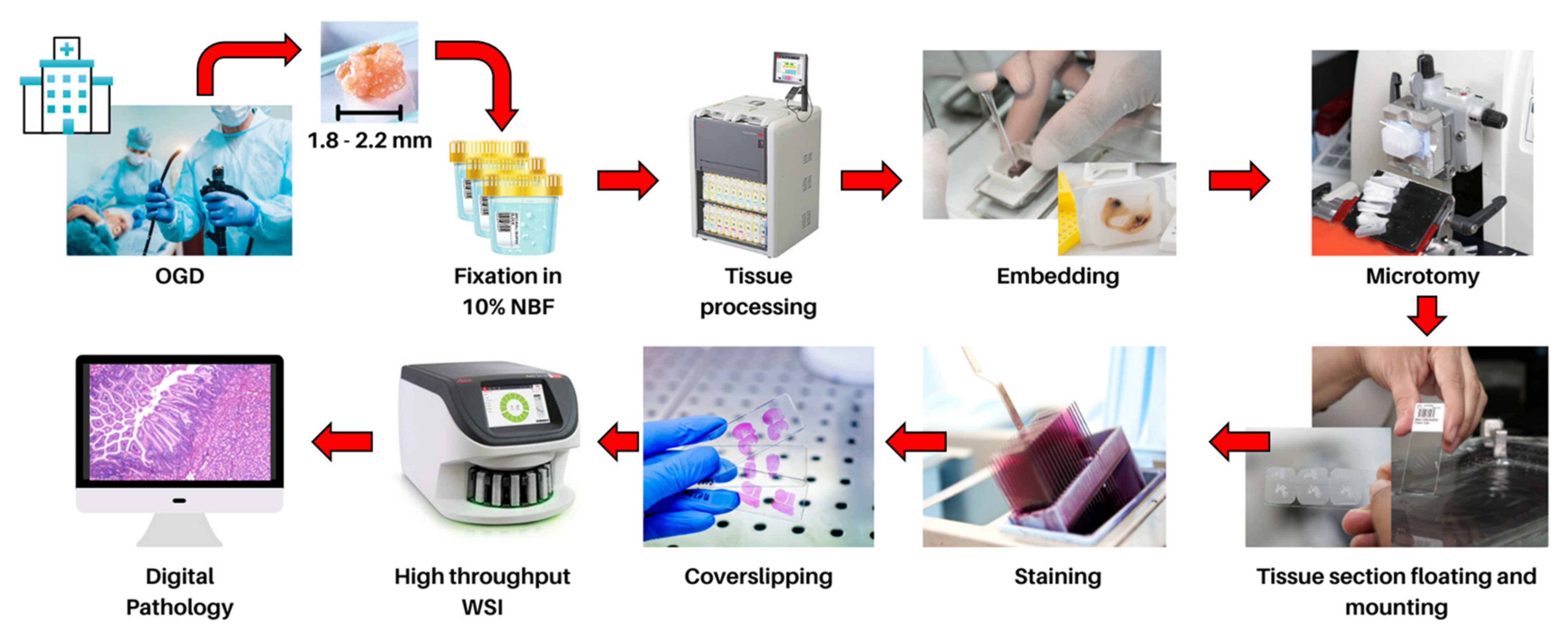
2. Current Histopathology Practices and Opportunities in Digital Pathology
3. Current Challenges of Algorithm Development in Computational Pathology
3.1. Colour Normalization
3.2. Pathologist Interpretation for Model Training
3.3. Model Transparency and Interpretability for Deployment of AI-Based Tools in Clinical Practice
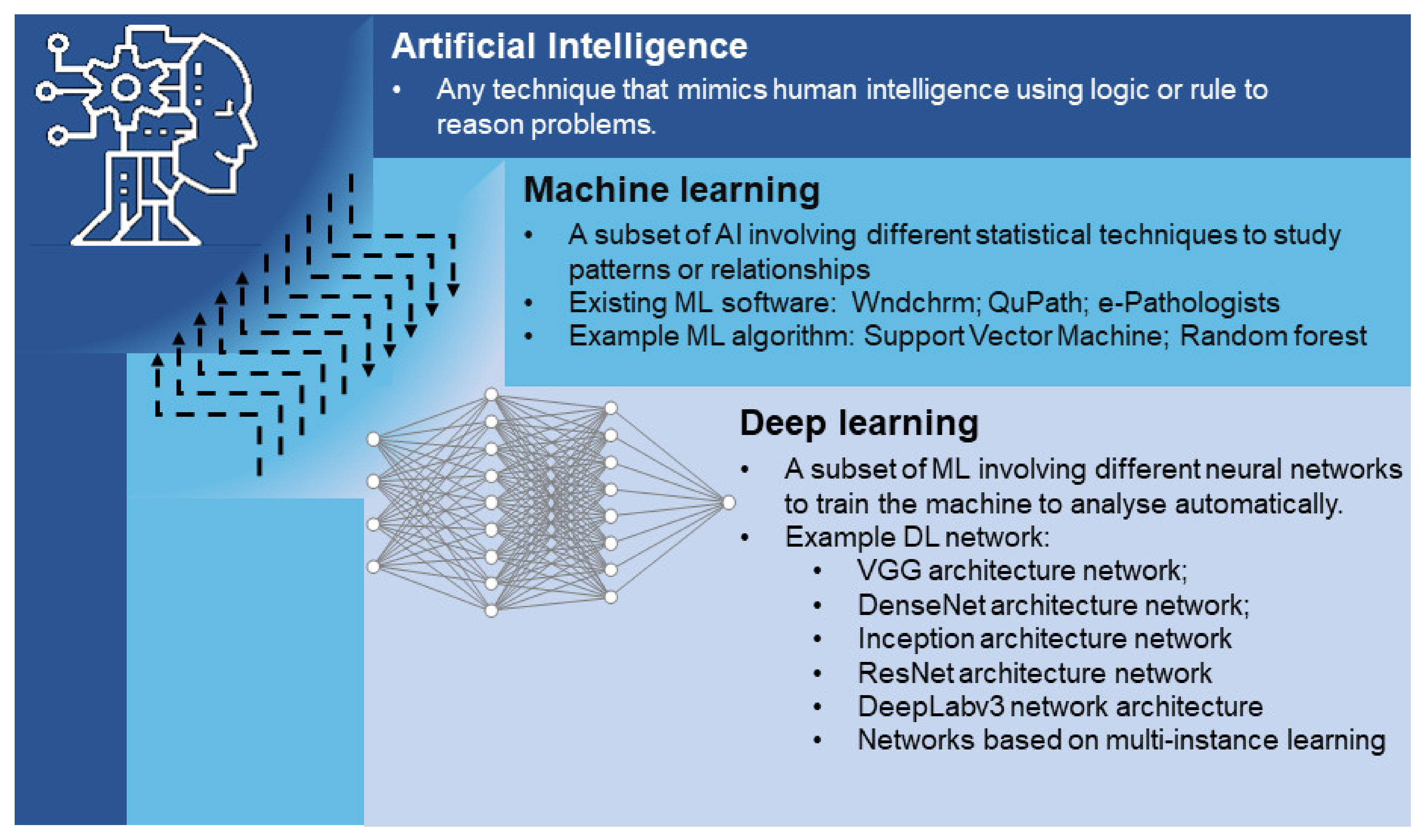
4. Machine Learning in GI Cancer Diagnosis
5. Deep Learning in GI Cancer Diagnosis
5.1. Fully Supervised Approach
5.1.1. Classification of GI Cancer
5.1.2. Segmentation in GI Cancer
5.1.3. Detection of Prognostic Markers and Prognosis in GI Cancer
5.2. Weakly Supervised Learning
| Author | Degree of Supervision | Task | Cancer Type | Type of WSI | Dataset | Algorithm/ Model | Performance | Clinical Application |
|---|---|---|---|---|---|---|---|---|
| Shen et al. [112] | Fully supervised | Classification | Gastric cancer | H&E | Training, validation and testing: 432 WSIs (TCGA-STAD cohort) + 460 WSIs (TCGA-COAD) + 171 WSIs (TCGA-READ) + 400 WSIs (Camelyon16) | DenseNet + Deformable Conditional Random Field model | Accuracy: 0.9398 (TCGA-STAD), 0.9337 (TCGA-COAD), 0.9294 (TCGA-READ), 0.9468 (Camelyon16) | Identification of suspected cancer area from histological imaging |
| Song et al. [30] | Fully supervised | Classification | Gastric cancer | H&E | Training: 2123 WSIs Validation: 300 WSIs Internal testing: 100 WSIs External validation: 3212 WSIs (daily gastric dataset) + 595 WSIs (PUMCH) + 987 WSIs (CHCAMS and Peking Union Medical College) | DeepLab v3 | Malignant vs. benign training AUC: 0.923 Internal testing AUC: 0.931 AUC: 0.995 (daily gastric dataset) AUC: 0.990 (PUMCH) AUC: 0.996 (CHCAMS and Peking Union Medical College) | Diagnosis of gastric cancer |
| Su et al. [28] | Fully supervised | Classification and detection | Gastric cancer | H&E | Training: 348 WSIs Testing: 88 WSIs External Validation: 31 WSIs | ResNet-18 | Poorly differentiated adenocarcinoma vs. well-differentiated adenocarcinoma and other normal tissue F1 score: 0.8615 Well-differentiated adenocarcinoma vs. poorly differentiated adenocarcinoma and other normal tissue F1 score: 0.8977 Patients with MSI vs. without MSI Accuracy: 0.7727 (95% CI 0.6857–0.8636) | Differentiation of cancer grade and diagnosis of MSI |
| Song et al. [29] | Fully supervised | Classification | Colorectal cancer | H&E | Training: 177 WSIs Validation: 40 WSIs Internal test: 194 WSIs External validation: 168 WSIs | Deep Lab v2 with ResNet34 | Adenomatous vs. normal AUC: 0.92 Accuracy: 0.904 | Diagnosis of colorectal adenomas |
| Sirinukunwattana et al. [114] | Fully supervised | Classification | Colorectal cancer | H&E | Training: 510 WSIs External validation: 431 WSIs (TCGA cohort) + 265 WSIs (GRAMPIAN cohort) | Inception V3 | Colorectal cancer consensus molecular subtypes 1 vs. 2 vs. 3 vs. 4 Training average accuracy: 70% Training AUC: 0.9 External validation accuracy: 0.64 (TCGA cohort) + 0.72 (GRAMPIAN cohort)External validation AUC:0.84 (TCGA cohort) + 0.85 (GRAMPIAN cohort) | Prediction of colorectal cancer molecular subtype |
| Popovici et al. [115] | Fully supervised | Classification | Colorectal cancer | H&E | Training: 100 WSIs Test: 200 WSIs | VGG-F | Molecular subtype A vs. B vs. C vs. D vs. EOverall accuracy: 0.84 (95% CI: 0.79−0.88)Overall recall: 0.85 (95% CI: 0.80−0.89)Overall precision: 0.84 (95% CI: 0.80−0.88) | Prediction of colorectal cancer molecular subtype |
| Korbar et al. [116] | Fully supervised | Classification | Colorectal cancer | H&E | Training: 458 WSIs Testing: 239 WSIs | ResNet-152 | Hyperplastic polyp vs. sessile serrated polyp vs. traditional serrated adenoma vs. tubular adenoma vs. tubulovillous/villous adenoma vs. normal Accuracy: 0.930 (95% CI: 0.890−0.959) Precision: 0.897 (95% CI: 0.852−0.932) Recall: 0.883 (95% CI: 0.836−0.921) F1 score: 0.888 (95% CI: 0.841−0.925) | Characterization of colorectal polyps |
| Wei et al. [118] | Fully supervised | Classification | Colorectal cancer | H&E | Training: 326 WSIs Validation: 25 WSIs Internet test: 157 WSIs External validation: 238 WSIs | Ensemble ResNet×5 | Hyperplastic polyp vs. sessile serrated adenoma vs. tubular adenoma vs. tubulovillous or villous adenoma. Internal test mean accuracy: 0.935 (95% CI: 0.896–0.974) External validation mean accuracy: 0.870 (95% CI: 0.827–0.913) | Colorectal polyp classification |
| Gupta et al. [119] | Fully supervised | Classification | Colorectal cancer | H&E | Training and testing: 303,012 normal WSI patches and approximately 1,000,000 abnormal WSI patches | Customized Inception-ResNet-v2 Type 5 (IR-v2 Type 5) model. | Abnormal region vs. normal region F-score: 0.99 AUC: 0.99 | Identification of suspected cancer area from histological imaging |
| Kather et al. [117] | Fully supervised | Classification and prognosis | Colorectal cancer | H&E | Training: 86 WSIs Testing: 25 WSIs External validation: 862 WSIs (TCGA cohort) + 409 WSIs (DACHS cohort) | VGG19 | Adipose tissue vs. background vs. lymphocytes vs. mucus vs. smooth muscle vs. normal colon mucosa vs. cancer-associated stroma vs. colorectal adenocarcinoma epithelium Internal testing Overall Accuracy: 0.99 External testing Overall accuracy: 0.943 High deep stroma score predicts shorter survival Hazard ratio: 1.99 (95% CI: 1.27–3.12) | Colorectal cancer detection and prediction of patient survival outcome |
| Zhu et al. [31] | Fully supervised | Classification and segmentation | Gastric and colorectal cancer | H&E | Training: 750 WSIs Testing: 250 WSIs | Adversarial CAC-UNet | Malignant region vs. benign region DSC: 0.8749 Recall: 0.9362 Precision: 0.9027 Accuracy: 0.8935 | Identification of suspected cancer area from histological imaging |
| Xu et al. [33] | Fully supervised | Segmentation | Colorectal cancer | H&E | Training: 750 WSIs Testing: 250 WSIs | CoUNet | Malignant region vs. benign region Dice: 0.746 AUC: 0.980 | Identification of suspected cancer area from histological imaging |
| Feng et al. [32] | Fully supervised | Segmentation | Colorectal cancer | H&E | Training: 750 WSIs Testing: 250 WSIs | U-Net-16 | Malignant region vs. benign region DSC: 0.7789 AUC:1 | Identification of suspected cancer area from histological imaging |
| Mahendra et al. [120] | Fully supervised | Segmentation | Colorectal cancer | H&E | Training: 270 WSIs (CAMELYON16) + 500 WSIs (CAMELYON17) + 660 WSIs (DigestPath) + 50 WSIs (PAIP) Testing: 129 WSIs (CAMELYON16) + 500 WSIs (CAMELYON17) + 212 WSIs (DigestPath) + 40 WSIs (PAIP) | DenseNet-121 + Inception-ResNet-V2 + DeeplabV3Plus | Malignant region vs. benign region Cohen kappa score: 0.9090 (CAMELYON17) DSC: 0.782 (DigestPath) | Identification of suspected cancer area from histological imaging |
| Gehrung et al. [34] | Fully supervised | Detection | Oesophageal cancer | H&E and TFF3 pathology slides | Training: 100 + 187 patients Validation: 187 patients External validation: 1519 patients | VGG-16 | Patients with Barrett’s oesophagus vs. no Barrett’s oesophagus AUC: 0.88 (95% CI: 0.85–0.91) Sensitivity: 0.7262 (95% CI: 0.6742–0.7821) Specificity: 0.9313 (95% CI: 0.9004–0.9613) Simulated realistic cohort workload reduction: 57% External validation cohort reduction: 57.41% | Detection of Barrett’s oesophagus |
| Kather et al. [122] | Fully supervised | Detection | Gastric and colorectal cancer | H&E | Training: 81 patients (UMM and NCT tissue bank) + 216 patients (TCGA-STAD) + 278 patients (TCGA-CRC-KR) + 260 patients (TCGA-CRC-DX) + 382 patients (UCEC) External validation: 99 patients (TCGA-STAD) + 109 patients (TCGA-CRC-KR) +100 patients (TCGA-CRC-DX) +110 patients (UCEC) + 185 patients (KCCH) | Resnet18 | Patients with MSI vs. no MSI Training AUC: >0.99 (UMM and NCT tissue bank) AUC: 0.81 (CI: 0.69–0.90) (TCGA-STAD) AUC: 0.84 (CI: 0.73–0.91) (TCGA-CRC-KR) AUC: 0.77 (CI: 0.62–0.87) (TCGA-CRC-DX) AUC: 0.75 (CI: 0.63–0.83) (UCEC) AUC: 0.69 (CI: 0.52–0.82) (KCCH) | Detection of MSI |
| Echle et al. [36] | Fully supervised | Detection | Colorectal cancer | H&E | Training: 6406 WSIs External validation: 771 WSIs | Shufflenet | Colorectal tumour sample with dMMR or MSI vs. no dMMR or MSI Mean AUC: 0.92 AUPRC: 0.93 Specificity: 0.67 Sensitivity: 0.95 External validation AUC without colour normalisation: 0.95 External validation AUC with colour normalisation: 0.96 | Detection of MSI |
| Cao et al. [121] | Fully supervised | Detection | Colorectal cancer | H&E | Training: 429 WSIs External validation: 785 WSIs | ResNet-18 | Colorectal cancer patients with MSI vs. no MSI AUC: 0.8848 (95% CI: 0.8185–0.9512) External validation AUC: 0.8504 (95% CI: 0.7591–0.9323) | Detection of MSI |
| Meier et al. [124] | Fully supervised | Prognosis | Gastric cancer | H&E IHC staining, including CD8, CD20, CD68 and Ki67 | Training and testing: 248 patients | GoogLeNet | Risk of the presence of Ki67&CD20 Hazard ratio = 1.47 (95% CI: 1.15–1.89) Risk of the presence of CD20&CD68 Hazard ratio = 1.33 (95% CI: 1.07–1.67) | Cancer prognosis based on various IHC markers to predict patient survival outcome |
| Bychkov et al. [123] | Fully supervised | Prognosis | Colorectal cancer | H&E | Training: 220 WSIs Validation: 60 WSIs Testing: 140 WSIs | VGG-16 | High-risk patients vs. low-risk patients Prediction with small tissue hazard ratio: 2.3 (95% CI: 1.79–3.03) | Survival analysis of colorectal cancer |
| Wang et al. [127] | Weakly supervised | Classification | Gastric cancer | H&E | Training: 408 WSIs Testing: 200 WSIs | recalibrated multi-instance deep learning | Cancer vs. dysplasia vs. normal Accuracy: 0.865 | Diagnosis of gastric cancer |
| Xu et al. [37] | Weakly supervised | Classification | Gastric cancer | H&E | Training, validation and testing: 185 WSIs (SRS dataset) + 2032 WSIs (Mars dataset) | multiple instance classification framework based on graph convolutional networks | Tumour vs. normal Recall: 0.904 (SRS dataset), 0.9824 (Mars dataset) Precision: 0.9116 (SRS dataset), 0.9826 (Mars dataset) F1-score: 0.9075 (SRS dataset), 0.9824 (Mars dataset) | Diagnosis of gastric cancer |
| Huang et al. [39] | Weakly supervised | Classification | Gastric cancer | H&E | Training and testing: 2333 WSIs External validation: 175 WSIs | GastroMIL | Gastric cancer vs. normal External validation accuracy: 0.92 GastroMIL risk score associated with patient overall survival Hazard ratio: 2.414 | Diagnosis of gastric cancer and prediction of patient survival outcome |
| Li et al. [131] | Weakly supervised | Classification | Gastric cancer | H&E | Training and testing: 10,894 WSIs | DLA34 + Otsu’s method | Tumour vs. normal Sensitivity: 1.0000 Specificity: 0.8932 AUC: 0.9906 | Diagnosis of gastric cancer |
| Chen et al. [133] | Weakly supervised | Classification | Colorectal cancer | H&E | Training and testing: 400 WSIs | CNN classifier | Normal (including hyperplastic polyp) vs. adenoma vs. adenocarcinoma vs. mucinous adenocarcinoma vs. signet ring cell carcinoma Overall accuracy: 0.76 | Prediction of colorectal cancer tumour grade |
| Ye et al. [38] | Weakly supervised | Classification | Colorectal cancer | H&E | Training and testing: 100 WSIs | Multiple-instance CNN | With epithelial cell nuclei vs. no epithelial cell nuclei Accuracy: 0.936 Precision: 0.922 Recall: 0.960 | Detection of colon cancer |
| Sharma et al. [129] | Weakly supervised | Classification | Gastrointestinal cancer | H&E | Training and testing: 413 WSIs | Cluster-to-Conquer framework | Celiac cancer vs. normal Accuracy: 0.862 Precision: 0.855 Recall: 0.922 F1-score: 0.887 | Detection of gastrointestinal cancer |
| Klein et al. [130] | Weakly supervised | Detection | Gastric cancer | H&E + Giemsa staining | Training: 191 H&E WSIs and 286 Giemsa-stained WSIs Validation: 71 H&E WSIs and 87 Giemsa-stained WSIs External validation: 364 H&E WSIs and 347 Giemsa-stained WSIs | VGG+ + active learning | H. pylori vs. no H. pylori External validation AUC: 0.81 (H&E) + 0.92 (Giemsa-stained) | Detection of H. pylori |
6. Clinical Insight for Selected AI Applications in Early Diagnosis and Monitoring of the Progression of GI Cancer
6.1. Detection of Early GI Premalignant Lesions
6.2. Tumour Subtyping Characterization and Estimation of Patient Outcome
6.3. Screening GI Cancer in Daily Clinical Operation
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Arnold, M.; Abnet, C.C.; Neale, R.E.; Vignat, J.; Giovannucci, E.L.; McGlynn, K.A.; Bray, F. Global Burden of 5 Major Types of Gastrointestinal Cancer. Gastroenterology 2020, 159, 335–349.e15. [Google Scholar] [CrossRef]
- Islami, F.; Goding Sauer, A.; Miller, K.D.; Siegel, R.L.; Fedewa, S.A.; Jacobs, E.J.; McCullough, M.L.; Patel, A.V.; Ma, J.; Soerjomataram, I.; et al. Proportion and number of cancer cases and deaths attributable to potentially modifiable risk factors in the United States. CA Cancer J. Clin. 2018, 68, 31–54. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Siegel, R.L.; Rosenberg, P.S.; Jemal, A. Emerging cancer trends among young adults in the USA: Analysis of a population-based cancer registry. Lancet Public Health 2019, 4, e137–e147. [Google Scholar] [CrossRef] [Green Version]
- Yan, H.; Sellick, K. Symptoms, psychological distress, social support, and quality of life of Chinese patients newly diagnosed with gastrointestinal cancer. Cancer Nurs. 2004, 27, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Kamel, H.M. Trends and challenges in pathology practice: Choices and necessities. Sultan Qaboos Univ. Med. J. 2011, 11, 38. [Google Scholar]
- Rao, G.G.; Crook, M.; Tillyer, M. Pathology tests: Is the time for demand management ripe at last? J. Clin. Pathol. 2003, 56, 243–248. [Google Scholar] [PubMed] [Green Version]
- Hassell, L.A.; Parwani, A.V.; Weiss, L.; Jones, M.A.; Ye, J. Challenges and opportunities in the adoption of College of American Pathologists checklists in electronic format: Perspectives and experience of Reporting Pathology Protocols Project (RPP2) participant laboratories. Arch. Pathol. Lab. Med. 2010, 134, 1152–1159. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, S.M.; Lewis, F.A.; Cao, Y.; Conrad, R.C.; Cronin, M.; Danenberg, K.D.; Goralski, T.J.; Langmore, J.P.; Raja, R.G.; Williams, P.M. Tissue handling and specimen preparation in surgical pathology: Issues concerning the recovery of nucleic acids from formalin-fixed, paraffin-embedded tissue. Arch. Pathol. Lab. Med. 2008, 132, 1929–1935. [Google Scholar] [CrossRef] [PubMed]
- Ribé, A.; Ribalta, T.; Lledó, R.; Torras, G.; Asenjo, M.A.; Cardesa, A. Evaluation of turnaround times as a component of quality assurance in surgical pathology. Int. J. Qual. Health Care 1998, 10, 241–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamil, S.M.; Srinivas, A. Evaluation of quality management systems implementation in medical diagnostic laboratories benchmarked for accreditation. J. Med. Lab. Diagn. 2015, 6, 27–35. [Google Scholar] [CrossRef] [Green Version]
- Peter, T.F.; Rotz, P.D.; Blair, D.H.; Khine, A.-A.; Freeman, R.R.; Murtagh, M.M. Impact of laboratory accreditation on patient care and the health system. Am. J. Clin. Pathol. 2010, 134, 550–555. [Google Scholar] [CrossRef] [Green Version]
- The Royal College of Pathologists. Digital Pathology. Available online: https://www.rcpath.org/profession/digital-pathology.html (accessed on 20 March 2022).
- Williams, B.J.; Hanby, A.; Millican-Slater, R.; Nijhawan, A.; Verghese, E.; Treanor, D. Digital pathology for the primary diagnosis of breast histopathological specimens: An innovative validation and concordance study on digital pathology validation and training. Histopathology 2018, 72, 662–671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, B.J.; Jayewardene, D.; Treanor, D. Digital immunohistochemistry implementation, training and validation: Experience and technical notes from a large clinical laboratory. J. Clin. Pathol. 2019, 72, 373–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bera, K.; Schalper, K.A.; Rimm, D.L.; Velcheti, V.; Madabhushi, A. Artificial intelligence in digital pathology—new tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 2019, 16, 703–715. [Google Scholar] [CrossRef] [PubMed]
- Van der Laak, J.; Litjens, G.; Ciompi, F. Deep learning in histopathology: The path to the clinic. Nat. Med. 2021, 27, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Tizhoosh, H.R.; Pantanowitz, L. Artificial Intelligence and Digital Pathology: Challenges and Opportunities. J. Pathol. Inform. 2018, 9, 38. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, A.; Khartchenko, A.F.; Pati, P.; Gabrani, M.; Schraml, P.; Kaigala, G.V. Quantitative microimmunohistochemistry for the grading of immunostains on tumour tissues. Nat. Biomed. Eng. 2019, 3, 478–490. [Google Scholar] [CrossRef]
- Arar, N.M.; Pati, P.; Kashyap, A.; Khartchenko, A.F.; Goksel, O.; Kaigala, G.V.; Gabrani, M. Computational immunohistochemistry: Recipes for standardization of immunostaining. In Medical Image Computing and Computer-Assisted Intervention−MICCAI 2017; Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D., Duchesne, S., Eds.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2017; pp. 48–55. [Google Scholar]
- Arar, N.M.; Pati, P.; Kashyap, A.; Khartchenko, A.F.; Goksel, O.; Kaigala, G.V.; Gabrani, M. High-quality immunohistochemical stains through computational assay parameter optimization. IEEE Trans. Biomed. Eng. 2019, 66, 2952–2963. [Google Scholar] [CrossRef] [Green Version]
- Srinivasu, P.N.; SivaSai, J.G.; Ijaz, M.F.; Bhoi, A.K.; Kim, W.; Kang, J.J. Classification of Skin Disease Using Deep Learning Neural Networks with MobileNet V2 and LSTM. Sensors 2021, 21, 2852. [Google Scholar] [CrossRef]
- Kelly, C.J.; Karthikesalingam, A.; Suleyman, M.; Corrado, G.; King, D. Key challenges for delivering clinical impact with artificial intelligence. BMC Med. 2019, 17, 195. [Google Scholar] [CrossRef] [Green Version]
- Vulli, A.; Srinivasu, P.N.; Sashank, M.S.K.; Shafi, J.; Choi, J.; Ijaz, M.F. Fine-Tuned DenseNet-169 for Breast Cancer Metastasis Prediction Using FastAI and 1-Cycle Policy. Sensors 2022, 22, 2988. [Google Scholar] [CrossRef]
- Yoshida, H.; Shimazu, T.; Kiyuna, T.; Marugame, A.; Yamashita, Y.; Cosatto, E.; Taniguchi, H.; Sekine, S.; Ochiai, A. Automated histological classification of whole-slide images of gastric biopsy specimens. Gastric Cancer 2017, 21, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, Y.; Tokunaga, K.; Koga, T.; Sakamoto, C.; Goldberg, I.G.; Saitoh, N.; Nakao, M. Computational analysis of morphological and molecular features in gastric cancer tissues. Cancer Med. 2020, 9, 2223–2234. [Google Scholar] [CrossRef] [PubMed]
- Cosatto, E.; Laquerre, P.-F.; Malon, C.; Graf, H.P.; Saito, A.; Kiyuna, T.; Marugame, A.; Kamijo, K. Automated gastric cancer diagnosis on H&E-stained sections; ltraining a classifier on a large scale with multiple instance machine learning. Proc. SPIE 2013, 8676, 867605. [Google Scholar] [CrossRef]
- Jiang, D.; Liao, J.; Duan, H.; Wu, Q.; Owen, G.; Shu, C.; Chen, L.; He, Y.; Wu, Z.; He, D.; et al. A machine learning-based prognostic predictor for stage III colon cancer. Sci. Rep. 2020, 10, 10333. [Google Scholar] [CrossRef] [PubMed]
- Su, F.; Li, J.; Zhao, X.; Wang, B.; Hu, Y.; Sun, Y.; Ji, J. Interpretable tumor differentiation grade and microsatellite instability recognition in gastric cancer using deep learning. Lab. Investig. 2022, 102, 641–649. [Google Scholar] [CrossRef]
- Song, Z.; Yu, C.; Zou, S.; Wang, W.; Huang, Y.; Ding, X.; Liu, J.; Shao, L.; Yuan, J.; Gou, X.; et al. Automatic deep learning-based colorectal adenoma detection system and its similarities with pathologists. BMJ Open 2020, 10, e036423. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Zou, S.; Zhou, W.; Huang, Y.; Shao, L.; Yuan, J.; Gou, X.; Jin, W.; Wang, Z.; Chen, X.; et al. Clinically applicable histopathological diagnosis system for gastric cancer detection using deep learning. Nat. Commun. 2020, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Mei, K.; Peng, T.; Luo, Y.; Liu, J.; Wang, Y.; Jin, M. Multi-level colonoscopy malignant tissue detection with adversarial CAC-UNet. Neurocomputing 2021, 438, 165–183. [Google Scholar] [CrossRef]
- Feng, R.; Liu, X.; Chen, J.; Chen, D.Z.; Gao, H.; Wu, J. A Deep Learning Approach for Colonoscopy Pathology WSI Analysis: Accurate Segmentation and Classification. IEEE J. Biomed. Health Inform. 2021, 25, 3700–3708. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Liu, H.; Wang, X.; Ouyang, H.; Qian, Y. CoUNet: An End-to-End Colonoscopy Lesion Image Segmentation and Classification Framework. In ICVIP 2020: 2020 The 4th International Conference on Video and Image Processing, 25–27 December 2020, Xi’an China; Association for Computing Machinery: New York, NY, USA, 2020; pp. 81–87. [Google Scholar]
- Gehrung, M.; Crispin-Ortuzar, M.; Berman, A.G.; O’Donovan, M.; Fitzgerald, R.C.; Markowetz, F. Triage-driven diagnosis of Barrett’s esophagus for early detection of esophageal adenocarcinoma using deep learning. Nat. Med. 2021, 27, 833–841. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Marklund, H.; Bláha, O.; Desai, M.; Martin, B.A.; Bingham, D.B.; Berry, G.J.; Gomulia, E.; Ng, A.; Shen, J. Deep learning assistance for the histopathologic diagnosis of Helicobacter pylori. Intell. Med. 2020, 1–2, 100004. [Google Scholar] [CrossRef]
- Echle, A.; Grabsch, H.I.; Quirke, P.; van den Brandt, P.A.; West, N.P.; Hutchins, G.G.A.; Heij, L.R.; Tan, X.; Richman, S.D.; Krause, J.; et al. Clinical-Grade Detection of Microsatellite Instability in Colorectal Tumors by Deep Learning. Gastroenterology 2020, 159, 1406–1416.e11. [Google Scholar] [CrossRef]
- Xiang, X.; Wu, X. Multiple Instance Classification for Gastric Cancer Pathological Images Based on Implicit Spatial Topological Structure Representation. Appl. Sci. 2021, 11, 10368. [Google Scholar] [CrossRef]
- Ye, T.; Lan, R.; Luo, X. Multiple-instance CNN Improved by S3TA for Colon Cancer Classification with Unannotated Histopathological Images. In Proceedings of the 2021 11th International Conference on Intelligent Control and Information Processing (ICICIP), Dali, China, 3–7 December 2021; pp. 444–448. [Google Scholar]
- Huang, B.; Tian, S.; Zhan, N.; Ma, J.; Huang, Z.; Zhang, C.; Zhang, H.; Ming, F.; Liao, F.; Ji, M.; et al. Accurate diagnosis and prognosis prediction of gastric cancer using deep learning on digital pathological images: A retrospective multicentre study. EBioMedicine 2021, 73, 103631. [Google Scholar] [CrossRef]
- Li, B.; Li, Y.; Eliceiri, K.W. Dual-stream Multiple Instance Learning Network for Whole Slide Image Classification with Self-supervised Contrastive Learning. In Proceedings of the 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Nashville, TN, USA, 20–25 June 2021; pp. 14313–14323. [Google Scholar]
- Tellez, D.; Litjens, G.; Bándi, P.; Bulten, W.; Bokhorst, J.-M.; Ciompi, F.; van der Laak, J. Quantifying the effects of data augmentation and stain color normalization in convolutional neural networks for computational pathology. Med. Image Anal. 2019, 58, 101544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Pantanowitz, L.; Schuffler, P.; Yarlagadda, D.V.K.; Ardon, O.; Reuter, V.E.; Hameed, M.; Klimstra, D.S.; Hanna, M.G. (Re) Defining the High-Power Field for Digital Pathology. J. Pathol. Inform. 2020, 11, 33. [Google Scholar] [CrossRef] [PubMed]
- Dun, X.-p.; Parkinson, D.B. Visualizing peripheral nerve regeneration by whole mount staining. PLoS ONE 2015, 10, e0119168. [Google Scholar]
- Heffner, S.; Colgan, O.; Doolan, C. Digital Pathology. Available online: https://www.leicabiosystems.com/en-br/knowledge-pathway/digital-pathology/ (accessed on 30 April 2022).
- Borowsky, A.D.; Glassy, E.F.; Wallace, W.D.; Kallichanda, N.S.; Behling, C.A.; Miller, D.V.; Oswal, H.N.; Feddersen, R.M.; Bakhtar, O.R.; Mendoza, A.E.; et al. Digital Whole Slide Imaging Compared with Light Microscopy for Primary Diagnosis in Surgical Pathology. Arch. Pathol. Lab. Med. 2020, 144, 1245–1253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snead, D.R.; Tsang, Y.W.; Meskiri, A.; Kimani, P.K.; Crossman, R.; Rajpoot, N.M.; Blessing, E.; Chen, K.; Gopalakrishnan, K.; Matthews, P.; et al. Validation of digital pathology imaging for primary histopathological diagnosis. Histopathology 2016, 68, 1063–1072. [Google Scholar] [CrossRef] [Green Version]
- Hanna, M.G.; Reuter, V.E.; Ardon, O.; Kim, D.; Sirintrapun, S.J.; Schüffler, P.J.; Busam, K.J.; Sauter, J.L.; Brogi, E.; Tan, L.K.; et al. Validation of a digital pathology system including remote review during the COVID-19 pandemic. Mod. Pathol. 2020, 33, 2115–2127. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.L.; Azhar, R.; Sng, S.H.; Chua, Y.Q.; Hwang, J.S.; Chin, J.P.; Seah, W.K.; Loke, J.C.; Ang, R.H.; Tan, P.H. Enabling digital pathology in the diagnostic setting: Navigating through the implementation journey in an academic medical centre. J. Clin. Pathol. 2016, 69, 784–792. [Google Scholar] [CrossRef] [PubMed]
- Aloqaily, A.; Polonia, A.; Campelos, S.; Alrefae, N.; Vale, J.; Caramelo, A.; Eloy, C. Digital Versus Optical Diagnosis of Follicular Patterned Thyroid Lesions. Head Neck Pathol. 2021, 15, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Salvi, M.; Acharya, U.R.; Molinari, F.; Meiburger, K.M. The impact of pre- and post-image processing techniques on deep learning frameworks: A comprehensive review for digital pathology image analysis. Comput. Biol. Med. 2021, 128, 104129. [Google Scholar] [CrossRef] [PubMed]
- Taqi, S.A.; Sami, S.A.; Sami, L.B.; Zaki, S.A. A review of artifacts in histopathology. J. Oral Maxillofac. Pathol. 2018, 22, 279. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef] [Green Version]
- Aubreville, M.; Bertram, C.; Klopfleisch, R.; Maier, A. SlideRunner. In Bildverarbeitung für die Medizin 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 309–314. [Google Scholar]
- Williams, B.; Hanby, A.; Millican-Slater, R.; Verghese, E.; Nijhawan, A.; Wilson, I.; Besusparis, J.; Clark, D.; Snead, D.; Rakha, E.; et al. Digital pathology for primary diagnosis of screen-detected breast lesions—Experimental data, validation and experience from four centres. Histopathology 2020, 76, 968–975. [Google Scholar] [CrossRef]
- Eloy, C.; Vale, J.; Curado, M.; Polonia, A.; Campelos, S.; Caramelo, A.; Sousa, R.; Sobrinho-Simoes, M. Digital Pathology Workflow Implementation at IPATIMUP. Diagnostics 2021, 11, 2111. [Google Scholar] [CrossRef]
- Abels, E.; Pantanowitz, L.; Aeffner, F.; Zarella, M.D.; van der Laak, J.; Bui, M.M.; Vemuri, V.N.; Parwani, A.V.; Gibbs, J.; Agosto-Arroyo, E.; et al. Computational pathology definitions, best practices, and recommendations for regulatory guidance: A white paper from the Digital Pathology Association. J. Pathol. 2019, 249, 286–294. [Google Scholar] [CrossRef]
- Hawkes, N. Cancer survival data emphasise importance of early diagnosis. BMJ 2019, 364, l408. [Google Scholar] [CrossRef]
- Schiffman, J.D.; Fisher, P.G.; Gibbs, P. Early detection of cancer: Past, present, and future. Am. Soc. Clin. Oncol. Educ. Book 2015, 35, 57–65. [Google Scholar] [CrossRef] [Green Version]
- Williams, B.J.; Bottoms, D.; Treanor, D. Future-proofing pathology: The case for clinical adoption of digital pathology. J. Clin. Pathol. 2017, 70, 1010–1018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maung, R. Pathologists’ workload and patient safety. Diagn. Histopathol. 2016, 22, 283–287. [Google Scholar] [CrossRef]
- Esteva, A.; Robicquet, A.; Ramsundar, B.; Kuleshov, V.; DePristo, M.; Chou, K.; Cui, C.; Corrado, G.; Thrun, S.; Dean, J. A guide to deep learning in healthcare. Nat. Med. 2019, 25, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Miotto, R.; Wang, F.; Wang, S.; Jiang, X.; Dudley, J.T. Deep learning for healthcare: Review, opportunities and challenges. Brief. Bioinform. 2018, 19, 1236–1246. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Jiang, Y.; Zhi, H.; Dong, Y.; Li, H.; Ma, S.; Wang, Y.; Dong, Q.; Shen, H.; Wang, Y. Artificial intelligence in healthcare: Past, present and future. Stroke Vasc. Neurol. 2017, 2, 230–243. [Google Scholar] [CrossRef] [PubMed]
- Holzinger, A.; Langs, G.; Denk, H.; Zatloukal, K.; Muller, H. Causability and explainability of artificial intelligence in medicine. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2019, 9, e1312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitney, J.; Corredor, G.; Janowczyk, A.; Ganesan, S.; Doyle, S.; Tomaszewski, J.; Feldman, M.; Gilmore, H.; Madabhushi, A. Quantitative nuclear histomorphometry predicts oncotype DX risk categories for early stage ER+ breast cancer. BMC Cancer 2018, 18, 610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinata, M.; Ushiku, T. Detecting immunotherapy-sensitive subtype in gastric cancer using histologic image-based deep learning. Sci. Rep. 2021, 11, 22636. [Google Scholar] [CrossRef]
- Rathore, S.; Iftikhar, M.A.; Chaddad, A.; Niazi, T.; Karasic, T.; Bilello, M. Segmentation and Grade Prediction of Colon Cancer Digital Pathology Images Across Multiple Institutions. Cancers 2019, 11, 1700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ehteshami Bejnordi, B.; Veta, M.; Johannes van Diest, P.; van Ginneken, B.; Karssemeijer, N.; Litjens, G.; van der Laak, J.; Hermsen, M.; Manson, Q.F.; Balkenhol, M.; et al. Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women with Breast Cancer. JAMA 2017, 318, 2199–2210. [Google Scholar] [CrossRef] [PubMed]
- Bandi, P.; Geessink, O.; Manson, Q.; Van Dijk, M.; Balkenhol, M.; Hermsen, M.; Ehteshami Bejnordi, B.; Lee, B.; Paeng, K.; Zhong, A.; et al. From Detection of Individual Metastases to Classification of Lymph Node Status at the Patient Level: The CAMELYON17 Challenge. IEEE Trans. Med. Imaging 2019, 38, 550–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Royal College of Pathologists of Australasia. Ageing Pathologists. Available online: https://www.rcpa.edu.au/getattachment/95c190e1-bdbe-4ab1-83e1-0a218c69ad82/Ageing-Pathologists.aspx (accessed on 1 May 2022).
- The Royal College of Pathologists of Australia. Becoming a Pathologist. Available online: https://www.rcpa.edu.au/Pathology-Careers/Becoming-a-Pathologist (accessed on 30 April 2022).
- Yagi, Y. Color standardization and optimization in whole slide imaging. Diagn. Pathol. 2011, 6 (Suppl. S1), S15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kothari, S.; Phan, J.H.; Moffitt, R.A.; Stokes, T.H.; Hassberger, S.E.; Chaudry, Q.; Young, A.N.; Wang, M.D. Automatic batch-invariant color segmentation of histological cancer images. In Proceedings of the 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Chicago, IL, USA, 30 March–2 April 2011; pp. 657–660. [Google Scholar]
- Tabesh, A.; Teverovskiy, M.; Pang, H.-Y.; Kumar, V.P.; Verbel, D.; Kotsianti, A.; Saidi, O. Multifeature prostate cancer diagnosis and Gleason grading of histological images. IEEE Trans. Med. Imaging 2007, 26, 1366–1378. [Google Scholar] [CrossRef] [PubMed]
- Reinhard, E.; Adhikhmin, M.; Gooch, B.; Shirley, P. Color transfer between images. IEEE Comput. Graph. Appl. 2001, 21, 34–41. [Google Scholar] [CrossRef]
- Abe, T.; Murakami, Y.; Yamaguchi, M.; Ohyama, N.; Yagi, Y. Color correction of pathological images based on dye amount quantification. Opt. Rev. 2005, 12, 293–300. [Google Scholar] [CrossRef]
- Magee, D.; Treanor, D.; Crellin, D.; Shires, M.; Smith, K.; Mohee, K.; Quirke, P. Colour normalisation in digital histopathology images. In Proceedings of the Optical Tissue Image analysis in Microscopy, Histopathology and Endoscopy (MICCAI Workshop), London, UK, 24 September 2009; Daniel Elson: London, UK, 2009; Volume 100, pp. 100–111. [Google Scholar]
- Macenko, M.; Niethammer, M.; Marron, J.S.; Borland, D.; Woosley, J.T.; Guan, X.; Schmitt, C.; Thomas, N.E. A method for normalizing histology slides for quantitative analysis. In Proceedings of the 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Boston, MA, USA, 28 June–1 July 2009; pp. 1107–1110. [Google Scholar]
- Tani, S.; Fukunaga, Y.; Shimizu, S.; Fukunishi, M.; Ishii, K.; Tamiya, K. Color standardization method and system for whole slide imaging based on spectral sensing. Anal. Cell. Pathol. 2012, 35, 107–115. [Google Scholar] [CrossRef]
- Niethammer, M.; Borland, D.; Marron, J.S.; Woosley, J.; Thomas, N.E. Appearance normalization of histology slides. In MLMI 2010: Machine Learning in Medical Imaging; Wang, F., Yan, P., Suzuki, K., Shen, D., Eds.; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2010; Volume 6357, pp. 58–66. [Google Scholar]
- Shaban, M.; Baur, C.; Navab, N.; Albarqouni, S. Staingan: Stain Style Transfer for Digital Histological Images. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 953–956. [Google Scholar]
- Kausar, T.; Kausar, A.; Ashraf, M.A.; Siddique, M.F.; Wang, M.; Sajid, M.; Siddique, M.Z.; Haq, A.U.; Riaz, I. SA-GAN: Stain Acclimation Generative Adversarial Network for Histopathology Image Analysis. Appl. Sci. 2022, 12, 288. [Google Scholar] [CrossRef]
- Cong, C.; Liu, S.; Di Ieva, A.; Pagnucco, M.; Berkovsky, S.; Song, Y. Texture Enhanced Generative Adversarial Network for Stain Normalisation in Histopathology Images. In Proceedings of the 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), Nice, France, 13–16 April 2021; pp. 1949–1952. [Google Scholar]
- Patil, A.; Talha, M.; Bhatia, A.; Kurian, N.C.; Mangale, S.; Patel, S.; Sethi, A. Fast, Self Supervised, Fully Convolutional Color Normalization of H&E Stained Images. In Proceedings of the 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), Nice, France, 13–16 April 2021; pp. 1563–1567. [Google Scholar]
- Bug, D.; Schneider, S.; Grote, A.; Oswald, E.; Feuerhake, F.; Schüler, J.; Merhof, D. Context-Based Normalization of Histological Stains Using Deep Convolutional Features. In Proceedings of the Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, Cham, Switzerland, 9 September 2017; pp. 135–142. [Google Scholar]
- Baba, A.I.; Câtoi, C. Comparative Oncology; The Publishing House of the Romanian Academy: Bucharest, Romania, 2007. [Google Scholar]
- Elmore, J.G.; Longton, G.M.; Carney, P.A.; Geller, B.M.; Onega, T.; Tosteson, A.N.; Nelson, H.D.; Pepe, M.S.; Allison, K.H.; Schnitt, S.J.; et al. Diagnostic concordance among pathologists interpreting breast biopsy specimens. JAMA 2015, 313, 1122–1132. [Google Scholar] [CrossRef]
- Ali, F.; Khan, P.; Riaz, K.; Kwak, D.; Abuhmed, T.; Park, D.; Kwak, K.S. A fuzzy ontology and SVM–based Web content classification system. IEEE Access 2017, 5, 25781–25797. [Google Scholar] [CrossRef]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin. 2017, 67, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Azam, A.S.; Miligy, I.M.; Kimani, P.K.U.; Maqbool, H.; Hewitt, K.; Rajpoot, N.M.; Snead, D.R.J. Diagnostic concordance and discordance in digital pathology: A systematic review and meta-analysis. J. Clin. Pathol. 2021, 74, 448. [Google Scholar] [CrossRef] [PubMed]
- Buck, T.P.; Dilorio, R.; Havrilla, L.; O’Neill, D.G. Validation of a whole slide imaging system for primary diagnosis in surgical pathology: A community hospital experience. J. Pathol. Inform. 2014, 5, 43. [Google Scholar] [CrossRef]
- Tabata, K.; Mori, I.; Sasaki, T.; Itoh, T.; Shiraishi, T.; Yoshimi, N.; Maeda, I.; Harada, O.; Taniyama, K.; Taniyama, D. Whole-slide imaging at primary pathological diagnosis: Validation of whole-slide imaging-based primary pathological diagnosis at twelve Japanese academic institutes. Pathol. Int. 2017, 67, 547–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cross, S.; Furness, P.; Igali, L.; Snead, D.; Treanor, D. Best Practice Recommendations for Implementing Digital Pathology January 2018; The Royal College of Pathologists: London, UK, 2018; Available online: https://www.rcpath.org/uploads/assets/f465d1b3-797b-4297-b7fedc00b4d77e51/Best-practice-recommendations-for-implementing-digital-pathology.pdf (accessed on 16 June 2022).
- Linardatos, P.; Papastefanopoulos, V.; Kotsiantis, S. Explainable AI: A Review of Machine Learning Interpretability Methods. Entropy 2020, 23, 18. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Ertosun, M.G.; Rubin, D.L. Automated Grading of Gliomas using Deep Learning in Digital Pathology Images: A modular approach with ensemble of convolutional neural networks. AMIA Annu. Symp. Proc. 2015, 2015, 1899–1908. [Google Scholar] [PubMed]
- Barker, J.; Hoogi, A.; Depeursinge, A.; Rubin, D.L. Automated classification of brain tumor type in whole-slide digital pathology images using local representative tiles. Med. Image Anal. 2016, 30, 60–71. [Google Scholar] [CrossRef] [Green Version]
- Langer, L.; Binenbaum, Y.; Gugel, L.; Amit, M.; Gil, Z.; Dekel, S. Computer-aided diagnostics in digital pathology: Automated evaluation of early-phase pancreatic cancer in mice. Int. J. Comput. Assist. Radiol. Surg. 2015, 10, 1043–1054. [Google Scholar] [CrossRef]
- Amann, J.; Blasimme, A.; Vayena, E.; Frey, D.; Madai, V.I. Explainability for artificial intelligence in healthcare: A multidisciplinary perspective. BMC Med. Inform. Decis. Mak. 2020, 20, 310. [Google Scholar] [CrossRef]
- Goldfarb, A.; Teodoridis, F. Why Is AI Adoption in Health Care Lagging? Available online: https://www.brookings.edu/research/why-is-ai-adoption-in-health-care-lagging/ (accessed on 30 April 2022).
- McKay, F.; Williams, B.J.; Prestwich, G.; Bansal, D.; Hallowell, N.; Treanor, D. The ethical challenges of artificial intelligence-driven digital pathology. J. Pathol. Clin. Res. 2022, 8, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Madabhushi, A.; Lee, G. Image analysis and machine learning in digital pathology: Challenges and opportunities. Med. Image Anal. 2016, 33, 170–175. [Google Scholar] [CrossRef] [Green Version]
- Tizhoosh, H.R.; Babaie, M. Representing Medical Images With Encoded Local Projections. IEEE Trans. Biomed. Eng. 2018, 65, 2267–2277. [Google Scholar] [CrossRef] [PubMed]
- Shamir, L.; Orlov, N.V.; Eckley, D.M.; Macura, T.J.; Johnston, J.; Goldberg, I.G. Wndchrm—An open source utility for biological image analysis-0. Source Code Biol. Med. 2008, 3, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Xie, J.-j.; Han, Z.; Liu, W.; Xi, S.; Huang, L.; Huang, W.; Lin, T.; Zhao, L.-y.; Hu, Y.; et al. Immunomarker Support Vector Machine Classifier for Prediction of Gastric Cancer Survival and Adjuvant Chemotherapeutic Benefit. Clin. Cancer Res. 2018, 24, 5574–5584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, H.; Zerbe, N.; Klempert, I.; Lohmann, S.; Lindequist, B.; Hellwich, O.; Hufnagl, P. Appearance-based necrosis detection using textural features and SVM with discriminative thresholding in histopathological whole slide images. In Proceedings of the 2015 IEEE 15th International Conference on Bioinformatics and Bioengineering (BIBE), Belgrade, Serbia, 2–4 November 2015; pp. 1–6. [Google Scholar]
- Wang, H.; Cruz-Roa, A.; Basavanhally, A.; Gilmore, H.; Shih, N.; Feldman, M.D.; Tomaszewski, J.E.; González, F.A.; Madabhushi, A. Mitosis detection in breast cancer pathology images by combining handcrafted and convolutional neural network features. J. Med. Imaging 2014, 1, 034003. [Google Scholar] [CrossRef] [PubMed]
- Geread, R.S.; Morreale, P.; Dony, R.D.; Brouwer, E.; Wood, G.A.; Androutsos, D.; Khademi, A. IHC Color Histograms for Unsupervised Ki67 Proliferation Index Calculation. Front. Bioeng. Biotechnol. 2019, 7, 226. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Samaras, D.; Kurç, T.M.; Gao, Y.; Davis, J.E.; Saltz, J.H. Patch-Based Convolutional Neural Network for Whole Slide Tissue Image Classification. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 2424–2433. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Gindra, R.H.; Green, E.J.; Burks, E.J.; Betke, M.; Beane, J.E.; Kolachalama, V.B. A graph-transformer for whole slide image classification. IEEE Trans. Med. Imag. 2022; Online ahead of print. [Google Scholar] [CrossRef]
- Shen, Y.; Ke, J. A Deformable CRF Model for Histopathology Whole-Slide Image Classification. In MICCAI 2020: Medical Image Computing and Computer Assisted Intervention—MICCAI 2020; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2020; Volume 12265, pp. 500–508. [Google Scholar]
- Iizuka, O.; Kanavati, F.; Kato, K.; Rambeau, M.; Arihiro, K.; Tsuneki, M. Deep Learning Models for Histopathological Classification of Gastric and Colonic Epithelial Tumours. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sirinukunwattana, K.; Domingo, E.; Richman, S.D.; Redmond, K.L.; Blake, A.; Verrill, C.; Leedham, S.J.; Chatzipli, A.; Hardy, C.W.; Whalley, C.M.; et al. Image-based consensus molecular subtype (imCMS) classification of colorectal cancer using deep learning. Gut 2020, 70, 544–554. [Google Scholar] [CrossRef] [PubMed]
- Popovici, V.; Budinská, E.; Dušek, L.; Kozubek, M.; Bosman, F.T. Image-based surrogate biomarkers for molecular subtypes of colorectal cancer. Bioinformatics 2017, 33, 2002–2009. [Google Scholar] [CrossRef] [Green Version]
- Korbar, B.; Olofson, A.M.; Miraflor, A.P.; Nicka, K.M.; Suriawinata, M.A.; Torresani, L.; Suriawinata, A.A.; Hassanpour, S. Deep Learning for Classification of Colorectal Polyps on Whole-slide Images. J. Pathol. Inform. 2017, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Kather, J.N.; Krisam, J.; Charoentong, P.; Luedde, T.; Herpel, E.; Weis, C.-A.; Gaiser, T.; Marx, A.; Valous, N.A.; Ferber, D.; et al. Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicenter study. PLoS Med. 2019, 16, e1002730. [Google Scholar] [CrossRef]
- Wei, J.; Suriawinata, A.A.; Vaickus, L.J.; Ren, B.; Liu, X.; Lisovsky, M.; Tomita, N.; Abdollahi, B.; Kim, A.S.; Snover, D.C.; et al. Evaluation of a Deep Neural Network for Automated Classification of Colorectal Polyps on Histopathologic Slides. JAMA Netw. Open 2020, 3, e203398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, P.; Huang, Y.; Sahoo, P.K.; You, J.-F.; Chiang, S.-F.; Onthoni, D.D.; Chern, Y.-J.; Chao, K.-Y.; Chiang, J.-M.; Yeh, C.-Y.; et al. Colon Tissues Classification and Localization in Whole Slide Images Using Deep Learning. Diagnostics 2021, 11, 1398. [Google Scholar] [CrossRef] [PubMed]
- Khened, M.; Kori, A.; Rajkumar, H.; Srinivasan, B.; Krishnamurthi, G. A generalized deep learning framework for whole-slide image segmentation and analysis. Sci. Rep. 2021, 11, 11579. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Yang, F.; Ma, S.-C.; Liu, L.; Zhao, Y.; Li, Y.; Wu, D.-H.; Wang, T.; Lu, W.-J.; Cai, W.-J.; et al. Development and interpretation of a pathomics-based model for the prediction of microsatellite instability in Colorectal Cancer. Theranostics 2020, 10, 11080–11091. [Google Scholar] [CrossRef] [PubMed]
- Kather, J.N.; Pearson, A.T.; Halama, N.; Jäger, D.; Krause, J.; Loosen, S.H.; Marx, A.; Boor, P.; Tacke, F.; Neumann, U.P.; et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 2019, 25, 1054–1056. [Google Scholar] [CrossRef] [PubMed]
- Bychkov, D.; Linder, N.; Turkki, R.; Nordling, S.; Kovanen, P.E.; Verrill, C.; Walliander, M.; Lundin, M.; Haglund, C.; Lundin, J. Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Meier, A.; Nekolla, K.; Hewitt, L.C.; Earle, S.; Yoshikawa, T.; Oshima, T.; Miyagi, Y.; Huss, R.; Schmidt, G.; Grabsch, H.I. Hypothesis-free deep survival learning applied to the tumour microenvironment in gastric cancer. J. Pathol. Clin. Res. 2020, 6, 273–282. [Google Scholar] [CrossRef]
- Lu, M.Y.; Williamson, D.F.K.; Chen, T.Y.; Chen, R.J.; Barbieri, M.; Mahmood, F. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat. Biomed. Eng. 2021, 5, 555–570. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.; Kruger, A.J.; Sha, L.; Kannan, M.; Khan, A.A.; Stumpe, M.C. Learning relevant H&E slide morphologies for prediction of colorectal cancer tumor mutation burden using weakly supervised deep learning. J. Clin. Oncol. 2020, 38, e15244. [Google Scholar] [CrossRef]
- Wang, S.; Zhu, Y.; Yu, L.; Chen, H.; Lin, H.; Wan, X.-B.; Fan, X.; Heng, P.-A. RMDL: Recalibrated multi-instance deep learning for whole slide gastric image classification. Med. Image Anal. 2019, 58, 101549. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.; Bian, H.; Chen, Y.; Wang, Y.; Zhang, J.; Ji, X.; Zhang, Y. TransMIL: Transformer based Correlated Multiple Instance Learning for Whole Slide Image Classication. Adv. Neural Inf. Process. Syst. 2021, 34, 2136–2147. [Google Scholar]
- Sharma, Y.; Shrivastava, A.; Ehsan, L.; Moskaluk, C.A.; Syed, S.; Brown, D.E. Cluster-to-Conquer: A Framework for End-to-End Multi-Instance Learning for Whole Slide Image Classification. Proc. Mach. Learn. Res. 2021, 143, 682–698. [Google Scholar]
- Klein, S.; Gildenblat, J.; Ihle, M.A.; Merkelbach-Bruse, S.; Noh, K.-W.; Peifer, M.; Quaas, A.M.; Büttner, R. Deep learning for sensitive detection of Helicobacter Pylori in gastric biopsies. BMC Gastroenterol. 2020, 20, 1–11. [Google Scholar] [CrossRef]
- Li, J.; Chen, W.; Huang, X.; Yang, S.; Hu, Z.; Duan, Q.; Metaxas, D.N.; Li, H.; Zhang, S. Hybrid Supervision Learning for Pathology Whole Slide Image Classification. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2021; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2021; Volume 12908, pp. 309–318. [Google Scholar]
- Adu, K.; Yu, Y.; Cai, J.; Owusu-Agyemang, K.; Twumasi, B.A.; Wang, X. DHS-CapsNet: Dual horizontal squash capsule networks for lung and colon cancer classification from whole slide histopathological images. Int. J. Imaging Syst. Technol. 2021, 31, 2075–2092. [Google Scholar] [CrossRef]
- Chen, H.; Han, X.; Fan, X.; Lou, X.; Liu, H.; Huang, J.; Yao, J. Rectified Cross-Entropy and Upper Transition Loss for Weakly Supervised Whole Slide Image Classifier. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2019; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2019; Volume 11764, pp. 351–359. [Google Scholar]
- Ren, J.; Hacihaliloglu, I.; Singer, E.A.; Foran, D.J.; Qi, X. Unsupervised Domain Adaptation for Classification of Histopathology Whole-Slide Images. Front. Bioeng. Biotechnol. 2019, 7, 102. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biscotti, C.V.; Dawson, A.E.; Dziura, B.; Galup, L.; Darragh, T.; Rahemtulla, A.; Wills-Frank, L. Assisted primary screening using the automated ThinPrep Imaging System. Am. J. Clin. Pathol. 2005, 123, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Vu, H.T.; Lopez, R.; Bennett, A.; Burke, C.A. Individuals with sessile serrated polyps express an aggressive colorectal phenotype. Dis. Colon Rectum 2011, 54, 1216–1223. [Google Scholar] [CrossRef]
- Osmond, A.; Li-Chang, H.; Kirsch, R.; Divaris, D.; Falck, V.; Liu, D.F.; Marginean, C.; Newell, K.; Parfitt, J.; Rudrick, B.; et al. Interobserver variability in assessing dysplasia and architecture in colorectal adenomas: A multicentre Canadian study. J. Clin. Pathol. 2014, 67, 781–786. [Google Scholar] [CrossRef]
- Foss, F.A.; Milkins, S.; McGregor, A.H. Inter-observer variability in the histological assessment of colorectal polyps detected through the NHS Bowel Cancer Screening Programme. Histopathology 2012, 61, 47–52. [Google Scholar] [CrossRef]
- Davidson, K.W.; Barry, M.J.; Mangione, C.M.; Cabana, M.; Caughey, A.B.; Davis, E.M.; Donahue, K.E.; Doubeni, C.A.; Krist, A.H.; Kubik, M.J.J. Screening for colorectal cancer: US Preventive Services Task Force recommendation statement. JAMA 2016, 315, 2564–2575. [Google Scholar] [CrossRef]
- Zhao, L.; Lee, V.; Ng, M.K.; Yan, H.; Bijlsma, M.F. Molecular subtyping of cancer: Current status and moving toward clinical applications. Briefings Bioinform. 2018, 20, 572–584. [Google Scholar] [CrossRef] [PubMed]
- De Smedt, L.; Lemahieu, J.; Palmans, S.; Govaere, O.; Tousseyn, T.; Van Cutsem, E.; Prenen, H.; Tejpar, S.; Spaepen, M.; Matthijs, G.; et al. Microsatellite instable vs stable colon carcinomas: Analysis of tumour heterogeneity, inflammation and angiogenesis. Br. J. Cancer 2015, 113, 500–509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baretti, M.; Le, D.T. DNA mismatch repair in cancer. Pharmacol. Ther. 2018, 189, 45–62. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Luo, H.; Huang, L.; Luo, H.; Zhu, X. Microsatellite instability: A review of what the oncologist should know. Cancer Cell Int. 2020, 20, 1–13. [Google Scholar] [CrossRef] [Green Version]
- André, T.; Shiu, K.-K.; Kim, T.W.; Jensen, B.V.; Jensen, L.H.; Punt, C.; Smith, D.; Garcia-Carbonero, R.; Benavides, M.; Gibbs, P.; et al. Pembrolizumab in Microsatellite-Instability–High Advanced Colorectal Cancer. N. Engl. J. Med. 2020, 383, 2207–2218. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Qian, J.; Zhao, Y.J.N. The loss of PTEN expression and microsatellite stability (MSS) were predictors of unfavorable prognosis in gastric cancer (GC). Neoplasma 2020, 67, 1359–1366. [Google Scholar] [CrossRef] [PubMed]
- Snowsill, T.; Coelho, H.; Huxley, N.; Jones-Hughes, T.; Briscoe, S.; Frayling, I.M.; Hyde, C. Molecular testing for Lynch syndrome in people with colorectal cancer: Systematic reviews and economic evaluation. Health Technol. Assess. 2017, 21, 1–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goss, P.E.; Lee, B.L.; Badovinac-Crnjevic, T.; Strasser-Weippl, K.; Chavarri-Guerra, Y.; St Louis, J.; Villarreal-Garza, C.; Unger-Saldaña, K.; Ferreyra, M.; Debiasi, M.; et al. Planning cancer control in Latin America and the Caribbean. Lancet Oncol. 2013, 14, 391–436. [Google Scholar] [CrossRef]
- Banatvala, J.J.L. COVID-19 testing delays and pathology services in the UK. Lancet 2020, 395, 1831. [Google Scholar] [CrossRef]
- Al-Shamsi, H.O.; Abu-Gheida, I.; Rana, S.K.; Nijhawan, N.; Abdulsamad, A.S.; Alrawi, S.; Abuhaleeqa, M.; Almansoori, T.M.; Alkasab, T.; Aleassa, E.M.; et al. Challenges for cancer patients returning home during SARS-COV-19 pandemic after medical tourism - a consensus report by the emirates oncology task force. BMC Cancer 2020, 20, 1–10. [Google Scholar] [CrossRef]
- Balasubramani, B.; Newsom, K.J.; Martinez, K.A.; Starostik, P.; Clare-Salzler, M.; Chamala, S. Pathology informatics and robotics strategies for improving efficiency of COVID-19 pooled testing. Acad. Pathol. 2021, 8, 23742895211020485. [Google Scholar] [CrossRef] [PubMed]
- Campanella, G.; Hanna, M.G.; Geneslaw, L.; Miraflor, A.; Silva, V.W.K.; Busam, K.J.; Brogi, E.; Reuter, V.E.; Klimstra, D.S.; Fuchs, T.J. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019, 25, 1301–1309. [Google Scholar] [CrossRef] [PubMed]
- Ianni, J.D.; Soans, R.E.; Sankarapandian, S.; Chamarthi, R.V.; Ayyagari, D.; Olsen, T.G.; Bonham, M.J.; Stavish, C.C.; Motaparthi, K.; Cockerell, C.J.; et al. Tailored for Real-World: A Whole Slide Image Classification System Validated on Uncurated Multi-Site Data Emulating the Prospective Pathology Workload. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef] [Green Version]
| Author | Task | Cancer Type | Type of WSI | Dataset | Algorithm/ Model | Performance | Clinical Application |
|---|---|---|---|---|---|---|---|
| Yoshida et al. [24] | Classification | Gastric cancer | H&E | Training and testing: 3062 WSIs | e-Pathologist | Positive for carcinoma or suspicion of carcinoma vs. caution for adenoma or suspicion of a neoplastic lesion vs. negative for a neoplastic lesion Overall concordance rate: 55.6% Kappa coefficient: 0.28 (95% CI: 0.26–0.30) Negative vs. non-negative Sensitivity: 89.5% (95% CI: 87.5–91.4%) Specificity: 50.7% (95% CI: 48.5–52.9%) Positive predictive value: 47.7% (95% CI: 45.4–49.9%) Negative predictive value: 90.6% (95% CI, 88.8–92.2%) | Differentiation and diagnosis gastric cancer grade |
| Yasuda et al. [25] | Classification | Gastric cancer | H&E | Training and testing: 66 WSIs | wndchrm | Noncancer vs. well-differentiated gastric cancer AUC: 0.99 Noncancer vs. moderately differentiated gastric cancer AUC: 0.98 Noncancer vs. poorly differentiated gastric cancer AUC: 0.99 | Differentiation and diagnosis gastric cancer grade |
| Jiang et al. [106] | Classification and prognosis | Gastric cancer | H&E | Training: 251 patients Internal validation: 248 patients External validation: 287 patients | Support vector machine | Patients might benefit more from postoperative adjuvant chemotherapy vs. patient might not postoperative adjuvant chemotherapy training cohort: 5-year overall survival AUC: 0.796 5-year disease-free survival AUC: 0.805 Internal validation cohort: 5-year overall survival AUC: 0.809 5-year disease-free survival AUC: 0.813 External validation cohort: 5-year overall survival AUC: 0.834 5-year disease-free survival AUC: 0.828 | Prognosis of gastric cancer patients and identification of patients who might benefit from adjuvant chemotherapy |
| Cosatto et al. [26] | Detection | Gastric cancer | H&E | Training set: 8558 patients Test set: 4168 patients | Semi-supervised multi-instance learning framework | Positive vs. negative AUC: 0.96 | Detection of gastric cancer |
| Jiang et al. [27] | Classification | Colon cancer | H&E | Training: 101 patients Internal validation: 67 patients External validation: 47 patients | InceptionResNetV2 + gradient-boosting decision tree machine classifier | High-risk recurrence vs. low-risk recurrence Internal validation hazard ratio: 8.9766 (95% CI: 2.824–28.528) External validation hazard ratio: 10.273 (95% CI: 2.177–48.472) Poor vs. good prognosis groups: Internal validation hazard ratio: 10.687 (95% CI: 2.908–39.272) External validation hazard ratio: 5.033 (95% CI: 1.792–14.132) | Prognosis of stage III colon cancer |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wong, A.N.N.; He, Z.; Leung, K.L.; To, C.C.K.; Wong, C.Y.; Wong, S.C.C.; Yoo, J.S.; Chan, C.K.R.; Chan, A.Z.; Lacambra, M.D.; et al. Current Developments of Artificial Intelligence in Digital Pathology and Its Future Clinical Applications in Gastrointestinal Cancers. Cancers 2022, 14, 3780. https://doi.org/10.3390/cancers14153780
Wong ANN, He Z, Leung KL, To CCK, Wong CY, Wong SCC, Yoo JS, Chan CKR, Chan AZ, Lacambra MD, et al. Current Developments of Artificial Intelligence in Digital Pathology and Its Future Clinical Applications in Gastrointestinal Cancers. Cancers. 2022; 14(15):3780. https://doi.org/10.3390/cancers14153780
Chicago/Turabian StyleWong, Alex Ngai Nick, Zebang He, Ka Long Leung, Curtis Chun Kit To, Chun Yin Wong, Sze Chuen Cesar Wong, Jung Sun Yoo, Cheong Kin Ronald Chan, Angela Zaneta Chan, Maribel D. Lacambra, and et al. 2022. "Current Developments of Artificial Intelligence in Digital Pathology and Its Future Clinical Applications in Gastrointestinal Cancers" Cancers 14, no. 15: 3780. https://doi.org/10.3390/cancers14153780
APA StyleWong, A. N. N., He, Z., Leung, K. L., To, C. C. K., Wong, C. Y., Wong, S. C. C., Yoo, J. S., Chan, C. K. R., Chan, A. Z., Lacambra, M. D., & Yeung, M. H. Y. (2022). Current Developments of Artificial Intelligence in Digital Pathology and Its Future Clinical Applications in Gastrointestinal Cancers. Cancers, 14(15), 3780. https://doi.org/10.3390/cancers14153780







