Machine Learning Improves the Prediction Rate of Non-Curative Resection of Endoscopic Submucosal Dissection in Patients with Early Gastric Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
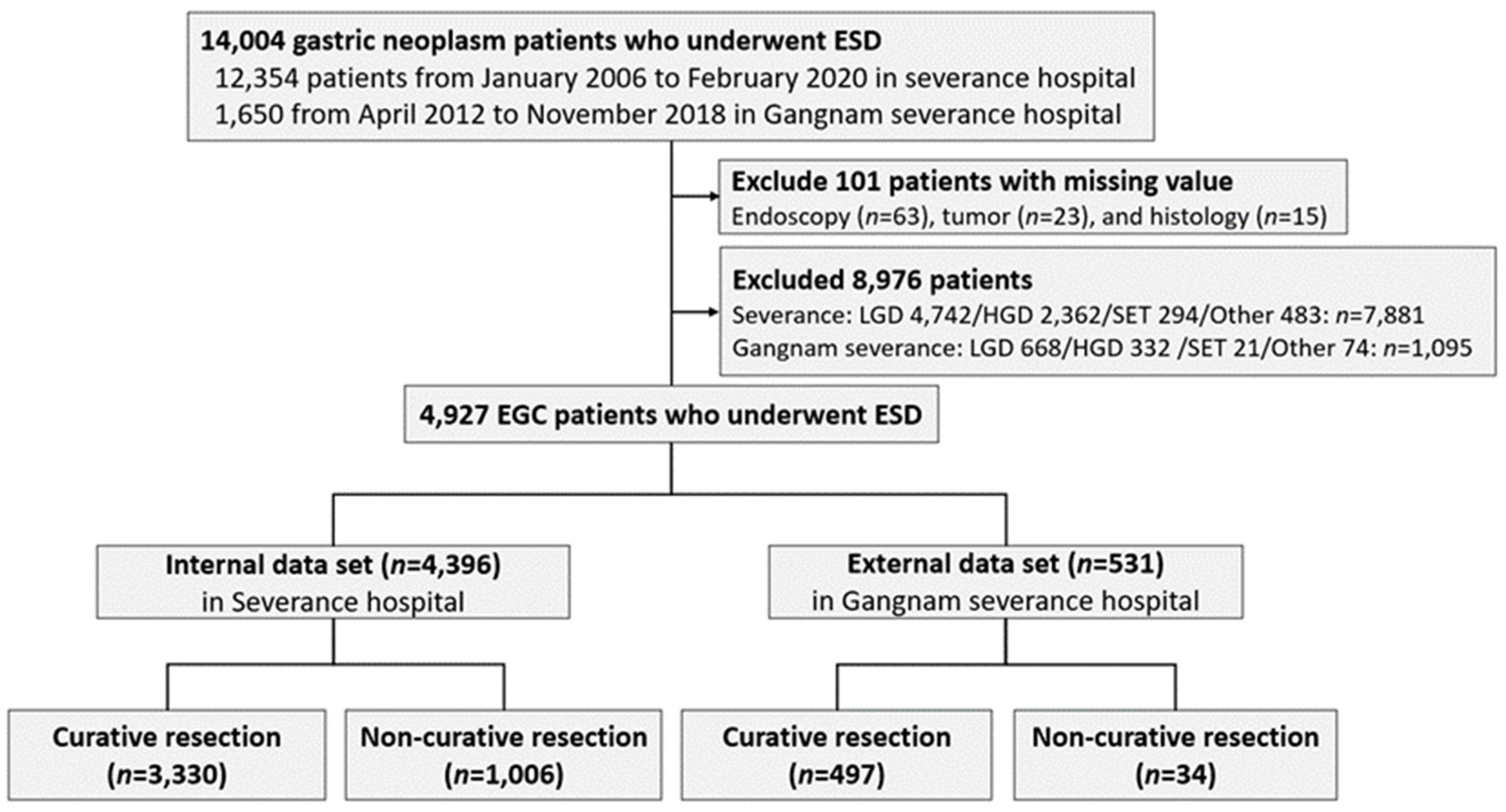
2.1. Data Collection and Study Population
2.2. Definitions
2.3. ESD Technique
2.4. Gross and Histopathologic Evaluation
2.5. Prediction Models
2.6. Statistical Analysis
3. Results
3.1. Baseline Characteristics of the Patients
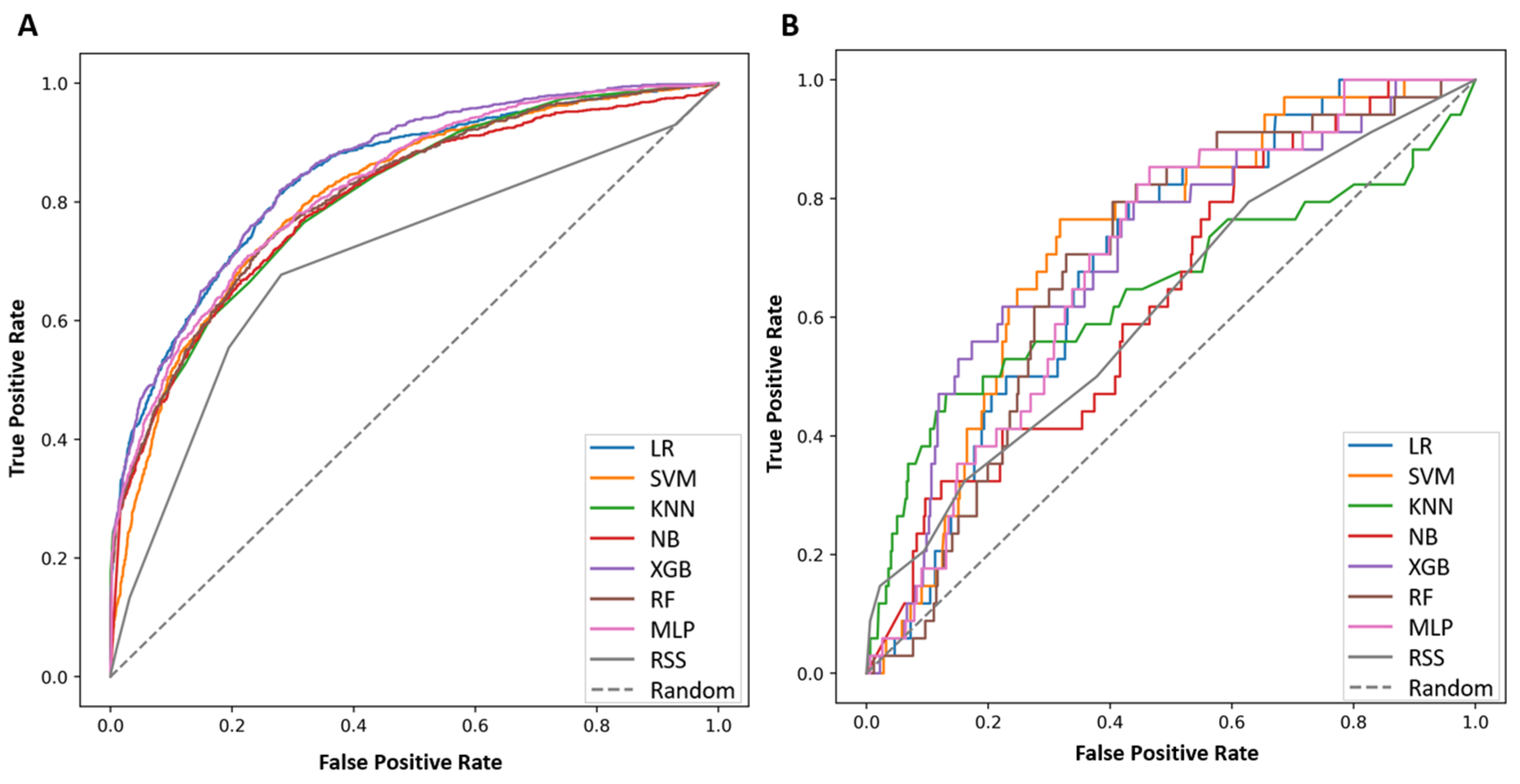
3.2. Performance of the ML Model for Prediction of NCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Japanese Gastric Cancer Association. Japanese gastric cancer treatment guidelines 2014 (ver. 4). Gastric Cancer 2017, 20, 1–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pimentel-Nunes, P.; Libanio, D.; Bastiaansen, B.A.J.; Bhandari, P.; Bisschops, R.; Bourke, M.J.; Esposito, G.; Lemmers, A.; Maselli, R.; Messmann, H.; et al. Endoscopic submucosal dissection for superficial gastrointestinal lesions: European Society of Gastrointestinal Endoscopy (ESGE) Guideline—Update 2022. Endoscopy 2022, 54, 591–622. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.R.; Lee, H.; Min, B.H.; Lee, J.H.; Rhee, P.L.; Kim, J.J.; Kim, K.M.; Kim, S. Effect of rescue surgery after non-curative endoscopic resection of early gastric cancer. Br. J. Surg. 2015, 102, 1394–1401. [Google Scholar] [CrossRef] [PubMed]
- Hatta, W.; Gotoda, T.; Oyama, T.; Kawata, N.; Takahashi, A.; Yoshifuku, Y.; Hoteya, S.; Nakagawa, M.; Hirano, M.; Esaki, M.; et al. A Scoring System to Stratify Curability after Endoscopic Submucosal Dissection for Early Gastric Cancer: “eCura system”. Am. J. Gastroenterol. 2017, 112, 874–881. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.W.; Kim, J.H.; Park, J.C.; Jeon, M.Y.; Lee, Y.C.; Lee, S.K.; Shin, S.K.; Chung, H.S.; Noh, S.H.; Kim, J.W.; et al. Additive endoscopic resection may be sufficient for patients with a positive lateral margin after endoscopic resection of early gastric cancer. Gastrointest. Endosc. 2017, 86, 849–856. [Google Scholar] [CrossRef]
- Japanese Gastric Cancer Association. Japanese gastric cancer treatment guidelines 2018 (5th edition). Gastric Cancer 2021, 24, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.H.; Park, J.C.; Song, I.J.; Kim, Y.J.; Joh, D.H.; Hahn, K.Y.; Lee, Y.K.; Kim, H.Y.; Chung, H.; Shin, S.K.; et al. Prediction model for non-curative resection of endoscopic submucosal dissection in patients with early gastric cancer. Gastrointest. Endosc. 2017, 85, 976–983. [Google Scholar] [CrossRef]
- Libanio, D.; Dinis-Ribeiro, M.; Pimentel-Nunes, P.; Dias, C.C.; Rodrigues, P.P. Predicting outcomes of gastric endoscopic submucosal dissection using a Bayesian approach: A step for individualized risk assessment. Endosc. Int. Open 2017, 5, E563–E572. [Google Scholar] [CrossRef] [Green Version]
- Nam, H.S.; Choi, C.W.; Kim, S.J.; Kang, D.H.; Kim, H.W.; Park, S.B.; Ryu, D.G.; Choi, J.S. Preprocedural prediction of non-curative endoscopic submucosal dissection for early gastric cancer. PLoS ONE 2018, 13, e0206179. [Google Scholar] [CrossRef]
- Kim, T.S.; Min, B.H.; Kim, K.M.; Yoo, H.; Kim, K.; Min, Y.W.; Lee, H.; Rhee, P.L.; Kim, J.J.; Lee, J.H. Risk-Scoring System for Prediction of Non-Curative Endoscopic Submucosal Dissection Requiring Additional Gastrectomy in Patients with Early Gastric Cancer. J. Gastric Cancer 2021, 21, 368–378. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, Q.; Zhu, S.; Zhang, S.; Sun, X. Risk Factors and Prediction Model for Non-curative Resection of Early Gastric Cancer With Endoscopic Resection and the Evaluation. Front. Med. (Lausanne) 2021, 8, 637875. [Google Scholar] [CrossRef] [PubMed]
- Kwan, J.L.; Lo, L.; Ferguson, J.; Goldberg, H.; Diaz-Martinez, J.P.; Tomlinson, G.; Grimshaw, J.M.; Shojania, K.G. Computerised clinical decision support systems and absolute improvements in care: Meta-analysis of controlled clinical trials. BMJ 2020, 370, m3216. [Google Scholar] [CrossRef] [PubMed]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Shiraishi, J.; Li, Q.; Appelbaum, D.; Doi, K. Computer-aided diagnosis and artificial intelligence in clinical imaging. Semin. Nucl. Med. 2011, 41, 449–462. [Google Scholar] [CrossRef] [PubMed]
- Bang, C.S.; Ahn, J.Y.; Kim, J.H.; Kim, Y.I.; Choi, I.J.; Shin, W.G. Establishing Machine Learning Models to Predict Curative Resection in Early Gastric Cancer with Undifferentiated Histology: Development and Usability Study. J. Med. Internet Res. 2021, 23, e25053. [Google Scholar] [CrossRef]
- Yao, K.; Uedo, N.; Kamada, T.; Hirasawa, T.; Nagahama, T.; Yoshinaga, S.; Oka, M.; Inoue, K.; Mabe, K.; Yao, T.; et al. Guidelines for endoscopic diagnosis of early gastric cancer. Dig. Endosc. 2020, 32, 663–698. [Google Scholar] [CrossRef]
- Kim, Y.I.; Kim, Y.W.; Choi, I.J.; Kim, C.G.; Lee, J.Y.; Cho, S.J.; Eom, B.W.; Yoon, H.M.; Ryu, K.W.; Kook, M.C. Long-term survival after endoscopic resection versus surgery in early gastric cancers. Endoscopy 2015, 47, 293–301. [Google Scholar] [CrossRef] [Green Version]
- Nakata, B.; Tendo, M.; Okuyama, M.; Nakahara, K.; Ishizu, H.; Masuda, G.; Lee, T.; Hori, T.; Ohsawa, M.; Sato, H.; et al. Additional surgical resection after endoscopic mucosal dissection for early gastric cancer: A medium-sized hospital’s experience. Int. J. Surg. 2016, 36, 335–341. [Google Scholar] [CrossRef]
- Choi, J.; Kim, S.G.; Im, J.P.; Kim, J.S.; Jung, H.C.; Song, I.S. Comparison of endoscopic ultrasonography and conventional endoscopy for prediction of depth of tumor invasion in early gastric cancer. Endoscopy 2010, 42, 705–713. [Google Scholar] [CrossRef]
- Lee, J.Y.; Choi, I.J.; Kim, C.G.; Cho, S.J.; Kook, M.C.; Ryu, K.W.; Kim, Y.W. Therapeutic Decision-Making Using Endoscopic Ultrasonography in Endoscopic Treatment of Early Gastric Cancer. Gut Liver 2016, 10, 42–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, C.H.; Park, J.C.; Chung, H.; Shin, S.K.; Lee, S.K.; Lee, Y.C. A specific role of endoscopic ultrasonography for therapeutic decision-making in patients with gastric cardia cancer. Surg. Endosc. 2016, 30, 4193–4199. [Google Scholar] [CrossRef]
- Kanesaka, T.; Sekikawa, A.; Tsumura, T.; Maruo, T.; Osaki, Y.; Wakasa, T.; Shintaku, M.; Yao, K. Absent microsurface pattern is characteristic of early gastric cancer of undifferentiated type: Magnifying endoscopy with narrow-band imaging. Gastrointest Endosc 2014, 80, 1194–1198.e1191. [Google Scholar] [CrossRef] [PubMed]
- Nagahama, T.; Yao, K.; Uedo, N.; Doyama, H.; Ueo, T.; Uchita, K.; Ishikawa, H.; Kanesaka, T.; Takeda, Y.; Wada, K.; et al. Delineation of the extent of early gastric cancer by magnifying narrow-band imaging and chromoendoscopy: A multicenter randomized controlled trial. Endoscopy 2018, 50, 566–576. [Google Scholar] [CrossRef] [PubMed]
- Horiuchi, Y.; Tokai, Y.; Yamamoto, N.; Yoshimizu, S.; Ishiyama, A.; Yoshio, T.; Hirasawa, T.; Yamamoto, Y.; Nagahama, M.; Takahashi, H.; et al. Additive Effect of Magnifying Endoscopy with Narrow-Band Imaging for Diagnosing Mixed-Type Early Gastric Cancers. Dig. Dis. Sci. 2020, 65, 591–599. [Google Scholar] [CrossRef] [PubMed]
- Shung, D.L.; Au, B.; Taylor, R.A.; Tay, J.K.; Laursen, S.B.; Stanley, A.J.; Dalton, H.R.; Ngu, J.; Schultz, M.; Laine, L. Validation of a Machine Learning Model That Outperforms Clinical Risk Scoring Systems for Upper Gastrointestinal Bleeding. Gastroenterology 2020, 158, 160–167. [Google Scholar] [CrossRef]
- Ogunleye, A.; Wang, Q.G. XGBoost Model for Chronic Kidney Disease Diagnosis. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020, 17, 2131–2140. [Google Scholar] [CrossRef]
- Huang, J.C.; Tsai, Y.C.; Wu, P.Y.; Lien, Y.H.; Chien, C.Y.; Kuo, C.F.; Hung, J.F.; Chen, S.C.; Kuo, C.H. Predictive modeling of blood pressure during hemodialysis: A comparison of linear model, random forest, support vector regression, XGBoost, LASSO regression and ensemble method. Comput. Methods Programs Biomed. 2020, 195, 105536. [Google Scholar] [CrossRef] [PubMed]
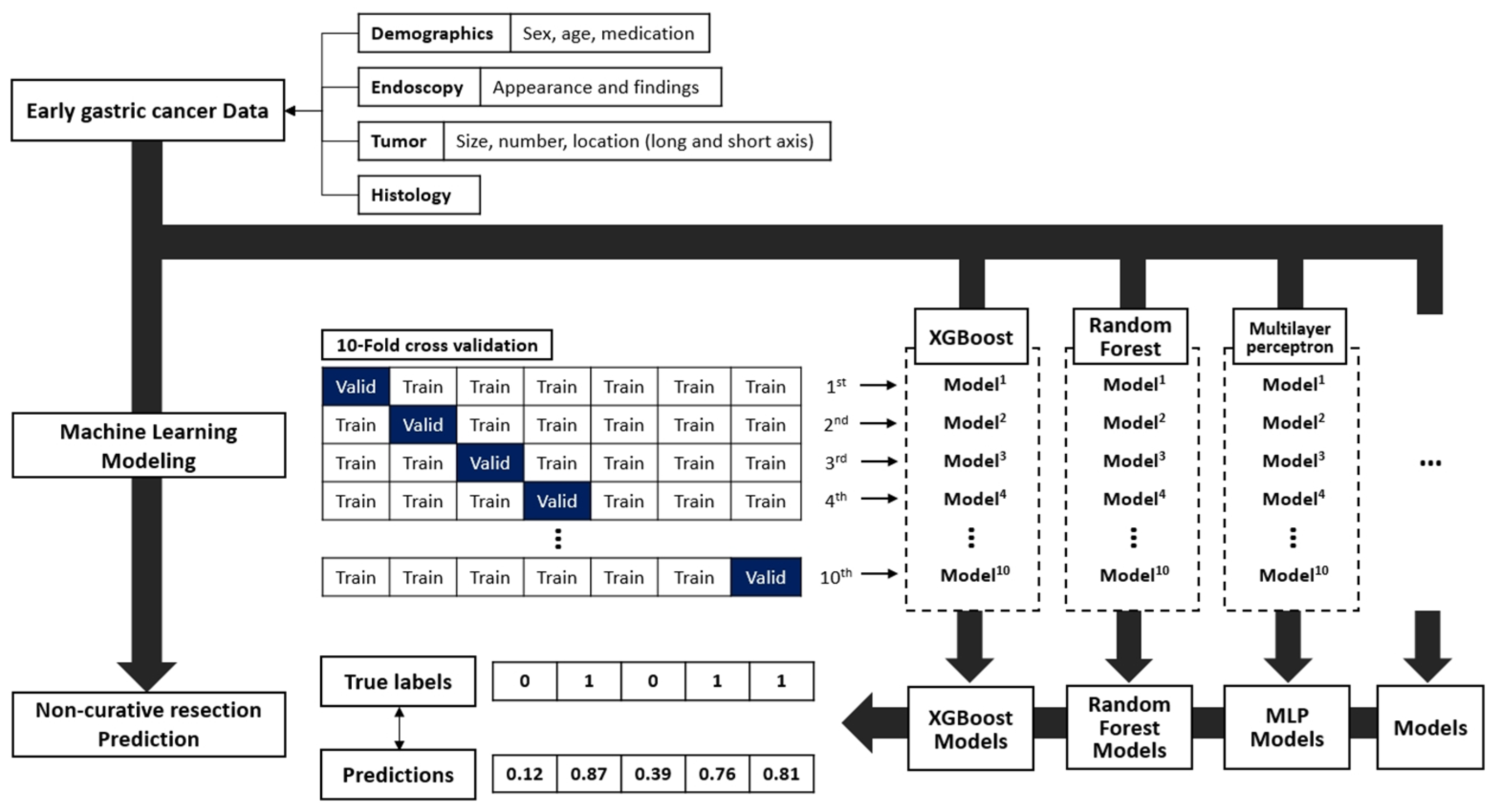
| Category | Variables | |
|---|---|---|
| Demographics | Sex, Age, Antithrombotics | |
| Endoscopy | Appearance | Elevated, Flat, Depressed |
| Finding | Ulcer, Fold, Erythema, Exudate, Whitish or atrophy, Nodularity or elevated, Spontaneous bleeding | |
| Tumor | Size | Size (mm) |
| Number | 1, 2, or >2 | |
| Location (long axis) | Upper, Middle, Lower | |
| Location (short axis) | Anterior wall, Posterior wall, Greater curvature, Lesser curvature | |
| Histology | Adenocarcinoma well-differentiated, Adenocarcinoma moderate-differentiated, Adenocarcinoma poorly differentiated, Signet-ring cell, Others (Mucinous, Carcinoma in situ, Squamous cell type, etc) | |
| Variables | Overall (n = 4927) | Internal Data Set (n = 4396) | External Data Set (n = 531) | p-Value |
|---|---|---|---|---|
| Demographics | ||||
| Age, years | 64.4 ± 10.2 | 64.7 ± 10.1 | 62.4 ± 11.2 | <0.001 |
| Male | 3620 (73.5) | 3240 (73.7) | 151 (71.6) | 0.29 |
| Medications | ||||
| Antithrombotics | 923 (18.7) | 830 (18.9) | 93 (17.5) | 0.45 |
| Histology | ||||
| AWD | 1565 (31.8) | 1425 (32.4) | 140 (26.4) | <0.001 |
| AMD | 1228 (24.9) | 1109 (25.2) | 119 (22.4) | 0.16 |
| APD | 145 (2.9) | 113 (2.6) | 32 (6.0) | <0.001 |
| SRC | 250 (5.1) | 210 (4.8) | 40 (7.5) | <0.001 |
| Other (Mucinous, CIS, SCC) | 1739 (35.2) | 1539 (35.0) | 200 (37.7) | 0.23 |
| Multiple lesions | ||||
| 1 | 4280 (86.9) | 3785 (86.1) | 495 (93.2) | 0.01 |
| 2 | 561 (11.4) | 532 (12.1) | 29 (5.5) | <0.001 |
| >2 | 647 (13.1) | 611 (13.9) | 36 (6.8) | <0.001 |
| Tumor location (long axis) | ||||
| Upper | 495 (10.0) | 441 (10.0) | 54 (10.2) | 0.92 |
| Mid | 1681 (34.1) | 1463 (33.3) | 218 (41.1) | <0.001 |
| Lower | 2635 (53.5) | 2369 (53.9) | 266 (50.1) | 0.10 |
| Tumor location (short axis) | ||||
| AW | 1050 (21.3) | 927 (21.1) | 123 (23.2) | 0.27 |
| PW | 1189 (24.1) | 1070 (24.3) | 119 (22.4) | 0.33 |
| LC | 1838 (37.3) | 1607 (36.6) | 231 (43.5) | <0.001 |
| GC | 1023 (20.8) | 904 (20.6) | 119 (22.4) | 0.32 |
| Tumor size (mm) | 13.1 ± 9.2 | 12.4 ± 8.4 | 20.0 ± 12.7 | <0.001 |
| Endoscopic appearance | ||||
| Elevated | 3253 (66.0) | 2976 (67.7) | 277 (52.2) | <0.001 |
| Flat | 1341 (27.2) | 1124 (25.6) | 217 (40.9) | <0.001 |
| Depressed | 2302 (46.7) | 2029 (46.2) | 273 (51.4) | 0.02 |
| Endoscopic finding | ||||
| Ulcer | 274 (5.6) | 225 (5.1) | 49 (9.2) | <0.001 |
| Fusion of fold, interruption, or smooth tapering of fold | 104 (2.1) | 70 (1.6) | 34 (6.4) | <0.001 |
| Erythema | 795 (16.1) | 534 (12.1) | 261 (49.2) | <0.001 |
| Exudate | 210 (4.3) | 102 (2.3) | 107 (20.2) | <0.001 |
| Whitish scar or atrophy | 269 (5.5) | 225 (5.1) | 44 (8.3) | 0.002 |
| Nodularity or elevated | 863 (17.5) | 596 (13.6) | 267 (50.3) | <0.001 |
| Spontaneous bleeding | 60 (1.2) | 38 (0.9) | 22 (4.1) | <0.001 |
| Risk Score | Precision | F1 Score | AUPRC (95%CI) | Sensitivity | Specificity | AUROC (95%CI) | p-Value a |
|---|---|---|---|---|---|---|---|
| Internal data | |||||||
| RSS | 0.636 | 0.777 | 0.463 (0.449–0.478) | 0.998 | 0.008 | 0.701 (0.683–0.720) | |
| LR | 0.735 | 0.547 | 0.691 (0.677–0.705) | 0.788 | 0.721 | 0.840 (0.825–0.854) | <0.001 |
| SVM | 0.700 | 0.460 | 0.596 (0.581–0.610) | 0.827 | 0.618 | 0.667 (0.647–0.687) | <0.001 |
| KNN | 0.835 | 0.436 | 0.652 (0.637–0.665) | 0.771 | 0.665 | 0.807 (0.792–0.822) | <0.001 |
| NB | 0.696 | 0.492 | 0.633 (0.619–0.647) | 0.946 | 0.380 | 0.799 (0.783–0.815) | <0.001 |
| XGB | 0.749 | 0.576 | 0.699 (0.685–0.713) | 0.785 | 0.732 | 0.851 (0.837–0.864) | <0.001 |
| RF | 0.925 | 0.326 | 0.647 (0.633–0.661) | 0.713 | 0.757 | 0.812 (0.797–0.827) | <0.001 |
| MLP | 0.718 | 0.527 | 0.676 (0.662–0.689) | 0.722 | 0.752 | 0.837 (0.823–0.850) | <0.001 |
| External data | |||||||
| RSS | 0.200 | 0.333 | 0.174 (0.163–0.186) | 0.977 | 0.147 | 0.616 (0.516–0.719) | |
| LR | 0.122 | 0.193 | 0.104 (0.095–0.113) | 0.561 | 0.794 | 0.693 (0.610–0.773) | 0.09 |
| SVM | 0.099 | 0.133 | 0.113 (0.104–0.122) | 0.563 | 0.794 | 0.693 (0.613–0.769) | 0.02 |
| KNN | 0.202 | 0.148 | 0.169 (0.159–0.181) | 0.829 | 0.470 | 0.645 (0.523–0.762) | 0.69 |
| NB | 0.096 | 0.147 | 0.151 (0.141–0.162) | 0.776 | 0.411 | 0.631 (0.540–0.722) | 0.74 |
| XGB | 0.187 | 0.274 | 0.125 (0.116–0.135) | 0.587 | 0.735 | 0.710 (0.612–0.803) | 0.02 |
| RF | 0.031 | 0.030 | 0.099 (0.090–0.108) | 0.394 | 0.911 | 0.688 (0.604–0.769) | 0.12 |
| MLP | 0.126 | 0.188 | 0.105 (0.096–0.114) | 0.551 | 0.823 | 0.691 (0.603–0.771) | 0.06 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yun, H.-R.; Huh, C.W.; Jung, D.H.; Lee, G.; Son, N.-H.; Kim, J.-H.; Youn, Y.H.; Park, J.C.; Shin, S.K.; Lee, S.K.; et al. Machine Learning Improves the Prediction Rate of Non-Curative Resection of Endoscopic Submucosal Dissection in Patients with Early Gastric Cancer. Cancers 2022, 14, 3742. https://doi.org/10.3390/cancers14153742
Yun H-R, Huh CW, Jung DH, Lee G, Son N-H, Kim J-H, Youn YH, Park JC, Shin SK, Lee SK, et al. Machine Learning Improves the Prediction Rate of Non-Curative Resection of Endoscopic Submucosal Dissection in Patients with Early Gastric Cancer. Cancers. 2022; 14(15):3742. https://doi.org/10.3390/cancers14153742
Chicago/Turabian StyleYun, Hae-Ryong, Cheal Wung Huh, Da Hyun Jung, Gyubok Lee, Nak-Hoon Son, Jie-Hyun Kim, Young Hoon Youn, Jun Chul Park, Sung Kwan Shin, Sang Kil Lee, and et al. 2022. "Machine Learning Improves the Prediction Rate of Non-Curative Resection of Endoscopic Submucosal Dissection in Patients with Early Gastric Cancer" Cancers 14, no. 15: 3742. https://doi.org/10.3390/cancers14153742
APA StyleYun, H.-R., Huh, C. W., Jung, D. H., Lee, G., Son, N.-H., Kim, J.-H., Youn, Y. H., Park, J. C., Shin, S. K., Lee, S. K., & Lee, Y. C. (2022). Machine Learning Improves the Prediction Rate of Non-Curative Resection of Endoscopic Submucosal Dissection in Patients with Early Gastric Cancer. Cancers, 14(15), 3742. https://doi.org/10.3390/cancers14153742







