Pan-Cancer Analysis Reveals SH3TC2 as an Oncogene for Colorectal Cancer and Promotes Tumorigenesis via the MAPK Pathway
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Sources
2.2. SH3TC2 Expression Pattern in Pan-Cancer
2.3. Prognostic Analysis of SH3TC2 in Different Tumors
2.4. SH3TC2 Expression Levels in CRC
2.5. Correlation between SH3TC2 Expression and Clinical Stage in Colorectal Cancer
2.6. Correlation between SH3TC2 Expression and Immune Cell Infiltration
2.7. The Co-Expressed Genes of SH3TC2 and Functional Enrichment Analysis
2.8. Clinical Specimens and Immunohistochemistry (IHC) Analysis
2.9. Cell Lines and Transfection
2.10. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.11. Western Blot
2.12. Transwell Analysis
2.13. Cell Viability Analysis
2.14. Cell Apoptosis Assays
2.15. In Vivo Analysis
2.16. Statistical Analysis
3. Results
3.1. Expression Levels and Prognostic Value of SH3TC2 in Human Cancers
3.2. Correlation between SH3TC2 Expression and Immune Cell Infiltration Levels
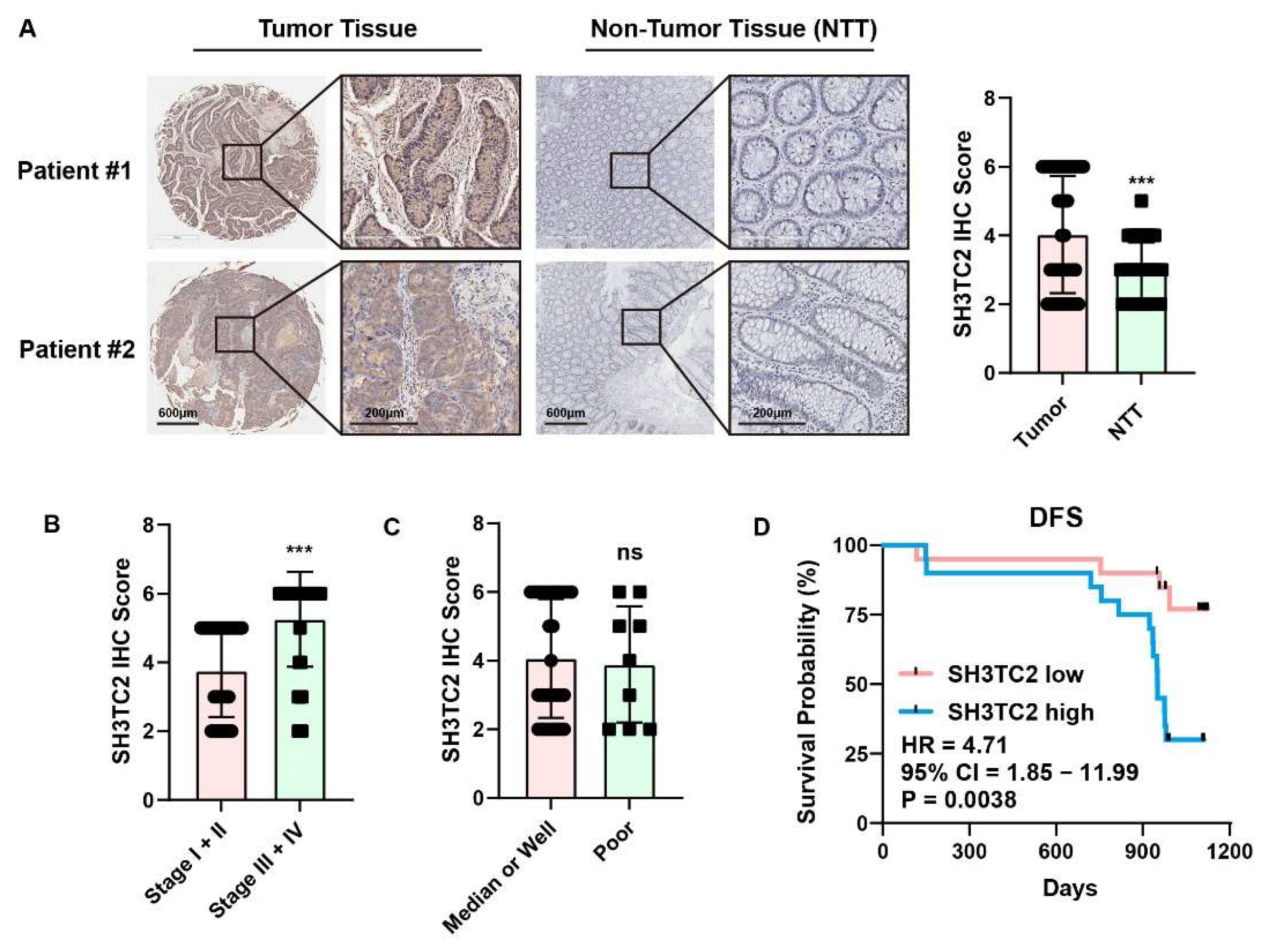
3.3. Clinical Specimen Analysis Revealed That Higher Expression of SH3TC2 Indicated Poor Prognosis
3.4. Downregulated SH3TC2 Expression Inhibits Tumorigenesis In Vitro
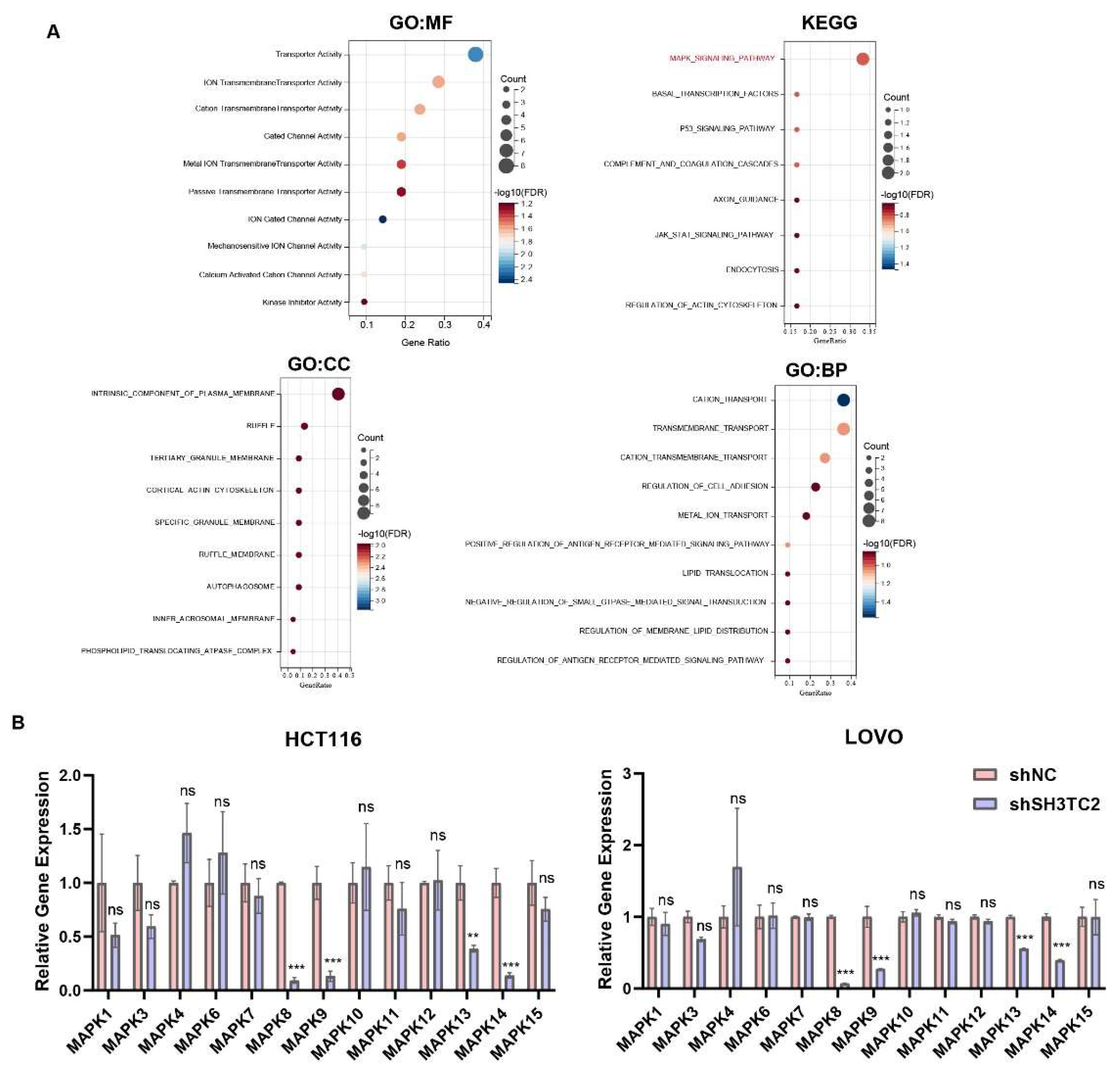
3.5. Coexpressed Genes of SH3TC2 and Functional Enrichment Analysis
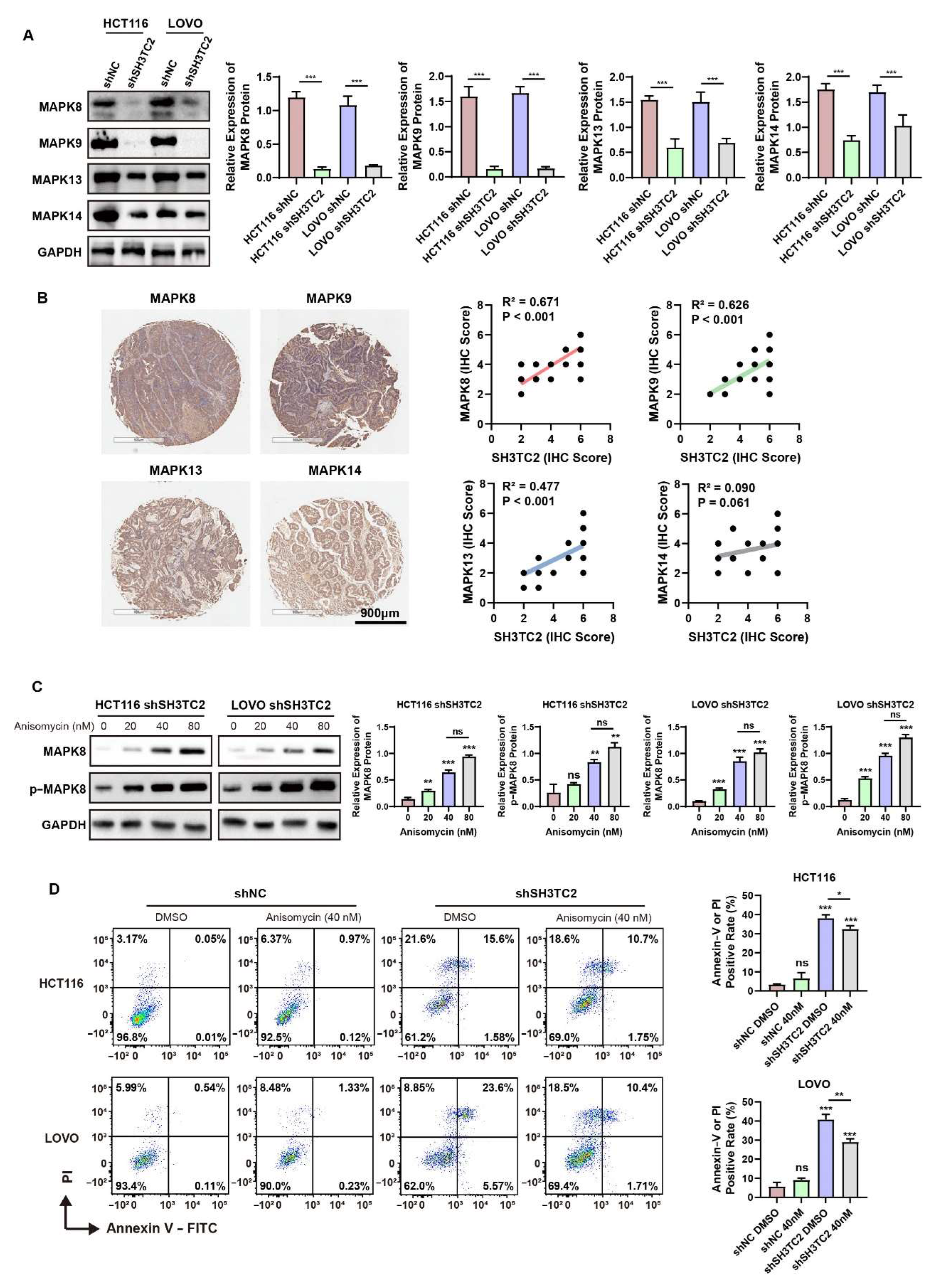
3.6. Downregulated SH3TC2 Inhibited MAPK Pathways In Vitro
3.7. Downregulation of SH3TC2 Inhibited Tumor Growth In Vivo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Brody, H. Colorectal cancer. Nature 2015, 521, S1. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.D.; Ortiz, A.P.; Pinheiro, P.S.; Bandi, P.; Minihan, A.; Fuchs, H.E.; Martinez Tyson, D.; Tortolero-Luna, G.; Fedewa, S.A.; Jemal, A.M.; et al. Cancer statistics for the US Hispanic/Latino population, 2021. CA Cancer J. Clin. 2021, 71, 466–487. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Bradner, J.E.; Hnisz, D.; Young, R.A. Transcriptional Addiction in Cancer. Cell 2017, 168, 629–643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hogg, S.J.; Beavis, P.A.; Dawson, M.A.; Johnstone, R.W. Targeting the epigenetic regulation of antitumour immunity. Nat. Rev. Drug Discov. 2020, 19, 776–800. [Google Scholar] [CrossRef]
- Park, J.W.; Han, J.W. Targeting epigenetics for cancer therapy. Arch. Pharm. Res. 2019, 42, 159–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Payer, L.M.; Burns, K.H. Transposable elements in human genetic disease. Nat. Rev. Genet. 2019, 20, 760–772. [Google Scholar] [CrossRef]
- Stendel, C.; Roos, A.; Kleine, H.; Arnaud, E.; Ozcelik, M.; Sidiropoulos, P.N.; Zenker, J.; Schupfer, F.; Lehmann, U.; Sobota, R.M.; et al. SH3TC2, a protein mutant in Charcot-Marie-Tooth neuropathy, links peripheral nerve myelination to endosomal recycling. Brain 2010, 133, 2462–2474. [Google Scholar] [CrossRef] [Green Version]
- Gouttenoire, E.A.; Lupo, V.; Calpena, E.; Bartesaghi, L.; Schupfer, F.; Medard, J.J.; Maurer, F.; Beckmann, J.S.; Senderek, J.; Palau, F.; et al. Sh3tc2 deficiency affects neuregulin-1/ErbB signaling. Glia 2013, 61, 1041–1051. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.J.; Nam, S.H.; Park, J.M.; Kanwal, S.; Choi, Y.J.; Lee, H.J.; Lee, K.S.; Lee, J.E.; Park, J.S.; Choi, B.O.; et al. Compound heterozygous mutations of SH3TC2 in Charcot-Marie-Tooth disease type 4C patients. J. Hum. Genet. 2019, 64, 961–965. [Google Scholar] [CrossRef] [PubMed]
- Schiza, N.; Georgiou, E.; Kagiava, A.; Medard, J.J.; Richter, J.; Tryfonos, C.; Sargiannidou, I.; Heslegrave, A.J.; Rossor, A.M.; Zetterberg, H.; et al. Gene replacement therapy in a model of Charcot-Marie-Tooth 4C neuropathy. Brain 2019, 142, 1227–1241. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; He, Z.Q.; Li, Y.R.; Bai, J.M.; Wang, H.R.; Wang, H.F.; Cui, F.; Yang, F.; Huang, X.S. Screening for SH3TC2 variants in Charcot-Marie-Tooth disease in a cohort of Chinese patients. Acta Neurol. Belg. 2021, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Guan, L.; Wang, Q.; Wang, L.; Wu, B.; Chen, Y.; Liu, F.; Ye, F.; Zhang, T.; Li, K.; Yan, B.; et al. Common variants on 17q25 and gene-gene interactions conferring risk of schizophrenia in Han Chinese population and regulating gene expressions in human brain. Mol. Psychiatry 2016, 21, 1244–1250. [Google Scholar] [CrossRef]
- Yu, P.; Lan, H.; Song, X.; Pan, Z. High Expression of the SH3TC2-DT/SH3TC2 Gene Pair Associated With FLT3 Mutation and Poor Survival in Acute Myeloid Leukemia: An Integrated TCGA Analysis. Front. Oncol. 2020, 10, 829. [Google Scholar] [CrossRef]
- Doecke, J.D.; Wang, Y.; Baggerly, K. Co-localized genomic regulation of miRNA and mRNA via DNA methylation affects survival in multiple tumor types. Cancer Genet. 2016, 209, 463–473. [Google Scholar] [CrossRef]
- Li, H.X.; Liang, J.H.; Wang, J.; Han, J.Y.; Li, S.; Huang, K.; Liu, C.Y. Mex3a promotes oncogenesis through the RAP1/MAPK signaling pathway in colorectal cancer and is inhibited by hsa-miR-6887-3p. Cancer Commun. 2021, 41, 472–491. [Google Scholar] [CrossRef]
- Morrison, D.K. MAP kinase pathways. Cold Spring Harb. Perspect. Biol. 2012, 4, a011254. [Google Scholar] [CrossRef]
- Steelman, L.S.; Franklin, R.A.; Abrams, S.L.; Chappell, W.; Kempf, C.R.; Basecke, J.; Stivala, F.; Donia, M.; Fagone, P.; Nicoletti, F.; et al. Roles of the Ras/Raf/MEK/ERK pathway in leukemia therapy. Leukemia 2011, 25, 1080–1094. [Google Scholar] [CrossRef] [Green Version]
- Wagner, E.F.; Nebreda, A.R. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat. Rev. Cancer 2009, 9, 537–549. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, H.T. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002, 12, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Coulthard, L.R.; White, D.E.; Jones, D.L.; McDermott, M.F.; Burchill, S.A. p38(MAPK): Stress responses from molecular mechanisms to therapeutics. Trends Mol. Med. 2009, 15, 369–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuadrado, A.; Nebreda, A.R. Mechanisms and functions of p38 MAPK signalling. Biochem. J. 2010, 429, 403–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Rawas, R.; Amaral, I.M.; Hofer, A. Is p38 MAPK Associated to Drugs of Abuse-Induced Abnormal Behaviors? Int. J. Mol. Sci. 2020, 21, 4833. [Google Scholar] [CrossRef]
- Falcicchia, C.; Tozzi, F.; Arancio, O.; Watterson, D.M.; Origlia, N. Involvement of p38 MAPK in Synaptic Function and Dysfunction. Int. J. Mol. Sci. 2020, 21, 5624. [Google Scholar] [CrossRef] [PubMed]
- Yong, H.Y.; Koh, M.S.; Moon, A. The p38 MAPK inhibitors for the treatment of inflammatory diseases and cancer. Expert Opin. Investig. Drugs 2009, 18, 1893–1905. [Google Scholar] [CrossRef]
- Cargnello, M.; Roux, P.P. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 2011, 75, 50–83. [Google Scholar] [CrossRef] [Green Version]
- Delire, B.; Starkel, P. The Ras/MAPK pathway and hepatocarcinoma: Pathogenesis and therapeutic implications. Eur. J. Clin. Investig. 2015, 45, 609–623. [Google Scholar] [CrossRef]
- Guo, Y.J.; Pan, W.W.; Liu, S.B.; Shen, Z.F.; Xu, Y.; Hu, L.L. ERK/MAPK signalling pathway and tumorigenesis. Exp. Ther. Med. 2020, 19, 1997–2007. [Google Scholar] [CrossRef] [Green Version]
- Molina, J.R.; Adjei, A.A. The Ras/Raf/MAPK pathway. J. Thorac. Oncol. 2006, 1, 7–9. [Google Scholar] [CrossRef] [Green Version]
- Santarpia, L.; Lippman, S.M.; El-Naggar, A.K. Targeting the MAPK-RAS-RAF signaling pathway in cancer therapy. Expert Opin. Ther. Targets 2012, 16, 103–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Mello, S.R. When Good Kinases Go Rogue: GSK3, p38 MAPK and CDKs as Therapeutic Targets for Alzheimer’s and Huntington’s Disease. Int. J. Mol. Sci. 2021, 22, 5911. [Google Scholar] [CrossRef] [PubMed]
- Dinda, B.; Dinda, M.; Kulsi, G.; Chakraborty, A.; Dinda, S. Therapeutic potentials of plant iridoids in Alzheimer’s and Parkinson’s diseases: A review. Eur. J. Med. Chem. 2019, 169, 185–199. [Google Scholar] [CrossRef]
- Lee, J.K.; Kim, N.J. Recent Advances in the Inhibition of p38 MAPK as a Potential Strategy for the Treatment of Alzheimer’s Disease. Molecules 2017, 22, 1287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Obergasteiger, J.; Frapporti, G.; Pramstaller, P.P.; Hicks, A.A.; Volta, M. A new hypothesis for Parkinson’s disease pathogenesis: GTPase-p38 MAPK signaling and autophagy as convergence points of etiology and genomics. Mol. Neurodegener. 2018, 13, 40. [Google Scholar] [CrossRef] [PubMed]
- Rai, S.N.; Dilnashin, H.; Birla, H.; Singh, S.S.; Zahra, W.; Rathore, A.S.; Singh, B.K.; Singh, S.P. The Role of PI3K/Akt and ERK in Neurodegenerative Disorders. Neurotox. Res. 2019, 35, 775–795. [Google Scholar] [CrossRef] [PubMed]
- Marmol, I.; Sanchez-de-Diego, C.; Pradilla Dieste, A.; Cerrada, E.; Rodriguez Yoldi, M.J. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int. J. Mol. Sci. 2017, 18, 197. [Google Scholar] [CrossRef] [Green Version]
- Stefani, C.; Miricescu, D.; Stanescu, S., II; Nica, R.I.; Greabu, M.; Totan, A.R.; Jinga, M. Growth Factors, PI3K/AKT/mTOR and MAPK Signaling Pathways in Colorectal Cancer Pathogenesis: Where Are We Now? Int. J. Mol. Sci. 2021, 22, 10260. [Google Scholar] [CrossRef] [PubMed]
- Grothey, A.; Fakih, M.; Tabernero, J. Management of BRAF-mutant metastatic colorectal cancer: A review of treatment options and evidence-based guidelines. Ann. Oncol. 2021, 32, 959–967. [Google Scholar] [CrossRef]
- Mauri, G.; Bonazzina, E.; Amatu, A.; Tosi, F.; Bencardino, K.; Gori, V.; Massihnia, D.; Cipani, T.; Spina, F.; Ghezzi, S.; et al. The Evolutionary Landscape of Treatment for BRAF(V600E) Mutant Metastatic Colorectal Cancer. Cancers 2021, 13, 137. [Google Scholar] [CrossRef]
- Rasanen, K.; Dang, K.X.; Mustonen, H.; Ho, T.H.; Lintula, S.; Koistinen, H.; Stenman, U.H.; Haglund, C.; Stenman, J. MAPK inhibitors induce serine peptidase inhibitor Kazal type 1 (SPINK1) secretion in BRAF V600E-mutant colorectal adenocarcinoma. Mol. Oncol. 2018, 12, 224–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ros, J.; Baraibar, I.; Sardo, E.; Mulet, N.; Salva, F.; Argiles, G.; Martini, G.; Ciardiello, D.; Cuadra, J.L.; Tabernero, J.; et al. BRAF, MEK and EGFR inhibition as treatment strategies in BRAF V600E metastatic colorectal cancer. Ther. Adv. Med. Oncol. 2021, 13, 1758835921992974. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.F.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartha, A.; Gyorffy, B. TNMplot.com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef]
- Zeng, D.Q.; Ye, Z.L.; Shen, R.F.; Yu, G.C.; Wu, J.N.; Xiong, Y.; Zhou, R.; Qiu, W.J.; Huang, N.; Sun, L.; et al. IOBR: Multi-Omics Immuno-Oncology Biological Research to Decode Tumor Microenvironment and Signatures. Front. Immunol. 2021, 12, 687975. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.C.; Wang, L.G.; Han, Y.Y.; He, Q.Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhou, Y.; Feng, X.; Gao, Y.; Huang, C.; Yao, X. Low-dose orlistat promotes the therapeutic effect of oxaliplatin in colorectal cancer. Biomed. Pharmacother. 2022, 153, 113426. [Google Scholar] [CrossRef]
- Huang, C.; Wang, M.; Wang, J.; Wu, D.; Gao, Y.; Huang, K.; Yao, X. Suppression MGP inhibits tumor proliferation and reverses oxaliplatin resistance in colorectal cancer. Biochem. Pharmacol. 2021, 189, 114390. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, Q.; Wang, M.; Huang, C.; Yao, X. Effective Delivery of siRNA-Loaded Nanoparticles for Overcoming Oxaliplatin Resistance in Colorectal Cancer. Front. Oncol. 2022, 12, 827891. [Google Scholar] [CrossRef]
- Naito, Y.; Yoshimura, J.; Morishita, S.; Ui-Tei, K. siDirect 2.0: Updated software for designing functional siRNA with reduced seed-dependent off-target effect. BMC Bioinform. 2009, 10, 392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnaud, E.; Zenker, J.; de Preux Charles, A.S.; Stendel, C.; Roos, A.; Medard, J.J.; Tricaud, N.; Kleine, H.; Luscher, B.; Weis, J.; et al. SH3TC2/KIAA1985 protein is required for proper myelination and the integrity of the node of Ranvier in the peripheral nervous system. Proc. Natl. Acad. Sci. USA 2009, 106, 17528–17533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, R.; Hu, J. The role of JNK in prostate cancer progression and therapeutic strategies. Biomed. Pharmacother. 2020, 121, 109679. [Google Scholar] [CrossRef]
- Bubici, C.; Papa, S. JNK signalling in cancer: In need of new, smarter therapeutic targets. Br. J. Pharmacol. 2014, 171, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, L.; Bai, J.; Li, L.; Fan, J.; Fu, Z.; Liu, J. Long noncoding RNA plasmacytoma variant translocation 1 promotes progression of colorectal cancer by sponging microRNA-152-3p and regulating E2F3/MAPK8 signaling. Cancer Sci. 2022, 113, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.Y.; Richardson, B.C. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 2005, 6, 322–327. [Google Scholar] [CrossRef]
- Drosten, M.; Barbacid, M. Targeting the MAPK Pathway in KRAS-Driven Tumors. Cancer Cell 2020, 37, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Pua, L.J.W.; Mai, C.W.; Chung, F.F.; Khoo, A.S.; Leong, C.O.; Lim, W.M.; Hii, L.W. Functional Roles of JNK and p38 MAPK Signaling in Nasopharyngeal Carcinoma. Int. J. Mol. Sci. 2022, 23, 1108. [Google Scholar] [CrossRef]
- Dong, Y.; Siegwart, D.J.; Anderson, D.G. Strategies, design, and chemistry in siRNA delivery systems. Adv. Drug Deliv. Rev. 2019, 144, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, J.A.; Witzigmann, D.; Chen, S.; Cullis, P.R.; van der Meel, R. Lipid Nanoparticle Technology for Clinical Translation of siRNA Therapeutics. Acc. Chem. Res. Hematol. 2019, 52, 2435–2444. [Google Scholar] [CrossRef] [PubMed]
- Young, S.W.; Stenzel, M.; Yang, J.L. Nanoparticle-siRNA: A potential cancer therapy? Crit. Rev. Oncol. 2016, 98, 159–169. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, C.; Yi, H.; Zhou, Y.; Zhang, Q.; Yao, X. Pan-Cancer Analysis Reveals SH3TC2 as an Oncogene for Colorectal Cancer and Promotes Tumorigenesis via the MAPK Pathway. Cancers 2022, 14, 3735. https://doi.org/10.3390/cancers14153735
Huang C, Yi H, Zhou Y, Zhang Q, Yao X. Pan-Cancer Analysis Reveals SH3TC2 as an Oncogene for Colorectal Cancer and Promotes Tumorigenesis via the MAPK Pathway. Cancers. 2022; 14(15):3735. https://doi.org/10.3390/cancers14153735
Chicago/Turabian StyleHuang, Chengzhi, Hui Yi, Yue Zhou, Qing Zhang, and Xueqing Yao. 2022. "Pan-Cancer Analysis Reveals SH3TC2 as an Oncogene for Colorectal Cancer and Promotes Tumorigenesis via the MAPK Pathway" Cancers 14, no. 15: 3735. https://doi.org/10.3390/cancers14153735
APA StyleHuang, C., Yi, H., Zhou, Y., Zhang, Q., & Yao, X. (2022). Pan-Cancer Analysis Reveals SH3TC2 as an Oncogene for Colorectal Cancer and Promotes Tumorigenesis via the MAPK Pathway. Cancers, 14(15), 3735. https://doi.org/10.3390/cancers14153735





