Deep Neural Network Models for Colon Cancer Screening
Abstract
:Simple Summary
Abstract
1. Introduction
2. Imaging Modalities
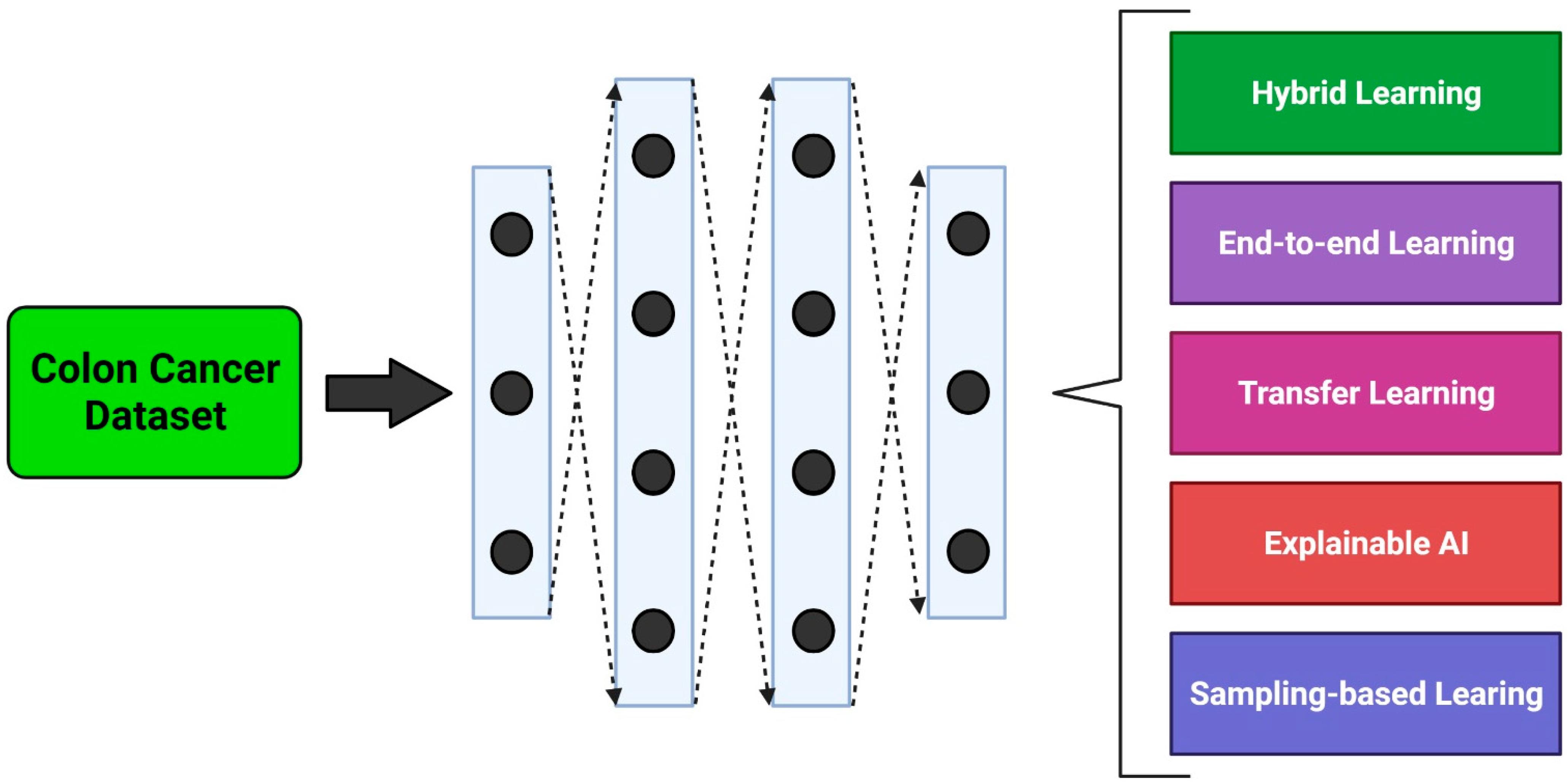
3. Methodological Approaches
3.1. Hybrid Learning Methods
3.2. End-to-End Learning Methods
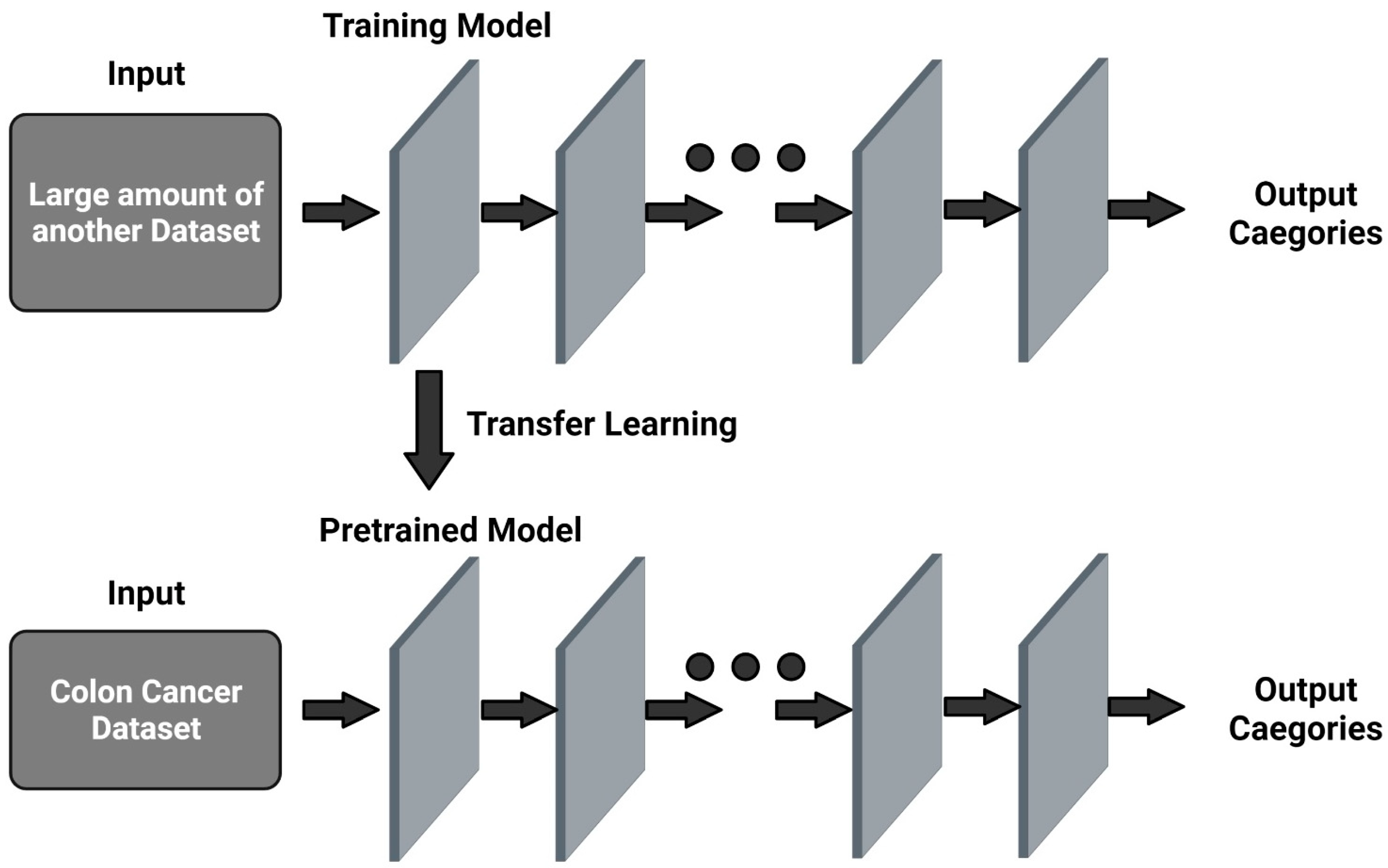
3.3. Transfer Learning Methods
3.4. Explainable Learning Methods
3.5. Sampling Methods
4. Results and Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424, Erratum in CA Cancer J. Clin. 2020, 70, 313. [Google Scholar] [CrossRef] [Green Version]
- Wong, M.C.S.; Huang, J.; Lok, V.; Wang, J.; Fung, F.; Ding, H.; Zheng, Z.J. Differences in Incidence and Mortality Trends of Colorectal Cancer Worldwide Based on Sex, Age, and Anatomic Location. Clin. Gastroenterol. Hepatol. 2021, 19, 955–966.e61. [Google Scholar] [CrossRef] [PubMed]
- Kahi, C.J. Reviewing the Evidence that Polypectomy Prevents Cancer. Gastrointest. Endosc. Clin. N. Am. 2019, 29, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Leggett, B.; Whitehall, V. Role of the serrated pathway in colorectal cancer pathogenesis. Gastroenterology 2010, 138, 2088–2100. [Google Scholar] [CrossRef]
- Bejnordi, B.E.; Zuidhof, G.; Balkenhol, M.; Hermsen, M.; Bult, P.; van Ginneken, B.; Karssemeijer, N.; Litjens, G.; van der Laak, J. Context-Aware stacked convolutionalneural networks for classification of breast carcinomas in whole-slide histopathology images. J. Med. Imaging 2017, 4, 044504. [Google Scholar] [CrossRef]
- Gschwantler, M.; Kriwanek, S.; Langner, E.; Goritzer, B.; SchrutkaKolbl, C.; Brownstone, E.; Feichtinger, H.; Weiss, W. High-grade dysplasia and invasive carcinoma in colorectal adenomas: A multivariate analysis of the impact of adenoma and patient characteristics. Eur. J. Gastroenterol. Hepatol. 2002, 14, 183188. [Google Scholar] [CrossRef]
- Lieberman, D.A.; Rex, D.K.; Winawer, S.J.; Giardiello, F.M.; Johnson, D.A.; Levin, T.R. Guidelines for colonoscopy surveillance after screening and polypectomy: A consensus update by the US Multi-Society Task Force on Colorectal Cancer. Gastroenterology 2012, 143, 844–857. [Google Scholar] [CrossRef] [Green Version]
- Vu, H.T.; Lopez, R.; Bennett, A.; Burke, C.A. Individuals with sessile serrated polyps express an aggressive colorectal phenotype. Dis. Colon Rectum 2011, 54, 1216–1223. [Google Scholar] [CrossRef]
- Miller, F.G.; Joffe, S. Equipoise and the dilemma of randomized clinical trials. N. Engl. J. Med. 2011, 364, 476–480. [Google Scholar] [CrossRef]
- Abdeljawad, K.; Vemulapalli, K.C.; Kahi, C.J.; Cummings, O.W.; Snover, D.C.; Rex, D.K. Sessile serrated polyp prevalence determined by a colonoscopist with a high lesion detection rate and an experienced pathologist. Gastrointest. Endosc. 2015, 81, 517–524. [Google Scholar] [CrossRef]
- Irshad, H.; Veillard, A.; Roux, L.; Racoceanu, D. Methods for nuclei detection, segmentation, and classification in digital histopathology: A review current status and future potential. IEEE Rev. Biomed. Eng. 2014, 7, 97–114. [Google Scholar] [CrossRef]
- Veta, M.; van Diest, P.J.; Willems, S.M.; Wang, H.; Madabhushi, A.; Cruz-Roa, A.; Gonzalez, F.; Larsen, A.B.L.; Vestergaard, J.S.; Dahl, A.B.; et al. Assessment of algorithms for mitosis detection in breast cancer histopathology images. Med. Image Anal. 2015, 20, 237–248. [Google Scholar] [CrossRef] [Green Version]
- Komura, D.; Ishikawa, S. Machine learning methods for histopathological image analysis. Comput. Struct. Biotechnol. J. 2018, 16, 34–42. [Google Scholar] [CrossRef]
- Malhi, A.; Kampik, T.; Pannu, H.; Madhikermi, M.; Främling, K. Explaining machine learning-based classifications of in-vivo gastral images. In Proceedings of the 2019 Digital Image Computing: Techniques and Applications, DICTA, Perth, Australia, 2–4 December 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1–7. [Google Scholar]
- Silva, J.; Histace, A.; Romain, O.; Dray, X.; Granado, B. Toward embedded detection of polyps in wce images for early diagnosis of colorectal cancer. Int. J. Comput. Assist. Radiol. Surg. 2014, 9, 283–293. [Google Scholar] [CrossRef]
- West, N.P.; Dattani, M.; McShane, P.; Hutchins, G.; Grabsch, J.; Mueller, W.; Treanor, D.; Quirke, P.; Grabsch, H. The proportion of tumour cells is an independent predictorfor survival in colorectal cancer patients. Br. J. Cancer 2010, 3, 1519–1523. [Google Scholar] [CrossRef] [Green Version]
- Rastogi, A.; Keighley, J.; Singh, V.; Callahan, P.; Bansal, A.; Wani, S.; Sharma, P. High accuracy of narrow band imaging without magnification for the real-time characterization of polyp histology and its comparison with high-definition white light colonoscopy: A prospective study. Am. J. Gastroenterol. 2009, 104, 2422–2430. [Google Scholar] [CrossRef]
- Lu, F.I.; de van Niekerk, W.; Owen, D.; Turbin, D.A.; Webber, D.L. Longitudinal outcome study of sessile serrated adenomas of the colorectum: An increased risk for subsequent right-sided colorectal carcinoma. Am. J. Surg. Pathol. 2010, 34, 927–934. [Google Scholar] [CrossRef]
- Tischendorf, J.J.; Gross, S.; Winograd, R.; Hecker, H.; Auer, R.; Behrens, A.; Trautwein, C.; Aach, T.; Stehle, T. Computer-aided classification of colorectal polyps based on vascular patterns: A pilot study. Endoscopy 2010, 42, 203–207. [Google Scholar] [CrossRef]
- Kitajima, K.; Fujimori, T.; Fujii, S.; Takeda, J.; Ohkura, Y.; Kawamata, H.; Kumamoto, T.; Ishiguro, S.; Kato, Y.; Shimoda, T.; et al. Correlations between lymph node metastasis and depth of submucosal invasion in submucosal invasive colorectal carcinoma: A Japanese collaborative study. J. Gastroenterol. 2004, 39, 534–543. [Google Scholar] [CrossRef]
- Sirinukunwattana, K.; Raza, S.E.A.; Tsang, Y.W.; Snead, D.R.; Cree, I.A.; Rajpoot, N.M. Locality sensitive deep learning for detection and classification of nuclei in routine colon cancer histology images. IEEE Trans. Med. Imaging 2016, 35, 1196–1206. [Google Scholar] [CrossRef] [Green Version]
- Mobadersany, P.; Yousefi, S.; Amgad, M.; Gutman, D.A.; Barnholtz-Sloan, J.S.; Velázquez Vega, J.E.; Brat, D.J.; Cooper, L.A. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc. Natl. Acad. Sci. USA 2018, 115, E2970–E2979. [Google Scholar] [CrossRef] [Green Version]
- Itoh, T.; Kawahira, H.; Nakashima, H.; Yata, N. Deep learning analyzes Helicobacter pylori infection by upper gastrointestinal endoscopyimages. Endosc. Int. Open 2018, 6, E139–E144. [Google Scholar]
- Korbar, B.; Olofson, A.M.; Miraflor, A.P.; Nicka, C.M.; Suriawinata, M.A.; Torresani, L.; Suriawinata, A.A.; Hassanpour, S. Deep learning for classification of colorectal polyps on whole-slide images. J Pathol. Inform. 2017, 8, 30. [Google Scholar] [CrossRef]
- Komeda, Y.; Handa, H.; Watanabe, T.; Nomura, T.; Kitahashi, M.; Sakurai, T.; Okamoto, A.; Minami, T.; Kono, M.; Arizumi, T.; et al. Computeraided diagnosis based on convolutional neural network system for colorectal polyp classification: Preliminary experience. Oncology 2017, 93 (Suppl. S1), 30–34. [Google Scholar] [CrossRef]
- Wimmer, G.; Tamaki, T.; Tischendorf, J.J.; Häfner, M.; Yoshida, S.; Tanaka, S.; Uhl, A. Directional wavelet based features for colonic polyp classification. Med. Image Anal. 2016, 31, 16–36. [Google Scholar] [CrossRef]
- Hafner, M.; Tamaki, T.; Tanaka, S.; Uhl, A.; Wimmer, G.; Yoshida, S. Local fractal dimension based approaches for colonic polyp classification. Med. Image Anal. 2015, 26, 92–107. [Google Scholar] [CrossRef]
- Okamoto, T.; Koide, T.; Sugi, K.; Shimizu, T.; Hoang, A.T.; Tamaki, T.; Raytchev, B.; Kaneda, K.; Kominami, Y.; Yoshida, S.; et al. Image segmentation of pyramid style identifier based on Support Vector Machine for colorectal endoscopic images. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; Volume 2015, pp. 2997–3000. [Google Scholar]
- Mori, Y.; Kudo, S.E.; Chiu, P.W.; Misawa, M.; Wakamura, K.; Kudo, T.; Hayashi, T.; Katagiri, A.; Miyachi, H.; Ishida, F.; et al. Impact of an automated system for endocytoscopic diagnosis of small colorectal lesions: An international web-based study. Endoscopy 2016, 48, 1110–1118. [Google Scholar] [CrossRef]
- Ştefănescu, D.; Streba, C.; Cârţână, E.T.; Săftoiu, A.; Gruionu, G.; Gruionu, L.G. Computer Aided Diagnosis for Confocal Laser Endomicroscopy in Advanced Colorectal Adenocarcinoma. PLoS ONE 2016, 11, e0154863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trebeschi, S.; van Griethuysen, J.J.M.; Lambregts, D.M.J.; Parmar, C.; Bakers, F.C.; Peters, N.H.; Beets-Tan, R.G.; Aerts, H.J. Deep Learning for Fully-Automated Localization and Segmentation of Rectal Cancer on Multiparametric MR. Sci. Rep. 2017, 7, 5301. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.P.; Xie, P.; Wang, M.; Li, W.; Pickhardt, P.J.; Xia, W.; Xiong, F.; Zhang, R.; Xie, Y.; Jian, J.; et al. Deep learning-based fully automated detection and segmentation of lymph nodes on multiparametric-mri for rectal cancer: A multicentre study. EBioMedicine 2020, 56, 102780. [Google Scholar] [CrossRef] [PubMed]
- Kainz, K.; Pfeiffer, M.; Urschler, M. Segmentation and classification of colon glands with deep convolutional neural networks and total variation regularization. PeerJ 2017, 5, e3874. [Google Scholar] [CrossRef]
- Urban, G.; Tripathi, P.; Alkayali, T.; Mittal, M.; Jalali, F.; Karnes, W.; Baldi, P. Deep learning localizes and identifies polyps in real time with 96% accuracy in screening colonoscopy. Gastroenterology 2018, 155, 1069–1078.e8. [Google Scholar] [CrossRef]
- Tajbakhsh, N.; Gurudu, S.R.; Liang, J. Automated polyp detection in colonoscopy videos using shape and context information. IEEE Trans. Med. Imaging 2016, 35, 630–644. [Google Scholar] [CrossRef]
- Misawa, M.; Kudo, S.E.; Mori, Y.; Cho, T.; Kataoka, S.; Yamauchi, A.; Ogawa, Y.; Maeda, Y.; Takeda, K.; Ichimasa, K. Artificial intelligence assisted polyp detection for colonoscopy: Initial experience. Gastroenterology 2018, 154, 2027–2029.e3. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Xiao, X.; Glissen Brown, J.R.; Berzin, T.M.; Tu, M.; Xiong, F.; Hu, X.; Liu, P.; Song, Y.; Zhang, D.; et al. Development and validation of a deep-learning algorithm for the detection of polyps during colonoscopy. Nat. Biomed. Eng. 2018, 2, 741–748. [Google Scholar] [CrossRef]
- Ding, H.; Pan, Z.; Cen, Q.; Li, Y.; Chen, S. Multi-scale fully convolutional network for gland segmentation using three-class classification. Neurocomputing 2020, 380, 150–161. [Google Scholar] [CrossRef]
- Yoon, H.; Lee, J.; Oh, J.E.; Kim, H.R.; Lee, S.; Chang, H.J.; Sohn, D.K. Tumor Identification in Colorectal Histology Images Using a Convolutional Neural Network. J. Digit. Imaging 2019, 32, 131–140. [Google Scholar] [CrossRef]
- Sena, P.; Fioresi, R.; Faglioni, F.; Losi, L.; Faglioni, G.; Roncucci, L. Deep learning techniques for detecting preneoplastic and neoplastic lesions in human colorectal histological images. Oncol. Lett. 2019, 18, 6101–6107. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Jia, Z.; Wang, L.B.; Ai, Y.; Zhang, F.; Lai, M.; Chang, E.I. Large scale tissue histopathology image classification, segmentation, and visualization via deep convolutional activation features. BMC Bioinform. 2017, 18, 281. [Google Scholar] [CrossRef] [Green Version]
- Korbar, B.; Olofson, A.M.; Miraflor, A.P.; Nicka, C.M.; Suriawinata, M.A.; Torresani, L.; Suriawinata, A.A.; Hassanpour, S. Looking under the hood: Deep neural network visualization to interpret whole-slide Image analysis outcomes for colorectal polyps. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Honolulu, HI, USA, 21–26 July 2017; pp. 69–75. [Google Scholar]
- Sabol, P.; Sinčák, P.; Hartono, P.; Kočan, P.; Benetinová, Z.; Blichárová, A.; Verbóová, Ľ.; Štammová, E.; Sabolová-Fabianová, A.; Jašková, A. Explainable classifier for improving the accountability in decision-making for colorectal cancer diagnosis from histopathological images. J. Biomed. Inform. 2020, 109, 103523. [Google Scholar] [CrossRef]
- Mary, S.; Hewitt, K.J.; Rajpoot, N. Deep Learning with Sampling in Colon Cancer Histology. Front. Bioeng. Biotechnol. 2019, 7, 52. [Google Scholar]
- Hong, L.T.T.; Thanh, N.C.; Long, T.Q. Polyp segmentation in colonoscopy Images using ensembles of U-Nets with efficientNet and asymmetric similarity loss function. In Proceedings of the 2020 International Conference on Computing and Communication Technologies (RIVF), Ho Chi Minh City, Vietnam, 14–15 October 2020; pp. 1–6. [Google Scholar] [CrossRef]
- Yamada, M.; Saito, Y.; Imaoka, H.; Saiko, M.; Yamada, S.; Kondo, H.; Takamaru, H.; Sakamoto, T.; Sese, J.; Kuchiba, A.; et al. Development of a real-time endoscopic image diagnosis support system using deep learning technology in colonoscopy. Sci. Rep. 2019, 9, 14465. [Google Scholar] [CrossRef] [Green Version]
- Ho, C.; Zhao, Z.; Chen, X.F.; Sauer, J.; Saraf, S.A.; Jialdasani, R.; Taghipour, K.; Sathe, A.; Khor, L.-Y.; Lim, K.-H.; et al. A promising deep learning assistive algorithm for histopathological screening of colorectal cancer. Sci. Rep. 2022, 12, 2222. [Google Scholar] [CrossRef]
- Xu, L.; Walker, B.; Liang, P.-I.; Tong, Y.; Xu, C.; Su, Y.C.; Karsan, A. Colorectal cancer detection based on deep learning. J. Pathol. Inform. 2020, 1, 28. [Google Scholar] [CrossRef]
- Buendgens, L.; Didem, C.; Laleh, N.G.; van Treeck, M.; Koenen, M.T.; Zimmermann, H.W.; Herbold, T.; Lux, T.J.; Hann, A.; Trautwein, C.; et al. Weakly supervised end-to-end artificial intelligence in gastrointestinal endoscopy. Sci. Rep. 2022, 12, 4829. [Google Scholar] [CrossRef]
- Takamatsu, M.; Yamamoto, N.; Kawachi, H.; Chino, A.; Saito, S.; Ueno, M.; Ishikawa, Y.; Takaawa, Y.; Takeuchi, K. Prediction of early colorectal cancer metastasis by machine learning using digital slide images. Comput. Methods Programs Biomed. 2019, 178, 155–161. [Google Scholar] [CrossRef]
- Ghosh, J.; Sharma, A.K.; Tomar, S. Feature extraction and classification of colon cancer using a hybrid approach of supervised and unsupervised learning. In Advanced Machine Learning Approaches in Cancer Prognosis; Springer: Cham, Switzerland, 2021; pp. 195–219. [Google Scholar]
- Thomaz, V.A.; Sierra-Franco, C.A.; Raposo, A.B. Training data enhancements for improving colonic polype detection using deep convolutional neural networks. Aritificial Intell. Med. 2020, 111, 101988. [Google Scholar] [CrossRef]
- Yu, G.; Sun, K.; Xu, C.; Shi, X.-H.; Wu, C.; Xie, T.; Meng, R.-Q.; Meng, X.-H.; Wang, K.-S.; Xiao, H.-M.; et al. Accurate recognition of colorectal cancer with semi-supervised deep learning on pathological images. Nat. Commun. 2021, 12, 6311. [Google Scholar] [CrossRef]
- Rączkowski, L.; Mozejko, M.; Zambonelli, J.; Szczurek, E. Ara: Accurate, reliable and active histopathological image classification framework with Bayesian deep learning. Sci. Rep. 2019, 9, 14347. [Google Scholar] [CrossRef] [Green Version]
- Mori, Y.; Kudo, S.-E.; Wakamura, K.; Misawa, M.; Ogawa, Y.; Kutsukawa, M.; Kudo, T.; Hayashi, T.; Miyachi, H.; Ishida, F.; et al. Novel computer-aided diagnostic system for colorectal lesions using endoscytoscopy. Gastrointest. Endosc. 2015, 81, 621–629. [Google Scholar] [CrossRef] [Green Version]
- Nartowt, B.J.; Hart, G.R.; Muhammad, W.; Liang, Y.; Stark, G.F.; Deng, J. Robust Machine Learning for Colorectal Cancer Risk Prediction and Stratification. Front. Big Data 2020, 3, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, J.-J.; Chen, B.-L.; Kong, Y.-X.; Ma, X.-G.; Yu, Y.-T. An early intestinal cancer prediction algorithm based on deep belief network. Sci. Rep. 2019, 9, 17418. [Google Scholar] [CrossRef] [PubMed]
- Ito, I.; Kawahira, H.; Nakashima, H.; Uesato, M.; Miyauchi, H.; Matsubara, H. Endoscopic diagnostic support system for cT1b colorectal cancer using deep learning. Oncology 2019, 96, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Tamai, N.; Saito, Y.; Sakamoto, T.; Nakajima, T.; Matsuda, T.; Sumiyama, K.; Tajiri, H.; Koyama, R.; Kido, S. Effectiveness of computer-aided diagnosis of colorectal lesions using novel software for magnifying narrow-band imaging: A pilot study. Endosc. Int. Open 2017, 5, E690–E694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Z.; Yu, C.; Zou, S.; Wang, W.; Huang, Y.; Ding, X.; Liu, J.; Shao, L.; Yuan, J.; Gou, X.; et al. Automatic deep learning-based colorectal adenoma detection system and its similarities with pathologists. BMJ Open 2020, 10, e036423. [Google Scholar] [CrossRef] [PubMed]
- Rathore, S.; Hussain, M.; Khan, A. Automated colon cancer detection using hybrid of novel geometric features and some traditional features. Comput. Biol. Med. 2015, 65, 279–296. [Google Scholar] [CrossRef] [PubMed]
- Nadimi, E.S.; Buijs, M.M.; Herp, J.; Kroijer, R.; Kobaek-Larsen, M.; Nielsen, E.; Pedersen, C.D.; Blanes-Vidal, V.; Baatrup, G. Application of deep learning for autonomous detection and localization of colorectal polyps in wireless colon capsule endoscopy. Comput. Electr. Eng. 2020, 81, 106531. [Google Scholar] [CrossRef]
- Glasmachers, T. Limits of end-to-end learning. Proc. Mach. Learn. Res. 2017, 77, 17–32. [Google Scholar]
- Graves, A.; Wayne, G.; Danihelka, I. Neural turing machines. arXiv 2014, arXiv:1410.54012014. [Google Scholar]
- Graves, G.; Wayne, M.; Reynolds, T.; Harley, I.; Danihelka, A.; Grabska-Barwi’nska, S.; Colmenarejo G’omez, E.; Grefenstette, T.R.; Agapiou, J. Hybrid computing using a neural network with dynamic external memory. Nature 2016, 538, 471–476. [Google Scholar] [CrossRef]
- Tamar, A.; Levine, S.; Abbeel, P.; Wu, Y.; Thomas, G. Value iteration networks. In Advances in Neural Information Processing Systems, Proceedings of the 30th Conference on Neural Information Processing Systems (NIPS 2016), Barcelona, Spain, 5–10 December 2016; Curran Associates: Red Hook, NY, USA, 2016; pp. 2146–2154. [Google Scholar]
- Mirowski, P.; Pascanu, R.; Viola, F.; Soyer, H.; Ballard, A.; Banino, A.; Denil, M.; Goroshin, R.; Sifre, L.; Kavukcuoglu, K. Learning to navigate in complex environments. arXiv 2016, arXiv:1611.03673. [Google Scholar]
- Iizuka, O.; Kanavati, F.; Kato, K.; Rambeau, M.; Arihiro, K.; Tsuneki, M. Deep learning models for histopathological classification of gastric and colonic epithelial tumours. Sci. Rep. 2020, 10, 1504. [Google Scholar] [CrossRef] [Green Version]
- Pinckaers, H.; Litjens, G. Neural ordinary differential equation for semantic segmentation of individual colon glands. arXiv 2019, arXiv:1910.10470v1. [Google Scholar]
- Olivas, E.S.; Guerrero, J.D.M.; Sober, M.M.; Benedito, J.R.M.; Lopez, A.J.S. Handbook of Research on Machine Learning Applications and Trends: Algorithms, Methods and Techniques; Information Science Reference; IGI Global: Hershey, PA, USA, 2009; pp. 1–834. [Google Scholar]
- Dilmegani, C. Transfer Learning in 2022: What It Is & How It Works. Artificial Intelligence Multiple, 2020. Available online: https://research.aimultiple.com/transfer-learning/ (accessed on 1 July 2022).
- Gessert, N.; Bengs, M.; Wittig, L.; Drömann, D.; Keck, T.; Schlaefer, A.; Ellebrecht, D.B. Deep transfer learning methods for colon cancer classification in confocal laser microscopy images. Int. J. Comput. Assist. Radiol. Surg. 2019, 14, 1837–1845. [Google Scholar] [CrossRef] [Green Version]
- Hamida, A.B.; Devanne, M.; Weber, J.; Truntzer, C.; Derangère, V.; Ghiringhelli, F.; Forestier, G.; Wemmert, C. Deep learning for colon cancer histopathological images analysis. In Computers in Biology and Medicine; Elsevier: Amsterdam, The Netherlands, 2021. [Google Scholar]
- Malik, J.; Kiranyaz, S.; Kunhoth, S.; Ince, T.; Al-Maadeed, S.; Hamila, R.; Gabbouj, M. Colorectal cancer diagnosis from histology images: A comparative study. arXiv 2019, arXiv:1903.11210. [Google Scholar]
- Kather, J.N.; Krisam, J.; Charoentong, P.; Luedde, T.; Herpel, E.; Weis, C.-A.; Gaiser, T.; Marx, A.; Valous, N.A.; Ferber, D.; et al. Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicentre study. PLoS Med. 2019, 16, e1002730. [Google Scholar] [CrossRef]
- Hägele, M.; Seegerer, P.; Lapuschkin, S.; Bockmayr, M.; Samek, W.; Klauschen, F.; Mülle, K.-R.; Binder, A. Resolving challenges in deep learning-based analyses of histopathological images using explanation methods. Sci. Rep. 2020, 10, 6423. [Google Scholar] [CrossRef] [Green Version]
- Kather, J.N.; Pearson, A.T.; Halama, N.; Jäger, D.; Krause, J.; Loosen, S.H.; Marx, A.; Boor, P.; Tacke, F.; Neumann, U.P.; et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 2019, 25, 1054–1056. [Google Scholar] [CrossRef]
- Bychkov, D.; Linder, N.; Turkki, R.; Nordling, S.; Kovanen, P.E.; Verrill, C.; Walliander, M.; Lundin, M.; Haglund, C.; Lundin, J. Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci. Rep. 2018, 8, 3395. [Google Scholar] [CrossRef]
- Ponzio, F.; Macii, E.; Ficarra, E.; Di Cataldo, S. Colorectal cancer classification using deep convolutional networks. In Proceedings of the 11th International Joint Conference on Biomedical Engineering Systems and Technologies 2018, Funchal, Portugal, 19–21 January 2018; Volume 2, pp. 58–66. [Google Scholar]
- Yao, J.; Zhu, X.; Jonnagaddala, J.; Hawkins, N.; Huang, J. Whole slide images based cancer survival prediction using attention guided deep multiple instance learning networks. Med. Image Anal. 2020, 65, 101789. [Google Scholar] [CrossRef]
- Koziarski, M. Two-Stage Resampling for Convolutional Neural Network Training in the Imbalanced Colorectal Cancer Image Classification. arXiv 2020, arXiv:2004.03332. [Google Scholar]
- Li, Y.; Eresen, A.; Shangguan, J.; Yang, J.; Lu, Y.; Chen, D.; Wang, J.; Velichko, Y.; Yaghmai, V.; Zhang, Z. Establishment of a new non-invasive imaging prediction model for liver metastasis in colon cancer. Am. J. Cancer Res. 2019, 9, 2482–2492. [Google Scholar]
- Wang, K.S.; Yu, G.; Xu, C.; Meng, X.H.; Zhou, J.; Zheng, C.; Deng, Z.; Shang, L.; Liu, R.; Su, S.; et al. Accurate diagnosis of colorectal cancer based on histopathology images using artificial intelligence. BMC Med. 2021, 19, 76. [Google Scholar] [CrossRef]
| References | Methods | Imaging Modality |
|---|---|---|
| Yao et al., 2020 [80] | Deep Attention Multiple Instance Survival Learning (DeepAttnMISL) with Multiple Instance Fully Convolutional Network (MI-FCN), DeepMISL, Finetuned-WSISA-LassoCOx, Finetuned-WSISA-MTLSA, WSISA-LassoCox, WSISA-MTLSA | Sample images of tissues taken through colonoscopy and turned into WSI clusters. |
| Sirinukunwattana et al., 2016 [21] | Spatially Constrained Convolutional Neural Network (SC-CNN) | 100 H&E-stained histology images of colorectal adenocarcinomas of 500 × 500 pixels were cropped from WSIs using Omnyx VLI20 scanner. |
| Sabol et al., 2020 [43] | Cumulative Fuzzy Class Membership Criterion (CFCMC) | H&E tissue slides cut into 5000 small sections of 150 × 150 pixels and annotation as one of eight tissue classes. |
| Korbar et al., 2017 [42] | ResNet | 176 H&E-stained WSIs collected from patients who underwent colorectal cancer screening. |
| Hägele et al., 2020 [76] | GoogLeNet from Caffe Model Zoo | H&E-stained images from TCGA Research Network were chosen and its WSIs were annotated. |
| Koziarski et al., 2020 [81] | MobileNet | Colorectal cancer histology image datasets divided into 5000 different types of tissues, each with a 150 × 150 pixels dimensionality. |
| Kainz et al., 2017 [33] | Object-Net and Separator-Net | 165 annotated, H&E-stained images of colorectal adenocarcinomas were collected at 20× magnification using a Zeiss MIRAX MIDI Scanner. |
| Hong et al., 2020 [45] | U-Net with EfficientNet B4 and EfficientNet B5 encoder | Datasets obtained from ETIS-Larib from the MICCAI 2015 polyp detection challenge, and CVCColonDB |
| Shapcott et al., 2019 [44] | Matconvnet, Tensorflow “cifar10” with Pycharm IDE | 142 H&E-stained, 40× magnification colorectal cancer images obtained from TCGA COAD data set from the Genomic Data Commons Portal 2018. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kavitha, M.S.; Gangadaran, P.; Jackson, A.; Venmathi Maran, B.A.; Kurita, T.; Ahn, B.-C. Deep Neural Network Models for Colon Cancer Screening. Cancers 2022, 14, 3707. https://doi.org/10.3390/cancers14153707
Kavitha MS, Gangadaran P, Jackson A, Venmathi Maran BA, Kurita T, Ahn B-C. Deep Neural Network Models for Colon Cancer Screening. Cancers. 2022; 14(15):3707. https://doi.org/10.3390/cancers14153707
Chicago/Turabian StyleKavitha, Muthu Subash, Prakash Gangadaran, Aurelia Jackson, Balu Alagar Venmathi Maran, Takio Kurita, and Byeong-Cheol Ahn. 2022. "Deep Neural Network Models for Colon Cancer Screening" Cancers 14, no. 15: 3707. https://doi.org/10.3390/cancers14153707
APA StyleKavitha, M. S., Gangadaran, P., Jackson, A., Venmathi Maran, B. A., Kurita, T., & Ahn, B.-C. (2022). Deep Neural Network Models for Colon Cancer Screening. Cancers, 14(15), 3707. https://doi.org/10.3390/cancers14153707









