Lung Cancer Organoids: The Rough Path to Personalized Medicine
Abstract
:Simple Summary
Abstract
1. Introduction
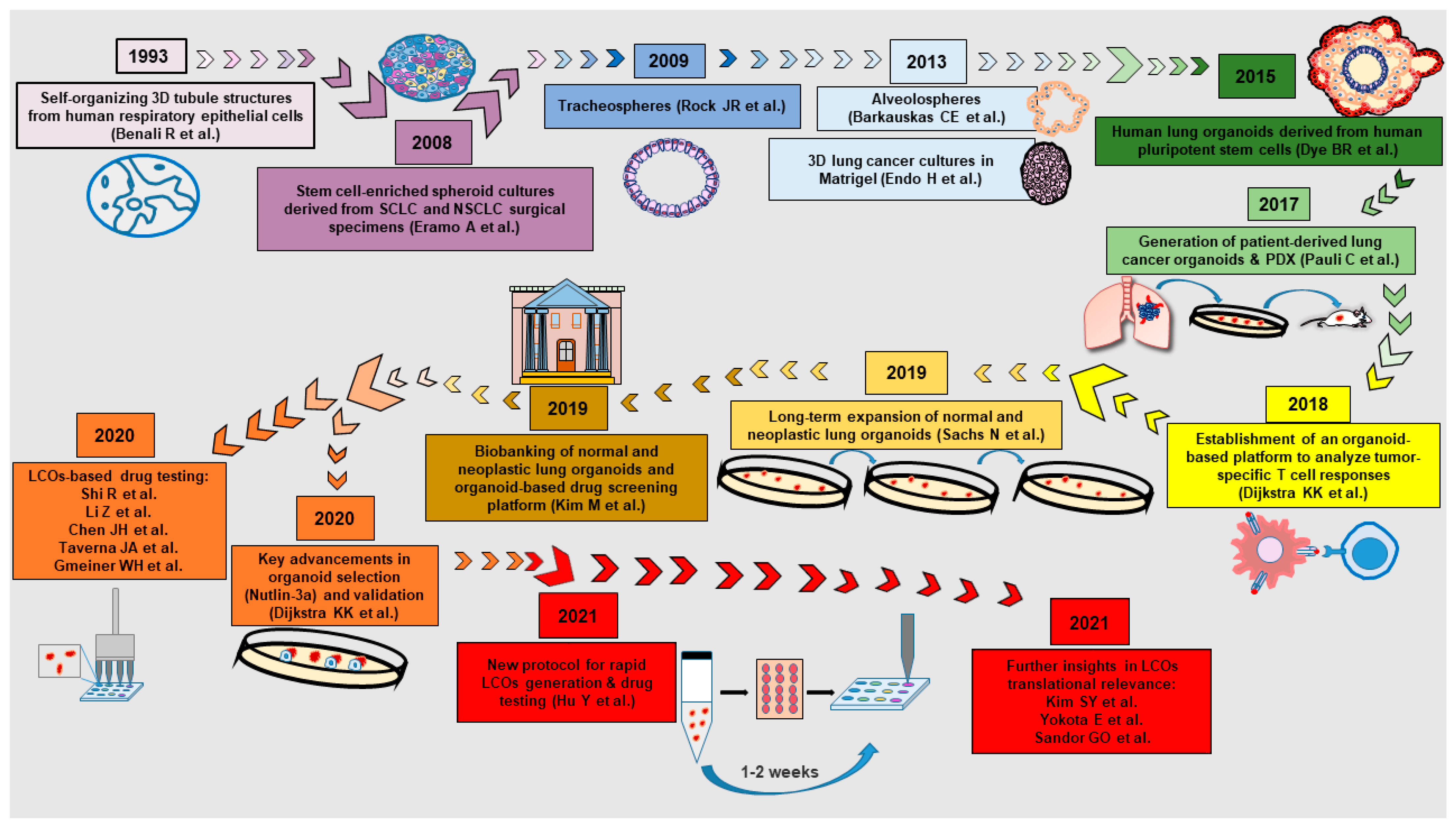
2. A Brief History of Normal and Neoplastic Lung Organoids
3. Problems and Solutions in the Generation of Lung Cancer Organoids
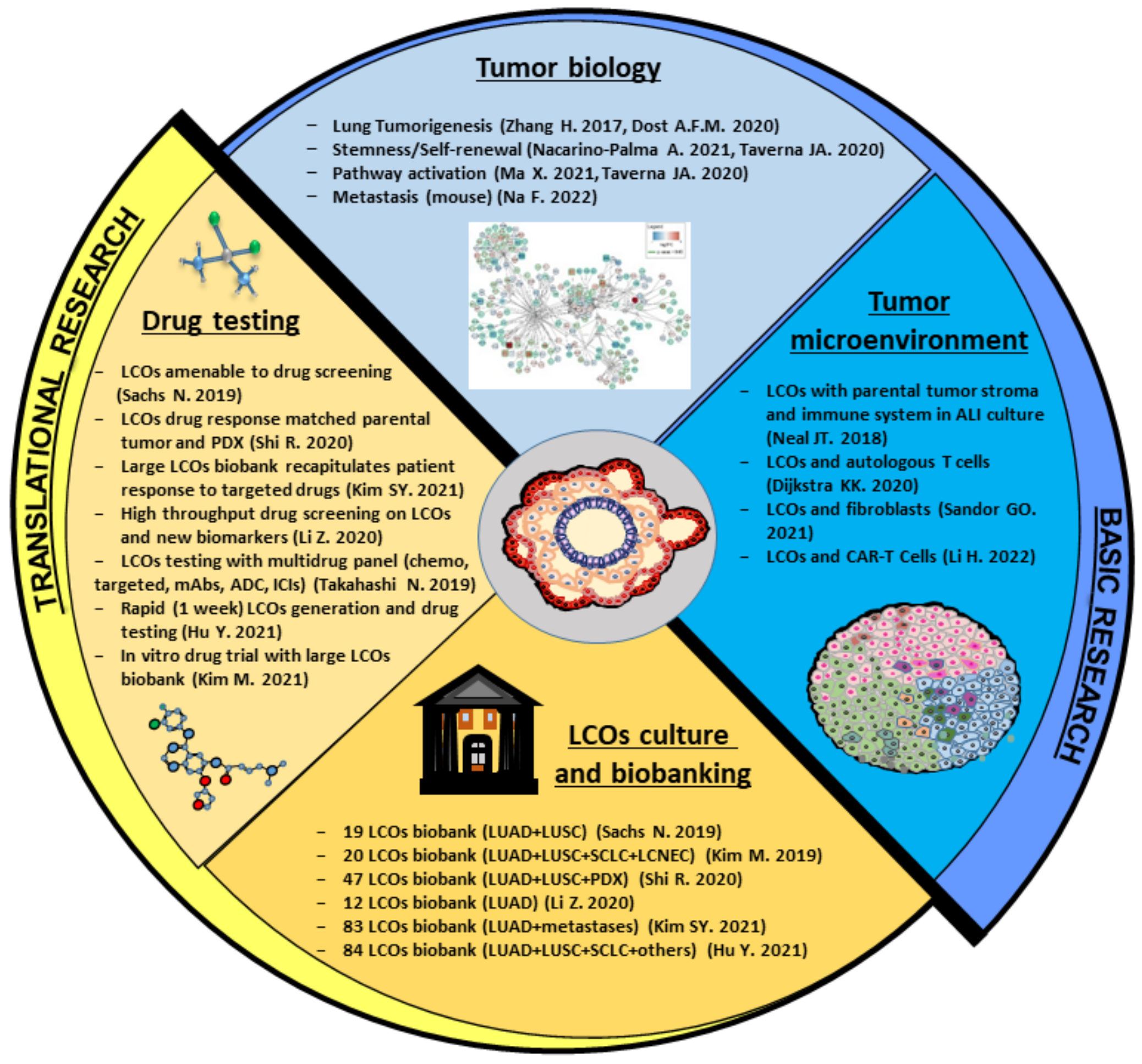
4. Preclinical Applications of Lung Cancer Organoids
4.1. LCOs in Basic Research: A Useful Tool to Understand Lung Cancer Biology
4.2. LCOs for Modeling the Tumor Microenvironment: Reconstituted or Holistic Co-Cultures?
4.3. LCOs in Translational Research: Applications for Personalized Medicine
5. Future Directions and Conclusions
5.1. Exploiting Biomaterial Technologies for an Improved Mimicking of the Lung ECM
5.2. Potential Applications of New 3D Technologies to LCO Cultures
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: Globocan Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Cancer Research, UK. Lung Cancer Statistics—Incidence. 2020. Available online: https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/lung-cancer/incidence#heading-Three (accessed on 19 June 2022).
- National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology, Non-Small Cell Lung Cancer—Version 2; National Comprehensive Cancer Network: Plymouth Meeting, PA, USA, 2022; p. 283. Available online: https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf (accessed on 19 June 2022).
- Hirano, T.; Yasuda, H.; Tani, T.; Hamamoto, J.; Oashi, A.; Ishioka, K.; Arai, D.; Nukaga, S.; Miyawaki, M.; Kawada, I.; et al. In vitro modeling to determine mutation specificity of EGFR tyrosine kinase inhibitors against clinically relevant EGFR mutants in non-small-cell lung cancer. Oncotarget 2015, 6, 38789–38803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huo, K.-G.; Arcangelo, E.D.; Tsao, M.-S. Patient-derived cell line, xenograft and organoid models in lung cancer therapy. Transl. Lung Cancer Res. 2020, 9, 2214–2232. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.Z.; Han, R.R.; Qiu, G.Z.; Ju, X.C.; Lou, G.; Jin, W.L. Organoids: An intermediate modeling platform in precision oncology. Cancer Lett. 2018, 414, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Rozenberg, J.M.; Filkov, G.I.; Trofimenko, A.V.; Karpulevich, E.A.; Parshin, V.D.; Royuk, V.V.; Sekacheva, M.I.; Durymanov, M.O. Biomedical Applications of Non-Small Cell Lung Cancer Spheroids. Front. Oncol. 2021, 11, 791069. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, P.; Liu, R.; Bi, H.; Xu, C.; Rosenholm, J.M.; Akerfelt, M. 3D Modeling of Epithelial Tumors-The Synergy between Materials Engineering, 3D Bioprinting, High-Content Imaging, and Nanotechnology. Int. J. Mol. Sci. 2021, 22, 6225. [Google Scholar] [CrossRef]
- Eramo, A.; Lotti, F.; Sette, G.; Pilozzi, E.; Biffoni, M.; Di Virgilio, A.; Conticello, C.; Ruco, L.; Peschle, C.; de Maria, R. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008, 15, 504–514. [Google Scholar] [CrossRef]
- Orienti, I.; Francescangeli, F.; de Angelis, M.L.; Fecchi, K.; Bongiorno-Borbone, L.; Signore, M.; Peschiaroli, A.; Boe, A.; Bruselles, A.; Costantino, A.; et al. A new bioavailable fenretinide formulation with antiproliferative, antimetabolic, and cytotoxic effects on solid tumors. Cell Death Dis. 2019, 10, 529. [Google Scholar] [CrossRef] [Green Version]
- Orienti, I.; Salvati, V.; Sette, G.; Zucchetti, M.; Bongiorno-Borbone, L.; Peschiaroli, A.; Zolla, L.; Francescangeli, F.; Ferrari, M.; Matteo, C.; et al. A novel oral micellar fenretinide formulation with enhanced bioavailability and antitumour activity against multiple tumours from cancer stem cells. J. Exp. Clin. Cancer Res. 2019, 38, 373. [Google Scholar] [CrossRef] [Green Version]
- Zeuner, A.; Francescangeli, F.; Contavalli, P.; Zapparelli, G.; Apuzzo, T.; Eramo, A.; Baiocchi, M.; De Angelis, M.L.; Biffoni, M.; Sette, G.; et al. Elimination of quiescent/slow-proliferating cancer stem cells by Bcl-XL inhibition in non-small cell lung cancer. Cell Death Differ. 2014, 21, 1877–1888. [Google Scholar] [CrossRef] [PubMed]
- Kamer, I.; Bab-Dinitz, E.; Zadok, O.; Ofek, E.; Gottfried, T.; Daniel-Meshulam, I.; Hout-Siloni, G.; Ben Nun, A.; Barshack, I.; Onn, A.; et al. Immunotherapy response modeling by ex-vivo organ culture for lung cancer. Cancer Immunol. Immunother. 2021, 70, 2223–2234. [Google Scholar] [CrossRef]
- Ivanova, E.; Kuraguchi, M.; Xu, M.; Portell, A.J.; Taus, L.; Diala, I.; Lalani, A.S.; Choi, J.; Chambers, E.S.; Li, S.; et al. Use of Ex Vivo Patient-Derived Tumor Organotypic Spheroids to Identify Combination Therapies for HER2 Mutant Non-Small Cell Lung Cancer. Clin. Cancer Res. 2020, 26, 2393–2403. [Google Scholar] [CrossRef] [Green Version]
- Weeber, F.; Ooft, S.N.; Dijkstra, K.K.; Voest, E.E. Tumor Organoids as a Pre-clinical Cancer Model for Drug Discovery. Cell Chem. Biol. 2017, 24, 1092–1100. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, K.K.; Monkhorst, K.; Schipper, L.J.; Hartemink, K.J.; Smit, E.F.; Kaing, S.; de Groot, R.; Wolkers, M.C.; Clevers, H.; Cuppen, E.; et al. Challenges in Establishing Pure Lung Cancer Organoids Limit Their Utility for Personalized Medicine. Cell Rep. 2020, 31, 107588. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Radulovich, N.; Ng, C.; Liu, N.; Notsuda, H.; Cabanero, M.; Martins-Filho, S.N.; Raghavan, V.; Li, Q.; Mer, A.S.; et al. Organoid Cultures as Preclinical Models of Non-Small Cell Lung Cancer. Clin. Cancer Res. 2020, 26, 1162–1174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Liello, R.; Ciaramella, V.; Barra, G.; Venditti, M.; Della Corte, C.M.; Papaccio, F.; Sparano, F.; Viscardi, G.; Iacovino, M.L.; Minucci, S.; et al. Ex vivo lung cancer spheroids resemble treatment response of a patient with NSCLC to chemotherapy and immunotherapy: Case report and translational study. ESMO Open 2019, 4, e000536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jenkins, R.W.; Aref, A.R.; Lizotte, P.H.; Ivanova, E.; Stinson, S.; Zhou, C.W.; Bowden, M.; Deng, J.; Liu, H.; Miao, D.; et al. Ex Vivo Profiling of PD-1 Blockade Using Organotypic Tumor Spheroids. Cancer Discov. 2018, 8, 196–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruppen, J.; Wildhaber, F.D.; Strub, C.; Hall, S.R.; Schmid, R.A.; Geiser, T.; Guenat, O.T. Towards personalized medicine: Chemosensitivity assays of patient lung cancer cell spheroids in a perfused microfluidic platform. Lab Chip 2015, 15, 3076–3085. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Wang, H.; Ding, Q.; Xing, Y.; Xu, Z.; Lu, C.; Luo, D.; Xu, L.; Xia, W.; Zhou, C.; et al. Establishment of patient-derived tumor spheroids for non-small cell lung cancer. PLoS ONE 2018, 13, e0194016. [Google Scholar] [CrossRef]
- Endo, H.; Okami, J.; Okuyama, H.; Kumagai, T.; Uchida, J.; Kondo, J.; Takehara, T.; Nishizawa, Y.; Imamura, F.; Higashiyama, M.; et al. Spheroid culture of primary lung cancer cells with neuregulin 1/HER3 pathway activation. J. Thorac. Oncol. 2013, 8, 131–139. [Google Scholar] [CrossRef] [Green Version]
- Benali, R.; Tournier, J.M.; Chevillard, M.; Zahm, J.M.; Klossek, J.M.; Hinnrasky, J.; Gaillard, D.; Maquart, F.X.; Puchelle, E. Tubule formation by human surface respiratory epithelial cells cultured in a three-dimensional collagen lattice. Am. J. Physiol. 1993, 264, L183–L192. [Google Scholar] [CrossRef] [PubMed]
- Barkauskas, C.E.; Cronce, M.J.; Rackley, C.R.; Bowie, E.J.; Keene, D.R.; Stripp, B.R.; Randell, S.H.; Noble, P.W.; Hogan, B.L. Type 2 alveolar cells are stem cells in adult lung. J. Clin. Investig. 2013, 123, 3025–3036. [Google Scholar] [CrossRef] [PubMed]
- Rock, J.R.; Onaitis, M.W.; Rawlins, E.L.; Lu, Y.; Clark, C.P.; Xue, Y.; Randell, S.H.; Hogan, B.L. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad. Sci. USA 2009, 106, 12771–12775. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.W.; Huang, S.X.; de Carvalho, A.; Ho, S.H.; Islam, M.N.; Volpi, S.; Notarangelo, L.D.; Ciancanelli, M.; Casanova, J.L.; Bhattacharya, J.; et al. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat. Cell Biol. 2017, 19, 542–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dye, B.R.; Hill, D.R.; Ferguson, M.A.; Tsai, Y.H.; Nagy, M.S.; Dyal, R.; Wells, J.M.; Mayhew, C.N.; Nattiv, R.; Klein, O.D.; et al. In vitro generation of human pluripotent stem cell derived lung organoids. eLife 2015, 4, e05098. [Google Scholar] [CrossRef]
- McCauley, K.B.; Hawkins, F.; Serra, M.; Thomas, D.C.; Jacob, A.; Kotton, D.N. Efficient Derivation of Functional Human Airway Epithelium from Pluripotent Stem Cells via Temporal Regulation of Wnt Signaling. Cell Stem Cell 2017, 20, 844–857.e6. [Google Scholar] [CrossRef] [Green Version]
- Tan, Q.; Choi, K.M.; Sicard, D.; Tschumperlin, D.J. Human airway organoid engineering as a step toward lung regeneration and disease modeling. Biomaterials 2017, 113, 118–132. [Google Scholar] [CrossRef] [Green Version]
- Nikolic, M.Z.; Caritg, O.; Jeng, Q.; Johnson, J.A.; Sun, D.; Howell, K.J.; Brady, J.L.; Laresgoiti, U.; Allen, G.; Butler, R.; et al. Human embryonic lung epithelial tips are multipotent progenitors that can be expanded in vitro as long-term self-renewing organoids. eLife 2017, 6, e26575. [Google Scholar] [CrossRef] [PubMed]
- Sette, G.; Lo Cicero, S.; Blacona, G.; Pierandrei, S.; Bruno, S.M.; Salvati, V.; Castelli, G.; Falchi, M.; Fabrizzi, B.; Cimino, G.; et al. Theratyping cystic fibrosis in vitro in ALI culture and organoid models generated from patient-derived nasal epithelial conditionally reprogrammed stem cells. Eur. Respir. J. 2021, 58, 2100908. [Google Scholar] [CrossRef]
- Pauli, C.; Hopkins, B.D.; Prandi, D.; Shaw, R.; Fedrizzi, T.; Sboner, A.; Sailer, V.; Augello, M.; Puca, L.; Rosati, R.; et al. Personalized In Vitro and In Vivo Cancer Models to Guide Precision Medicine. Cancer Discov. 2017, 7, 462–477. [Google Scholar] [CrossRef] [Green Version]
- Finnberg, N.K.; Gokare, P.; Lev, A.; Grivennikov, S.I.; MacFarlane, A.W.T.; Campbell, K.S.; Winters, R.M.; Kaputa, K.; Farma, J.M.; Abbas, A.E.; et al. Application of 3D tumoroid systems to define immune and cytotoxic therapeutic responses based on tumoroid and tissue slice culture molecular signatures. Oncotarget 2017, 8, 66747–66757. [Google Scholar] [CrossRef] [Green Version]
- Sachs, N.; Papaspyropoulos, A.; Zomer-van Ommen, D.D.; Heo, I.; Bottinger, L.; Klay, D.; Weeber, F.; Huelsz-Prince, G.; Iakobachvili, N.; Amatngalim, G.D.; et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 2019, 38, e100300. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Kim, S.M.; Lim, S.; Lee, J.Y.; Choi, S.J.; Yang, S.D.; Yun, M.R.; Kim, C.G.; Gu, S.R.; Park, C.; et al. Modeling Clinical Responses to Targeted Therapies by Patient-Derived Organoids of Advanced Lung Adenocarcinoma. Clin. Cancer Res. 2021, 27, 4397–4409. [Google Scholar] [CrossRef] [PubMed]
- Mazzocchi, A.; Devarasetty, M.; Herberg, S.; Petty, W.J.; Marini, F.; Miller, L.; Kucera, G.; Dukes, D.K.; Ruiz, J.; Skardal, A.; et al. Pleural Effusion Aspirate for use in 3D Lung Cancer Modeling and Chemotherapy Screening. ACS Biomater. Sci. Eng. 2019, 5, 1937–1943. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.H.; Chu, X.P.; Zhang, J.T.; Nie, Q.; Tang, W.F.; Su, J.; Yan, H.H.; Zheng, H.P.; Chen, Z.X.; Chen, X.; et al. Genomic characteristics and drug screening among organoids derived from non-small cell lung cancer patients. Thorac. Cancer 2020, 11, 2279–2290. [Google Scholar] [CrossRef]
- Gmeiner, W.H.; Miller, L.D.; Chou, J.W.; Dominijanni, A.; Mutkus, L.; Marini, F.; Ruiz, J.; Dotson, T.; Thomas, K.W.; Parks, G.; et al. Dysregulated Pyrimidine Biosynthesis Contributes to 5-FU Resistance in SCLC Patient-Derived Organoids but Response to a Novel Polymeric Fluoropyrimidine, CF10. Cancers 2020, 12, 788. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Sui, X.; Song, F.; Li, Y.; Li, K.; Chen, Z.; Yang, F.; Chen, X.; Zhang, Y.; Wang, X.; et al. Lung cancer organoids analyzed on microwell arrays predict drug responses of patients within a week. Nat. Commun. 2021, 12, 2581. [Google Scholar] [CrossRef]
- Kim, M.; Mun, H.; Sung, C.O.; Cho, E.J.; Jeon, H.J.; Chun, S.M.; Jung, D.J.; Shin, T.H.; Jeong, G.S.; Kim, D.K.; et al. Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat. Commun. 2019, 10, 3991. [Google Scholar] [CrossRef]
- Takahashi, N.; Hoshi, H.; Higa, A.; Hiyama, G.; Tamura, H.; Ogawa, M.; Takagi, K.; Goda, K.; Okabe, N.; Muto, S.; et al. An In Vitro System for Evaluating Molecular Targeted Drugs Using Lung Patient-Derived Tumor Organoids. Cells 2019, 8, 481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokota, E.; Iwai, M.; Yukawa, T.; Yoshida, M.; Naomoto, Y.; Haisa, M.; Monobe, Y.; Takigawa, N.; Guo, M.; Maeda, Y.; et al. Clinical application of a lung cancer organoid (tumoroid) culture system. NPJ Precis. Oncol. 2021, 5, 29. [Google Scholar] [CrossRef] [PubMed]
- Cattaneo, C.M.; Dijkstra, K.K.; Fanchi, L.F.; Kelderman, S.; Kaing, S.; van Rooij, N.; van den Brink, S.; Schumacher, T.N.; Voest, E.E. Tumor organoid-T-cell coculture systems. Nat. Protoc. 2020, 15, 15–39. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, K.K.; Cattaneo, C.M.; Weeber, F.; Chalabi, M.; van de Haar, J.; Fanchi, L.F.; Slagter, M.; van der Velden, D.L.; Kaing, S.; Kelderman, S.; et al. Generation of Tumor-Reactive T Cells by Co-culture of Peripheral Blood Lymphocytes and Tumor Organoids. Cell 2018, 174, 1586–1598.e12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Harrison, E.B.; Li, H.; Hirabayashi, K.; Chen, J.; Li, Q.X.; Gunn, J.; Weiss, J.; Savoldo, B.; Parker, J.S.; et al. Targeting brain lesions of non-small cell lung cancer by enhancing CCL2-mediated CAR-T cell migration. Nat. Commun. 2022, 13, 2154. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Fillmore Brainson, C.; Koyama, S.; Redig, A.J.; Chen, T.; Li, S.; Gupta, M.; Garcia-de-Alba, C.; Paschini, M.; Herter-Sprie, G.S.; et al. Lkb1 inactivation drives lung cancer lineage switching governed by Polycomb Repressive Complex 2. Nat. Commun. 2017, 8, 14922. [Google Scholar] [CrossRef]
- Neal, J.T.; Li, X.; Zhu, J.; Giangarra, V.; Grzeskowiak, C.L.; Ju, J.; Liu, I.H.; Chiou, S.H.; Salahudeen, A.A.; Smith, A.R.; et al. Organoid Modeling of the Tumor Immune Microenvironment. Cell 2018, 175, 1972–1988.e16. [Google Scholar] [CrossRef] [Green Version]
- Seo, J.H.; Chae, Y.C.; Kossenkov, A.V.; Lee, Y.G.; Tang, H.Y.; Agarwal, E.; Gabrilovich, D.I.; Languino, L.R.; Speicher, D.W.; Shastrula, P.K.; et al. MFF Regulation of Mitochondrial Cell Death Is a Therapeutic Target in Cancer. Cancer Res. 2019, 79, 6215–6226. [Google Scholar] [CrossRef] [Green Version]
- Jung, D.J.; Shin, T.H.; Kim, M.; Sung, C.O.; Jang, S.J.; Jeong, G.S. A one-stop microfluidic-based lung cancer organoid culture platform for testing drug sensitivity. Lab Chip 2019, 19, 2854–2865. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, T.; Qin, Z.; Jiang, J.; Wang, Q.; Yang, S.; Rivard, C.; Gao, G.; Ng, T.L.; Tu, M.M.; et al. HER2 exon 20 insertions in non-small-cell lung cancer are sensitive to the irreversible pan-HER receptor tyrosine kinase inhibitor pyrotinib. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2019, 30, 447–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Qian, Y.; Li, W.; Liu, L.; Yu, L.; Liu, X.; Wu, G.; Wang, Y.; Luo, W.; Fang, F.; et al. Human Lung Adenocarcinoma-Derived Organoid Models for Drug Screening. iScience 2020, 23, 101411. [Google Scholar] [CrossRef]
- Taverna, J.A.; Hung, C.N.; DeArmond, D.T.; Chen, M.; Lin, C.L.; Osmulski, P.A.; Gaczynska, M.E.; Wang, C.M.; Lucio, N.D.; Chou, C.W.; et al. Single-Cell Proteomic Profiling Identifies Combined AXL and JAK1 Inhibition as a Novel Therapeutic Strategy for Lung Cancer. Cancer Res. 2020, 80, 1551–1563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Yu, L.; Chen, D.; Meng, Z.; Chen, W.; Huang, W. Protocol for generation of lung adenocarcinoma organoids from clinical samples. STAR Protoc. 2021, 2, 100239. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Lewin Mejia, D.; Chiang, B.; Luker, K.E.; Luker, G.D. Hybrid collagen alginate hydrogel as a platform for 3D tumor spheroid invasion. Acta Biomater. 2018, 75, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.Y.; Cho, Y.-H.; Kim, D.-S.; Ji, W.; Choi, C.-M.; Lee, J.C.; Rho, J.K.; Jeong, G.S. Establishment and Long-Term Expansion of Small Cell Lung Cancer Patient-Derived Tumor Organoids. Int. J. Mol. Sci. 2021, 22, 1349. [Google Scholar] [CrossRef]
- Sandor, G.O.; Soos, A.A.; Lorincz, P.; Rojko, L.; Harko, T.; Bogyo, L.; Tolgyes, T.; Bursics, A.; Buzas, E.I.; Moldvay, J.; et al. Wnt Activity and Cell Proliferation Are Coupled to Extracellular Vesicle Release in Multiple Organoid Models. Front. Cell Dev. Biol. 2021, 9, 670825. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Yang, S.; Jiang, H.; Wang, Y.; Xiang, Z. Transcriptomic analysis of tumor tissues and organoids reveals the crucial genes regulating the proliferation of lung adenocarcinoma. J. Transl. Med. 2021, 19, 368. [Google Scholar] [CrossRef]
- Fujii, M.; Shimokawa, M.; Date, S.; Takano, A.; Matano, M.; Nanki, K.; Ohta, Y.; Toshimitsu, K.; Nakazato, Y.; Kawasaki, K.; et al. A Colorectal Tumor Organoid Library Demonstrates Progressive Loss of Niche Factor Requirements during Tumorigenesis. Cell Stem Cell 2016, 18, 827–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sette, G.; Salvati, V.; Giordani, I.; Pilozzi, E.; Quacquarini, D.; Duranti, E.; de Nicola, F.; Pallocca, M.; Fanciulli, M.; Falchi, M.; et al. Conditionally reprogrammed cells (CRC) methodology does not allow the in vitro expansion of patient-derived primary and metastatic lung cancer cells. Int. J. Cancer 2018, 143, 88–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blakely, C.M.; Watkins, T.B.K.; Wu, W.; Gini, B.; Chabon, J.J.; McCoach, C.E.; McGranahan, N.; Wilson, G.A.; Birkbak, N.J.; Olivas, V.R.; et al. Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat. Genet. 2017, 49, 1693–1704. [Google Scholar] [CrossRef]
- Lo, Y.H.; Karlsson, K.; Kuo, C.J. Applications of Organoids for Cancer Biology and Precision Medicine. Nat. Cancer 2020, 1, 761–773. [Google Scholar] [CrossRef]
- De Angelis, M.L.; Francescangeli, F.; Nicolazzo, C.; Signore, M.; Giuliani, A.; Colace, L.; Boe, A.; Magri, V.; Baiocchi, M.; Ciardi, A.; et al. An organoid model of colorectal circulating tumor cells with stem cell features, hybrid EMT state and distinctive therapy response profile. J. Exp. Clin. Cancer Res. 2022, 41, 86. [Google Scholar] [CrossRef]
- Bolhaqueiro, A.C.F.; Ponsioen, B.; Bakker, B.; Klaasen, S.J.; Kucukkose, E.; van Jaarsveld, R.H.; Vivie, J.; Verlaan-Klink, I.; Hami, N.; Spierings, D.C.J.; et al. Ongoing chromosomal instability and karyotype evolution in human colorectal cancer organoids. Nat. Genet. 2019, 51, 824–834. [Google Scholar] [CrossRef] [PubMed]
- Drost, J.; van Boxtel, R.; Blokzijl, F.; Mizutani, T.; Sasaki, N.; Sasselli, V.; de Ligt, J.; Behjati, S.; Grolleman, J.E.; van Wezel, T.; et al. Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science 2017, 358, 234–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drost, J.; van Jaarsveld, R.H.; Ponsioen, B.; Zimberlin, C.; van Boxtel, R.; Buijs, A.; Sachs, N.; Overmeer, R.M.; Offerhaus, G.J.; Begthel, H.; et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 2015, 521, 43–47. [Google Scholar] [CrossRef]
- Li, X.; Nadauld, L.; Ootani, A.; Corney, D.C.; Pai, R.K.; Gevaert, O.; Cantrell, M.A.; Rack, P.G.; Neal, J.T.; Chan, C.W.; et al. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat. Med. 2014, 20, 769–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dost, A.F.M.; Moye, A.L.; Vedaie, M.; Tran, L.M.; Fung, E.; Heinze, D.; Villacorta-Martin, C.; Huang, J.; Hekman, R.; Kwan, J.H.; et al. Organoids Model Transcriptional Hallmarks of Oncogenic KRAS Activation in Lung Epithelial Progenitor Cells. Cell Stem Cell 2020, 27, 663–678.e8. [Google Scholar] [CrossRef] [PubMed]
- Semba, T.; Sato, R.; Kasuga, A.; Suina, K.; Shibata, T.; Kohno, T.; Suzuki, M.; Saya, H.; Arima, Y. Lung Adenocarcinoma Mouse Models Based on Orthotopic Transplantation of Syngeneic Tumor-Initiating Cells Expressing EpCAM, SCA-1, and Ly6d. Cancers 2020, 12, 3805. [Google Scholar] [CrossRef] [PubMed]
- Na, F.; Pan, X.; Chen, J.; Chen, X.; Wang, M.; Chi, P.; You, L.; Zhang, L.; Zhong, A.; Zhao, L.; et al. KMT2C deficiency promotes small cell lung cancer metastasis through DNMT3A-mediated epigenetic reprogramming. Nat. Cancer 2022, 3, 753–767. [Google Scholar] [CrossRef] [PubMed]
- Miura, A.; Yamada, D.; Nakamura, M.; Tomida, S.; Shimizu, D.; Jiang, Y.; Takao, T.; Yamamoto, H.; Suzawa, K.; Shien, K.; et al. Oncogenic potential of human pluripotent stem cell-derived lung organoids with HER2 overexpression. Int. J. Cancer 2021, 149, 1593–1604. [Google Scholar] [CrossRef]
- Nacarino-Palma, A.; Rejano-Gordillo, C.M.; Gonzalez-Rico, F.J.; Ordiales-Talavero, A.; Roman, A.C.; Cuadrado, M.; Bustelo, X.R.; Merino, J.M.; Fernandez-Salguero, P.M. Loss of Aryl Hydrocarbon Receptor Favors K-Ras(G12D)-Driven Non-Small Cell Lung Cancer. Cancers 2021, 13, 4071. [Google Scholar] [CrossRef]
- Fiorini, E.; Veghini, L.; Corbo, V. Modeling Cell Communication in Cancer With Organoids: Making the Complex Simple. Front. Cell Dev. Biol. 2020, 8, 166. [Google Scholar] [CrossRef] [Green Version]
- Ho, T.; Msallam, R. Tissues and Tumor Microenvironment (TME) in 3D: Models to Shed Light on Immunosuppression in Cancer. Cells 2021, 10, 831. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorthy, P.; Thomas, S.M.; Kaushik, G.; Subramaniam, D.; Chastain, K.M.; Dhar, A.; Tawfik, O.; Kasi, A.; Sun, W.; Ramalingam, S.; et al. Metastatic Tumor-in-a-Dish, a Novel Multicellular Organoid to Study Lung Colonization and Predict Therapeutic Response. Cancer Res. 2019, 79, 1681–1695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vazquez-Armendariz, A.I.; Heiner, M.; El Agha, E.; Salwig, I.; Hoek, A.; Hessler, M.C.; Shalashova, I.; Shrestha, A.; Carraro, G.; Mengel, J.P.; et al. Multilineage murine stem cells generate complex organoids to model distal lung development and disease. EMBO J. 2020, 39, e103476. [Google Scholar] [CrossRef] [PubMed]
- Yuki, K.; Cheng, N.; Nakano, M.; Kuo, C.J. Organoid Models of Tumor Immunology. Trends Immunol. 2020, 41, 652–664. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Giannakou, A.; Wyman, S.; Gruzas, J.; Golas, J.; Zhong, W.; Loreth, C.; Sridharan, L.; Yamin, T.T.; Damelin, M.; et al. Cancer-associated fibroblasts suppress SOX2-induced dysplasia in a lung squamous cancer coculture. Proc. Natl. Acad. Sci. USA 2018, 115, E11671–E11680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Bufalo, F.; Manzo, T.; Hoyos, V.; Yagyu, S.; Caruana, I.; Jacot, J.; Benavides, O.; Rosen, D.; Brenner, M.K. 3D modeling of human cancer: A PEG-fibrin hydrogel system to study the role of tumor microenvironment and recapitulate the in vivo effect of oncolytic adenovirus. Biomaterials 2016, 84, 76–85. [Google Scholar] [CrossRef]
- Yu, T.; Guo, Z.; Fan, H.; Song, J.; Liu, Y.; Gao, Z.; Wang, Q. Cancer-associated fibroblasts promote non-small cell lung cancer cell invasion by upregulation of glucose-regulated protein 78 (GRP78) expression in an integrated bionic microfluidic device. Oncotarget 2016, 7, 25593–25603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, H.; Sugano, M.; Miyashita, T.; Hashimoto, H.; Ochiai, A.; Suzuki, K.; Tsuboi, M.; Ishii, G. Organoid culture containing cancer cells and stromal cells reveals that podoplanin-positive cancer-associated fibroblasts enhance proliferation of lung cancer cells. Lung Cancer 2019, 134, 100–107. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014, 511, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Skoulidis, F.; Heymach, J.V. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat. Rev. Cancer 2019, 19, 495–509. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Cong, L.; Cong, X. Patient-Derived Organoids in Precision Medicine: Drug Screening, Organoid-on-a-Chip and Living Organoid Biobank. Front. Oncol. 2021, 11, 762184. [Google Scholar] [CrossRef]
- Ooft, S.N.; Weeber, F.; Dijkstra, K.K.; McLean, C.M.; Kaing, S.; van Werkhoven, E.; Schipper, L.; Hoes, L.; Vis, D.J.; van de Haar, J.; et al. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci. Transl. Med. 2019, 11, eaay2574. [Google Scholar] [CrossRef] [PubMed]
- Ooft, S.N.; Weeber, F.; Schipper, L.; Dijkstra, K.K.; McLean, C.M.; Kaing, S.; van de Haar, J.; Prevoo, W.; van Werkhoven, E.; Snaebjornsson, P.; et al. Prospective experimental treatment of colorectal cancer patients based on organoid drug responses. ESMO Open 2021, 6, 100103. [Google Scholar] [CrossRef] [PubMed]
- Burgstaller, G.; Oehrle, B.; Gerckens, M.; White, E.S.; Schiller, H.B.; Eickelberg, O. The instructive extracellular matrix of the lung: Basic composition and alterations in chronic lung disease. Eur. Respir. J. 2017, 50, 1601805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sullivan, K.M.; Ko, E.; Kim, E.M.; Ballance, W.C.; Ito, J.D.; Chalifoux, M.; Kim, Y.J.; Bashir, R.; Kong, H. Extracellular Microenvironmental Control for Organoid Assembly. Tissue Eng. Part B Rev. 2022. ahead of print. [Google Scholar] [CrossRef]
- Pickup, M.W.; Mouw, J.K.; Weaver, V.M. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014, 15, 1243–1253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popova, N.V.; Jucker, M. The Functional Role of Extracellular Matrix Proteins in Cancer. Cancers 2022, 14, 238. [Google Scholar] [CrossRef]
- Talbot, N.C.; Caperna, T.J. Proteome array identification of bioactive soluble proteins/peptides in Matrigel: Relevance to stem cell responses. Cytotechnology 2015, 67, 873–883. [Google Scholar] [CrossRef] [Green Version]
- Hughes, C.S.; Postovit, L.M.; Lajoie, G.A. Matrigel: A complex protein mixture required for optimal growth of cell culture. Proteomics 2010, 10, 1886–1890. [Google Scholar] [CrossRef]
- Kaur, S.; Kaur, I.; Rawal, P.; Tripathi, D.M.; Vasudevan, A. Non-matrigel scaffolds for organoid cultures. Cancer Lett. 2021, 504, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Suki, B.; Ito, S.; Stamenovic, D.; Lutchen, K.R.; Ingenito, E.P. Biomechanics of the lung parenchyma: Critical roles of collagen and mechanical forces. J. Appl. Physiol. 2005, 98, 1892–1899. [Google Scholar] [CrossRef] [PubMed]
- Hautefort, I.; Poletti, M.; Papp, D.; Korcsmaros, T. Everything You Always Wanted to Know About Organoid-Based Models (and Never Dared to Ask). Cell. Mol. Gastroenterol. Hepatol. 2022, 14, 311–331. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Zhou, X.; Mandal, K.; He, N.; Wennerberg, W.; Qu, M.; Jiang, X.; Sun, W.; Khademhosseini, A. Reconstructing the tumor architecture into organoids. Adv. Drug Deliv. Rev. 2021, 176, 113839. [Google Scholar] [CrossRef] [PubMed]
- Nii, T.; Makino, K.; Tabata, Y. Three-Dimensional Culture System of Cancer Cells Combined with Biomaterials for Drug Screening. Cancers 2020, 12, 2754. [Google Scholar] [CrossRef] [PubMed]
- DelNero, P.; Lane, M.; Verbridge, S.S.; Kwee, B.; Kermani, P.; Hempstead, B.; Stroock, A.; Fischbach, C. 3D culture broadly regulates tumor cell hypoxia response and angiogenesis via pro-inflammatory pathways. Biomaterials 2015, 55, 110–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhiman, H.K.; Ray, A.R.; Panda, A.K. Characterization and evaluation of chitosan matrix for in vitro growth of MCF-7 breast cancer cell lines. Biomaterials 2004, 25, 5147–5154. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhou, Y.; Chen, W.; Yuan, Z.; You, B.; Liu, Y.; Yang, S.; Li, F.; Qu, C.; Zhang, X. A Novel 3D in Vitro Tumor Model Based on Silk Fibroin/Chitosan Scaffolds To Mimic the Tumor Microenvironment. ACS Appl. Mater. Interfaces 2018, 10, 36641–36651. [Google Scholar] [CrossRef]
- Engel, B.J.; Constantinou, P.E.; Sablatura, L.K.; Doty, N.J.; Carson, D.D.; Farach-Carson, M.C.; Harrington, D.A.; Zarembinski, T.I. Multilayered, Hyaluronic Acid-Based Hydrogel Formulations Suitable for Automated 3D High Throughput Drug Screening of Cancer-Stromal Cell Cocultures. Adv. Healthc. Mater. 2015, 4, 1664–1674. [Google Scholar] [CrossRef]
- Gill, B.J.; Gibbons, D.L.; Roudsari, L.C.; Saik, J.E.; Rizvi, Z.H.; Roybal, J.D.; Kurie, J.M.; West, J.L. A synthetic matrix with independently tunable biochemistry and mechanical properties to study epithelial morphogenesis and EMT in a lung adenocarcinoma model. Cancer Res. 2012, 72, 6013–6023. [Google Scholar] [CrossRef] [Green Version]
- Cruz-Acuna, R.; Vunjak-Novakovic, G.; Burdick, J.A.; Rustgi, A.K. Emerging technologies provide insights on cancer extracellular matrix biology and therapeutics. iScience 2021, 24, 102475. [Google Scholar] [CrossRef]
- Yi, S.A.; Zhang, Y.; Rathnam, C.; Pongkulapa, T.; Lee, K.B. Bioengineering Approaches for the Advanced Organoid Research. Adv. Mater. 2021, 33, e2007949. [Google Scholar] [CrossRef] [PubMed]
- Loebel, C.; Weiner, A.I.; Eiken, M.K.; Katzen, J.B.; Morley, M.P.; Bala, V.; Cardenas-Diaz, F.L.; Davidson, M.D.; Shiraishi, K.; Basil, M.C.; et al. Microstructured Hydrogels to Guide Self-Assembly and Function of Lung Alveolospheres. Adv. Mater. 2022, 34, e2202992. [Google Scholar] [CrossRef] [PubMed]
- Hofer, M.; Lutolf, M.P. Engineering organoids. Nat. Rev. Mater. 2021, 6, 402–420. [Google Scholar] [CrossRef] [PubMed]
- Spiller, E.R.; Ung, N.; Kim, S.; Patsch, K.; Lau, R.; Strelez, C.; Doshi, C.; Choung, S.; Choi, B.; Juarez Rosales, E.F.; et al. Imaging-Based Machine Learning Analysis of Patient-Derived Tumor Organoid Drug Response. Front. Oncol. 2021, 11, 771173. [Google Scholar] [CrossRef] [PubMed]
- Hahn, W.C.; Bader, J.S.; Braun, T.P.; Califano, A.; Clemons, P.A.; Druker, B.J.; Ewald, A.J.; Fu, H.; Jagu, S.; Kemp, C.J.; et al. An expanded universe of cancer targets. Cell 2021, 184, 1142–1155. [Google Scholar] [CrossRef] [PubMed]
| SPHEROIDS | ||||||
| YEAR | REFERENCES | ECM/SUPPLEMENTS | SOURCE | N | SUCCESS RATE | APPLICATIONS |
| 2008 | [9] | DMEM/F12 medium, EGF and bFGF | Resection | 7 lines of stem cell-enriched tumor-derived spheroids (TDS) | 7/19 (36.8%) | Identification and characterization of lung cancer stem cells; generation of xenografts recapitulating the histology of parental tumors |
| 2013 | [22] | StemPro hESC medium, Matrigel GFR, NRG1, Long-IGF1, bFGF, Activin A, EGF | Resection/pleural effusion | 108 TDS | Total: 108/143 (75.5%) 100/125 (80%) surgical samples, 8/18 (44.4%) pleural effusions | Method to expand patient-derived lung tumor cells |
| 2018 | [46] | Advanced DMEM/F-12, N2, Noggin, B27 | Resection | 3 TDS | 100% | Method to expand patient-derived lung tumor cells |
| 2020 | [14] | Collagen hydrogels in a 3D microfluidic culture system | Core needle biopsy/surgical biopsy/pleural effusion | 2 PDX-derived spheroids | 2/2 (100%) | Drug testing |
| ORGANOIDS | ||||||
| YEAR | REFERENCES | ECM/SUPPLEMENTS | SOURCE | N | SUCCESS RATE | APPLICATIONS |
| 2017 | [32] | Matrigel GFR, B27, N-acetylcisteine, R-spondin-1, Noggin, FGF10, FGF2, EGF, A83-01, Y-2763, SB202190, Nicotinamide, Prostaglandin E2 | Resection/biopsy | 1 PDO | 1/2 (50%) LUAD | Biobanking |
| 2017 | [33] | Advanced DMEM/F-12, Matrigel, B27, N-acetylcysteine, Gastrin, Nicotinamide, EGF, Noggin, Wnt-3a, R-Spondin-1, A83-01, SB202190 and Y-27632 | Resection/biopsy | 3 PDOs | 3/3 (100%) NSCLC | Evaluation of immune cell populations infiltrating cultured tissues; drug testing |
| 2019 | [34] | Cultrex Basal Membrane Extract GFR, R-Spondin-1, FGF7, FGF10, Noggin, A83-01, Y-27632, SB202190, B27, Nutlin 3a | Resection/biopsy | 19 PDOs | Total: 19/34 (55.8%) 14/16 (87.5%) primary NSCLC 5/18 (27.8%) metastatic NSCLC | Long-term expansion of LCOs, validation, and drug testing |
| 2018 | [47] | ALI | Resection/biopsy | 9 PDOs | 9/20 (45%) NSCLC | New method for preserving endogenous tumor-infiltrating lymphocytes, suitable for immuno-oncology investigations and personalized immunotherapy testing |
| 2018 | [44] | Geltrex Free Reduced growth factor basement membrane matrix, B27, N-acetylcisteine, R-Spondin-1, Noggin, FGF10, FGF7, A83-01, Y-27632, SB202190, Nicotinamide, Nutlin-3a | Resection/biopsy | 6 PDOs | 6/6 (100%) NSCLC | Development of a platform to analyze tumor-specific T cell responses in a personalized manner |
| 2019 | [40] | Matrigel GFR, B27, N2, FGF2, EGF, Y-27632 | Resection/biopsy | 20 PDOs | Total: 20/23 (87%) 12/14 (85.7%) LUAD 5/6 (83.3%) LUSC 2/2 (100%) SCLC 1/1 (100%) LCNEC | Biobanking, drug testing |
| 2019 | [48] | ECM base medium without supplements | Resection/biopsy | 1 PDO | NA | Investigation and inhibition of mitochondrial fission regulators in multiple tumor organoids |
| 2019 | [49] | Matrigel, B27, N2, FGF2, EGF, Y-27632 | Resection/biopsy | NA | NA | Microfluidic platform-enabling LCO culturing and drug sensitivity tests |
| 2019 | [41] | ECM base medium without supplements | Resection/biopsy | 3 PDOs | Total: 3/3 (100%) 2/2 (100%) LUSC 1/1 (100%) LUAD | Broad-spectrum drug testing |
| 2019 | [36] | ECM base medium without supplements | Pleural effusion | 5 PDOs | 5/5 (100%) LUAD | Establishment of an LCO culture system from pleural effusions; drug testing |
| 2019 | [50] | NA | Resection/biopsy | 1 PDO | 100% | Drug testing |
| 2020 | [17] | Matrigel, B27, N-acetylcysteine, Noggin, FGF10, FGF4, EGF, A83-01, Y-27632, CHIR 99021, SAG | Resection/PDX | 19 PDOs 28 XDOs | Total: 47/65 (72.3%) 13/16 (81%) LUAD PDOs 9/13 (69%) LUAD XDOs 6/14 (43%) LUSC PDOs 19/22 (86%) LUSC XDOs (results are referred to short term LCO cultures) | Platform for LCO expansion and validation; drug testing |
| 2020 | [43] | Geltrex, B27, N-acetylcysteine, R-Spondin-1, Noggin, FGF10, FGF7, A83-01, Y-27632, SB202190, Nicotinamide | Resection/biopsy Autologous PBLs | NA | NA | Protocol for co-culture LCOs and autologous PBLs for the individualized testing of T-cell-based immunotherapy |
| 2020 | [16] | Geltrex, B27, N-acetylcysteine, R-Spondin-1, Noggin, FGF10, FGF7, A83-01, Y-27632, SB202190, Nicotinamide | Resection/biopsy | 10 PDOs | 10/58 (17%) (4 from primary tumor; 6 from metastasis) | Evaluation of several methods to identify tumor purity of organoids established from intrapulmonary tumors |
| 2020 | [51] | Matrigel, B27, N-acetylcysteine, R-Spondin-1, Noggin, FGF10, FGF7, A83-01, Y-27632, SB202190, Nicotinamide | Resection/biopsy | 12 PDOs | 12/15 (80%) | LCO biobanking and characterization; drug testing |
| 2020 | [37] | MBM + Matrigel (1:3 ratio), B27, N2, FGF2, EGF, Y-27632 | Resection/biopsy | 7 PDOs | Total: 7/7 (100%) 6/6 (100%) LUAD 1/1 (100%) LUSC | LCOs biobanking and characterization; drug testing |
| 2020 | [38] | ECM base medium without supplements | PDX derived from biopsies | 4 XDOs | 4/4 (100%) SCLC | Organoid generation from PDXs obtained from SCLC biopsies; drug testing |
| 2020 | [52] | Matrigel, B27, N2, R-Spondin-1, Noggin, FGF10, FGF2, EGF, A83-01, Y-27632, SB202190, Nicotinamide, Prostaglandin E2, HGF | Resection/biopsy | 6 PDOs | 6/11 (54.5%) LUAD | Testing of pathway inhibitors identified by single-cell proteomics |
| 2021 | [53] | Matrigel, B27, GlutaMAX, Noggin, FGF10, FGF7, SB202190, Nicotinamide, N-acetylcysteine, R-Spondin-1, Y-27632, A83-01 | Resection/biopsy | 12 PDOs | 12/15 (80%) LUAD | Protocol for LCO generation from LUAD with high success rate |
| 2021 | [42] | Matrigel, B27, N-acetylcysteine, R-Spondin-1, Noggin, FGF10, FGF7, A83-01, Y-27632, SB202190, Nicotinamide, Nutlin-3a | Resection/pleural effusion | 3 PDOs | Total: 3/41 (7%) 3/30 (10%) LUAD 0/7 (0%) LUSC 0/2 (0%) SCLC 0/2 (0%) Pleomorphic Carcinoma | LCO generation and characterization; targeted drug testing |
| 2021 | [35] | Matrigel, B27, N-acetylcysteine, R-Spondin-1, Noggin, FGF10, FGF7, A83-01, Y-27632, SB202190, Nicotinamide | Metastasis/pleural effusion | 83 PDOs | 83/100 (83%) LUAD | LCO generation and characterization; targeted drug testing |
| 2021 | [54] | Matrigel, B27, N2, Nicotinamide, N-acetylcysteine, Y-27632, EGF, SB202190, A83-01, Forskolin, Dexamethasone | Resection/biopsy | Refers to [39] | Method for on-chip LCO cryopreservation and drug testing | |
| 2021 | [55] | Matrigel, B27, Y-27632, R-Spondin-1, Noggin, A83-01, Wnt-3a, EGF, FGF | Resection/biopsy | 8 PDOs | 8/10 (80%) SCLC | Generation and characterization of SCLC LCOs |
| 2021 | [56] | Matrigel, B27, N-acetylcysteine, R-Spondin-1, Noggin, FGF10, FGF7, A83-01, Y-27632, SB202190, Nicotinamide, Nutlin-3a, Heregulin-β1 | Resection | 6 PDOs | 6/6 (100%) LUAD | Studies on cancer microniche and role of extracellular vesicles |
| 2021 | [39] | Matrigel, B27, N2, Nicotinamide, N-acetylcysteine, Y-27632, EGF, SB202190, A83-01, Forskolin, Dexamethasone | Resection/biopsy | 84 PDOs | Total: 84/109 (77%) Resection: 55/71 (77.4%) LUAD 18/23 (78.2%) LUSC 4/4 (100%) SCLC 4/5 (80%) others Biopsy: 3/6 (50%) LUAD | Rapid LCO generation and drug testing by using a super-hydrophobic microwell array chip; consistency of in vitro results with clinical response |
| 2021 | [57] | OmaStem Lung Cancer Medium | Resection/pleural effusion | 2 PDOs | 2/6 (33.3%) LUAD | Differential gene expression analysis, prognostic analysis, and gene co-expression network analysis |
| 2021 | [45] | NA | Resection/biopsy | 2 PDOs | NA | Drug testing (cisplatin sensitization by halofuginone) |
| MAIN PROBLEMS | POSSIBLE SOLUTIONS |
|---|---|
| LOW SUCCESS RATE (overgrowth of normal airway organoids) |
|
| LOW YIELD OF LCO CULTURES |
|
| CULTURE EXTINCTION |
|
| LCO GENERATION TOO LONG FOR CLINICAL APPLICATIONS |
|
| LCO PRODUCTION IS PARTICULARLY DIFFICULT FOR EARLY STAGE TUMORS |
|
| LIMITED HETEROGENEITY |
| LCOs VALIDATIONS | |||||
|---|---|---|---|---|---|
| HISTOMORPHOLOGY | IHC | GENETIC | XENOGRAFT FORMATION | ||
| NSCLC | ADENOCARCINOMA | Solid or cystic | H&E NAPSIN TTF-1 CK7 | COPY NUMBER LUNG CANCER-RELATED MUTATIONS WHOLE EXOME SEQUENCING  |  |
| ADENOSQUAMOUS CARCINOMA | H&E CK7 CK5/6 p63 | ||||
| SQUAMOUS CELL CARCINOMA | H&E p63 TTF-1 CK5/6 | ||||
| SCLC | SMALL CELLCARCINOMA | H&E CD56 TTF-1 Synaptophysin | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rossi, R.; De Angelis, M.L.; Xhelili, E.; Sette, G.; Eramo, A.; De Maria, R.; Cesta Incani, U.; Francescangeli, F.; Zeuner, A. Lung Cancer Organoids: The Rough Path to Personalized Medicine. Cancers 2022, 14, 3703. https://doi.org/10.3390/cancers14153703
Rossi R, De Angelis ML, Xhelili E, Sette G, Eramo A, De Maria R, Cesta Incani U, Francescangeli F, Zeuner A. Lung Cancer Organoids: The Rough Path to Personalized Medicine. Cancers. 2022; 14(15):3703. https://doi.org/10.3390/cancers14153703
Chicago/Turabian StyleRossi, Rachele, Maria Laura De Angelis, Eljona Xhelili, Giovanni Sette, Adriana Eramo, Ruggero De Maria, Ursula Cesta Incani, Federica Francescangeli, and Ann Zeuner. 2022. "Lung Cancer Organoids: The Rough Path to Personalized Medicine" Cancers 14, no. 15: 3703. https://doi.org/10.3390/cancers14153703
APA StyleRossi, R., De Angelis, M. L., Xhelili, E., Sette, G., Eramo, A., De Maria, R., Cesta Incani, U., Francescangeli, F., & Zeuner, A. (2022). Lung Cancer Organoids: The Rough Path to Personalized Medicine. Cancers, 14(15), 3703. https://doi.org/10.3390/cancers14153703





