Prognostic and Clinicopathological Significance of Telomerase Reverse Transcriptase Upregulation in Oral Cancer: A Systematic Review and Meta-Analysis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Protocol
2.2. Search Strategy
2.3. Eligibility Criteria
2.4. Study-Selection Process
2.5. Data Extraction
2.6. Evaluation of the Methodological Quality and Risk of Bias across Primary-Level Studies
2.7. Statistical Analysis
2.8. Validation of Methodological Quality
3. Results
3.1. Results of the Literature Search
3.2. Study Characteristics
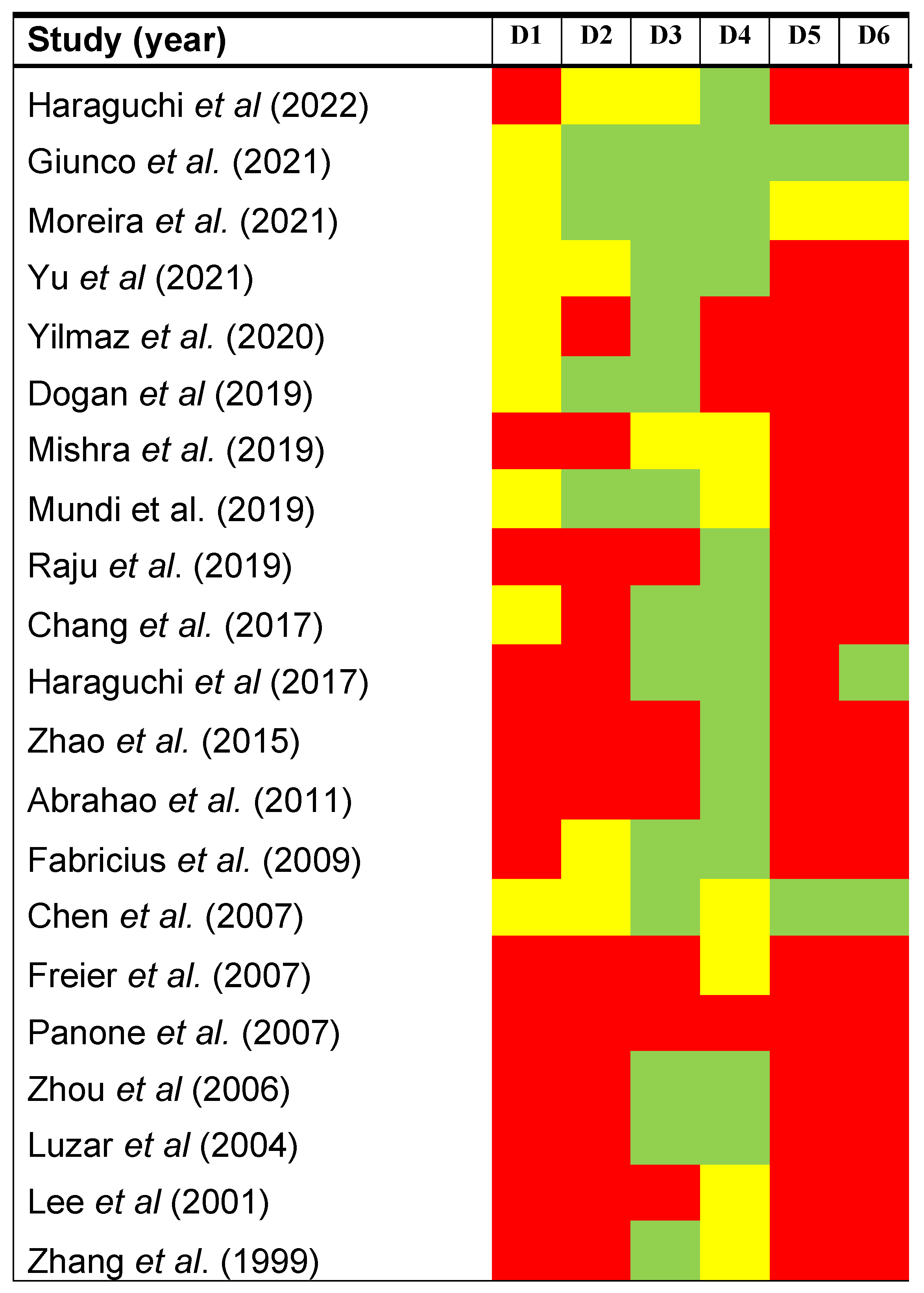
3.3. Qualitative Evaluation
3.4. Quantitative Evaluation (Meta-Analysis)
3.4.1. Association between TERT Upregulation and Prognostic Variables
3.4.2. Association between TERT Upregulation and Clinicopathological Variables
3.4.3. Association between TERT Upregulation and Clinicopathological Variables Not Included in Meta-Analysis
3.5. Quantitative Evaluation (Secondary Analyses)
3.5.1. Meta-Regression Analysis
3.5.2. Sensitivity Analysis
3.5.3. Analysis of Small-Study Effects
3.6. Validation of Methodological Quality
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. The Hallmarks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Arndt, G.M.; MacKenzie, K.L. New prospects for targeting telomerase beyond the telomere. Nat. Rev. Cancer 2016, 16, 508–524. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.W.; Prowse, K.R.; Harley, C.B.; West, M.D.; Ho, P.L.C.; Coviello, G.M.; Weinrich, S.L.; Piatyszek, M.A.; Wright, W.E.; Shay, J.W. Specific Association of Human Telomerase Activity with Immortal Cells and Cancer. Science 1994, 266, 2011–2015. [Google Scholar] [CrossRef]
- Shay, J.W.; Bacchetti, S. A survey of telomerase activity in human cancer. Eur. J. Cancer Part A 1997, 33, 787–791. [Google Scholar] [CrossRef]
- Harley, B.C.; Futcher, B.A.; Greider, W.C. Telomeres shorten during ageing of human fibroblasts. Nature 1990, 345, 458–460. [Google Scholar] [CrossRef]
- Reaper, P.M.; Clay-Farrace, L.; Fiegler, H.; Carr, P.; Von Zglinicki, T.; Saretzki, G. A DNA damage checkpoint response in telomere-initiated senescence. Nature 2003, 426, 194–198. [Google Scholar]
- Greider, C.W.; Blackburn, E.H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 1985, 43, 405–413. [Google Scholar] [CrossRef]
- Bodnar, A.G.; Ouellette, M.; Frolkis, M.; Holt, S.E.; Chiu, C.P.; Morin, G.B.; Harley, C.B.; Shay, J.W.; Lichtsteiner, S.; Wright, W.E. Extension of life-span by introduction of telomerase into normal human cells. Science 1998, 279, 349–352. [Google Scholar] [CrossRef]
- Feng, J.; Funk, W.D.; Wang, S.S.; Weinrich, S.L.; Avilion, A.A.; Chiu, C.P.; Adams, R.R.; Chang, E.; Allsopp, R.C.; Yu, J.; et al. The RNA component of human telomerase. Science 1995, 269, 1236–1241. [Google Scholar] [CrossRef]
- Meyerson, M.; Counter, C.M.; Eaton, E.N.; Ellisen, L.W.; Steiner, P.; Caddle, S.D.; Ziaugra, L.; Beijersbergen, R.L.; Davidoff, M.J.; Qingyun, L.; et al. hEST2, the putative human telomerase catalytic subunit gene, is up- regulated in tumor cells and during immortalization. Cell 1997, 90, 785–795. [Google Scholar] [CrossRef]
- Weinrich, S.L.; Pruzan, R.; Ma, L.; Ouellette, M.; Tesmer, V.M.; Holt, S.E.; Bodnar, A.G.; Lichtsteiner, S.; Kim, N.W.; Trager, J.B.; et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 1997, 17, 498–502. [Google Scholar] [CrossRef]
- Nakamura, T.M.; Morin, G.B.; Chapman, K.B.; Weinrich, S.L.; Andrews, W.H.; Lingner, J.; Harley, C.B.; Cech, T.R. Telomerase catalytic subunit homologs from fission yeast and human. Science 1997, 277, 955–959. [Google Scholar] [CrossRef]
- Riley, R.D.; Ridley, G.; Williams, K.; Altman, D.G.; Hayden, J.; de Vet, H.C.W. Prognosis research: Toward evidence-based results and a Cochrane methods group. J. Clin. Epidemiol. 2007, 60, 863–865. [Google Scholar] [CrossRef]
- Higgins, J.P.; Green, S. (Eds.) Cochrane Handbook for Systematic Reviews of Interventions; John Wiley & Sons, Ltd: Chichester, UK, 2008; ISBN 9780470712184. [Google Scholar]
- Shea, B.J.; Reeves, B.C.; Wells, G.; Thuku, M.; Hamel, C.; Moran, J.; Moher, D.; Tugwell, P.; Welch, V.; Kristjansson, E.; et al. AMSTAR 2: A critical appraisal tool for systematic reviews that include randomised or non-randomised studies of healthcare interventions, or both. BMJ 2017, 358, j4008. [Google Scholar] [CrossRef]
- Shamseer, L.; Moher, D.; Clarke, M.; Ghersi, D.; Liberati, A.; Petticrew, M.; Shekelle, P.; Stewart, L.A. PRISMA-P Group Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015: Elaboration and explanation. BMJ 2015, 350, g7647. [Google Scholar] [CrossRef]
- Luo, D.; Wan, X.; Liu, J.; Tong, T. Optimally estimating the sample mean from the sample size, median, mid-range, and/or mid-quartile range. Stat. Methods Med. Res. 2018, 27, 1785–1805. [Google Scholar] [CrossRef]
- Wan, X.; Wang, W.; Liu, J.; Tong, T. Estimating the sample mean and standard deviation from the sample size, median, range and/or interquartile range. BMC Med. Res. Methodol. 2014, 14, 135. [Google Scholar] [CrossRef]
- Hayden, J.A.; Côté, P.; Bombardier, C. Evaluation of the quality of prognosis studies in systematic reviews. Ann. Intern. Med. 2006, 144, 427–437. [Google Scholar] [CrossRef]
- González-Moles, M.Á.; Ayén, Á.; González-Ruiz, I.; de Porras-Carrique, T.; González-Ruiz, L.; Ruiz-Ávila, I.; Ramos-García, P. Prognostic and Clinicopathological Significance of FADD Upregulation in Head and Neck Squamous Cell Carcinoma: A Systematic Review and Meta-Analysis. Cancers 2020, 12, 2393. [Google Scholar] [CrossRef]
- Ramos-García, P.; González-Moles, M.Á. Prognostic and Clinicopathological Significance of the Aberrant Expression of β-Catenin in Oral Squamous Cell Carcinoma: A Systematic Review and Meta-Analysis. Cancers 2022, 14, 479. [Google Scholar] [CrossRef]
- Ramos-García, P.; González-Moles, M.Á.; Warnakulasuriya, S. Significance of p53 overexpression in the prediction of the malignant transformation risk of oral potentially malignant disorders: A systematic review and meta-analysis. Oral Oncol. 2022, 126, 105734. [Google Scholar] [CrossRef]
- Tierney, J.F.; Stewart, L.A.; Ghersi, D.; Burdett, S.; Sydes, M.R. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 2007, 8, 16. [Google Scholar] [CrossRef]
- Parmar, M.K.; Torri, V.; Stewart, L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat. Med. 1998, 17, 2815–2834. [Google Scholar] [CrossRef]
- Anzures-Cabrera, J.; Sarpatwari, A.; Higgins, J.P. Expressing findings from meta-analyses of continuous outcomes in terms of risks. Stat. Med. 2011, 30, 2967–2985. [Google Scholar] [CrossRef]
- Higgins, J.P.T.; Thompson, S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002, 21, 1539–1558. [Google Scholar] [CrossRef]
- Higgins, J.P.T.; Thompson, S.G.; Deeks, J.J.; Altman, D.G. Measuring inconsistency in meta-analyses. BMJ 2003, 327, 557–560. [Google Scholar] [CrossRef]
- Thompson, S.G.; Higgins, J.P.T. How should meta-regression analyses be undertaken and interpreted? Stat. Med. 2002, 21, 1559–1573. [Google Scholar] [CrossRef]
- Higgins, J.P.T.; Thompson, S.G. Controlling the risk of spurious findings from meta-regression. Stat. Med. 2004, 23, 1663–1682. [Google Scholar] [CrossRef]
- Manly, B.F.J. Randomization, Bootstrap and Monte Carlo Methods in Biology; Taylor & Francis: Oxfordshire, UK, 2006; Volume 53. [Google Scholar]
- Viechtbauer, W.; Cheung, M.W.-L. Outlier and influence diagnostics for meta-analysis. Res. Synth. Methods 2010, 1, 112–125. [Google Scholar] [CrossRef]
- Egger, M.; Davey Smith, G.; Schneider, M.; Minder, C. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997, 315, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Harrison, S.; Jones, H.E.; Martin, R.M.; Lewis, S.J.; Higgins, J.P.T. The albatross plot: A novel graphical tool for presenting results of diversely reported studies in a systematic review. Res. Synth. Methods 2017, 8, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Haraguchi, K.; Habu, M.; Yada, N.; Sasaguri, M.; Yoshioka, I.; Tominaga, K. Human telomerase reverse transcriptase protein expression is associated with survival in patients with oral squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 2022, 15, 29–37. [Google Scholar] [PubMed]
- Moreira, A.; Poulet, A.; Masliah-Planchon, J.; Lecerf, C.; Vacher, S.; Larbi Chérif, L.; Dupain, C.; Marret, G.; Girard, E.; Syx, L.; et al. Prognostic value of tumor mutational burden in patients with oral cavity squamous cell carcinoma treated with upfront surgery. ESMO Open 2021, 6, 100178. [Google Scholar] [CrossRef]
- Chang, K.-P.; Wang, C.-I.; Pickering, C.R.; Huang, Y.; Tsai, C.-N.; Tsang, N.-M.; Kao, H.-K.; Cheng, M.-H.; Myers, J.N. Prevalence of promoter mutations in the TERT gene in oral cavity squamous cell carcinoma. Head Neck 2017, 39, 1131–1137. [Google Scholar] [CrossRef]
- Zhao, T.; Hu, F.; Qiao, B.; Chen, Z.; Tao, Q. Telomerase reverse transcriptase potentially promotes the progression of oral squamous cell carcinoma through induction of epithelial-mesenchymal transition. Int. J. Oncol. 2015, 46, 2205–2215. [Google Scholar] [CrossRef]
- Abrahao, A.C.; Bonelli, B.V.; Nunes, F.D.; Dias, E.P.; Cabral, M.G. Immunohistochemical expression of p53, p16 and hTERT in oral squamous cell carcinoma and potentially malignant disorders. Braz. Oral Res. 2011, 25, 34–41. [Google Scholar] [CrossRef]
- Fabricius, E.-M.; Kruse-Boitschenko, U.; Khoury, R.; Wildner, G.-P.; Raguse, J.-D.; Klein, M. Immunohistochemical determination of the appropriate anti-hTERT antibodies for in situ detection of telomerase activity in frozen sections of head and neck squamous cell carcinomas and tumor margin tissues. Int. J. Oncol. 2009, 34, 1257–1279. [Google Scholar] [CrossRef][Green Version]
- Chen, H.-H.; Yu, C.-H.; Wang, J.-T.; Liu, B.-Y.; Wang, Y.-P.; Sun, A.; Tsai, T.-C.; Chiang, C.-P. Expression of human telomerase reverse transcriptase (hTERT) protein is significantly associated with the progression, recurrence and prognosis of oral squamous cell carcinoma in Taiwan. Oral Oncol. 2007, 43, 122–129. [Google Scholar] [CrossRef]
- Pannone, G.; De Maria, S.; Zamparese, R.; Metafora, S.; Serpico, R.; Morelli, F.; Rubini, C.; Farina, E.; Carteni, M.; Staibano, S.; et al. Prognostic value of human telomerase reverse transcriptase gene expression in oral carcinogenesis. Int. J. Oncol. 2007, 30, 1349–1357. [Google Scholar] [CrossRef]
- Freier, K.; Pungs, S.; Flechtenmacher, C.; Bosch, F.X.; Lichter, P.; Joos, S.; Hofele, C. Frequent high telomerase reverse transcriptase expression in primary oral squamous cell carcinoma. J. Oral Pathol. Med. 2007, 36, 267–272. [Google Scholar] [CrossRef]
- Luzar, B.; Poljak, M.; Marin, I.J.; Eberlinc, A.; Klopcic, U.; Gale, N. Human telomerase catalytic subunit gene re-expression is an early event in oral carcinogenesis. Histopathology 2004, 45, 13–19. [Google Scholar] [CrossRef]
- Lee, B.K.; Diebel, E.; Neukam, F.W.; Wiltfang, J.; Ries, J. Diagnostic and prognostic relevance of expression of human telomerase subunits in oral cancer. Int. J. Oncol. 2001, 19, 1063–1068. [Google Scholar] [CrossRef]
- Zhou, Q.; Wang, X.; Jiang, Y.; Li, R.; Zhong, M. Expression of telomerase genes hTRTmRNA in oral squamous cell carcinomas. Shanghai Kou Qiang Yi Xue 2006, 15, 259–262. [Google Scholar]
- Yu, Y.; Fan, D.; Song, X.; Zakeri, K.; Chen, L.; Kang, J.; McBride, S.; Tsai, C.J.; Dunn, L.; Sherman, E.; et al. TERT Promoter Mutations Are Enriched in Oral Cavity Cancers and Associated With Locoregional Recurrence. JCO Precis. Oncol. 2021, 5, PO.20.00515. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, W. Telomerase hTR and hTRT gene expression in oral precancerous lesions and squamous cell carcinomas. Chin. J. Dent. Res. 1999, 2, 43–48. [Google Scholar]
- Yilmaz, I.; Erkul, B.E.; Ozturk Sari, S.; Issin, G.; Tural, E.; Terzi Kaya Terzi, N.; Karatay, H.; Celik, M.; Ulusan, M.; Bilgic, B. Promoter region mutations of the telomerase reverse transcriptase (TERT) gene in head and neck squamous cell carcinoma. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2020, 130, 63–70. [Google Scholar] [CrossRef]
- Giunco, S.; Boscolo-Rizzo, P.; Rampazzo, E.; Tirelli, G.; Alessandrini, L.; Di Carlo, R.; Rossi, M.; Nicolai, P.; Menegaldo, A.; Carraro, V.; et al. TERT Promoter Mutations and rs2853669 Polymorphism: Useful Markers for Clinical Outcome Stratification of Patients With Oral Cavity Squamous Cell Carcinoma. Front. Oncol. 2021, 11, 782658. [Google Scholar] [CrossRef]
- Raju, K.L.; Haragannavar, V.C.; Patil, S.; Rao, R.S.; Nagaraj, T.; Augustine, D.; Venkatesiah, S.S.; Nambiar, S. Expression of hTERT in Oral Submucous Fibrosis and Oral Squamous Cell Carcinoma—An Immunohistochemical Analysis. Pathol. Oncol. Res. 2020, 26, 1573–1582. [Google Scholar] [CrossRef]
- Mundi, N.; Prokopec, S.D.; Ghasemi, F.; Warner, A.; Patel, K.; MacNeil, D.; Howlett, C.; Stecho, W.; Plantinga, P.; Pinto, N.; et al. Genomic and human papillomavirus profiling of an oral cancer cohort identifies TP53 as a predictor of overall survival. Cancers Head Neck 2019, 4, 5. [Google Scholar] [CrossRef]
- Dogan, S.; Xu, B.; Middha, S.; Vanderbilt, C.M.; Bowman, A.S.; Migliacci, J.; Morris, L.G.T.; Seshan, V.E.; Ganly, I. Identification of prognostic molecular biomarkers in 157 HPV-positive and HPV-negative squamous cell carcinomas of the oropharynx. Int. J. Cancer 2019, 145, 3152–3162. [Google Scholar] [CrossRef]
- Mishra, N.; Tandon, N.; Fatima, N.; Srivastava, A.N.; Lal, N.; Kumar, V. Immunohistochemical expression of human telomerase reverse transcriptase in oral cancer and precancer: A case-control study. J. Oral Maxillofac. Pathol. 2019, 23, 412–417. [Google Scholar] [CrossRef]
- Haraguchi, K.; Yada, N.; Sato, S.; Habu, M.; Hayakawa, M.; Takahashi, O.; Sasaguri, M.; Takenaka, S.; Yoshioka, I.; Matsuo, K.; et al. The methylation status and expression of human telomerase reverse transcriptase is significantly high in oral carcinogenesis. APMIS 2017, 125, 797–807. [Google Scholar] [CrossRef]
- Deng, W.; Peng, W.; Wang, T.; Chen, J.; Zhu, S. Overexpression of MMPs Functions as a Prognostic Biomarker for Oral Cancer Patients: A Systematic Review and Meta-analysis. Oral Health Prev. Dent. 2019, 17, 505–514. [Google Scholar] [CrossRef]
- Stewart, S.A.; Hahn, W.C.; O’Connor, B.F.; Banner, E.N.; Lundberg, A.S.; Modha, P.; Mizuno, H.; Brooks, M.W.; Fleming, M.; Zimonjic, D.B.; et al. Telomerase contributes to tumorigenesis by a telomere length-independent mechanism. Proc. Natl. Acad. Sci. USA 2002, 99, 12606–12611. [Google Scholar] [CrossRef] [PubMed]
- Fleisig, H.B.; Wong, J.M.Y. Telomerase promotes efficient cell cycle kinetics and confers growth advantage to telomerase-negative transformed human cells. Oncogene 2012, 31, 954–965. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.G.; Gupta, A.; Wang, H.; Scherthan, H.; Dhar, S.; Gandhi, V.; Iliakis, G.; Shay, J.W.; Young, C.S.H.; Pandita, T.K. hTERT associates with human telomeres and enhances genomic stability and DNA repair. Oncogene 2003, 22, 131–146. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, O.G.; Assfalg, R.; Koch, S.; Schelling, A.; Meena, J.K.; Kraus, J.; Lechel, A.; Katz, S.F.; Benes, V.; Scharffetter-Kochanek, K.; et al. Telomerase stimulates ribosomal DNA transcription under hyperproliferative conditions. Nat. Commun. 2014, 5, 4599. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.L.; Coller, H.A.; Roberts, J.M. Telomerase modulates expression of growth-controlling genes and enhances cell proliferation. Nat. Cell Biol. 2003, 5, 474–479. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Firpo, E.J.; Wang, Y.; Roberts, J.M. Separation of telomerase functions by reverse genetics. Proc. Natl. Acad. Sci. USA 2011, 108, E1363–E1371. [Google Scholar] [CrossRef]
- Topoisomerase, I.; Ludwig, A.; Saretzki, G.; Holm, P.S.; Tiemann, F.; Lorenz, M.; Emrich, T.; Harley, C.B.; Zglinicki, T. Von Ribozyme cleavage of telomerase mRNA sensitizes breast epithelial cells to inhibitors of topoisomerase. Cancer Res. 2001, 61, 3053–3061. [Google Scholar]
- Cao, Y.; Li, H.; Deb, S.; Liu, J.P. TERT regulates cell survival independent of telomerase enzymatic activity. Oncogene 2002, 21, 3130–3138. [Google Scholar] [CrossRef]
- Luiten, R.M.; Pène, J.; Yssel, H.; Spits, H. Ectopic hTERT expression extends the life span of human CD4+ helper and regulatory T-cell clones and confers resistance to oxidative stress-induced apoptosis. Blood 2003, 101, 4512–4519. [Google Scholar] [CrossRef]
- Dudognon, C.; Pendino, F.; Hillion, J.; Saumet, A.; Lanotte, M.; Ségal-Bendirdjian, E. Death receptor signaling regulatory function for telomerase: hTERT abolishes TRAIL-induced apoptosis, independently of telomere maintenance. Oncogene 2004, 23, 7469–7474. [Google Scholar] [CrossRef][Green Version]
- Saretzki, G.; Ludwig, A.; Von Zglinicki, T.; Runnebaum, I.B. Ribozyme-mediated telomerase inhibition induces immediate cell loss but not telomere shortening in ovarian cancer cells. Cancer Gene Ther. 2001, 8, 827–834. [Google Scholar] [CrossRef]
- Kraemer, K.; Fuessel, S.; Schmidt, U.; Kotzsch, M.; Schwenzer, B.; Wirth, M.P.; Meye, A. Antisense-mediated hTERT inhibition specifically reduces the growth of human bladder cancer cells. Clin. Cancer Res. 2003, 9, 3794–3800. [Google Scholar]
- Park, J.I.; Venteicher, A.S.; Hong, J.Y.; Choi, J.; Jun, S.; Shkreli, M.; Chang, W.; Meng, Z.; Cheung, P.; Ji, H.; et al. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature 2009, 460, 66–72. [Google Scholar] [CrossRef]
- González-moles, M.A.; Ruiz-ávila, I.; Gil-montoya, J.A.; Plaza-campillo, J.; Scully, C. β-Catenin in oral cancer: An update on current knowledge. Oral Oncol. 2014, 50, 818–824. [Google Scholar] [CrossRef]
- Koh, C.M.; Khattar, E.; Leow, S.C.; Liu, C.Y.; Muller, J.; Ang, W.X.; Li, Y.; Franzoso, G.; Li, S.; Guccione, E.; et al. Telomerase regulates MYC-driven oncogenesis independent of its reverse transcriptase activity. J. Clin. Investig. 2015, 125, 2109–2122. [Google Scholar] [CrossRef]
- Luo, J.; Solimini, N.L.; Elledge, S.J. Principles of Cancer Therapy: Oncogene and Non-oncogene Addiction. Cell 2009, 136, 823–837. [Google Scholar] [CrossRef]
- Shin, K.H.; Kang, M.K.; Dicterow, E.; Kameta, A.; Baluda, M.A.; Park, N.H. Introduction of Human Telomerase Reverse Transcriptase to Normal Human Fibroblasts Enhances DNA Repair Capacity. Clin. Cancer Res. 2004, 10, 2551–2560. [Google Scholar] [CrossRef]
- Masutomi, K.; Possemato, R.; Wong, J.M.Y.; Currier, J.L.; Tothova, Z.; Manola, J.B.; Ganesan, S.; Lansdorp, P.M.; Collins, K.; Hahn, W.C. The telomerase reverse transcriptase regulates chromatin state and DNA damage responses. Proc. Natl. Acad. Sci. USA 2005, 102, 8222–8227. [Google Scholar] [CrossRef]
- Hayden, J.A.; van der Windt, D.A.; Cartwright, J.L.; Côté, P.; Bombardier, C. Assessing bias in studies of prognostic factors. Ann. Intern. Med. 2013, 158, 280–286. [Google Scholar] [CrossRef]
- Kyzas, P.A.; Denaxa-Kyza, D.; Ioannidis, J.P.A. Almost all articles on cancer prognostic markers report statistically significant results. Eur. J. Cancer 2007, 43, 2559–2579. [Google Scholar] [CrossRef]


| Total Sample Size | 21 Studies |
|---|---|
| Total patients (range) | 1698 (30–218) |
| Year of publication | 1999–2022 |
| Study design | |
| Retrospective cohort | 21 studies |
| TERT upregulation analysis | |
| Gene mutation | 7 studies (959 patients) |
| mRNA expression | 4 studies (174 patients) |
| protein overexpression | 10 studies (565 patients) |
| Study continent | |
| Asia | 8 studies (523 patients) |
| Europe | 8 studies (769 patients) |
| North America | 3 studies (361 patients) |
| South America | 1 study (30 patients) |
| Pooled Data | Heterogeneity | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Meta-Analyses | Studies, n | Patients, n | Stat. Model | Wt | ES (95% CI) | p-Value | Phet | I2 (%) | Supplementary Materials a |
| Overall Survival | |||||||||
| TERT upregulation (all) b | 9 | 1068 | REM | D-L | HR = 1.40 (0.95–2.07) | 0.09 | 0.02 | 55.0 | Manuscript, Figure 3 |
| Subgroup analysis by alteration c | 0.001 d | ||||||||
| TERT mutations | 6 | 892 | REM | D-L | HR = 1.03 (0.80–1.31) | 0.83 | 0.43 | 0.0 | |
| TERT protein overexpression | 3 | 176 | REM | D-L | HR = 3.01 (1.70–5.35) | <0.001 | 0.48 | 0.0 | |
| Subgroup analysis by geographical area c | 0.27 d | Figure S1 | |||||||
| Asian | 3 | 336 | REM | D-L | HR = 2.09 (0.86–5.08) | 0.11 | 0.04 | 68.4 | |
| Non-Asian | 6 | 732 | REM | D-L | HR = 1.19 (0.77–1.84) | 0.43 | 0.11 | 45.0 | |
| Univariable meta-regressions by study design and patients characteristics e | |||||||||
| Follow-up (months) | 9 | 1068 | random-effects meta-regression | Coef = −0.002 (−0.010 to 0.006) | 0.58 ± 0.005 f | hetexplained = −24.83%g | Figure S2 | ||
| Sex (proportion of males, %) | 8 | 1027 | random-effects meta-regression | Coef = −0.005 (−0.052 to 0.043) | 0.13 ± 0.003 f | hetexplained = −80.22% g | Figure S3 | ||
| Age (years, mean) | 8 | 911 | random-effects meta-regression | Coef = −0.003 (−0.122 to 0.115) | 0.92 ± 0.003 f | hetexplained = -−43.25%g | Figure S4 | ||
| Clinical stage (proportion of stage-III/IV patients,%) | 2 | 295 | — | — | — | — | — | ||
| Tobacco consumption (proportion of smokers, %) | 7 | 974 | random-effects meta-regression | Coef = 0.005 (−0.030 to 0.040) | 0.71 ± 0.005 f | hetexplained = −74.01% g | Figure S5 | ||
| Areca nut/betel quid consumption (proportion of chewers, %) | 2 | 283 | — | — | — | — | — | ||
| Alcohol consumption (% of patients with positive habit) | 6 | 817 | random-effects meta-regression | Coef = 0.008 (−0.019 to 0.035) | 0.44 ± 0.005 f | hetexplained = −41.06% g | Figure S6 | ||
| Disease-free survival | |||||||||
| TERT upregulation (all) b | 8 | 967 | REM | D-L | HR = 1.64 (1.06–2.54) | 0.03 | 0.07 | 46.4 | Figure S7 |
| Subgroup analysis by alteration c | 0.006 d | ||||||||
| TERT mutations | 5 | 790 | REM | D-L | HR = 1.13 (0.81–1.59) | 0.46 | 0.68 | 0.0 | |
| TERT mRNA overexpression | 1 | 42 | — | — | HR = 3.79 (1.03–13.98) | 0.05 | — | 0.0 | |
| TERT protein overexpression | 2 | 135 | REM | D-L | HR = 4.03 (1.80–9.05) | 0.001 | 0.49 | 0.0 | |
| T status | |||||||||
| TERT upregulation (all) b | 11 | 1055 | REM | D-L | OR = 1.15 (0.66–2.03) | 0.62 | 0.001 | 65.4 | Figure S8 |
| Subgroup analysis by alteration c | 0.22 d | ||||||||
| TERT mutations | 4 | 569 | REM | D-L | OR = 0.89 (0.56–1.40) | 0.61 | 0.41 | 0.0 | |
| TERT mRNA overexpression | 1 | 42 | — | — | OR = 0.40 (0.09–1.84) | 0.24 | — | 0.0 | |
| TERT protein overexpression | 6 | 444 | REM | D-L | OR = 1.81 (0.69–4.73) | 0.23 | 0.001 | 77.3 | |
| N status | |||||||||
| TERT upregulation (all) b | 10 | 1013 | REM | D-L | OR = 1.25 (0.62–2.50) | 0.54 | <0.001 | 75.4 | Figure S9 |
| Subgroup analysis by alteration c | 0.21 d | ||||||||
| TERT mutations | 4 | 569 | REM | D-L | OR = 0.80 (0.50–1.29) | 0.36 | 0.78 | 0.0 | |
| TERT protein overexpression | 6 | 444 | REM | D-L | OR = 1.82 (0.56–5.90) | 0.32 | <0.001 | 84.5 | |
| Clinical stage | |||||||||
| TERT upregulation (all) b | 7 | 526 | REM | D-L | OR = 1.33 (0.41–4.34) | 0.64 | <0.001 | 86.4 | Figure S10 |
| Subgroup analysis by alteration c | 0.24 d | ||||||||
| TERT mutations | 2 | 295 | REM | D-L | OR = 0.66 (0.38–1.17) | 0.16 | 0.88 | 0.0 | |
| TERT mRNA overexpression | 1 | 42 | — | — | OR = 0.14 (0.01–2.64) | 0.19 | — | 0.0 | |
| TERT protein overexpression | 4 | 189 | REM | D-L | OR = 2.75 (0.34–22.61) | 0.64 | <0.001 | 91.6 | |
| Histological grade | |||||||||
| TERT upregulation (all) b | 13 | 630 | REM | D-L | OR = 1.94 (1.14–3.30) | 0.01 | 0.21 | 23.0 | Figure S11 |
| Subgroup analysis by alteration c | 0.02 d | ||||||||
| TERT mutations | 1 | 144 | — | — | OR = 0.42 (0.09–1.98) | 0.28 | — | 0.0 | |
| TERT mRNA overexpression | 4 | 174 | REM | D-L | OR = 1.16 (0.48–2.76) | 0.74 | 0.44 | 0.0 | |
| TERT protein overexpression | 8 | 312 | REM | D-L | OR = 3.20 (1.83–5.62) | <0.001 | 0.69 | 0.0 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-Moles, M.Á.; Moya-González, E.; García-Ferrera, A.; Nieto-Casado, P.; Ramos-García, P. Prognostic and Clinicopathological Significance of Telomerase Reverse Transcriptase Upregulation in Oral Cancer: A Systematic Review and Meta-Analysis. Cancers 2022, 14, 3673. https://doi.org/10.3390/cancers14153673
González-Moles MÁ, Moya-González E, García-Ferrera A, Nieto-Casado P, Ramos-García P. Prognostic and Clinicopathological Significance of Telomerase Reverse Transcriptase Upregulation in Oral Cancer: A Systematic Review and Meta-Analysis. Cancers. 2022; 14(15):3673. https://doi.org/10.3390/cancers14153673
Chicago/Turabian StyleGonzález-Moles, Miguel Ángel, Eloísa Moya-González, Alberto García-Ferrera, Paola Nieto-Casado, and Pablo Ramos-García. 2022. "Prognostic and Clinicopathological Significance of Telomerase Reverse Transcriptase Upregulation in Oral Cancer: A Systematic Review and Meta-Analysis" Cancers 14, no. 15: 3673. https://doi.org/10.3390/cancers14153673
APA StyleGonzález-Moles, M. Á., Moya-González, E., García-Ferrera, A., Nieto-Casado, P., & Ramos-García, P. (2022). Prognostic and Clinicopathological Significance of Telomerase Reverse Transcriptase Upregulation in Oral Cancer: A Systematic Review and Meta-Analysis. Cancers, 14(15), 3673. https://doi.org/10.3390/cancers14153673








