Integrated Clinical and Genomic Models to Predict Optimal Cytoreduction in High-Grade Serous Ovarian Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Data
2.2. Genomic Analysis
2.3. Statistical Analysis
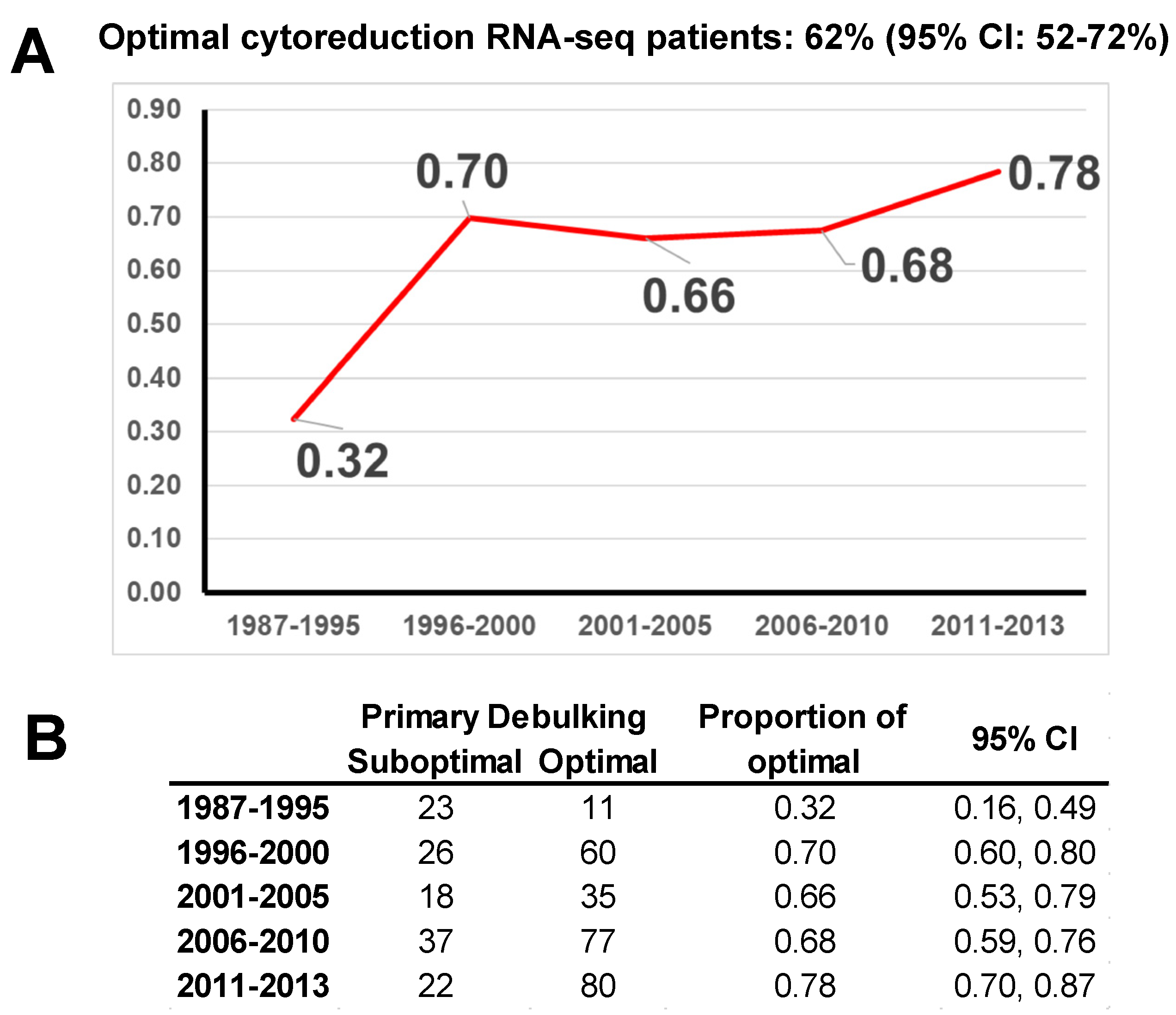
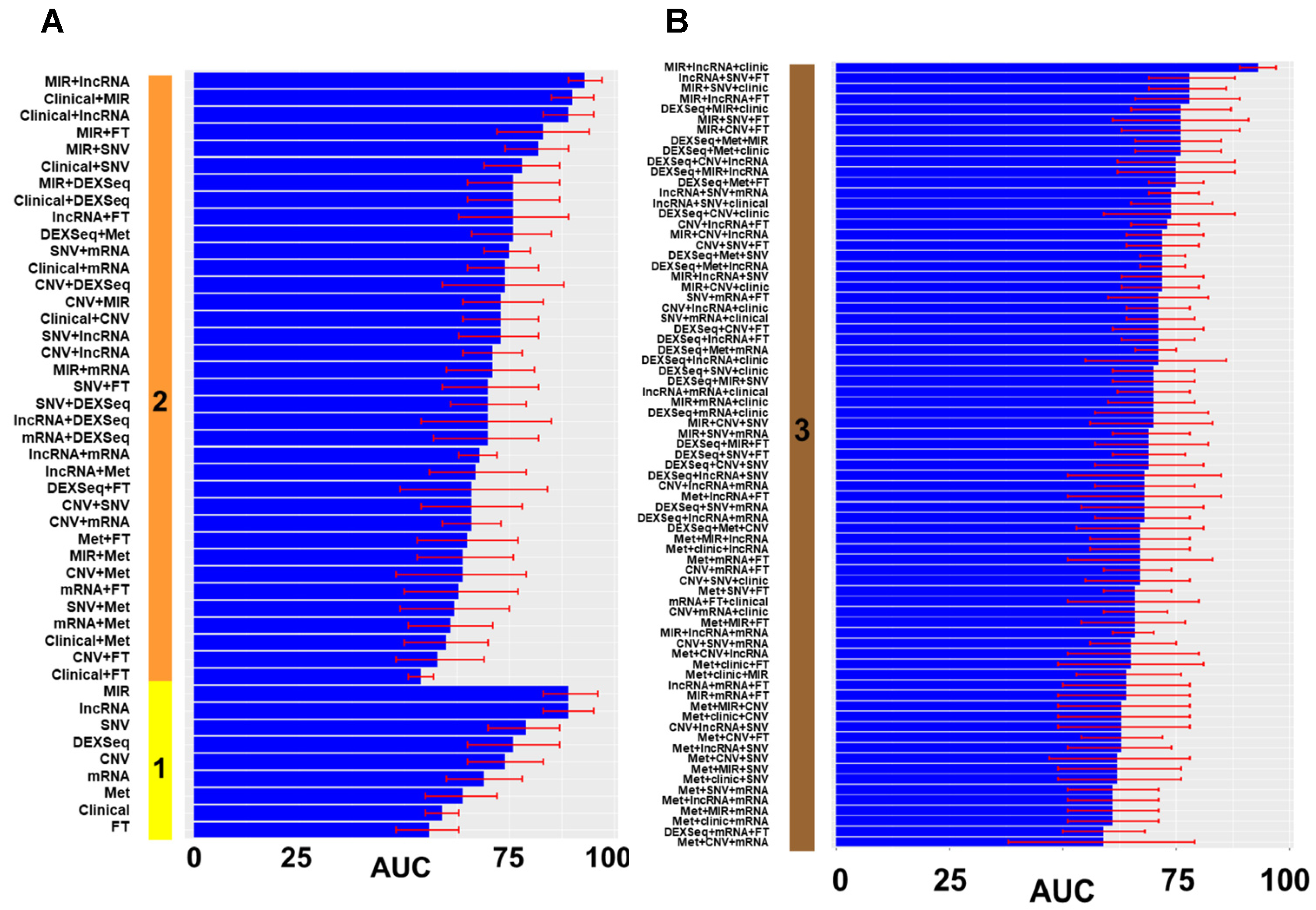
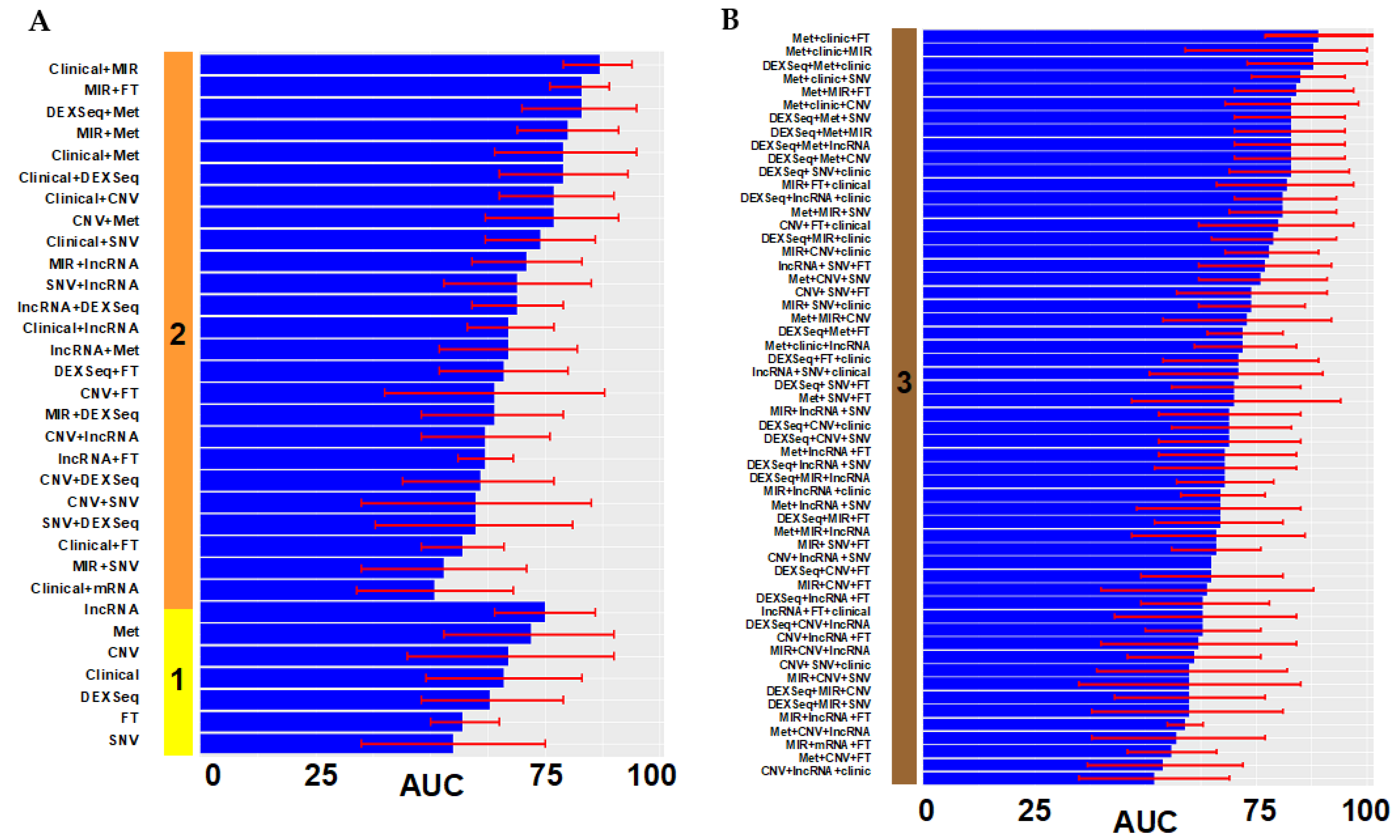
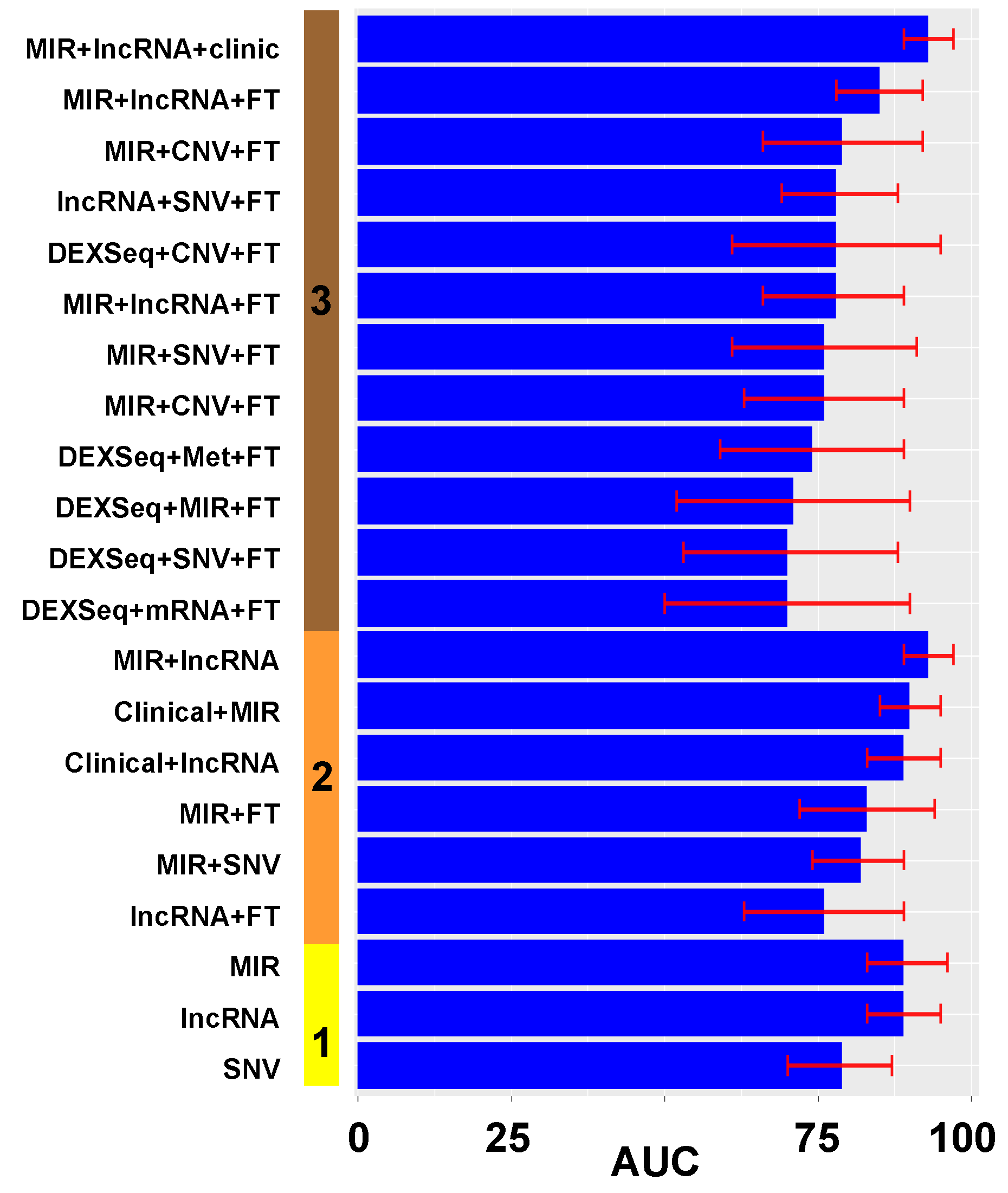
3. Results
3.1. Model Construction
3.2. Model Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Howlader, N.; Noone, A.; Krapcho, M. SEER Cancer Statistics Review 1975–2016. National Cancer Institute. 2019. Available online: https://seer.cancer.gov/archive/csr/1975_2016/ (accessed on 17 July 2022).
- Morgan, R.J., Jr.; Copeland, L.; Gershenson, D.; Locker, G.; McIntosh, D.; Ozols, R.; Teng, N. NCCN Ovarian Cancer Practice Guidelines. The National Comprehensive Cancer Network. Oncology 1996, 10, 293–310. [Google Scholar] [PubMed]
- McGuire, W.P.; Hoskins, W.J.; Brady, M.F.; Kucera, P.R.; Partridge, E.E.; Look, K.Y.; Clarke-Pearson, D.L.; Davidson, M. Cyclophosphamide and cisplatin compared with paclitaxel and cisplatin in patients with stage III and stage IV ovarian cancer. N. Engl. J. Med. 1996, 334, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Ozols, R.F.; Bundy, B.N.; Greer, B.E.; Fowler, J.M.; Clarke-Pearson, D.; Burger, R.A.; Mannel, R.S.; DeGeest, K.; Hartenbach, E.M.; Baergen, R.; et al. Phase III trial of carboplatin and paclitaxel compared with cisplatin and paclitaxel in patients with optimally resected stage III ovarian cancer: A Gynecologic Oncology Group study. J. Clin. Oncol. 2003, 21, 3194–3200. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, D.K.; Bundy, B.; Wenzel, L.; Huang, H.Q.; Baergen, R.; Lele, S.; Copeland, L.J.; Walker, J.L.; Burger, R.A.; Gynecologic Oncology, G. Intraperitoneal cisplatin and paclitaxel in ovarian cancer. N. Engl. J. Med. 2006, 354, 34–43. [Google Scholar] [CrossRef] [Green Version]
- Bookman, M.A.; Brady, M.F.; McGuire, W.P.; Harper, P.G.; Alberts, D.S.; Friedlander, M.; Colombo, N.; Fowler, J.M.; Argenta, P.A.; De Geest, K.; et al. Evaluation of new platinum-based treatment regimens in advanced-stage ovarian cancer: A Phase III Trial of the Gynecologic Cancer Intergroup. J. Clin. Oncol. 2009, 27, 1419–1425. [Google Scholar] [CrossRef]
- Burger, R.A.; Brady, M.F.; Bookman, M.A.; Fleming, G.F.; Monk, B.J.; Huang, H.; Mannel, R.S.; Homesley, H.D.; Fowler, J.; Greer, B.E.; et al. Incorporation of bevacizumab in the primary treatment of ovarian cancer. N. Engl. J. Med. 2011, 365, 2473–2483. [Google Scholar] [CrossRef] [Green Version]
- Walker, J.L.; Brady, M.F.; Wenzel, L.; Fleming, G.F.; Huang, H.Q.; DiSilvestro, P.A.; Fujiwara, K.; Alberts, D.S.; Zheng, W.; Tewari, K.S.; et al. Randomized Trial of Intravenous Versus Intraperitoneal Chemotherapy Plus Bevacizumab in Advanced Ovarian Carcinoma: An NRG Oncology/Gynecologic Oncology Group Study. J. Clin. Oncol. 2019, 37, 1380–1390. [Google Scholar] [CrossRef]
- Bristow, R.E.; Montz, F.J.; Lagasse, L.D.; Leuchter, R.S.; Karlan, B.Y. Survival impact of surgical cytoreduction in stage IV epithelial ovarian cancer. Gynecol. Oncol. 1999, 72, 278–287. [Google Scholar] [CrossRef]
- Winter, W.E.; Maxwell, G.L.; Tian, C.; Sundborg, M.J.; Rose, G.S.; Rose, P.G.; Rubin, S.C.; Muggia, F.; McGuire, W.P.; Gynecologic Oncology, G. Tumor residual after surgical cytoreduction in prediction of clinical outcome in stage IV epithelial ovarian cancer: A Gynecologic Oncology Group Study. J. Clin. Oncol. 2008, 26, 83–89. [Google Scholar] [CrossRef]
- Bristow, R.E.; Tomacruz, R.S.; Armstrong, D.K.; Trimble, E.L.; Montz, F.J. Survival effect of maximal cytoreductive surgery for advanced ovarian carcinoma during the platinum era: A meta-analysis. J. Clin. Oncol. 2002, 20, 1248–1259. [Google Scholar] [CrossRef]
- Chang, S.J.; Hodeib, M.; Chang, J.; Bristow, R.E. Survival impact of complete cytoreduction to no gross residual disease for advanced-stage ovarian cancer: A meta-analysis. Gynecol. Oncol. 2013, 130, 493–498. [Google Scholar] [CrossRef]
- Chi, D.S.; Eisenhauer, E.L.; Zivanovic, O.; Sonoda, Y.; Abu-Rustum, N.R.; Levine, D.A.; Guile, M.W.; Bristow, R.E.; Aghajanian, C.; Barakat, R.R. Improved progression-free and overall survival in advanced ovarian cancer as a result of a change in surgical paradigm. Gynecol. Oncol. 2009, 114, 26–31. [Google Scholar] [CrossRef]
- Armstrong, D.K.; Alvarez, R.D.; Bakkum-Gamez, J.N.; Barroilhet, L.; Behbakht, K.; Berchuck, A.; Chen, L.M.; Cristea, M.; DeRosa, M.; Eisenhauer, E.L.; et al. Ovarian Cancer, Version 2.2020, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 2021, 19, 191–226. [Google Scholar] [CrossRef]
- Wright, A.A.; Bohlke, K.; Armstrong, D.K.; Bookman, M.A.; Cliby, W.A.; Coleman, R.L.; Dizon, D.S.; Kash, J.J.; Meyer, L.A.; Moore, K.N.; et al. Neoadjuvant Chemotherapy for Newly Diagnosed, Advanced Ovarian Cancer: Society of Gynecologic Oncology and American Society of Clinical Oncology Clinical Practice Guideline. J. Clin. Oncol. 2016, 34, 3460–3473. [Google Scholar] [CrossRef]
- Kehoe, S.; Hook, J.; Nankivell, M.; Jayson, G.C.; Kitchener, H.; Lopes, T.; Luesley, D.; Perren, T.; Bannoo, S.; Mascarenhas, M.; et al. Primary chemotherapy versus primary surgery for newly diagnosed advanced ovarian cancer (CHORUS): An open-label, randomised, controlled, non-inferiority trial. Lancet 2015, 386, 249–257. [Google Scholar] [CrossRef]
- Fagotti, A.; Scambia, G. Neoadjuvant chemotherapy versus upfront debulking surgery in advanced tubo-ovarian cancer. Lancet Oncol. 2018, 19, 1558–1560. [Google Scholar] [CrossRef]
- Mulligan, K.M.; Glennon, K.; Donohoe, F.; O’Brien, Y.; Mc Donnell, B.C.; Bartels, H.C.; Vermeulen, C.; Walsh, T.; Shields, C.; McCormack, O.; et al. Multidisciplinary Surgical Approach to Increase Complete Cytoreduction Rates for Advanced Ovarian Cancer in a Tertiary Gynecologic Oncology Center. Ann. Surg. Oncol. 2021, 28, 4553–4560. [Google Scholar] [CrossRef]
- Feng, L.Y.; Liao, S.B.; Li, L. Preoperative serum levels of HE4 and CA125 predict primary optimal cytoreduction in advanced epithelial ovarian cancer: A preliminary model study. J. Ovarian Res. 2020, 13, 17. [Google Scholar] [CrossRef] [Green Version]
- Suidan, R.S.; Ramirez, P.T.; Sarasohn, D.M.; Teitcher, J.B.; Iyer, R.B.; Zhou, Q.; Iasonos, A.; Denesopolis, J.; Zivanovic, O.; Long Roche, K.C.; et al. A multicenter assessment of the ability of preoperative computed tomography scan and CA-125 to predict gross residual disease at primary debulking for advanced epithelial ovarian cancer. Gynecol. Oncol. 2017, 145, 27–31. [Google Scholar] [CrossRef] [Green Version]
- Ferrandina, G.; Sallustio, G.; Fagotti, A.; Vizzielli, G.; Paglia, A.; Cucci, E.; Margariti, A.; Aquilani, L.; Garganese, G.; Scambia, G. Role of CT scan-based and clinical evaluation in the preoperative prediction of optimal cytoreduction in advanced ovarian cancer: A prospective trial. Br. J. Cancer 2009, 101, 1066–1073. [Google Scholar] [CrossRef] [Green Version]
- Fleming, N.D.; Nick, A.M.; Coleman, R.L.; Westin, S.N.; Ramirez, P.T.; Soliman, P.T.; Fellman, B.; Meyer, L.A.; Schmeler, K.M.; Lu, K.H.; et al. Laparoscopic Surgical Algorithm to Triage the Timing of Tumor Reductive Surgery in Advanced Ovarian Cancer. Obstet. Gynecol. 2018, 132, 545–554. [Google Scholar] [CrossRef]
- Torres, D.; Wang, C.; Kumar, A.; Bakkum-Gamez, J.N.; Weaver, A.L.; McGree, M.E.; Konecny, G.E.; Goode, E.L.; Cliby, W.A. Factors that influence survival in high-grade serous ovarian cancer: A complex relationship between molecular subtype, disease dissemination, and operability. Gynecol. Oncol. 2018, 150, 227–232. [Google Scholar] [CrossRef] [Green Version]
- Riester, M.; Wei, W.; Waldron, L.; Culhane, A.C.; Trippa, L.; Oliva, E.; Kim, S.H.; Michor, F.; Huttenhower, C.; Parmigiani, G.; et al. Risk prediction for late-stage ovarian cancer by meta-analysis of 1525 patient samples. J. Natl. Cancer Inst. 2014, 106, dju048. [Google Scholar] [CrossRef] [Green Version]
- Heitz, F.; Kommoss, S.; Tourani, R.; Grandelis, A.; Uppendahl, L.; Aliferis, C.; Burges, A.; Wang, C.; Canzler, U.; Wang, J.; et al. Dilution of Molecular-Pathologic Gene Signatures by Medically Associated Factors Might Prevent Prediction of Resection Status After Debulking Surgery in Patients With Advanced Ovarian Cancer. Clin. Cancer Res. 2020, 26, 213–219. [Google Scholar] [CrossRef] [Green Version]
- Charlson, M.E.; Pompei, P.; Ales, K.L.; MacKenzie, C.R. A new method of classifying prognostic comorbidity in longitudinal studies: Development and validation. J. Chronic Dis. 1987, 40, 373–383. [Google Scholar] [CrossRef]
- Aletti, G.D.; Dowdy, S.C.; Podratz, K.C.; Cliby, W.A. Relationship among surgical complexity, short-term morbidity, and overall survival in primary surgery for advanced ovarian cancer. Am. J. Obstet. Gynecol. 2007, 197, 676-e1–676-e7. [Google Scholar] [CrossRef]
- Gonzalez Bosquet, J.; Devor, E.J.; Newtson, A.M.; Smith, B.J.; Bender, D.P.; Goodheart, M.J.; McDonald, M.E.; Braun, T.A.; Thiel, K.W.; Leslie, K.K. Creation and validation of models to predict response to primary treatment in serous ovarian cancer. Sci. Rep. 2021, 11, 5957. [Google Scholar] [CrossRef]
- Miller, M.D.; Salinas, E.A.; Newtson, A.M.; Sharma, D.; Keeney, M.E.; Warrier, A.; Smith, B.J.; Bender, D.P.; Goodheart, M.J.; Thiel, K.W.; et al. An integrated prediction model of recurrence in endometrial endometrioid cancers. Cancer Manag. Res. 2019, 11, 5301–5315. [Google Scholar] [CrossRef] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, S.; Reyes, A.; Huber, W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012, 22, 2008–2017. [Google Scholar] [CrossRef] [PubMed]
- Flensburg, C.; Sargeant, T.; Oshlack, A.; Majewski, I.J. SuperFreq: Integrated mutation detection and clonal tracking in cancer. PLoS Comput. Biol. 2020, 16, e1007603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardillo, N.; Russo, D.; Newtson, A.; Reyes, H.; Lyons, Y.; Devor, E.; Bender, D.; Goodheart, M.J.; Gonzalez-Bosquet, J. Identification of Novel lncRNAs in Ovarian Cancer and Their Impact on Overall Survival. Int. J. Mol. Sci. 2021, 22, 1079. [Google Scholar] [CrossRef]
- Sun, Z.; Nair, A.; Chen, X.; Prodduturi, N.; Wang, J.; Kocher, J.P. Author Correction: UClncR: Ultrafast and comprehensive long non-coding RNA detection from RNA-seq. Sci. Rep. 2018, 8, 5124. [Google Scholar] [CrossRef] [Green Version]
- Haas, B.J.; Dobin, A.; Stransky, N.; Li, B.; Yang, X.; Tickle, T.; Bankapur, A.; Ganote, C.; Doak, T.G.; Pochet, N.; et al. STAR-Fusion: Fast and Accurate Fusion Transcript Detection from RNA-Seq. Biorxiv 2017, 10.1101, 120295. [Google Scholar] [CrossRef] [Green Version]
- Reyes, H.D.; Devor, E.J.; Warrier, A.; Newtson, A.M.; Mattson, J.; Wagner, V.; Duncan, G.N.; Leslie, K.K.; Gonzalez-Bosquet, J. Differential DNA methylation in high-grade serous ovarian cancer (HGSOC) is associated with tumor behavior. Sci. Rep. 2019, 9, 17996. [Google Scholar] [CrossRef]
- Aryee, M.J.; Jaffe, A.E.; Corrada-Bravo, H.; Ladd-Acosta, C.; Feinberg, A.P.; Hansen, K.D.; Irizarry, R.A. Minfi: A flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014, 30, 1363–1369. [Google Scholar] [CrossRef] [Green Version]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Kuhn, M. Building Predictive Models in R Using the caret Package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Cancer Genome Atlas Research. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef]
- Sing, T.; Sander, O.; Beerenwinkel, N.; Lengauer, T. ROCR: Visualizing classifier performance in R. Bioinformatics 2005, 21, 3940–3941. [Google Scholar] [CrossRef]
- TensorFlow Developers. TensorFlow. Available online: https://doi.org/10.5281/zenodo.5949169 (accessed on 17 July 2022).
- Mohammad, N.; Muad, A.M.; Ahmad, R.; Yusof, M. Accuracy of advanced deep learning with tensorflow and keras for classifying teeth developmental stages in digital panoramic imaging. BMC Med. Imaging 2022, 22, 66. [Google Scholar] [CrossRef]
- Abadi, A.A.M.; Barham, P.; Brevdo, E. TensorFlow: Large-Scale Machine Learning on Heterogeneous Systems. 2015. Available online: https://www.tensorflow.org/tutorials (accessed on 5 September 2021).
- Wong, D.H.; Mardock, A.L.; Manrriquez, E.N.; Lai, T.S.; Sanaiha, Y.; Sinno, A.K.; Benharash, P.; Cohen, J.G. Trends in extent of surgical cytoreduction for patients with ovarian cancer. PLoS ONE 2021, 16, e0260255. [Google Scholar] [CrossRef]
- Lyons, Y.A.; Reyes, H.D.; McDonald, M.E.; Newtson, A.; Devor, E.; Bender, D.P.; Goodheart, M.J.; Gonzalez Bosquet, J. Interval debulking surgery is not worth the wait: A National Cancer Database study comparing primary cytoreductive surgery versus neoadjuvant chemotherapy. Int. J. Gynecol. Cancer 2020, 30, 845–852. [Google Scholar] [CrossRef]
- Fagotti, A.; Ferrandina, G.; Fanfani, F.; Ercoli, A.; Lorusso, D.; Rossi, M.; Scambia, G. A laparoscopy-based score to predict surgical outcome in patients with advanced ovarian carcinoma: A pilot study. Ann. Surg. Oncol. 2006, 13, 1156–1161. [Google Scholar] [CrossRef]
- Veeraraghavan, H.; Vargas, H.A.; Jimenez-Sanchez, A.; Micco, M.; Mema, E.; Lakhman, Y.; Crispin-Ortuzar, M.; Huang, E.P.; Levine, D.A.; Grisham, R.N.; et al. Integrated Multi-Tumor Radio-Genomic Marker of Outcomes in Patients with High Serous Ovarian Carcinoma. Cancers 2020, 12, 3403. [Google Scholar] [CrossRef]
- Boehm, K.M.; Aherne, E.A.; Ellenson, L.; Nikolovski, I.; Alghamdi, M.; Vazquez-Garcia, I.; Zamarin, D.; Roche, K.L.; Liu, Y.; Patel, D.; et al. Multimodal data integration using machine learning improves risk stratification of high-grade serous ovarian cancer. Nat. Cancer 2022, 3, 723–733. [Google Scholar] [CrossRef]
- Lu, H.; Arshad, M.; Thornton, A.; Avesani, G.; Cunnea, P.; Curry, E.; Kanavati, F.; Liang, J.; Nixon, K.; Williams, S.T.; et al. A mathematical-descriptor of tumor-mesoscopic-structure from computed-tomography images annotates prognostic- and molecular-phenotypes of epithelial ovarian cancer. Nat. Commun. 2019, 10, 764. [Google Scholar] [CrossRef]
- Narasimhulu, D.M.; Kumar, A.; Weaver, A.L.; Langstraat, C.L.; Cliby, W.A. Less guessing, more evidence in identifying patients least fit for cytoreductive surgery in advanced ovarian cancer: A triage algorithm to individualize surgical management. Gynecol. Oncol. 2020, 157, 572–577. [Google Scholar] [CrossRef]
- Cardillo, N.; Devor, E.; Calma, C.; Pedra Nobre, S.; Gabrilovich, S.; Bender, D.P.; Goodheart, M.; Gonzalez-Bosquet, J. Investigating the effect of optimal cytoreduction in the context of platinum sensitivity in high-grade serous ovarian cancer. Acta Obstet. Gynecol. Scand. 2022. [Google Scholar] [CrossRef]
- Miller, M.D.; Devor, E.J.; Salinas, E.A.; Newtson, A.M.; Goodheart, M.J.; Leslie, K.K.; Gonzalez-Bosquet, J. Population Substructure Has Implications in Validating Next-Generation Cancer Genomics Studies with TCGA. Int. J. Mol. Sci. 2019, 20, 1192. [Google Scholar] [CrossRef] [Green Version]
- Simon, R. Roadmap for developing and validating therapeutically relevant genomic classifiers. J. Clin. Oncol. 2005, 23, 7332–7341. [Google Scholar] [CrossRef] [Green Version]
- Stelloo, E.; Nout, R.A.; Naves, L.C.; Ter Haar, N.T.; Creutzberg, C.L.; Smit, V.T.; Bosse, T. High concordance of molecular tumor alterations between pre-operative curettage and hysterectomy specimens in patients with endometrial carcinoma. Gynecol. Oncol. 2014, 133, 197–204. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [Green Version]
- National Cancer Institute. Genomic Data Commons Data Portal. 2019. Available online: https://portal.gdc.cancer.gov/ (accessed on 17 July 2022).
- Bioconductor. Open Source Software for Bioinformatics. 2021. Available online: http://bioconductor.org/ (accessed on 17 July 2022).

| All UI HGSC Population | Samples with RNA-seq and DNA Methylation | ||||||
|---|---|---|---|---|---|---|---|
| Optimal (n = 273) | Suboptimal (n = 132) | p-Value | Optimal (n = 52) | Suboptimal (n = 31) | p-Value | ||
| Preoperative characteristics | Age (mean) | 61 | 61 | 0.742 | 62 | 58 | 0.204 |
| BMI (mean) | 28.3 | 28.1 | 0.772 | 25.5 | 27.8 | 0.149 | |
| Charlson Morbidity Index ** | 0.604 | ||||||
| Low (1–3) | 44 | 19 | Ref | 8 | 6 | Ref | |
| Medium (4–6) | 182 | 87 | 0.738 | 31 | 16 | 0.548 | |
| High (>6) | 37 | 19 | 0.660 | 5 | 3 | 0.806 | |
| Preop CA-125 (mean) | 1613 | 2067 | 0.224 | 2695 | 3030 | 0.808 | |
| Disease in Upper abdomen (Other than Omentum) by Imaging | 203 | 101 | 0.638 | 34 | 20 | 0.936 | |
| Large bowel | 14 | 8 | 0.698 | 1 | 1 | 0.711 | |
| Spleen | 5 | 0 | 0.982 | 0 | 0 | - | |
| Mesenteric LN | 15 | 7 | 0.936 | 2 | 1 | 0.884 | |
| Porta/Hepatis | 21 | 17 | 0.097 | 2 | 1 | 0.884 | |
| Ascites (upper abdomen) | 115 | 56 | 0.954 | 20 | 8 | 0.241 | |
| Other | 90 | 52 | 0.204 | 12 | 11 | 0.225 | |
| Disease in the Chest by Imaging | 25 | 29 | <0.001 * | 1 | 4 | 0.077 | |
| Tumor | 7 | 19 | 0.026 * | 1 | 2 | 0.313 | |
| Pleural effusion | 25 | 27 | 0.002* | 1 | 2 | 0.313 | |
| Neoadjuvant chemotherapy | 22 | 9 | 0.700 | 10 | 4 | 0.450 | |
| Operative characteristics | FIGO Stage | 0.001 * | 0.225 | ||||
| III | 224 | 89 | 40 | 20 | |||
| IV | 49 | 43 | 12 | 11 | |||
| Hysterectomy | 210 (77%) | 79 (60%) | <0.001 * | 38 (73%) | 18 (58%) | 0.161 | |
| Surgery to remove cervix | 188 (69%) | 63 (48%) | <0.001 * | 32 (62%) | 13 (42%) | 0.085 | |
| Adnexectomy | 263 (96%) | 125 (95%) | 0.443 | 51 (98%) | 29 (94%) | 0.313 | |
| Omentectomy | 260 (95%) | 118 (89%) | 0.031 * | 49 (94%) | 29 (94%) | 0.894 | |
| Surgery large bowel | 96 (35%) | 35 (27%) | 0.082 | 16 (31%) | 9 (29%) | 0.868 | |
| Surgery small bowel | 5 (2%) | 3 (2%) | 0.765 | 0 (0%) | 1 (3%) | 0.991 | |
| Splenectomy | 4 (1%) | 0 (0%) | 0.984 | 1 (2%) | 0 (0%) | 0.992 | |
| Diaphragmatic stripping | 8 (3%) | 0 (0%) | 0.977 | 5 (10%) | 0 (0%) | 0.992 | |
| Para-aortic lymphadenectomy | 56 (10%) | 13 (21%) | 0.009 * | 3 (6%) | 3 (10%) | 0.510 | |
| Residual disease | 0.981 | 0.988 | |||||
| Microscopic | 71 | 0 | 11 | 0 | |||
| Macroscopic | 202 | 132 | 41 | 31 | |||
| Surgical complexity index # | |||||||
| Low | 126 | 91 | Ref | 30 | 20 | Ref | |
| Intermediate | 140 | 41 | <0.001 * | 20 | 11 | 0.685 | |
| High | 7 | 0 | 0.978 | 2 | 0 | 0.992 | |
| Outcomes | 30-day mortality | 4 (1.5%) | 3 (2.3%) | 0.558 | 0 (0%) | 0 (0%) | N/A |
| 90-day mortality | 8 (2.9%) | 9 (6.8%) | 0.071 | 0 (0%) | 1 (1%) | 0.991 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cardillo, N.; Devor, E.J.; Pedra Nobre, S.; Newtson, A.; Leslie, K.; Bender, D.P.; Smith, B.J.; Goodheart, M.J.; Gonzalez-Bosquet, J. Integrated Clinical and Genomic Models to Predict Optimal Cytoreduction in High-Grade Serous Ovarian Cancer. Cancers 2022, 14, 3554. https://doi.org/10.3390/cancers14143554
Cardillo N, Devor EJ, Pedra Nobre S, Newtson A, Leslie K, Bender DP, Smith BJ, Goodheart MJ, Gonzalez-Bosquet J. Integrated Clinical and Genomic Models to Predict Optimal Cytoreduction in High-Grade Serous Ovarian Cancer. Cancers. 2022; 14(14):3554. https://doi.org/10.3390/cancers14143554
Chicago/Turabian StyleCardillo, Nicholas, Eric J. Devor, Silvana Pedra Nobre, Andreea Newtson, Kimberly Leslie, David P. Bender, Brian J. Smith, Michael J. Goodheart, and Jesus Gonzalez-Bosquet. 2022. "Integrated Clinical and Genomic Models to Predict Optimal Cytoreduction in High-Grade Serous Ovarian Cancer" Cancers 14, no. 14: 3554. https://doi.org/10.3390/cancers14143554
APA StyleCardillo, N., Devor, E. J., Pedra Nobre, S., Newtson, A., Leslie, K., Bender, D. P., Smith, B. J., Goodheart, M. J., & Gonzalez-Bosquet, J. (2022). Integrated Clinical and Genomic Models to Predict Optimal Cytoreduction in High-Grade Serous Ovarian Cancer. Cancers, 14(14), 3554. https://doi.org/10.3390/cancers14143554







