ZEB2/TWIST1/PRMT5/NuRD Multicomplex Contributes to the Epigenetic Regulation of EMT and Metastasis in Colorectal Carcinoma
Abstract
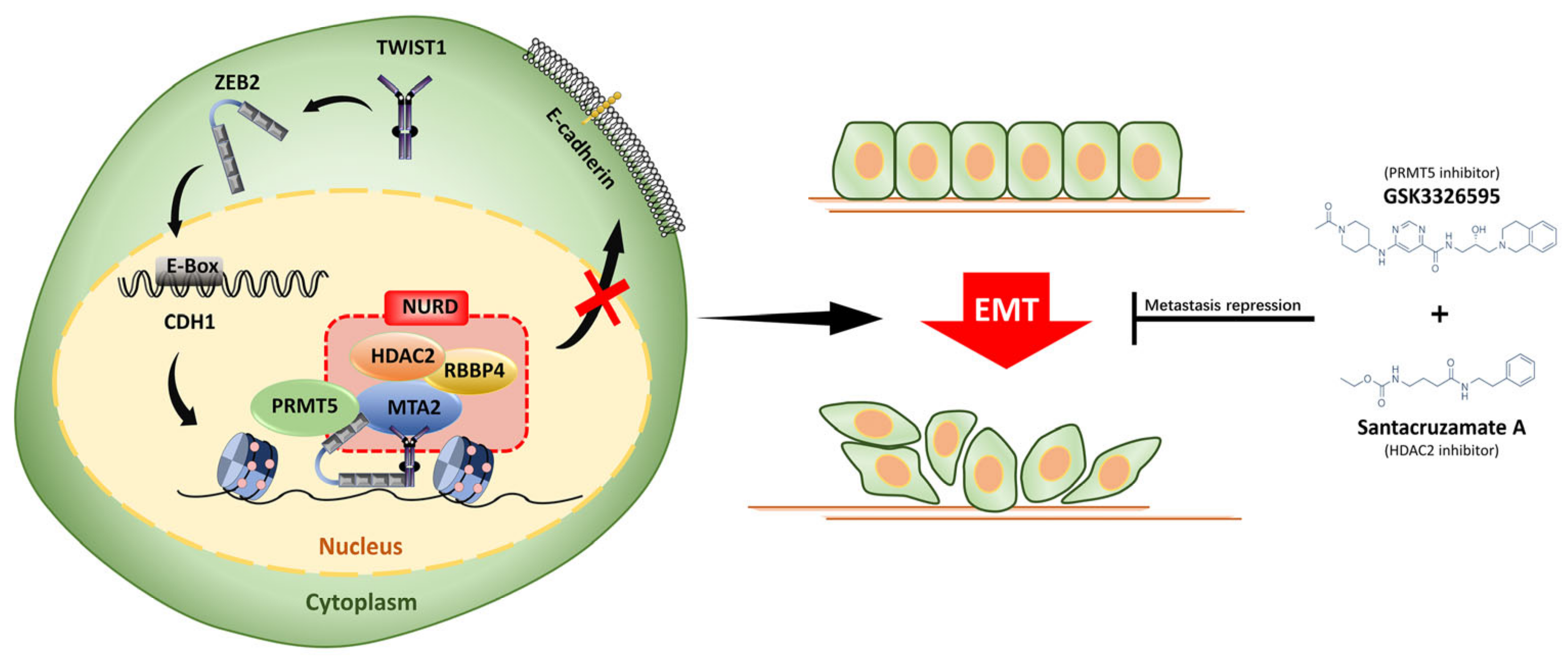
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue Samples
2.2. Database of Cancer Patients
2.3. Antibodies and Reagents
2.4. Cell Cultures and Stable Cell Lines
2.5. Western Blotting and Immunoprecipitation
2.6. Mass Spectrometry
2.7. Chromatin Immunoprecipitation Assay
2.8. Quantitative Real-Time PCR
2.9. Luciferase Assay
2.10. Cell Migration and Invasion
2.11. Liver Metastasis Model
2.12. Statistical Analysis
3. Results
3.1. ZEB2 and TWIST1 Are Significantly Upregulated during TGF-β-Induced EMT in CRC Cells and Patient Tissues
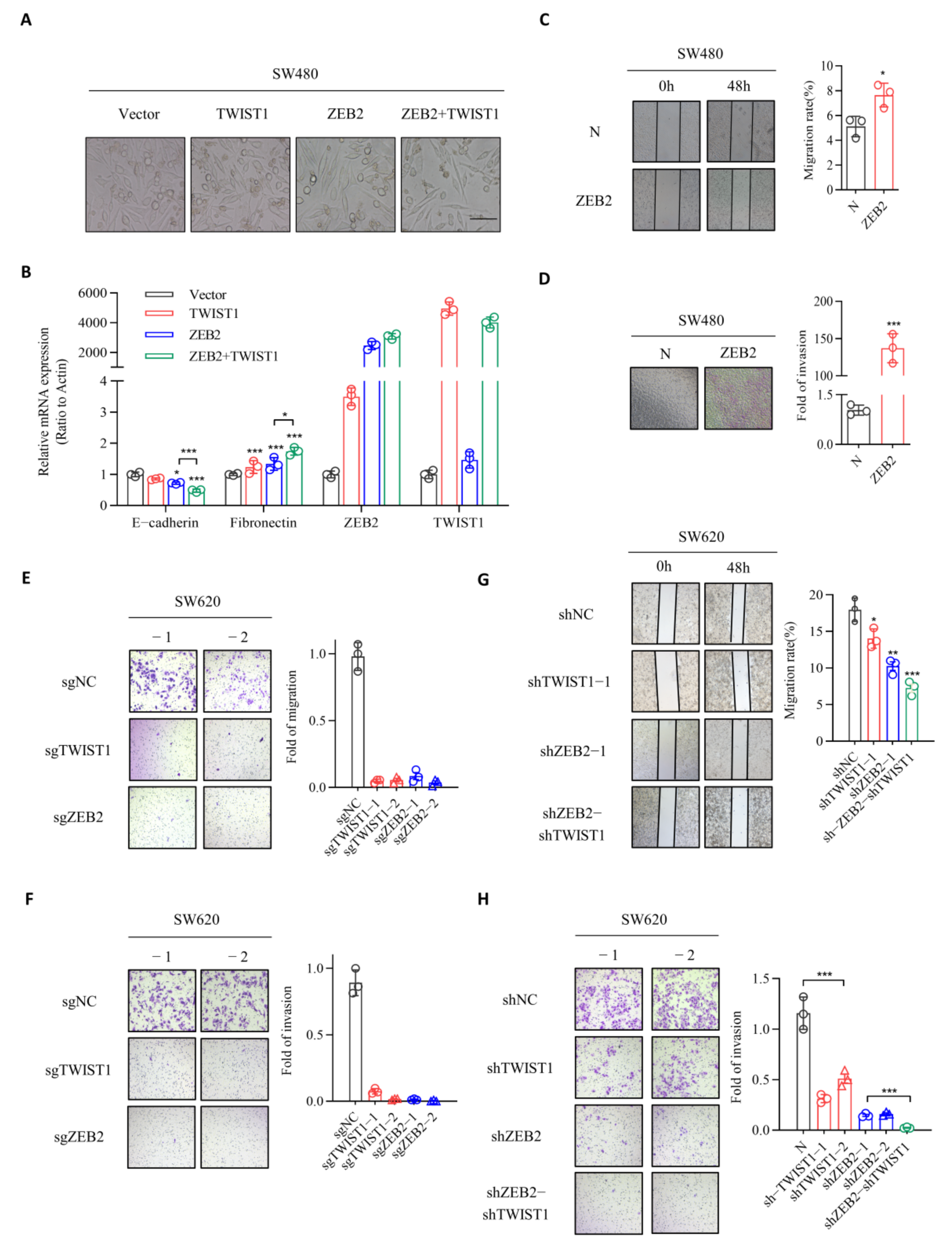
3.2. ZEB2 and TWIST1 Contribute to the EMT, Migration, and Invasion Processes in CRC
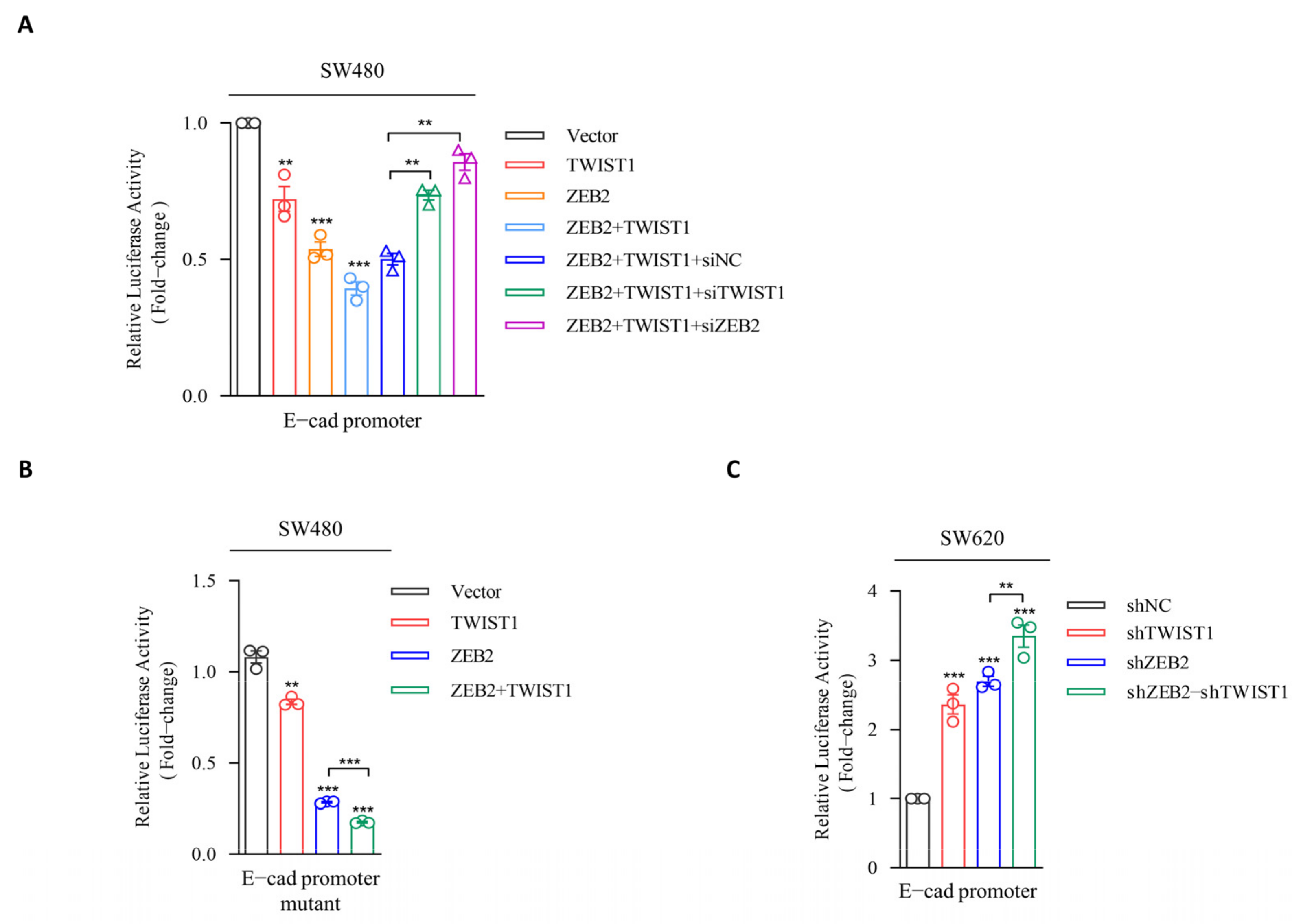
3.3. ZEB2 Recruits TWIST1 to the Promoter of E-cadherin and Synergistically Represses the Transcription
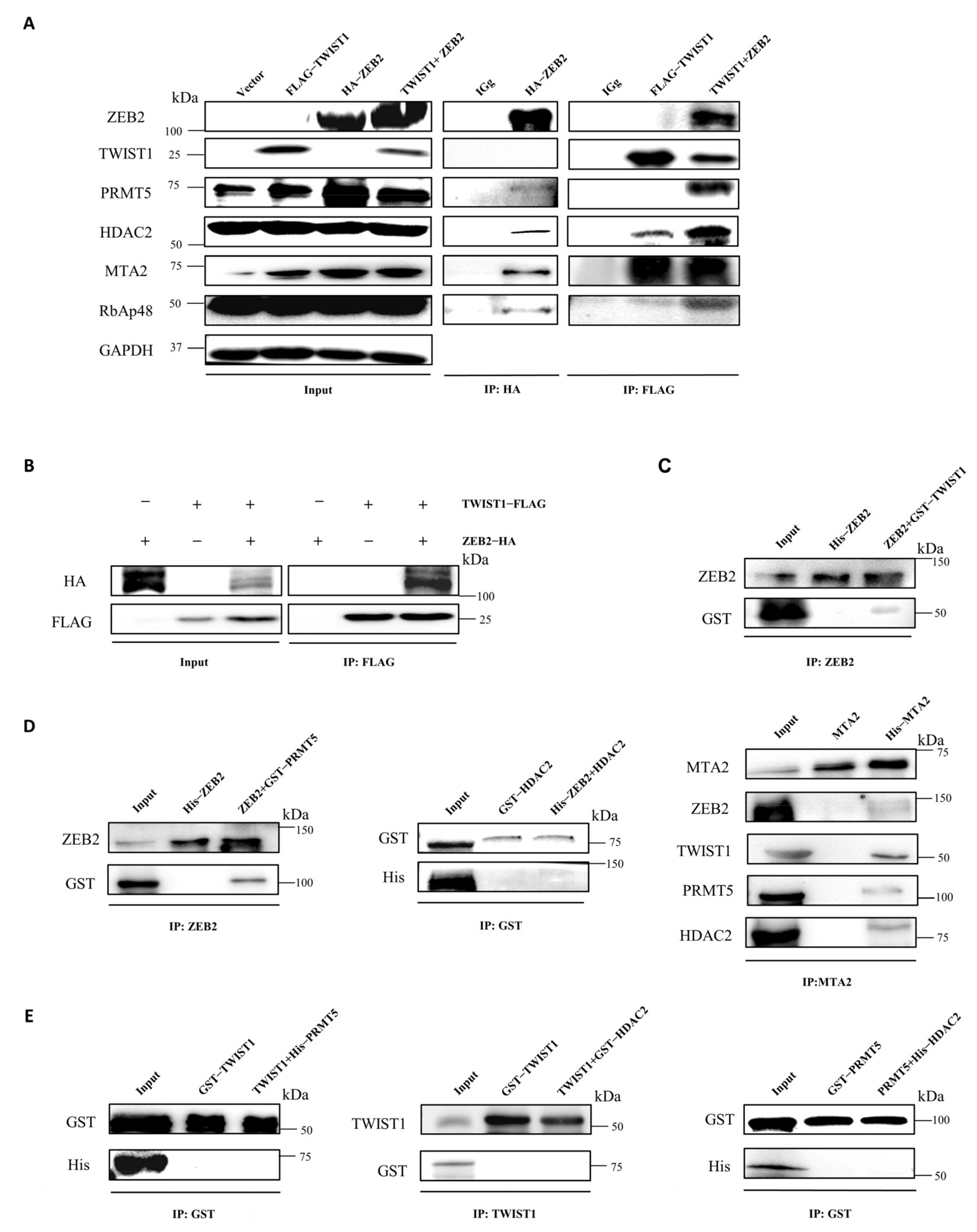
3.4. ZEB2 Interacts with TWIST1, PRMT5, and NuRD Complex to Form a Functional Multicomplex
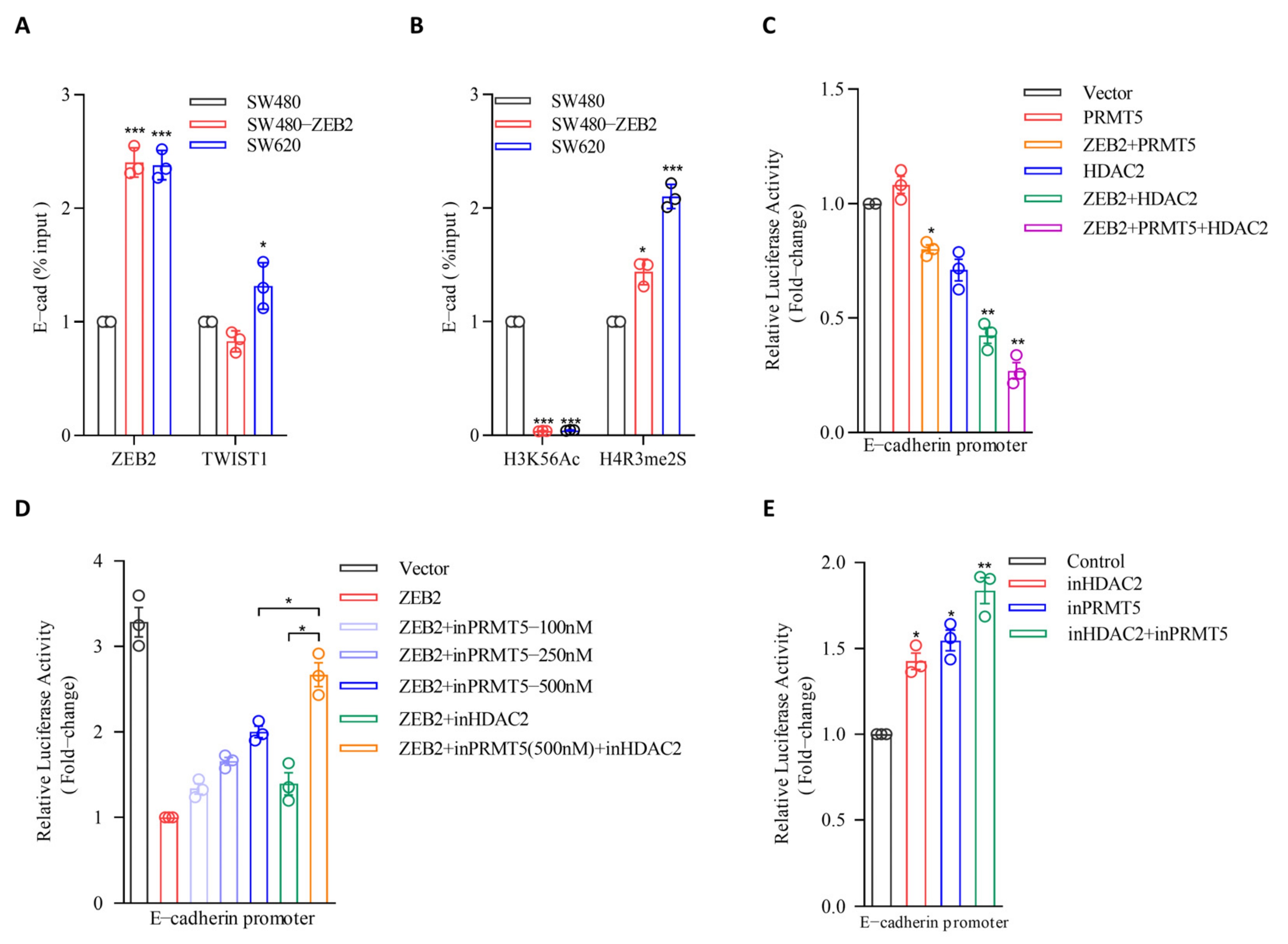
3.5. PRMT5 and HDAC2 Are Responsible for the ZEB2-Mediated E-cadherin Repression in CRC
3.6. PRMT5 and HDAC2 Mediate the Migration-, Invasion-, and Metastasis-Promoting Properties of ZEB2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CRC | colorectal carcinoma |
| EMT | epithelial to mesenchymal transition |
| NuRD | nucleosome remodeling and deacetylase |
| PRMT5 | Protein arginine methyltransferase 5 |
| HDAC | Histone deacetylases |
| Co-IP | co-immunoprecipitation |
| inHDAC2 | Santacruzamate A |
| inPRMT5 | GSK3326595 |
References
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683–691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nanduri, L.K.; Hissa, B.; Weitz, J.; Scholch, S.; Bork, U. The prognostic role of circulating tumor cells in colorectal cancer. Expert Rev. Anticancer Ther. 2019, 19, 1077–1088. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Xu, E.; Liu, H.; Wan, L.; Lai, M. Epithelial-mesenchymal transition in colorectal cancer metastasis: A system review. Pathol. Res. Pract. 2015, 211, 557–569. [Google Scholar] [CrossRef] [PubMed]
- Nieto, M.A.; Huang, R.Y.; Jackson, R.A.; Thiery, J.P. EMT: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef] [Green Version]
- Pena, C.; Garcia, J.M.; Silva, J.; Garcia, V.; Rodriguez, R.; Alonso, I.; Millan, I.; Salas, C.; de Herreros, A.G.; Munoz, A.; et al. E-cadherin and vitamin D receptor regulation by SNAIL and ZEB1 in colon cancer: Clinicopathological correlations. Hum. Mol. Genet. 2005, 14, 3361–3370. [Google Scholar] [CrossRef]
- He, X.; Chen, Z.; Jia, M.; Zhao, X. Downregulated E-cadherin expression indicates worse prognosis in Asian patients with colorectal cancer: Evidence from meta-analysis. PLoS ONE 2013, 8, e70858. [Google Scholar] [CrossRef]
- Toiyama, Y.; Yasuda, H.; Saigusa, S.; Tanaka, K.; Inoue, Y.; Goel, A.; Kusunoki, M. Increased expression of Slug and Vimentin as novel predictive biomarkers for lymph node metastasis and poor prognosis in colorectal cancer. Carcinogenesis 2013, 34, 2548–2557. [Google Scholar] [CrossRef] [Green Version]
- Peinado, H.; Olmeda, D.; Cano, A. Snail, Zeb and bHLH factors in tumour progression: An alliance against the epithelial phenotype? Nat. Rev. Cancer 2007, 7, 415–428. [Google Scholar] [CrossRef]
- Gomez, I.; Pena, C.; Herrera, M.; Munoz, C.; Larriba, M.J.; Garcia, V.; Dominguez, G.; Silva, J.; Rodriguez, R.; Garcia, D.H.A.; et al. TWIST1 is expressed in colorectal carcinomas and predicts patient survival. PLoS ONE 2011, 6, e18023. [Google Scholar] [CrossRef] [Green Version]
- Li, M.Z.; Wang, J.J.; Yang, S.B.; Li, W.F.; Xiao, L.B.; He, Y.L.; Song, X.M. ZEB2 promotes tumor metastasis and correlates with poor prognosis of human colorectal cancer. Am. J. Transl. Res. 2017, 9, 2838–2851. [Google Scholar]
- Kong, Y.H.; Syed, Z.S.; Lau, S.H.; Ramanathan, A.; Kallarakkal, T.G.; Vincent-Chong, V.K.; Wan, M.W.; Abraham, M.T.; Abdul, R.Z.; Zain, R.B.; et al. Co-Expression of TWIST1 and ZEB2 in oral squamous cell carcinoma is associated with poor survival. PLoS ONE 2015, 10, e134045. [Google Scholar] [CrossRef] [Green Version]
- Larriba, M.J.; Martin-Villar, E.; Garcia, J.M.; Pereira, F.; Pena, C.; de Herreros, A.G.; Bonilla, F.; Munoz, A. Snail2 cooperates with Snail1 in the repression of vitamin D receptor in colon cancer. Carcinogenesis 2009, 30, 1459–1468. [Google Scholar] [CrossRef] [Green Version]
- Serrano-Gomez, S.J.; Maziveyi, M.; Alahari, S.K. Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Mol. Cancer 2016, 15, 18. [Google Scholar] [CrossRef] [Green Version]
- Dong, C.; Wu, Y.; Yao, J.; Wang, Y.; Yu, Y.; Rychahou, P.G.; Evers, B.M.; Zhou, B.P. G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J. Clin. Investig. 2012, 122, 1469–1486. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Zheng, Y.; Dai, M.; Wang, X.; Wu, J.; Yu, B.; Zhang, H.; Cui, Y.; Kong, W.; Wu, H.; et al. G9a and histone deacetylases are crucial for Snail2-mediated E-cadherin repression and metastasis in hepatocellular carcinoma. Cancer Sci. 2019, 110, 3442–3452. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Dai, M.; Zheng, Y.; Wu, J.; Yu, B.; Zhang, H.; Kong, W.; Wu, H.; Yu, X. Epigenetic suppression of E-cadherin expression by Snail2 during the metastasis of colorectal cancer. Clin. Epigenet. 2018, 10, 154. [Google Scholar] [CrossRef]
- Aggarwal, P.; Vaites, L.P.; Kim, J.K.; Mellert, H.; Gurung, B.; Nakagawa, H.; Herlyn, M.; Hua, X.; Rustgi, A.K.; Mcmahon, S.B.; et al. Nuclear cyclin D1/CDK4 kinase regulates CUL4 expression and triggers neoplastic growth via activation of the PRMT5 methyltransferase. Cancer Cell 2010, 18, 329–340. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Pal, S.; Sif, S. Protein arginine methyltransferase 5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells. Mol. Cell. Biol. 2008, 28, 6262–6277. [Google Scholar] [CrossRef] [Green Version]
- Hu, D.; Gur, M.; Zhou, Z.; Gamper, A.; Hung, M.C.; Fujita, N.; Lan, L.; Bahar, I.; Wan, Y. Interplay between arginine methylation and ubiquitylation regulates KLF4-mediated genome stability and carcinogenesis. Nat. Commun. 2015, 6, 8419. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.; Gao, J.; Yang, Y.; Qiu, R.; Zheng, Y.; Huang, W.; Zeng, Y.; Hou, Y.; Wang, S.; Leng, S.; et al. PHD finger protein 1 (PHF1) is a novel reader for histone H4R3 symmetric dimethylation and coordinates with PRMT5-WDR77/CRL4B complex to promote tumorigenesis. Nucleic Acids Res. 2018, 46, 6608–6626. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Liu, R.; Feng, D.; Huang, W.; Huo, M.; Zhang, J.; Leng, S.; Yang, Y.; Yang, T.; Yin, X.; et al. Snail/PRMT5/NuRD complex contributes to DNA hypermethylation in cervical cancer by TET1 inhibition. Cell Death Differ. 2021, 28, 2818–2836. [Google Scholar] [CrossRef] [PubMed]
- Peinado, H.; Ballestar, E.; Esteller, M.; Cano, A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol. Cell. Biol. 2004, 24, 306–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aghdassi, A.; Sendler, M.; Guenther, A.; Mayerle, J.; Behn, C.O.; Heidecke, C.D.; Friess, H.; Buchler, M.; Evert, M.; Lerch, M.M.; et al. Recruitment of histone deacetylases HDAC1 and HDAC2 by the transcriptional repressor ZEB1 downregulates E-cadherin expression in pancreatic cancer. Gut 2012, 61, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Qin, L.; He, T.; Qin, J.; Hong, J.; Wong, J.; Liao, L.; Xu, J. The TWIST/Mi2/NuRD protein complex and its essential role in cancer metastasis. Cell Res. 2011, 21, 275–289. [Google Scholar] [CrossRef] [Green Version]
- Boyer, L.A.; Plath, K.; Zeitlinger, J.; Brambrink, T.; Medeiros, L.A.; Lee, T.I.; Levine, S.S.; Wernig, M.; Tajonar, A.; Ray, M.K.; et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 2006, 441, 349–353. [Google Scholar] [CrossRef]
- Hossan, T.; Nagarajan, S.; Baumgart, S.J.; Xie, W.; Magallanes, R.T.; Hernandez, C.; Chiaroni, P.M.; Indenbirken, D.; Spitzner, M.; Thomas-Chollier, M.; et al. Histone chaperone SSRP1 is essential for wnt signaling pathway activity during osteoblast differentiation. Stem Cells 2016, 34, 1369–1376. [Google Scholar] [CrossRef] [Green Version]
- Shimada, H.; Abe, S.; Kohno, T.; Satohisa, S.; Konno, T.; Takahashi, S.; Hatakeyama, T.; Arimoto, C.; Kakuki, T.; Kaneko, Y.; et al. Loss of tricellular tight junction protein LSR promotes cell invasion and migration via upregulation of TEAD1/AREG in human endometrial cancer. Sci. Rep. 2017, 7, 37049. [Google Scholar] [CrossRef] [Green Version]
- Lu, D.; Yao, Q.; Zhan, C.; Le-Meng, Z.; Liu, H.; Cai, Y.; Tu, C.; Li, X.; Zou, Y.; Zhang, S. MicroRNA-146a promote cell migration and invasion in human colorectal cancer via carboxypeptidase M/src-FAK pathway. Oncotarget 2017, 8, 22674–22684. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, A.; Pattnaik, N.; Mohanty, J.A.; Dixit, M. Increased FSHD region gene1 expression reduces in vitro cell migration, invasion, and angiogenesis, ex vivo supported by reduced expression in tumors. Biosci. Rep. 2017, 37, BSR20171062. [Google Scholar] [CrossRef] [Green Version]
- Kahlert, C.; Lahes, S.; Radhakrishnan, P.; Dutta, S.; Mogler, C.; Herpel, E.; Brand, K.; Steinert, G.; Schneider, M.; Mollenhauer, M.; et al. Overexpression of ZEB2 at the invasion front of colorectal cancer is an independent prognostic marker and regulates tumor invasion in vitro. Clin. Cancer Res. 2011, 17, 7654–7663. [Google Scholar] [CrossRef] [Green Version]
- Shioiri, M.; Shida, T.; Koda, K.; Oda, K.; Seike, K.; Nishimura, M.; Takano, S.; Miyazaki, M. Slug expression is an independent prognostic parameter for poor survival in colorectal carcinoma patients. Br. J. Cancer 2006, 94, 1816–1822. [Google Scholar] [CrossRef] [Green Version]
- Kroepil, F.; Fluegen, G.; Vallbohmer, D.; Baldus, S.E.; Dizdar, L.; Raffel, A.M.; Hafner, D.; Stoecklein, N.H.; Knoefel, W.T. Snail1 expression in colorectal cancer and its correlation with clinical and pathological parameters. BMC Cancer 2013, 13, 145. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Bedford, M.T. Protein arginine methyltransferases and cancer. Nat. Rev. Cancer 2013, 13, 37–50. [Google Scholar] [CrossRef]
- Powers, M.A.; Fay, M.M.; Factor, R.E.; Welm, A.L.; Ullman, K.S. Protein arginine methyltransferase 5 accelerates tumor growth by arginine methylation of the tumor suppressor programmed cell death 4. Cancer Res. 2011, 71, 5579–5587. [Google Scholar] [CrossRef] [Green Version]
- Bao, X.; Zhao, S.; Liu, T.; Liu, Y.; Liu, Y.; Yang, X. Overexpression of PRMT5 promotes tumor cell growth and is associated with poor disease prognosis in epithelial ovarian cancer. J. Histochem. Cytochem. 2013, 61, 206–217. [Google Scholar] [CrossRef] [Green Version]
- Benard, A.; Goossens-Beumer, I.J.; van Hoesel, A.Q.; Horati, H.; de Graaf, W.; Putter, H.; Zeestraten, E.C.; Liefers, G.J.; van de Velde, C.J.; Kuppen, P.J. Nuclear expression of histone deacetylases and their histone modifications predicts clinical outcome in colorectal cancer. Histopathology 2015, 66, 270–282. [Google Scholar] [CrossRef]
- Qi, Z.P.; Yalikong, A.; Zhang, J.W.; Cai, S.L.; Li, B.; Di, S.; Lv, Z.T.; Xu, E.P.; Zhong, Y.S.; Zhou, P.H. HDAC2 promotes the EMT of colorectal cancer cells and via the modular scaffold function of ENSG00000274093.1. J. Cell. Mol. Med. 2021, 25, 1190–1197. [Google Scholar] [CrossRef]
- Lai, A.Y.; Wade, P.A. Cancer biology and NuRD: A multifaceted chromatin remodelling complex. Nat. Rev. Cancer 2011, 11, 588–596. [Google Scholar] [CrossRef]
- Manshouri, R.; Coyaud, E.; Kundu, S.T.; Peng, D.H.; Stratton, S.A.; Alton, K.; Bajaj, R.; Fradette, J.J.; Minelli, R.; Peoples, M.D.; et al. ZEB1/NuRD complex suppresses TBC1D2b to stimulate E-cadherin internalization and promote metastasis in lung cancer. Nat. Commun. 2019, 10, 5125. [Google Scholar] [CrossRef]


| Accession a | Peptide Count b | Confidence Score c | p-Valued d | q-Valued d | Protein Description |
|---|---|---|---|---|---|
| A0JP08 | 43 (43) | 1595.43 | <0.0001 | <0.0001 | Zinc finger E-box-binding homeobox 2 |
| B4DV00 | 5 (5) | 119.52 | <0.0001 | <0.0001 | Protein arginine N-methyltransferase 5 |
| Q09028 | 2 (2) | 57.93 | 0.00073 | 0.00169 | Histone-binding protein RBBP4 |
| O94776 | 1 (1) | 54.81 | 0.00040 | 0.00025 | Metastasis-associated protein MTA2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Y.; Dai, M.; Dong, Y.; Yu, H.; Liu, T.; Feng, X.; Yu, B.; Zhang, H.; Wu, J.; Kong, W.; et al. ZEB2/TWIST1/PRMT5/NuRD Multicomplex Contributes to the Epigenetic Regulation of EMT and Metastasis in Colorectal Carcinoma. Cancers 2022, 14, 3426. https://doi.org/10.3390/cancers14143426
Zheng Y, Dai M, Dong Y, Yu H, Liu T, Feng X, Yu B, Zhang H, Wu J, Kong W, et al. ZEB2/TWIST1/PRMT5/NuRD Multicomplex Contributes to the Epigenetic Regulation of EMT and Metastasis in Colorectal Carcinoma. Cancers. 2022; 14(14):3426. https://doi.org/10.3390/cancers14143426
Chicago/Turabian StyleZheng, Yayuan, Mingrui Dai, Yue Dong, Hanqiao Yu, Tianfu Liu, Xuejian Feng, Bin Yu, Haihong Zhang, Jiaxin Wu, Wei Kong, and et al. 2022. "ZEB2/TWIST1/PRMT5/NuRD Multicomplex Contributes to the Epigenetic Regulation of EMT and Metastasis in Colorectal Carcinoma" Cancers 14, no. 14: 3426. https://doi.org/10.3390/cancers14143426






