Towards Personalized Sampling in Clear Cell Renal Cell Carcinomas
Abstract
:Simple Summary
Abstract
1. Introduction
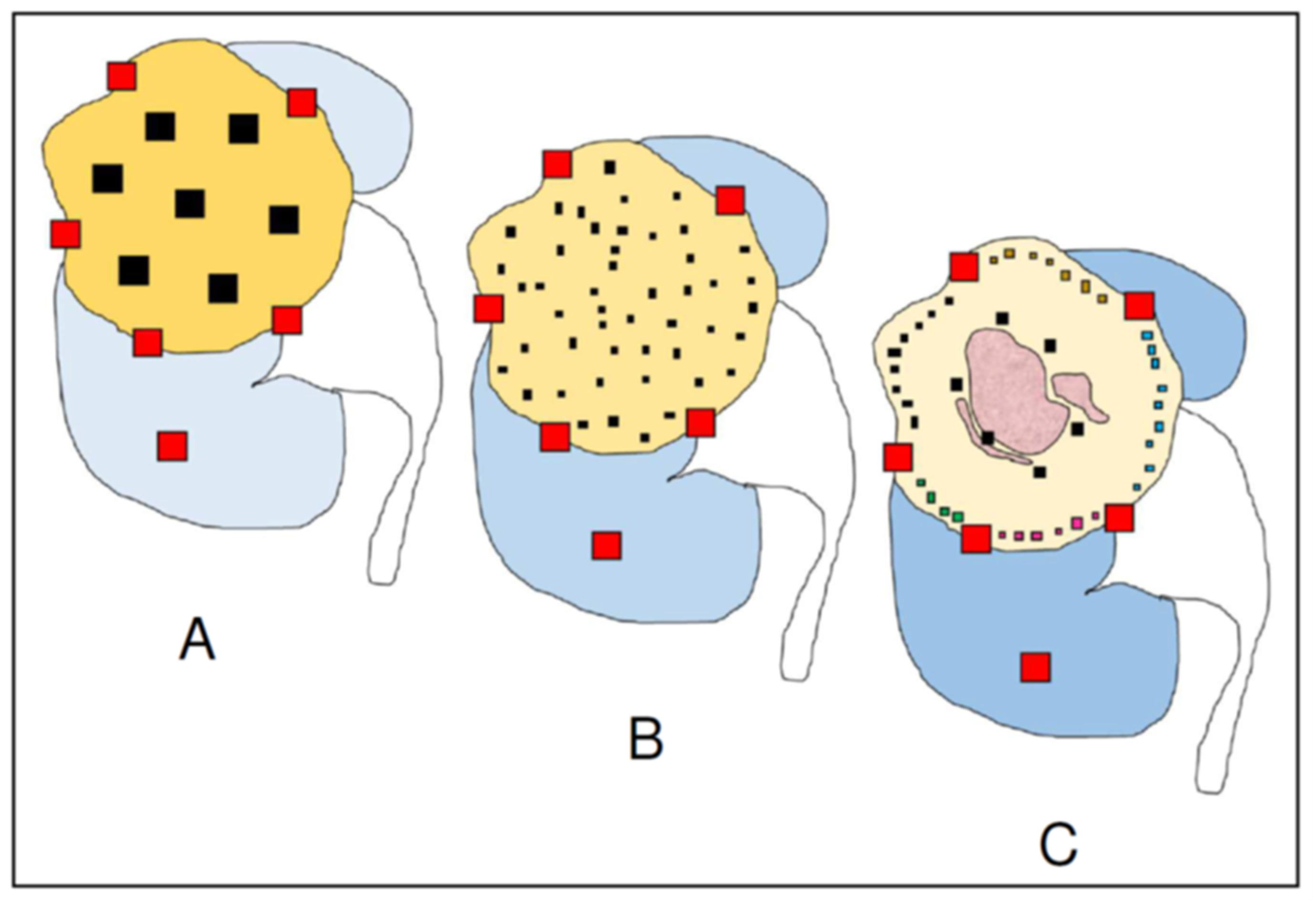
2. Classic Sampling Protocols
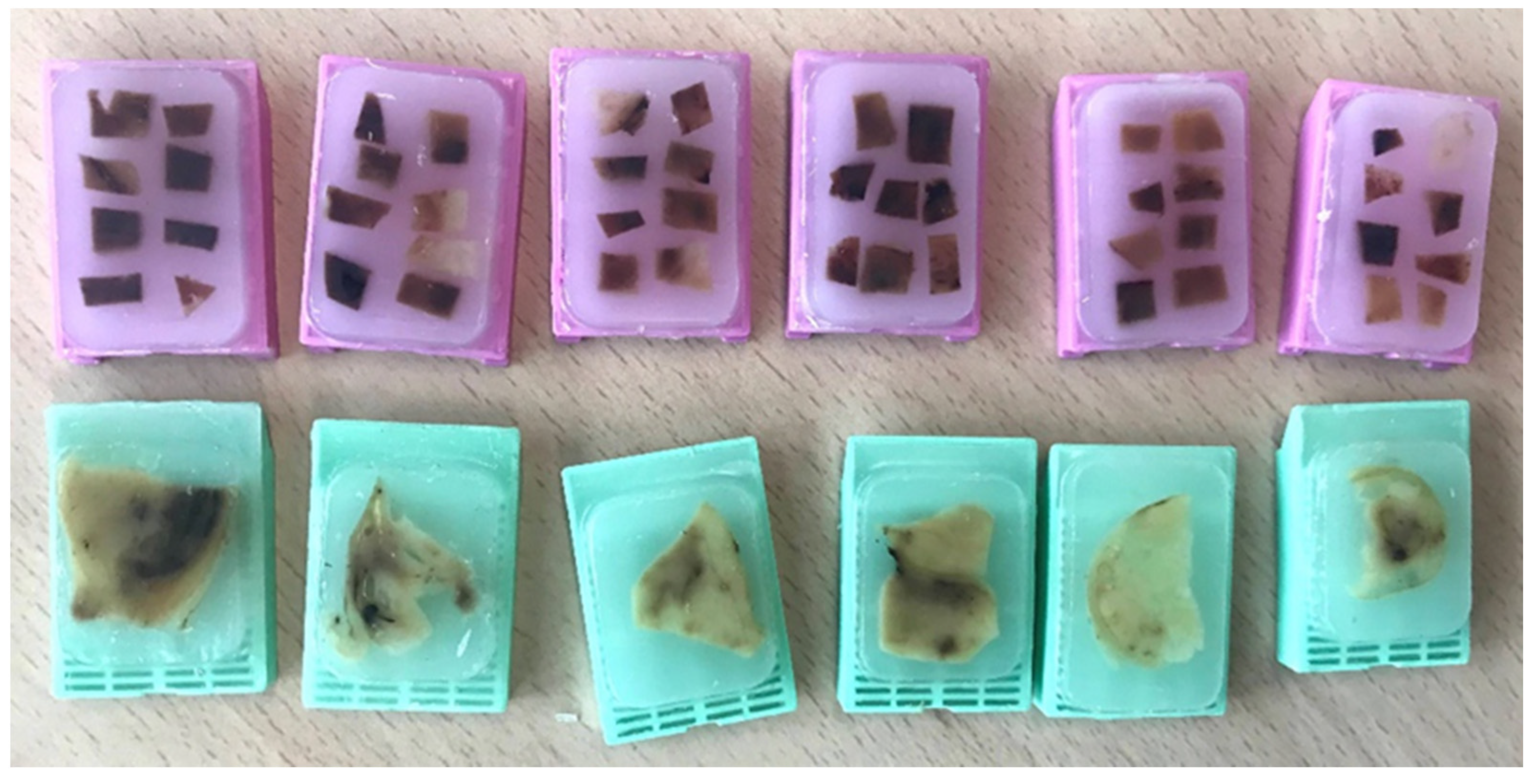
3. Multisite Tumor Sampling (MSTS)
4. Homogenization of the Residual Tumor Tissue
5. Personalized Multisite Tumor Sampling (pMSTS)
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Soultati, A.; Stares, M.; Swanton, C.; Larkin, J.; Turajlic, S. How should clinicians address intratumour heterogeneity in clear cell renal cell carcinoma? Curr. Opin. Urol. 2015, 25, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Manini, C.; López-Fernández, E.; López, J.I. Precision sampling fuels precision oncology: An evolutionary perspective. Trends Cancer 2021, 7, 978–981. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fusch, H.E.; Jemal, A. Cancer statistics, 2022. CA A Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Math, M.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef] [Green Version]
- Turajlic, S.; Xu, H.; Litchfield, K.; Rowan, A.; Horswell, S.; Chambers, T.; O’Brien, T.; Lopez, J.I.; Watkins, T.B.K.; Nicol, D.; et al. Deterministic evolutionary trajectories influence primary tumor growth: TRACERx Renal. Cell 2018, 173, 595–610. [Google Scholar] [CrossRef] [Green Version]
- Turajlic, S.; Xu, H.; Litchfield, K.; Rowan, A.; Chambers, T.; López, J.I.; Nicol, D.; O’Brien, T.; Larkin, J.; Horswell, S.; et al. Tracking cancer evolution reveals constrained routes to metastases: TRACERx Renal. Cell 2018, 173, 581–594. [Google Scholar] [CrossRef] [Green Version]
- Davis, A.; Gao, R.; Navin, N. Tumor evolution: Linear, branching, neutral or punctuated? Biochim. Biophys. Acta 2017, 1867, 151–161. [Google Scholar] [CrossRef] [Green Version]
- Nunes-Xavier, C.E.; Angulo, J.C.; Pulido, R.; López, J.I. A critical insight into the clinical translation of PD-1/PD-L1 blockade therapy in clear cell renal cell carcinoma. Curr. Urol. Rep. 2019, 20, 1–10. [Google Scholar] [CrossRef]
- Khagi, Y.; Kurzrock, R.; Patel, S.P. Next generation predictive biomarkers for immune checkpoint inhibition. Cancer Metastasis Rev. 2017, 36, 179–190. [Google Scholar] [CrossRef] [Green Version]
- Trpkov, K.; Grignon, D.J.; Bonsib, S.M.; Amin, M.B.; Billis, A.; Lopez-Beltran, A.; Samaratunga, H.; Tamboli, P.; Delahunt, B.; Egevad, L.; et al. Handling and staging or renal cell carcinoma: The International Society of Urological Pathology Consensus (ISUP) conference recommendations. Am. J. Surg. Pathol. 2013, 37, 1505–1517. [Google Scholar] [CrossRef]
- Sankin, A.; Hakimi, A.A.; Mikkilineni, N.; Ostrovnaya, I.; Silk, M.T.; Liang, Y.; Mano, R.; Chevinski, M.; Motzer, R.J.; Solomon, S.B.; et al. The impact of genetic heterogeneity on biomarker development in kidney cancer assessed by multiregional sampling. Cancer Med. 2014, 3, 1485–1492. [Google Scholar] [CrossRef] [PubMed]
- López, J.I.; Cortés, J.M. Multisite tumor sampling: A new tumor selection method to enhance intratumor heterogeneity detection. Hum. Pathol. 2017, 64, 1–6. [Google Scholar] [CrossRef] [PubMed]
- López, J.I.; Cortés, J.M. A divide-and-conquer strategy in tumor sampling enhances detection of intratumor heterogeneity in routine pathology: A modeling approach in clear cell renal cell carcinoma. F1000Reserch 2016, 5, 385. [Google Scholar] [CrossRef] [Green Version]
- Erramuzpe, A.; Cortés, J.M.; López, J.I. Multisite tumor sampling enhances the detection of intratumor heterogeneity at all different temporal stages of tumor evolution. Virchows Arch. 2018, 472, 187–194. [Google Scholar] [CrossRef]
- Yoshida, T.; Ohe, C.; Ikeda, J.; Atsumi, N.; Ohsugi, H.; Sugi, M.; Higasa, K.; Saito, R.; Tsuta, K.; Matsuda, T.; et al. Eosinophilic features in clear cell renal cell carcinoma correlate with outcomes of immune checkpoint and angiogenesis blockade. J. Immun. Ther. Cancer 2021, 9, e002922. [Google Scholar] [CrossRef]
- Guarch, R.; Cortés, J.M.; Lawrie, C.H.; López, J.I. Multi-site tumor sampling (MSTS) improves the performance of histological detection of intratumor heterogeneity in clear cell renal cell carcinoma (CCRCC). F1000Reserch 2016, 5, 2020. [Google Scholar] [CrossRef] [Green Version]
- Lakis, S.; Kotoula, V.; Koliou, G.; Efstratiou, I.; Chrisafi, S.; Papanikolaou, A.; Zebekakis, P.; Fountzilas, G. Multisite tumor sampling reveals extensive heterogeneity of tumor and host immune response in ovarian cancer. Cancer Genom. Proteom. 2020, 17, 529–541. [Google Scholar] [CrossRef]
- Meiller, C.; Montagne, F.; Hirsch, T.Z.; Caruso, S.; de Wolf, J.; Bayard, Q.; Assié, J.B.; Meunier, L.; Blum, Y.; Quetel, L.; et al. Multi-site tumor sampling highlights molecular intra-tumor heterogeneity in malignant pleural mesothelioma. Genome Med. 2021, 13, 1–16. [Google Scholar] [CrossRef]
- Jie, W.; Bai, J.; Yan, J.; Chi, Y.; Li, B.B. Multi-site tumour sampling improves the detection of intra-tumour heterogeneity in oral and oropharyngeal squamous cell carcinoma. Front. Med. 2021, 8, 670305. [Google Scholar] [CrossRef]
- Brunelli, M.; Martignoni, G.; Malpeli, G.; Volpe, A.; Cima, L.; Raspollini, M.R.; Barbareschi, M.; Tafuri, A.; Masi, G.; Barzon, L.; et al. Validation of a novel three-dimensional (3D fusion) gross sampling protocol for clear cell renal cell carcinoma to overcome intratumoral heterogeneity: The Meet-Uro 18 study. J. Pers. Med. 2022, 12, 727. [Google Scholar] [CrossRef]
- Cortés, J.M.; de Petris, G.; López, J.I. Detection of intratumor heterogeneity in modern pathology: A multisite tumor sampling perspective. Front. Med. 2017, 4, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallegos, L.L.; Gilchrist, A.; Spain, L.; Stanislaw, S.; Hill, S.M.; Primus, V.; Jones, C.; Agrawal, S.; Tippu, Z.; Barhoumi, A.; et al. A protocol for representative sampling of solid tumors to improve the accuracy of sequencing results. STAR Protoc. 2021, 2, 100624. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Fu, X.; López, J.I.; Rowan, A.; Au, L.; Fendler, A.; Hazell, S.; Xu, H.; Horswell, S.; Shepherd, S.T.C.; et al. Selection of metastasis competent subclones in the tumour interior. Nat. Ecol. Evol. 2021, 5, 1033–1045. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Zhao, Y.; López, J.I.; Rowan, A.; Au, L.; Fendler, A.; Hazell, S.; Xu, H.; Horswell, S.; Shepherd, S.T.C.; et al. Spatial patterns of tumour growth impact clonal diversification in a computational model and the TRACERx Renal study. Nat. Ecol. Evol. 2022, 6, 88–102. [Google Scholar] [CrossRef]
- Manini, C.; López-Fernández, E.; Lawrie, C.H.; Laruelle, A.; Angulo, J.C.; López, J.I. Clear cell renal cell carcinomas with aggressive behavior display low intratumor heterogeneity at the histological level. Curr. Urol. Rep. 2022, 23, 93–97. [Google Scholar] [CrossRef]
- Petrova, V.; Annicchiarico-Petruzzelli, M.; Melino, G.; Amelio, I. The hypoxic tumour microenvironment. Oncogenesis 2018, 7, 10. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manini, C.; López-Fernández, E.; López, J.I. Towards Personalized Sampling in Clear Cell Renal Cell Carcinomas. Cancers 2022, 14, 3381. https://doi.org/10.3390/cancers14143381
Manini C, López-Fernández E, López JI. Towards Personalized Sampling in Clear Cell Renal Cell Carcinomas. Cancers. 2022; 14(14):3381. https://doi.org/10.3390/cancers14143381
Chicago/Turabian StyleManini, Claudia, Estíbaliz López-Fernández, and José I. López. 2022. "Towards Personalized Sampling in Clear Cell Renal Cell Carcinomas" Cancers 14, no. 14: 3381. https://doi.org/10.3390/cancers14143381
APA StyleManini, C., López-Fernández, E., & López, J. I. (2022). Towards Personalized Sampling in Clear Cell Renal Cell Carcinomas. Cancers, 14(14), 3381. https://doi.org/10.3390/cancers14143381







