Increased Soluble PD-1 Predicts Response to Nivolumab plus Ipilimumab in Melanoma
Abstract
Simple Summary
Abstract
1. Background
2. Material and Methods
2.1. Patients and Treatment
2.2. Disease Characteristics and Response Assessment
2.3. Sample Collection and Processing
2.4. Plasma Cytokine Analysis
2.5. Statistical Analysis
3. Results
3.1. Patient Characteristics
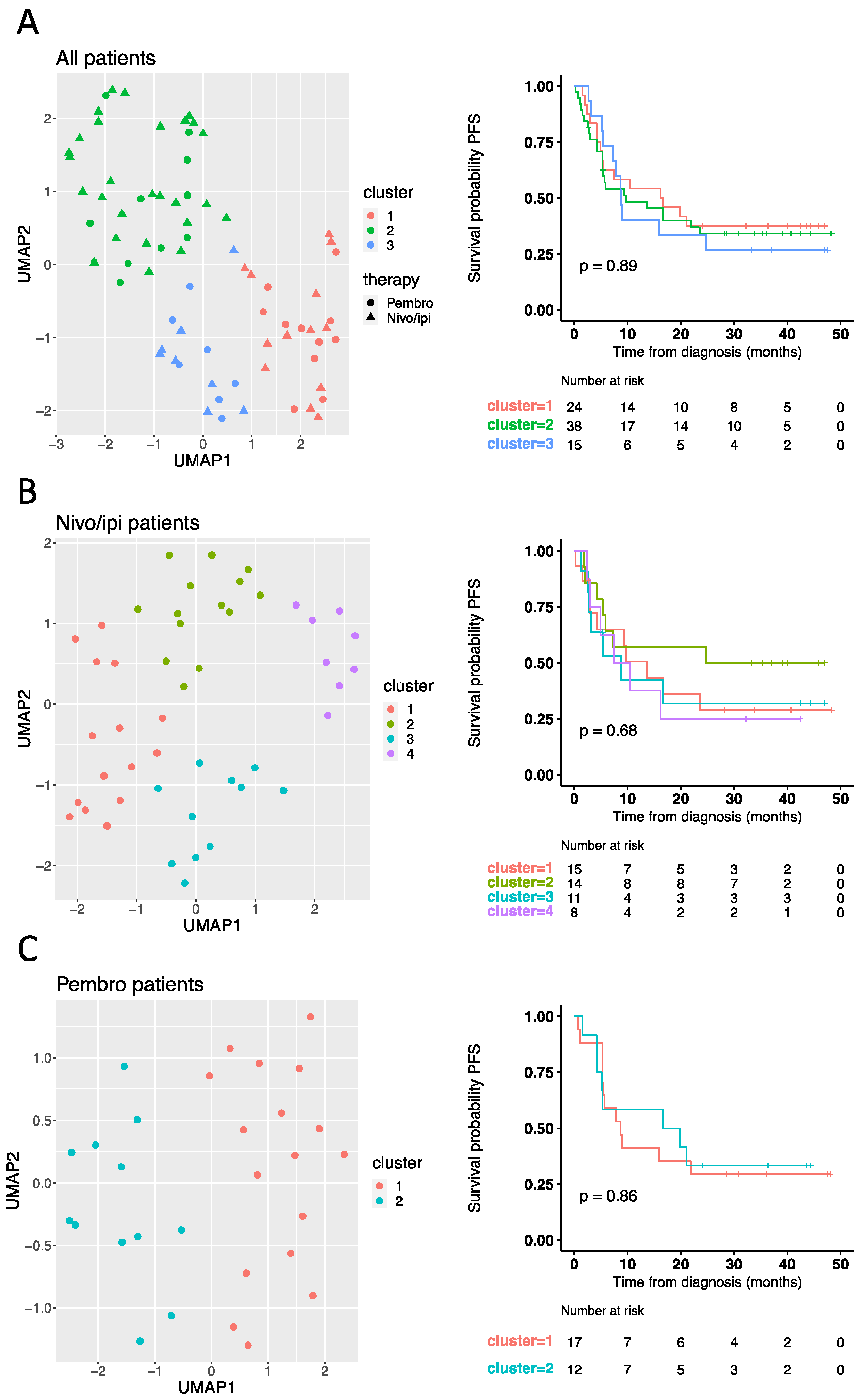
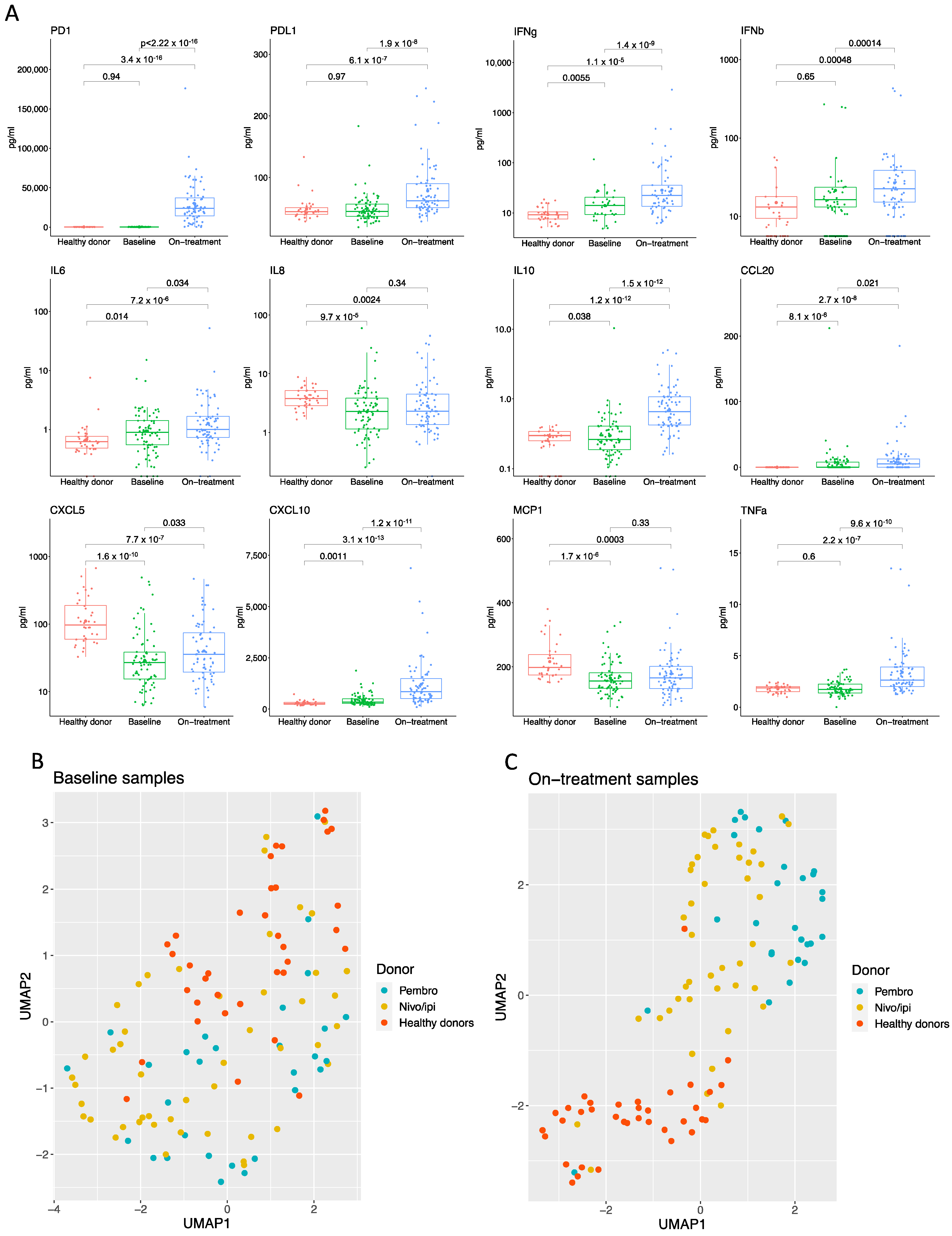
3.2. Baseline Cytokine Profile Does Not Predict Response to Checkpoint Inhibitors
3.3. Checkpoint Inhibitor Treatment Induces an Inflammatory Cytokine Profile Distinct from Baseline and Healthy Donors
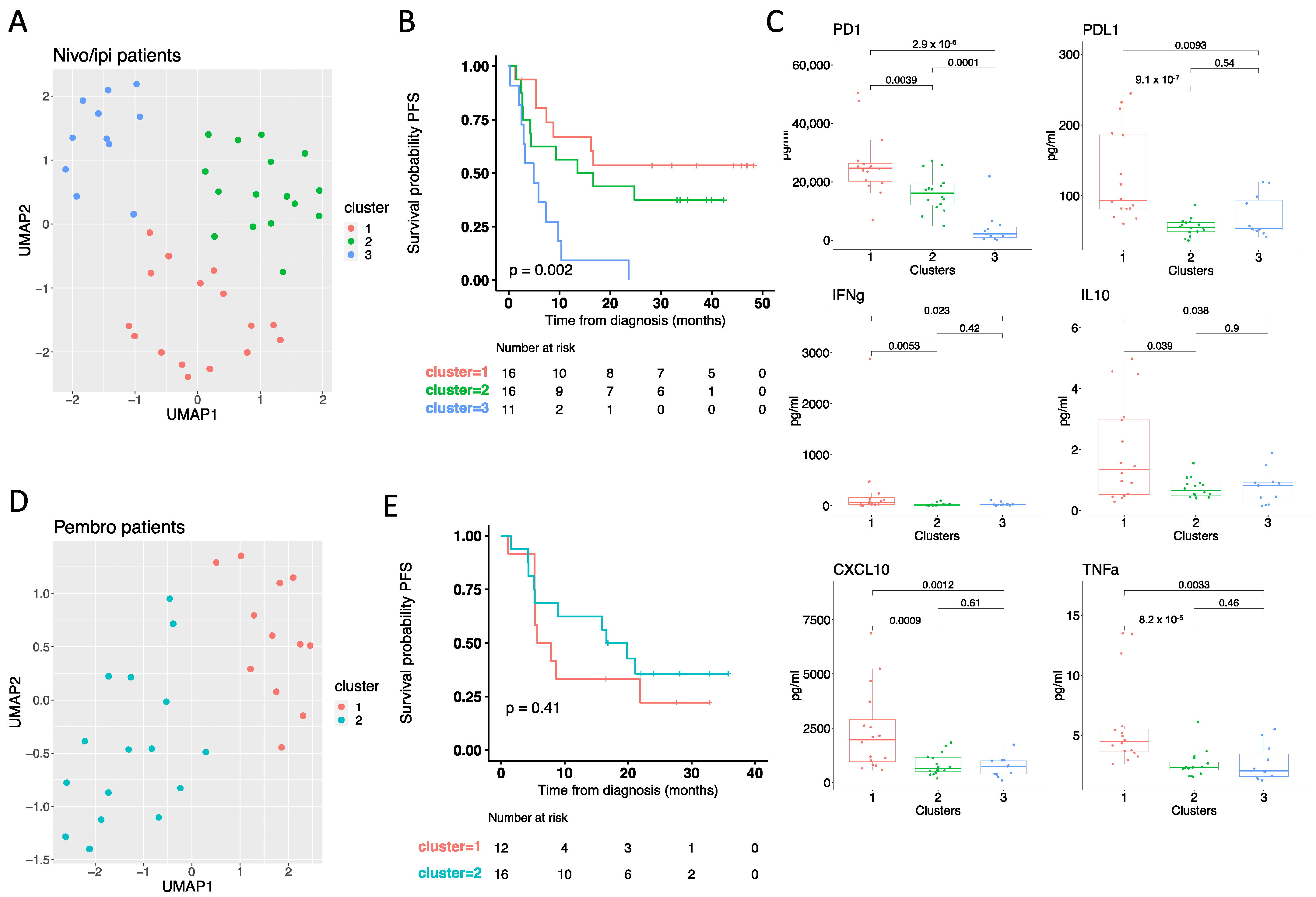
3.4. On-Treatment Cytokine Profile Predicts Response to Checkpoint Inhibitors
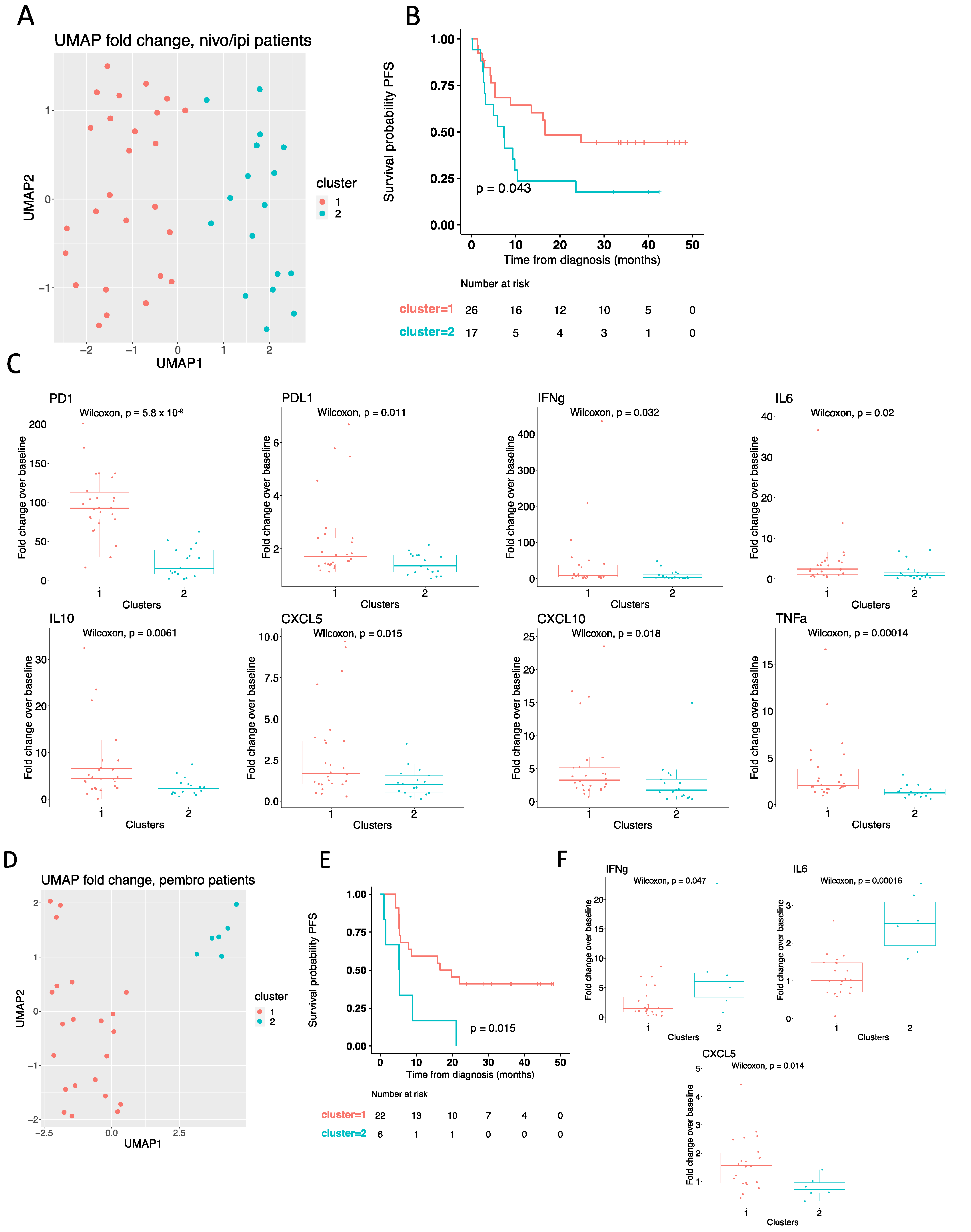
3.5. High PD-1 Increment Predicts Response to Nivolumab plus Ipilimumab
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| OS | Overall survival |
| PFS | Progression-free survival |
| HR | Hazard ratio |
| CI | Confidence interval |
| Pembro | Pembrolizumab |
| Nivo/ipi | Nivolumab plus ipilimumab |
| MM | Malignant melanoma |
| ECOG | Eastern Cooperative Oncology Group |
| U/L | Units/liter |
| LHD | Lactate dehydrogenase |
| CRP | C-reactive protein |
| PD-L1 | Programmed death ligand 1 |
| EDTA | Ethylenediaminetetraacetic acid |
| IFN | Interferon |
| IL | Interleukin |
| CXCL10 | C-X-C motif chemokine ligand 10 |
| CCL20 | Chemokine (C-C-motif) ligand 20 |
| TNF | Tumor Necrosis factor |
| MCP | Monocyte Chemoattractant Protein 1 |
| PD-1 | Programmed cell death protein 1 |
| CTLA-4 | Cytotoxic T-lymphocyte-associated protein 4 |
| UMAP | Uniform Manifold Approximation and Projection |
| CD8 | Cluster of differentiation 8 |
| TGF-β | Transforming growth factor β |
| PET/CT | Position emission tomography/computed tomography |
| MRI | Magnetic resonance imaging |
| NSCLC | Non-small cell lung cancer |
References
- Robert, C.; Ribas, A.; Schachter, J.; Arance, A.; Grob, J.-J.; Mortier, L.; Daud, A.; Carlino, M.S.; McNeil, C.M.; Lotem, M.; et al. Pembrolizumab versus ipilimumab in advanced melanoma (KEYNOTE-006): Post-hoc 5-year results from an open-label, multicentre, randomised, controlled, phase 3 study. Lancet Oncol. 2019, 20, 1239–1251. [Google Scholar] [CrossRef]
- Wolchok, J.D.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.-J.; Rutkowski, P.; Lao, C.D.; Cowey, C.L.; Schadendorf, D.; Wagstaff, J.; Dummer, R.; et al. Long-Term Outcomes With Nivolumab Plus Ipilimumab or Nivolumab Alone Versus Ipilimumab in Patients With Advanced Melanoma. J. Clin. Oncol. 2022, 40, 127–137. [Google Scholar] [CrossRef]
- Larkin, J.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.J.; Cowey, C.L.; Lao, C.D.; Schadendorf, D.; Dummer, R.; Smylie, M.; Rutkowski, P.; et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N. Engl. J. Med. 2015, 373, 23–34. [Google Scholar] [CrossRef]
- Schachter, J.; Ribas, A.; Long, G.V.; Arance, A.; Grob, J.-J.; Mortier, L.; Daud, A.; Carlino, M.S.; McNeil, C.; Lotem, M.; et al. Pembrolizumab versus ipilimumab for advanced melanoma: Final overall survival results of a multicentre, randomised, open-label phase 3 study (KEYNOTE-006). Lancet 2017, 390, 1853–1862. [Google Scholar] [CrossRef]
- Gide, T.N.; Wilmott, J.S.; Scolyer, R.A.; Long, G.V. Primary and Acquired Resistance to Immune Checkpoint Inhibitors in Metastatic Melanoma. Clin. Cancer Res. 2018, 24, 1260–1270. [Google Scholar] [CrossRef] [PubMed]
- Tumeh, P.C.; Harview, C.L.; Yearley, J.H.; Shintaku, I.P.; Taylor, E.J.M.; Robert, L. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014, 515, 568–571. [Google Scholar] [CrossRef] [PubMed]
- Wong, P.F.; Wei, W.; Smithy, J.W.; Acs, B.; Toki, M.I.; Blenman, K.R.M.; Zelterman, D.; Kluger, H.M.; Rimm, D.L. Multiplex Quantitative Analysis of Tumor-Infiltrating Lymphocytes and Immunotherapy Outcome in Metastatic Melanoma. Clin. Cancer Res. 2019, 25, 2442–2449. [Google Scholar] [CrossRef] [PubMed]
- Ayers, M.; Lunceford, J.; Nebozhyn, M.; Murphy, E.; Loboda, A.; Kaufman, D.R.; Albright, A.; Cheng, J.D.; Kang, S.P.; Shankaran, V.; et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Investig. 2017, 127, 2930–2940. [Google Scholar] [CrossRef]
- Van Allen, E.M.; Miao, D.; Schilling, B.; Shukla, S.A.; Blank, C.; Zimmer, L.; Sucker, A.; Hillen, U.; Foppen, M.H.G.; Goldinger, S.M.; et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015, 350, 207–211. [Google Scholar] [CrossRef]
- Snyder, A.; Makarov, V.; Merghoub, T.; Yuan, J.; Zaretsky, J.M.; Desrichard, A.; Walsh, L.A.; Postow, M.A.; Wong, P.; Ho, T.S.; et al. Genetic Basis for Clinical Response to CTLA-4 Blockade in Melanoma. N. Engl. J. Med. 2014, 371, 2189–2199. [Google Scholar] [CrossRef]
- Hodi, F.S.; Wolchok, J.D.; Schadendorf, D.; Larkin, J.; Long, G.V.; Qian, X.; Saci, A.; Young, T.C.; Srinivasan, S.; Chang, H.; et al. TMB and Inflammatory Gene Expression Associated with Clinical Outcomes following Immunotherapy in Advanced Melanoma. Cancer Immunol. Res. 2021, 9, 1202–1213. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, N.; Kiyohara, Y.; Uhara, H.; Iizuka, H.; Uehara, J.; Otsuka, F.; Fujisawa, Y.; Takenouchi, T.; Isei, T.; Iwatsuki, K.; et al. Cytokine biomarkers to predict antitumor responses to nivolumab suggested in a phase 2 study for advanced melanoma. Cancer Sci. 2017, 108, 1022–1031. [Google Scholar] [CrossRef] [PubMed]
- Nonomura, Y.; Otsuka, A.; Nakashima, C.; Seidel, J.; Kitoh, A.; Dainichi, T.; Nakajima, S.; Sawada, Y.; Matsushita, S.; Aoki, M.; et al. Peripheral blood Th9 cells are a possible pharmacodynamic biomarker of nivolumab treatment efficacy in metastatic melanoma patients. OncoImmunology 2016, 5, e1248327. [Google Scholar] [CrossRef] [PubMed]
- Schalper, K.A.; Carleton, M.; Zhou, M.; Chen, T.; Feng, Y.; Huang, S.-P.; Walsh, A.M.; Baxi, V.; Pandya, D.; Baradet, T.; et al. Elevated serum interleukin-8 is associated with enhanced intratumor neutrophils and reduced clinical benefit of immune-checkpoint inhibitors. Nat. Med. 2020, 26, 688–692. [Google Scholar] [CrossRef] [PubMed]
- Sanmamed, M.F.; Perez-Gracia, J.L.; Schalper, K.A.; Fusco, J.P.; Gonzalez, A.; Rodriguez-Ruiz, M.E.; Oñate, C.; Perez, G.; Alfaro, C.; Martín-Algarra, S.; et al. Changes in serum interleukin-8 (IL-8) levels reflect and predict response to anti-PD-1 treatment in melanoma and non-small-cell lung cancer patients. Ann. Oncol. 2017, 28, 1988–1995. [Google Scholar] [CrossRef]
- Zhou, J.; Mahoney, K.M.; Giobbie-Hurder, A.; Zhao, F.; Lee, S.; Liao, X.; Rodig, S.; Li, J.; Wu, X.; Butterfield, L.H.; et al. Soluble PD-L1 as a Biomarker in Malignant Melanoma Treated with Checkpoint Blockade. Cancer Immunol. Res. 2017, 5, 480–492. [Google Scholar] [CrossRef] [PubMed]
- Machiraju, D.; Wiecken, M.; Lang, N.; Hülsmeyer, I.; Roth, J.; Schank, T.E.; Eurich, R.; Halama, N.; Enk, A.; Hassel, J.C. Soluble immune checkpoints and T-cell subsets in blood as biomarkers for resistance to immunotherapy in melanoma patients. OncoImmunology 2021, 10, 1926762. [Google Scholar] [CrossRef]
- Ugurel, S.; Schadendorf, D.; Horny, K.; Sucker, A.; Schramm, S.; Utikal, J.; Pföhler, C.; Herbst, R.; Schilling, B.; Blank, C.; et al. Elevated baseline serum PD-1 or PD-L1 predicts poor outcome of PD-1 inhibition therapy in metastatic melanoma. Ann. Oncol. 2020, 31, 144–152. [Google Scholar] [CrossRef]
- McInnes, L.; Healy, J.; Melville, J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv 2018, arXiv:1802.03426. [Google Scholar]
- Daud, A.I.; Wolchok, J.D.; Robert, C.; Hwu, W.-J.; Weber, J.S.; Ribas, A.; Hodi, F.S.; Joshua, A.; Kefford, R.; Hersey, P.; et al. Programmed Death-Ligand 1 Expression and Response to the Anti–Programmed Death 1 Antibody Pembrolizumab in Melanoma. J. Clin. Oncol. 2016, 34, 4102–4109. [Google Scholar] [CrossRef]
- Koguchi, Y.; Hoen, H.M.; Bambina, S.A.; Rynning, M.D.; Fuerstenberg, R.K.; Curti, B.D.; Urba, W.J.; Milburn, C.; Bahjat, F.R.; Korman, A.J.; et al. Serum Immunoregulatory Proteins as Predictors of Overall Survival of Metastatic Melanoma Patients Treated with Ipilimumab. Cancer Res. 2015, 75, 5084–5092. [Google Scholar] [CrossRef] [PubMed]
- Bjoern, J.; Juul Nitschke, N.; Zeeberg Iversen, T.; Schmidt, H.; Fode, K.; Svane, I.M. Immunological correlates of treatment and response in stage IV malignant melanoma patients treated with Ipilimumab. Oncoimmunology 2016, 5, e1100788. [Google Scholar] [CrossRef]
- Biancotto, A.; Feng, X.; Langweiler, M.; Young, N.S.; Philip McCoy, J. Effect of anticoagulants on multiplexed measurement of cytokine/chemokines in healthy subjects. Cytokine 2012, 60, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Parkitny, L.; McAuley, J.H.; Kelly, P.J.; Di Pietro, F.; Cameron, B.; Moseley, G.L. Multiplex cytokine concentration measurement: How much do the medium and handling matter? Mediat. Inflamm. 2013, 2013, 890706. [Google Scholar] [CrossRef] [PubMed]
- Babačić, H.; Lehtiö, J.; Coaña YP de Pernemalm, M.; Eriksson, H. In-depth plasma proteomics reveals increase in circulating PD-1 during anti-PD-1 immunotherapy in patients with metastatic cutaneous melanoma. J. Immunother. Cancer 2020, 8, e000204. [Google Scholar] [CrossRef]
- Sorensen, S.F.; Demuth, C.; Weber, B.; Sorensen, B.S.; Meldgaard, P. Increase in soluble PD-1 is associated with prolonged survival in patients with advanced EGFR-mutated non-small cell lung cancer treated with erlotinib. Lung Cancer 2016, 100, 77–84. [Google Scholar] [CrossRef]
- Tiako Meyo, M.; Jouinot, A.; Giroux-Leprieur, E.; Fabre, E.; Wislez, M.; Alifano, M.; Leroy, K.; Boudou-Rouquette, P.; Tlemsani, C.; Khoudour, N.; et al. Predictive Value of Soluble PD-1, PD-L1, VEGFA, CD40 Ligand and CD44 for Nivolumab Therapy in Advanced Non-Small Cell Lung Cancer: A Case-Control Study. Cancers 2020, 12, 473. [Google Scholar] [CrossRef]
- Zhang, A.; Sun, Y.; Wang, S.; Du, J.; Gao, X.; Yuan, Y.; Zhao, L.; Yang, Y.; Xu, L.; Lei, Y.; et al. Secretion of human soluble programmed cell death protein 1 by chimeric antigen receptor-modified T cells enhances anti-tumor efficacy. Cytotherapy 2020, 22, 734–743. [Google Scholar] [CrossRef]
- Shin, S.-P.; Seo, H.-H.; Shin, J.-H.; Park, H.-B.; Lim, D.-P.; Eom, H.-S.; Bae, Y.-S.; Kim, I.-H.; Choi, K.; Lee, S.-J. Adenovirus Expressing Both Thymidine Kinase and Soluble PD1 Enhances Antitumor Immunity by Strengthening CD8 T-cell Response. Mol. Ther. 2013, 21, 688–695. [Google Scholar] [CrossRef]
- Xiao, H.; Huang, B.; Yuan, Y.; Li, D.; Han, L.-F.; Liu, Y.; Gong, W.; Wu, F.-H.; Zhang, G.-M.; Feng, Z.-H. Soluble PD-1 Facilitates 4-1BBL–Triggered Antitumor Immunity against Murine H22 Hepatocarcinoma In Vivo. Clin. Cancer Res. 2007, 13, 1823–1830. [Google Scholar] [CrossRef][Green Version]
- Song, M.Y.; Park, S.H.; Nam, H.J.; Choi, D.H.; Sung, Y.C. Enhancement of vaccine-induced primary and memory CD8+ t-cell responses by soluble PD-1. J. Immunother. 2011, 34, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Music, M.; Iafolla, M.A.J.; Ren, A.H.; Soosaipillai, A.; Prassas, I.; Diamandis, E.P. Serum PD-1 is elevated after pembrolizumab treatment but has no predictive value. Mol. Cancer Ther. 2019, 18, 1844–1851. [Google Scholar] [CrossRef] [PubMed]
- Das, R.; Verma, R.; Sznol, M.; Boddupalli, C.S.; Gettinger, S.N.; Kluger, H.; Callahan, M.; Wolchok, J.D.; Halaban, R.; Dhodapkar, M.V.; et al. Combination Therapy with Anti–CTLA-4 and Anti–PD-1 Leads to Distinct Immunologic Changes In Vivo. J. Immunol. 2014, 194, 950–959. [Google Scholar] [CrossRef] [PubMed]
| Parameter | Pembrolizumab (anti-PD1) | Nivolumab/Ipilimumab (anti-PD1/CTLA-4) | p |
|---|---|---|---|
| Total | 29 | 48 | |
| Age, year mean (range) | 68 (31–89) | 60 (30–85) | |
| Sex, n (%) - Male - Female | 18 (62) 11 (38) | 29 (60.4) 19 (39.6) | 0.89 |
| AJCC stage, n (%) - Stage III - Stage IV | 3 (10.3) 26 (89.7) | 5 (10.4) 43 (89.6) | 0.99 |
| BRAF status, n (%) - Wild type - Mutated | 16 (55.2) 13 (44.8) | 26 (54.2) 22 (45.8) | 0.93 |
| Tumor PD-L1 expression, n (%) - < 1% - ≥ 1% - N/A | 4 (13.8) 22 (75.9) 3 (10.3) | 43 (89.6) 2 (4.2) 3 (6.2) | <0.0001 |
| Serum LDH, n (%) - Normal - Elevated - N/A | 21 (72.4) 6 (20.7) 2 (6.9) | 32 (66.7) 14 (29.2) 2 (4.1) | 0.65 |
| Progression, n (%) - No - Yes | 9 (31) 20 (69) | 18 (37.5) 30 (62.5) | 0.56 |
| Alive, n (%) - No - Yes | 13 (44.8) 16 (55.2) | 24 (50) 24 (50) | 0.81 |
| Parameters | PFS | |
|---|---|---|
| Hazard Ratio (95% Confidence Interval) | p | |
| Univariate Cox regression analysis | ||
| Age | 1 (0.97–1.03) | 0.92 |
| Gender (Female vs male) | 1.13 (0.54–2.48) | 0.75 |
| LDH (normal vs elevated) | 0.56 (0.24–1.32) | 0.18 |
| Performance status (ECOG) (0 vs 1 or 2) | 2.19 (1.06–4.53) | 0.034 |
| PD-1 fold change (above vs below median) | 0.35 (0.16–0.77) | 0.0086 |
| CXCL10 fold change (above vs below median) | 1.15 (0.55–2.41) | 0.71 |
| TNFα fold change (above vs below median) | 1.09 (0.52–2.29) | 0.82 |
| PD-L1 fold change (above vs below median) | 1.27 (0.60–2.66) | 0.53 |
| IFNγ fold change (above vs below median) | 0.98 (0.47–2.07) | 0.97 |
| IL10 fold change (above vs below median) | 0.94 (0.45–1.98) | 0.87 |
| Multivariable Cox regression analysis | ||
| Age | 0.98 (0.95–1.026) | 0.49 |
| Gender (Female vs male) | 1.042 (0.43–2.5) | 0.93 |
| LDH (normal vs elevated) | 0.16 (0.041–0.64) | 0.0096 |
| Performance status (ECOG) (0 vs 1 or 2) | 2.41 (0.81–7.19) | 0.12 |
| PD-1 fold change (above vs below median) | 0.13 (0.0034–0.49) | 0.0026 |
| CXCL10 fold change (above vs below median) | 2.43 (0.68–8.71) | 0.17 |
| TNFα fold change (above vs below median) | 0.81 (0.18–3.69) | 0.79 |
| PD-L1 fold change (above vs below median) | 0.46 (0.14–1.5) | 0.20 |
| IFNγ fold change (above vs below median) | 1.42 (0.53–3.82) | 0.48 |
| IL10 fold change (above vs below median) | 2.095 (0.51–8.53) | 0.30 |
| Parameters | PFS | |
|---|---|---|
| Hazard Ratio (95% Confidence Interval) | p | |
| Univariate Cox regression analysis | ||
| Age | 1 (0.97–1.28) | 0.89 |
| Gender (Female vs male) | 1.84 (0.71–4.80) | 0.21 |
| LDH (normal vs elevated) | 2.17 (0.77–6.079) | 0.14 |
| Performance status (ECOG) (0 vs 1 or 2) | 1.72 (0.69–4.31) | 0.24 |
| IFNγ fold change (above vs below median) | 1.74 (0.69–4.34) | 0.24 |
| CXCL5 fold change (above vs below median) | 0.64 (0.26–1.6) | 0.34 |
| IL6 fold change (above vs below median) | 0.64 (0.26–1.6) | 0.34 |
| Multivariable Cox regression analysis | ||
| Age | 0.99 (0.95–1.024) | 0.47 |
| Gender (Female vs male) | 2.21 (0.67–7.24) | 0.19 |
| LDH (normal vs elevated) | 1.74 (0.50–6.052) | 0.38 |
| Performance status (ECOG) (0 vs 1 or 2) | 0.91 (0.3–2.81) | 0.87 |
| IFNγ fold change (above vs below median) | 1.91 (0.45–8.032) | 0.38 |
| CXCL5 fold change (above vs below median) | 0.8 (0.23–2.75) | 0.72 |
| IL6 fold change (above vs below median) | 0.66 (0.21–2.067) | 0.48 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pedersen, J.G.; Sokac, M.; Sørensen, B.S.; Luczak, A.A.; Aggerholm-Pedersen, N.; Birkbak, N.J.; Øllegaard, T.H.; Jakobsen, M.R. Increased Soluble PD-1 Predicts Response to Nivolumab plus Ipilimumab in Melanoma. Cancers 2022, 14, 3342. https://doi.org/10.3390/cancers14143342
Pedersen JG, Sokac M, Sørensen BS, Luczak AA, Aggerholm-Pedersen N, Birkbak NJ, Øllegaard TH, Jakobsen MR. Increased Soluble PD-1 Predicts Response to Nivolumab plus Ipilimumab in Melanoma. Cancers. 2022; 14(14):3342. https://doi.org/10.3390/cancers14143342
Chicago/Turabian StylePedersen, Jesper Geert, Mateo Sokac, Boe Sandahl Sørensen, Adam Andrzej Luczak, Ninna Aggerholm-Pedersen, Nicolai Juul Birkbak, Trine Heide Øllegaard, and Martin Roelsgaard Jakobsen. 2022. "Increased Soluble PD-1 Predicts Response to Nivolumab plus Ipilimumab in Melanoma" Cancers 14, no. 14: 3342. https://doi.org/10.3390/cancers14143342
APA StylePedersen, J. G., Sokac, M., Sørensen, B. S., Luczak, A. A., Aggerholm-Pedersen, N., Birkbak, N. J., Øllegaard, T. H., & Jakobsen, M. R. (2022). Increased Soluble PD-1 Predicts Response to Nivolumab plus Ipilimumab in Melanoma. Cancers, 14(14), 3342. https://doi.org/10.3390/cancers14143342






