Accurate Screening for Early-Stage Breast Cancer by Detection and Profiling of Circulating Tumor Cells
Abstract
:Simple Summary
Abstract
1. Background
2. Methods
2.1. Study Participants and Samples
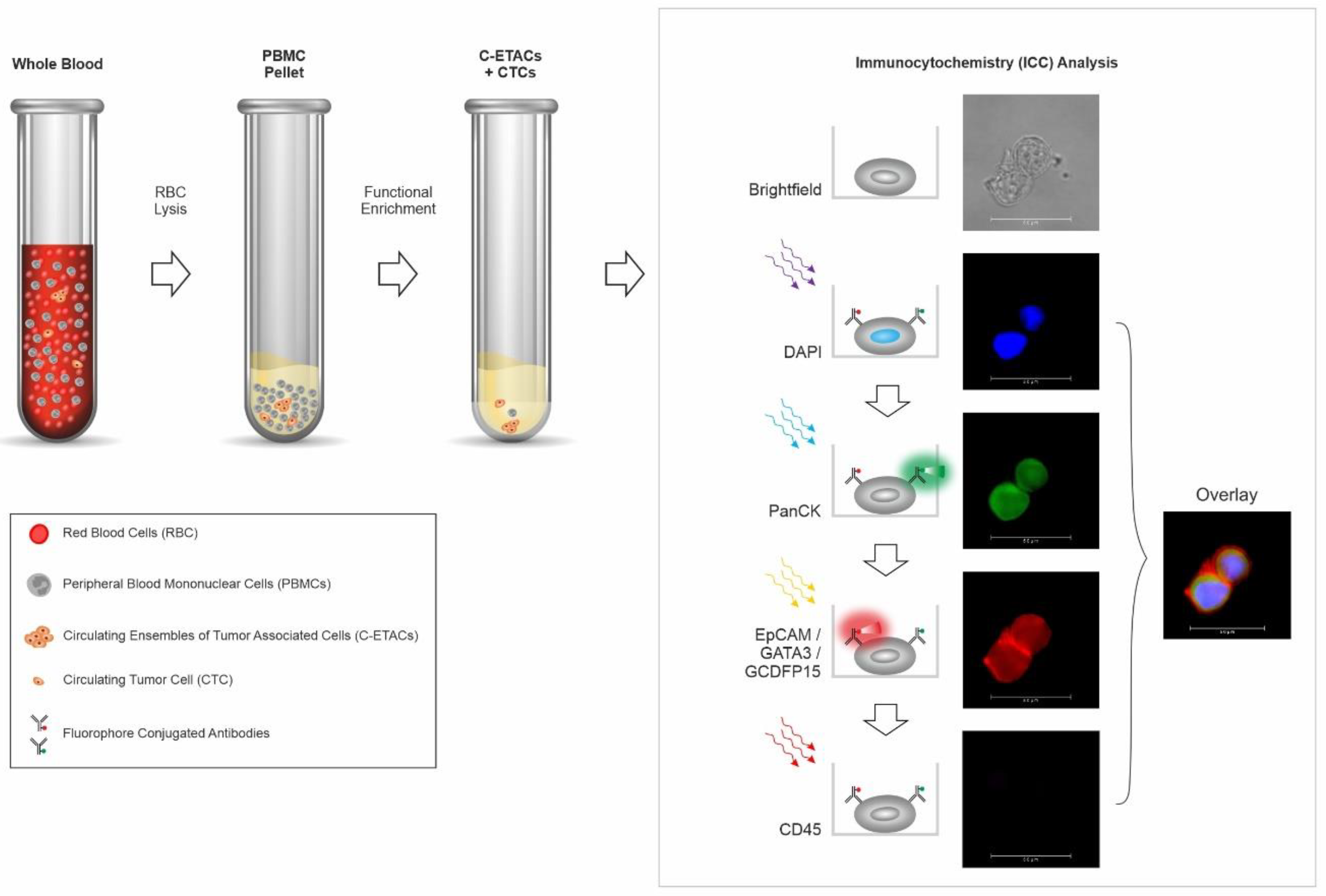
2.2. Enrichment of Circulating Tumor Cells from Peripheral Blood
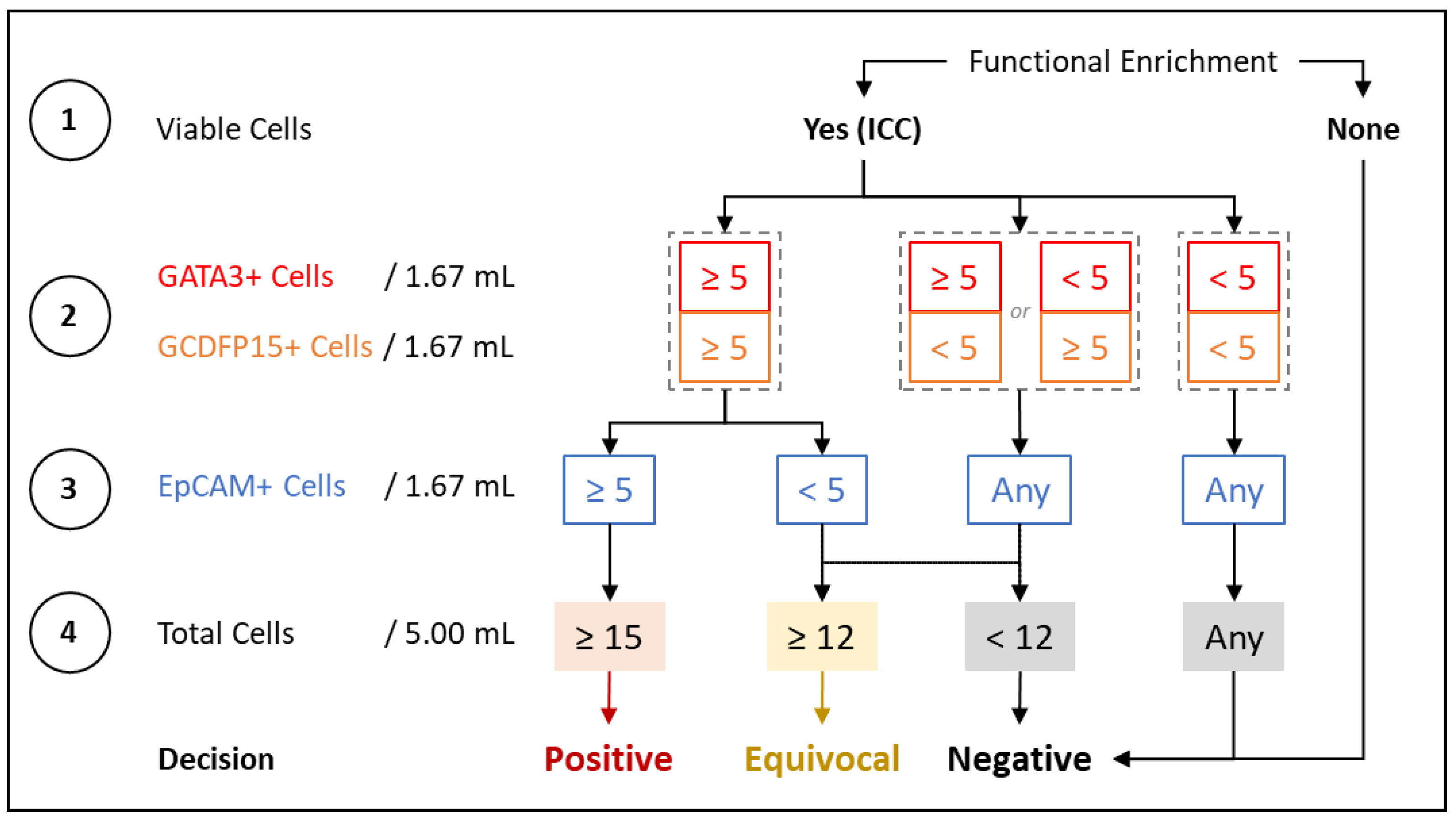
2.3. Immunocytochemistry Profiling of CTCs
2.4. Method Development and Optimization
2.5. Analytical Validation
2.6. Case–Control Clinical Study
2.7. Prospective Clinical Study
2.8. Molecular Concordance Study
3. Results
3.1. Method Development and Optimization
3.2. Analytical Validation
3.3. Case–Control Clinical Study
3.4. Prospective Clinical Study
3.5. Molecular Concordance Study
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Guidi, A.J.; Fischer, L.; Harris, J.R.; Schnitt, S.J. Microvessel density and distribution in ductal carcinoma in situ of the breast. J. Natl. Cancer Inst. 1994, 86, 614–619. [Google Scholar] [CrossRef]
- Teo, N.B.; Shoker, B.S.; Jarvis, C.; Martin, L.; Sloane, J.P.; Holcombe, C. Vascular density and phenotype around ductal carcinoma in situ (DCIS) of the breast. Br. J. Cancer 2002, 86, 905–911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heffelfinger, S.C.; Yassin, R.; Miller, M.A.; Lower, E. Vascularity of proliferative breast disease and carcinoma in situ correlates with histological features. Clin. Cancer Res. 1996, 2, 1873–1878. [Google Scholar]
- Sänger, N.; Effenberger, K.E.; Riethdorf, S.; Van Haasteren, V.; Gauwerky, J.; Wiegratz, I.; Strebhardt, K.; Kaufmann, M.; Pantel, K. Disseminated tumor cells in the bone marrow of patients with ductal carcinoma in situ. Int. J. Cancer 2011, 129, 2522–2526. [Google Scholar] [CrossRef] [PubMed]
- Gruber, I.V.; Hartkopf, A.D.; Hahn, M.; Taran, F.-A.; Staebler, A.; Wallwiener, D.; Brucker, S.Y.; Hanke, J.; Fehm, T. Relationship Between Hematogenous Tumor Cell Dissemination and Cellular Immunity in DCIS Patients. Anticancer Res. 2016, 36, 2345–2351. [Google Scholar]
- Hosseini, H.; Obradović, M.M.S.; Hoffmann, M.; Harper, K.L.; Sosa, M.S.; Werner-Klein, M.; Nanduri, L.K.; Werno, C.; Ehrl, C.; Maneck, M.; et al. Early dissemination seeds metastasis in breast cancer. Nature 2016, 540, 552–558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krol, I.; Schwab, F.D.; Carbone, R.; Ritter, M.; Picocci, S.; De Marni, M.L.; Stepien, G.; Franchi, G.M.; Zanardi, A.; Rissoglio, M.D.; et al. Detection of clustered circulating tumour cells in early breast cancer. Br. J. Cancer 2021, 125, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Reduzzi, C.; Di Cosimo, S.; Gerratana, L.; Motta, R.; Martinetti, A.; Vingiani, A.; D’Amico, P.; Zhang, Y.; Vismara, M.; Depretto, C.; et al. Circulating Tumor Cell Clusters Are Frequently Detected in Women with Early-Stage Breast Cancer. Cancers 2021, 13, 2356. [Google Scholar] [CrossRef]
- Jin, L.; Zhao, W.; Zhang, J.; Chen, W.; Xie, T.; Wang, L.; Fan, W.; Xie, S.; Shen, J.; Zheng, H.; et al. Evaluation of the diagnostic value of circulating tumor cells with CytoSorter® CTC capture system in patients with breast cancer. Cancer Med. 2020, 9, 1638–1647. [Google Scholar] [CrossRef] [Green Version]
- Fina, E.; Reduzzi, C.; Motta, R.; Di Cosimo, S.; Bianchi, G.; Martinetti, A.; Wechsler, J.; Cappelletti, V.; Daidone, M.G. Did circulating tumor cells tell us all they could? The missed circulating tumor cell message in breast cancer. Int. J. Biol. Markers 2015, 30, e429–e433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fina, E.; Cleris, L.; Dugo, M.; Lecchi, M.; Ciniselli, C.M.; Lecis, D.; Bianchi, G.V.; Verderio, P.; Daidone, M.G.; Cappelletti, V. Gene signatures of circulating breast cancer cell models are a source of novel molecular determinants of metastasis and improve circulating tumor cell detection in patients. J. Exp. Clin. Cancer Res. 2022, 25, 78. [Google Scholar] [CrossRef] [PubMed]
- Spizzo, G.; Fong, D.; Wurm, M.; Ensinger, C.; Obrist, P.; Hofer, C.; Mazzoleni, G.; Gastl, G.; Went, P. EpCAM expression in primary tumour tissues and metastases: An immunohistochemical analysis. J. Clin. Pathol. 2011, 64, 415–420. [Google Scholar] [CrossRef] [Green Version]
- Soysal, S.D.; Muenst, S.; Barbie, T.; Fleming, T.; Gao, F.; Spizzo, G.; Oertli, D.; Viehl, C.T.; Obermann, E.C.; Gillanders, W.E. EpCAM expression varies significantly and is differentially associated with prognosis in the luminal B HER2+, basal-like, and HER2 intrinsic subtypes of breast cancer. Br. J. Cancer 2013, 108, 1480–1487. [Google Scholar] [CrossRef] [PubMed]
- Rao, C.G.; Chianese, D.; Doyle, G.V.; Miller, M.C.; Russell, T.; Sanders, R.A.J.; Terstappen, L.W.M.M. Expression of epithelial cell adhesion molecule in carcinoma cells present in blood and primary and metastatic tumors. Int. J. Oncol. 2005, 27, 49–57. [Google Scholar] [CrossRef] [PubMed]
- de Wit, S.; Manicone, M.; Rossi, E.; Lampignano, R.; Yang, L.; Zill, B.; Rengel-Puertas, A.; Ouhlen, M.; Crespo, M.; Berghuis, A.M.S.; et al. EpCAM(high) and EpCAM(low) circulating tumor cells in metastatic prostate and breast cancer patients. Oncotarget 2018, 9, 35705–35716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, D.L.; Stefansson, S.; Haudenschild, C.; Martin, S.S.; Charpentier, M.; Chumsri, S.; Cristofanilli, M.; Tang, C.-M.; Alpaugh, R.K. Cytometric characterization of circulating tumor cells captured by microfiltration and their correlation to the CellSearch® CTC test. Cytometry A 2015, 87, 137–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, G.; Herrler, M.; Burgess, D.; Manna, E.; Krag, D.; Burke, J.F. Enrichment with anti-cytokeratin alone or combined with anti-EpCAM antibodies significantly increases the sensitivity for circulating tumor cell detection in metastatic breast cancer patients. Breast Cancer Res. 2008, 10, R69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheng, Y.; Wang, T.; Li, H.; Zhang, Z.; Chen, J.; He, C.; Li, Y.; Lv, Y.; Zhang, J.; Xu, C.; et al. Comparison of analytic performances of Cellsearch and iFISH approach in detecting circulating tumor cells. Oncotarget 2017, 8, 8801–8806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akolkar, D.; Patil, D.; Crook, T.; Limaye, S.; Page, R.; Datta, V.; Patil, R.; Sims, C.; Ranade, A.; Fulmali, P.; et al. Circulating ensembles of tumor-associated cells: A redoubtable new systemic hallmark of cancer. Int. J. Cancer 2020, 146, 3485–3494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaya, A.; Crook, T.; Plowman, N.; Ranade, A.; Limaye, S.; Bhatt, A.; Page, R.; Patil, R.; Fulmali, P.; Datta, V.; et al. Evaluation of circulating tumor cell clusters for pan-cancer noninvasive diagnostic triaging. Cancer Cytopathol. 2021, 129, 226–238. [Google Scholar] [CrossRef] [PubMed]
- von Elm, E.; Altman, D.G.; Egger, M.; Pocock, S.J.; Gøtzsche, P.C.; Vandenbroucke, J.P. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) Statement: Guidelines for reporting observational studies. Int. J. Surg. 2014, 12, 1495–1499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crook, T.; Gaya, A.; Page, R.; Limaye, S.; Ranade, A.; Bhatt, A.; Patil, S.; Kumar, P.; Patil, D.; Akolkar, D. Clinical utility of circulating tumor-associated cells to predict and monitor chemo-response in solid tumors. Cancer Chemother. Pharmacol. 2021, 87, 197–205. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Position Paper on Mammography Screening; World Health Organization: Geneva, Switzerland, 2014; ISBN 978-92-4-150793-6. [Google Scholar]
- Siu, A.L. Screening for Breast Cancer: U.S. Preventive Services Task Force Recommendation Statement. Ann. Intern. Med. 2016, 164, 279–296. [Google Scholar] [CrossRef] [Green Version]
- NCCN Guidelines: Breast Cancer Screening and Diagnosis version 1. 2021. Available online: https://www.nccn.org/professionals/physician_gls/pdf/breast-screening.pdf (accessed on 25 July 2021).
- Cardoso, F.; Kyriakides, S.; Ohno, S.; Penault-Llorca, F.; Poortmans, P.; Rubio, I.T.; Zackrisson, S.; Senkus, E. Early breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2019, 30, 1194–1220. [Google Scholar] [CrossRef] [Green Version]
- National Guideline Alliance (UK). Early and Locally Advanced Breast Cancer: Diagnosis and Management; National Guideline Alliance: London, UK, 2018; ISBN 978-1-4731-3008-1. [Google Scholar]
- Wöckel, A.; Festl, J.; Stüber, T.; Brust, K.; Stangl, S.; Heuschmann, P.U.; Albert, U.-S.; Budach, W.; Follmann, M.; Janni, W.; et al. Interdisciplinary Screening, Diagnosis, Therapy and Follow-up of Breast Cancer. Guideline of the DGGG and the DKG (S3-Level, AWMF Registry Number 032/045OL, December 2017)-Part 1 with Recommendations for the Screening, Diagnosis and Therapy of Breast C. Geburtshilfe Frauenheilkd 2018, 78, 927–948. [Google Scholar] [CrossRef] [Green Version]
- Schünemann, H.J.; Lerda, D.; Quinn, C.; Follmann, M.; Alonso-Coello, P.; Rossi, P.G.; Lebeau, A.; Nyström, L.; Broeders, M.; Ioannidou-Mouzaka, L.; et al. Breast Cancer Screening and Diagnosis: A Synopsis of the European Breast Guidelines. Ann. Intern. Med. 2020, 172, 46–56. [Google Scholar] [CrossRef] [Green Version]
- Abdullah, P.; Alabousi, M.; Ramadan, S.; Zawawi, I.; Zawawi, M.; Bhogadi, Y.; Freitas, V.; Patlas, M.N.; Alabousi, A. Synthetic 2D Mammography Versus Standard 2D Digital Mammography: A Diagnostic Test Accuracy Systematic Review and Meta-Analysis. Am. J. Roentgenol. 2021, 217, 314–325. [Google Scholar] [CrossRef]
- Salim, M.; Dembrower, K.; Eklund, M.; Lindholm, P.; Strand, F. Range of Radiologist Performance in a Population-based Screening Cohort of 1 Million Digital Mammography Examinations. Radiology 2020, 297, 33–39. [Google Scholar] [CrossRef]
- Lehman, C.D.; Wellman, R.D.; Buist, D.S.M.; Kerlikowske, K.; Tosteson, A.N.A.; Miglioretti, D.L. Diagnostic Accuracy of Digital Screening Mammography With and Without Computer-Aided Detection. JAMA Intern. Med. 2015, 175, 1828–1837. [Google Scholar] [CrossRef]
- McNeil, C. Screening Mammograms in Younger Women Have Low Accuracy and Detect Few Cancers. JNCI J. Natl. Cancer Inst. 2010, 102, 841–842. [Google Scholar] [CrossRef] [Green Version]
- Nelson, H.D.; Fu, R.; Cantor, A.; Pappas, M.; Daeges, M.; Humphrey, L. Effectiveness of Breast Cancer Screening: Systematic Review and Meta-analysis to Update the 2009 U.S. Preventive Services Task Force Recommendation. Ann. Intern. Med. 2016, 164, 244–255. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, R.A.; Kerlikowske, K.; Flowers, C.I.; Yankaskas, B.C.; Zhu, W.; Miglioretti, D.L. Cumulative probability of false-positive recall or biopsy recommendation after 10 years of screening mammography: A cohort study. Ann. Intern. Med. 2011, 155, 481–492. [Google Scholar] [CrossRef] [PubMed]
- Goldfrank, D.; Chuai, S.; Bernstein, J.L.; Ramon, Y.; Cajal, T.; Lee, J.B.; Alonso, M.C.; Diez, O.; Baiget, M.; Kauff, N.D.; et al. Effect of mammography on breast cancer risk in women with mutations in BRCA1 or BRCA2. Cancer Epidemiol. Biomark. Prev. 2006, 15, 2311–2313. [Google Scholar] [CrossRef] [Green Version]
- Niraula, S.; Biswanger, N.; Hu, P.; Lambert, P.; Decker, K. Incidence, Characteristics, and Outcomes of Interval Breast Cancers Compared With Screening-Detected Breast Cancers. JAMA Netw. Open 2020, 3, e2018179. [Google Scholar] [CrossRef]
- Health, United States, 2019—Data Finder. Available online: https://www.cdc.gov/nchs/data/hus/2019/033-508.pdf (accessed on 26 July 2021).
- Locke, W.J.; Guanzon, D.; Ma, C.; Liew, Y.J.; Duesing, K.R.; Fung, K.Y.C.; Ross, J.P. DNA Methylation Cancer Biomarkers: Translation to the Clinic. Front. Genet. 2019, 10, 1150. [Google Scholar] [CrossRef]
- Liu, M.C.; Oxnard, G.R.; Klein, E.A.; Swanton, C.; Seiden, M. V Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 2020, 31, 745–759. [Google Scholar] [CrossRef]
- Klein, E.A.; Richards, D.; Cohn, A.; Tummala, M.; Lapham, R.; Cosgrove, D.; Chung, G.; Clement, J.; Gao, J.; Hunkapiller, N.; et al. Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann. Oncol. Off. 2021, 32, 1167–1177. [Google Scholar] [CrossRef]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef] [Green Version]
- Sieuwerts, A.M.; Kraan, J.; Bolt, J.; van der Spoel, P.; Elstrodt, F.; Schutte, M.; Martens, J.W.M.; Gratama, J.-W.; Sleijfer, S.; Foekens, J.A. Anti-epithelial cell adhesion molecule antibodies and the detection of circulating normal-like breast tumor cells. J. Natl. Cancer Inst. 2009, 101, 61–66. [Google Scholar] [CrossRef]
- Aktas, B.; Tewes, M.; Fehm, T.; Hauch, S.; Kimmig, R.; Kasimir-Bauer, S. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. 2009, 11, R46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krawczyk, N.; Meier-Stiegen, F.; Banys, M.; Neubauer, H.; Ruckhaeberle, E.; Fehm, T. Expression of stem cell and epithelial-mesenchymal transition markers in circulating tumor cells of breast cancer patients. BioMed Res. Int. 2014, 2014, 415721. [Google Scholar] [CrossRef] [PubMed]
- MQSA National Statistics. Available online: https://www.fda.gov/radiation-emitting-products/mqsa-insights/mqsa-national-statistics (accessed on 17 September 2021).
- John, M. Eisenberg Center for Clinical Decisions and Communications Science Core-Needle Biopsy for Breast Abnormalities. Available online: https://www.ncbi.nlm.nih.gov/books/NBK368367/ (accessed on 7 July 2022).
- Cancer Stat Facts: Female Breast Cancer. Available online: https://seer.cancer.gov/statfacts/html/breast.html (accessed on 17 September 2021).
- Peintinger, F. National Breast Screening Programs across Europe. Breast Care 2019, 14, 354–358. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Evaluation of Linearity of Quantitative Measurement Procedures-Table 3. Deviations from True Ratios for Different Percentages of ADL; EP06Ed2. 2020. Available online: https://clsi.org/standards/products/method-evaluation/documents/ep06/ (accessed on 24 May 2022).
| EpCAM, PanCK, CD45 | GATA3, PanCK, CD45 | GCDFP15, PanCK, CD45 | Overall | |
|---|---|---|---|---|
| Analyte stability | 48 h | |||
| Recovery 1 | 94.6% | 86.4% | 88.6% | 89.9% |
| Limit of detection | 1 cell/mL | |||
| Linear range | 1–64 cells/mL | |||
| Linearity | R2 ≥ 0.98 | R2 ≥ 0.98 | R2 ≥ 0.98 | R2 ≥ 0.98 |
| Sensitivity | 96.0% (86.3%–99.5%) | 98.0% (89.4%–99.9%) | 94.0% (83.5%–98.8%) | 94.0% (83.5%–98.8%) |
| Specificity | 100.0% (88.4%–100.0%) | 100.0% (88.4%–100.0%) | 100.0% (88.4%–100.0%) | 100.0% (88.4%–100.0%) |
| Accuracy | 97.5% (91.3% to 99.7%) | 98.8% (93.2% to 99.9%) | 96.3% (89.4%–99.2%) | 96.3% (89.4%–99.2%) |
| Precision | CV = 4.6% | CV = 3.9% | CV = 3.8% | CV = 4.1% |
| Robustness | CV < 5% | |||
| Case–Control Study, Cancer vs. Asymptomatic Specificity: 100.00% (95% CI: 99.87%–100.00%) | Prospective Study, Cancer vs. Benign Specificity: 93.10% (95% CI: 77.23%–99.15%) | |||
|---|---|---|---|---|
| Sensitivity | Accuracy | Sensitivity | Accuracy | |
| Cumulative | 92.07% 95% CI: 91.12%–93.03% | 99.57% 95% CI: 99.34%–99.81% | 94.64% 95% CI: 88.70%–98.01% | 94.33% 95% CI: 89.13%–97.52% |
| Stage 0 | 70.00% 95% CI: 34.75%–93.33% | 99.90% 95% CI: 99.70%–99.98% | 87.50% 95% CI: 67.64%–97.34% | 90.57% 95% CI: 79.34%–96.87% |
| Stage I | 89.36% 95% CI: 76.90%–96.45% | 99.81% 95% CI: 99.60%–99.94% | 95.83% 95% CI: 78.88%–99.89% | 94.34% 95% CI: 84.34%–98.82% |
| Stage II | 95.74% 95% CI: 85.46%–99.48% | 99.91% 95% CI: 99.75%–99.99% | 95.83% 95% CI: 78.88%–99.89% | 94.34% 95% CI: 84.34%–98.82% |
| Stage III | 100.0% 95% CI: 88.43%–100.00% | 100.0% 95% CI: 99.87%–100.00% | 95.00% 95% CI: 75.13%–99.87% | 93.88% 95% CI: 83.13%–98.72% |
| Stage IV | 100.0% 95% CI: 88.43%–100.00% | 100.0% 95% CI: 99.87%–100.00% | 100.00% 95% CI: 83.16%–100.00% | 95.92% 95% CI: 86.02%–99.50% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Crook, T.; Leonard, R.; Mokbel, K.; Thompson, A.; Michell, M.; Page, R.; Vaid, A.; Mehrotra, R.; Ranade, A.; Limaye, S.; et al. Accurate Screening for Early-Stage Breast Cancer by Detection and Profiling of Circulating Tumor Cells. Cancers 2022, 14, 3341. https://doi.org/10.3390/cancers14143341
Crook T, Leonard R, Mokbel K, Thompson A, Michell M, Page R, Vaid A, Mehrotra R, Ranade A, Limaye S, et al. Accurate Screening for Early-Stage Breast Cancer by Detection and Profiling of Circulating Tumor Cells. Cancers. 2022; 14(14):3341. https://doi.org/10.3390/cancers14143341
Chicago/Turabian StyleCrook, Timothy, Robert Leonard, Kefah Mokbel, Alastair Thompson, Michael Michell, Raymond Page, Ashok Vaid, Ravi Mehrotra, Anantbhushan Ranade, Sewanti Limaye, and et al. 2022. "Accurate Screening for Early-Stage Breast Cancer by Detection and Profiling of Circulating Tumor Cells" Cancers 14, no. 14: 3341. https://doi.org/10.3390/cancers14143341
APA StyleCrook, T., Leonard, R., Mokbel, K., Thompson, A., Michell, M., Page, R., Vaid, A., Mehrotra, R., Ranade, A., Limaye, S., Patil, D., Akolkar, D., Datta, V., Fulmali, P., Apurwa, S., Schuster, S., Srinivasan, A., & Datar, R. (2022). Accurate Screening for Early-Stage Breast Cancer by Detection and Profiling of Circulating Tumor Cells. Cancers, 14(14), 3341. https://doi.org/10.3390/cancers14143341








