Selective CDK9 Inhibition by Natural Compound Toyocamycin in Cancer Cells
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. GFP Cell-Based System, Natural Drug Library, and Drugs
2.2. Drug Screening Conditions
2.3. Flow Cytometry for GFP Expression and Data Analysis
2.4. DNA Methylation Analysis by Pyrosequencing
2.5. Histone Acetylation Analysis by Mass Spectrometry
2.6. Anchorage-Dependent Clonogenic Assays
2.7. RNA Sequencing and Transcriptomic Analysis
2.8. Cell Viability, Proliferation Assays, and Cell Cycle Analysis
2.9. Cyclin-Dependent Kinase Inhibition Assays
2.10. Western Blot and Plasmid Constructs
2.11. Molecular Docking Simulations
2.12. Statistics
3. Results
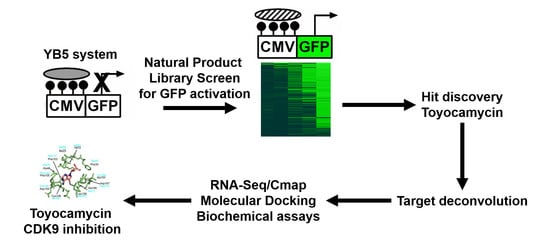
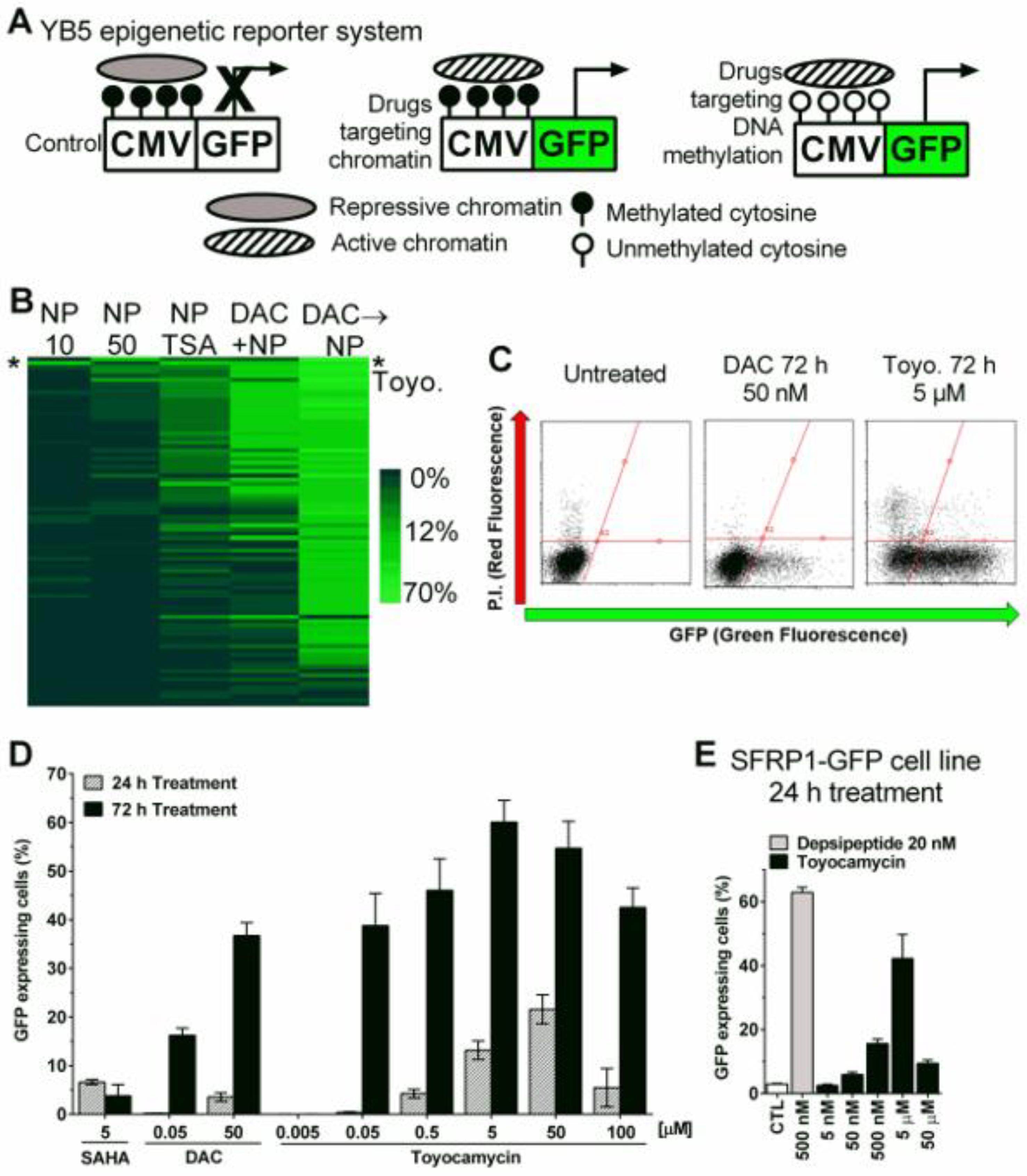
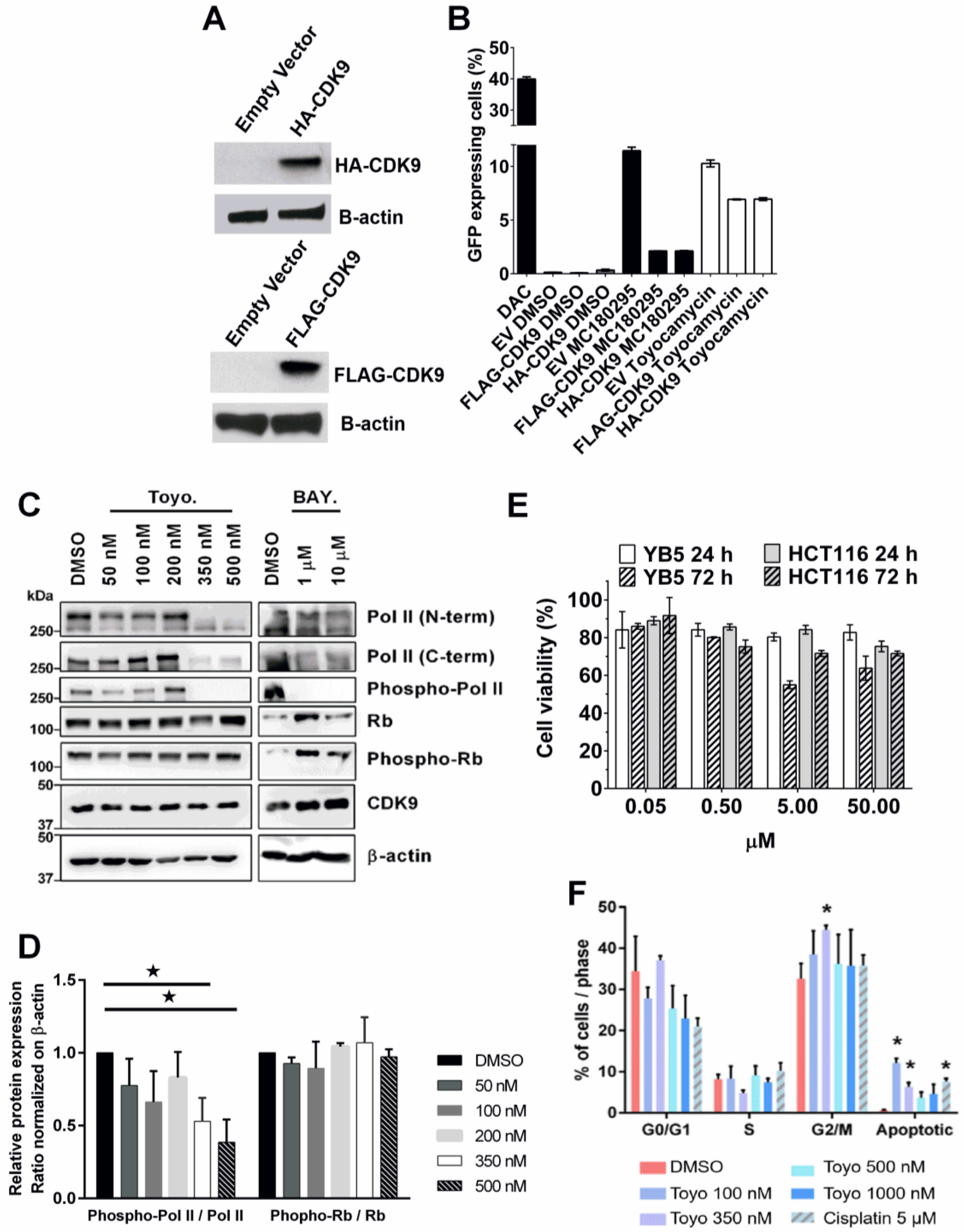
3.1. Epigenetic Drug Screening with the Natural Product Library
3.2. Toyocamycin Induced GFP Expression without Global Changes in DNA Methylation or Histone Acetylation Levels
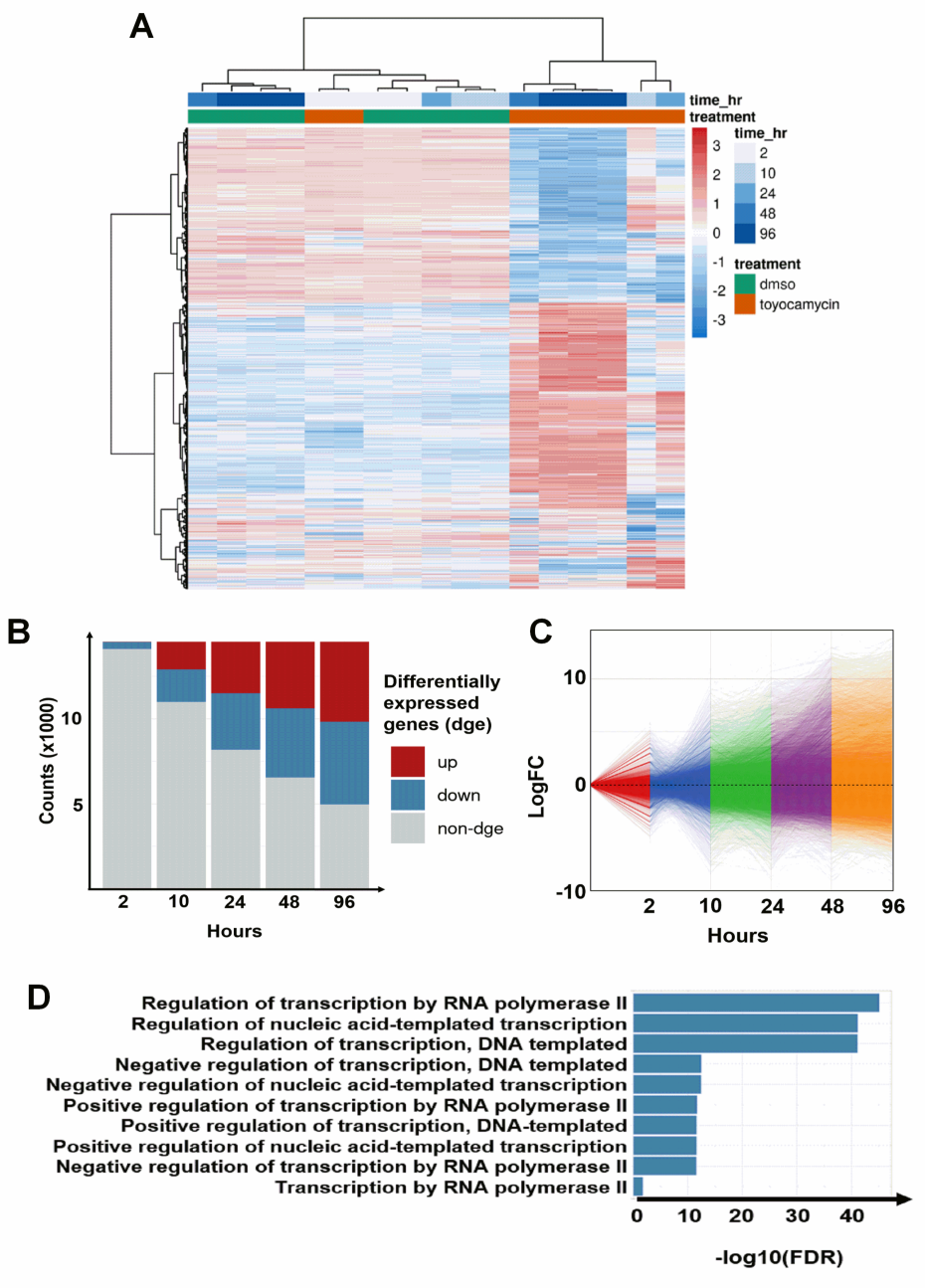
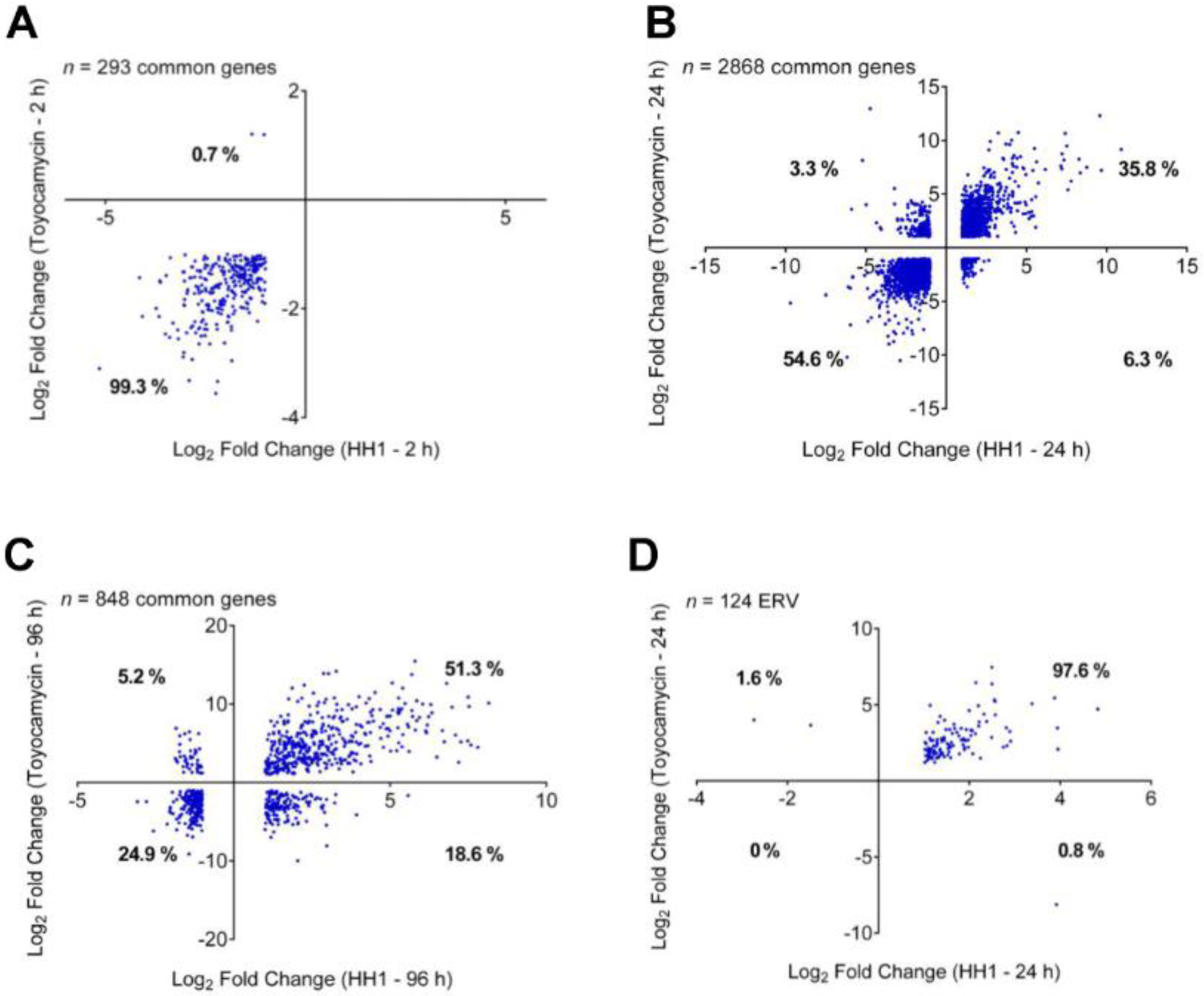
3.3. Regulation of Transcription by RNA Polymerase II Is Altered by Toyocamycin Treatment
3.4. Cyclin-Dependent Kinase 9 Inhibition by Toyocamycin
3.5. Toyocamycin Modulates CDK9 Enzymatic Activity in Colon Cancer Cell Lines
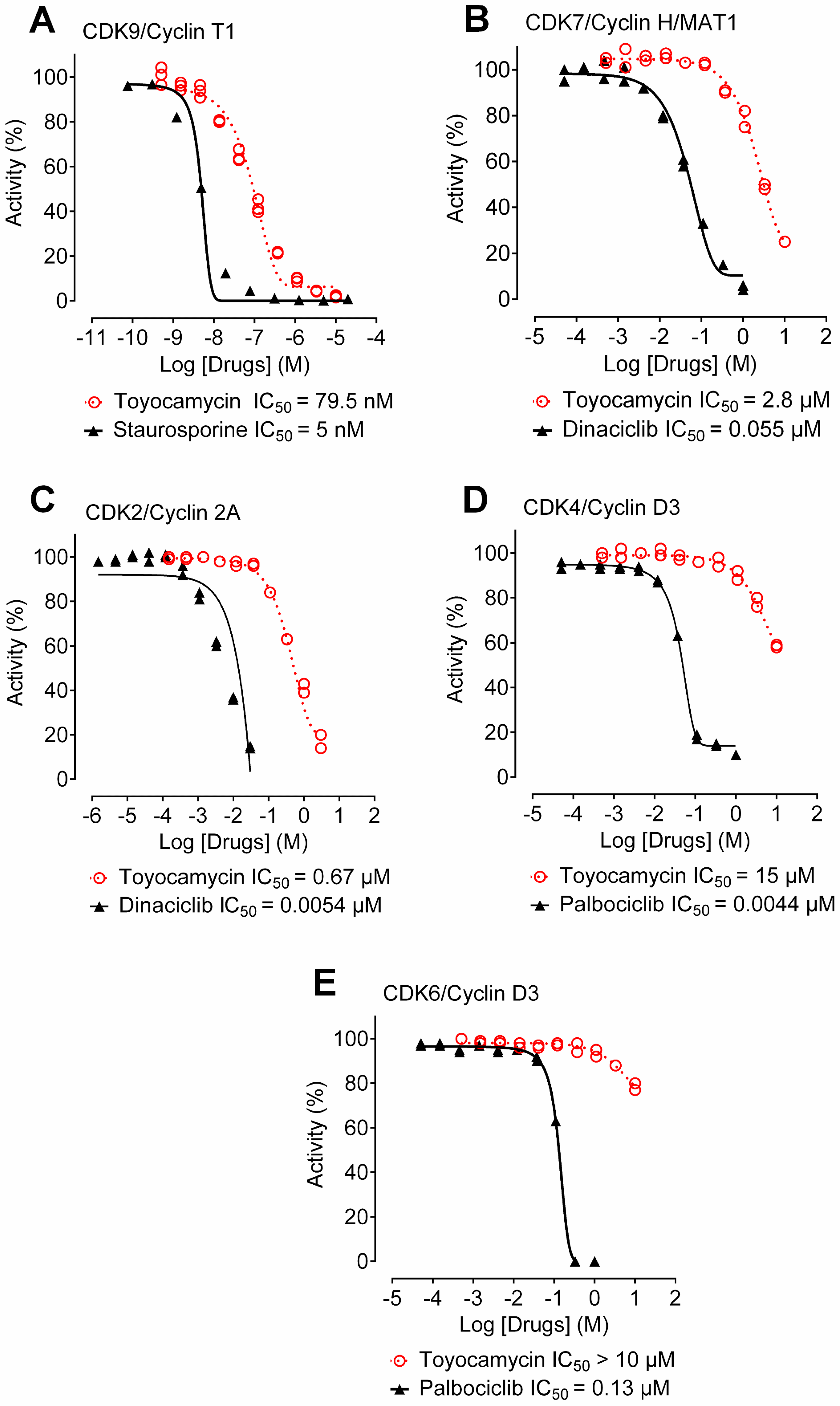
3.6. Toyocamycin Shows CDK9 Inhibition Selectivity in Kinase Assays
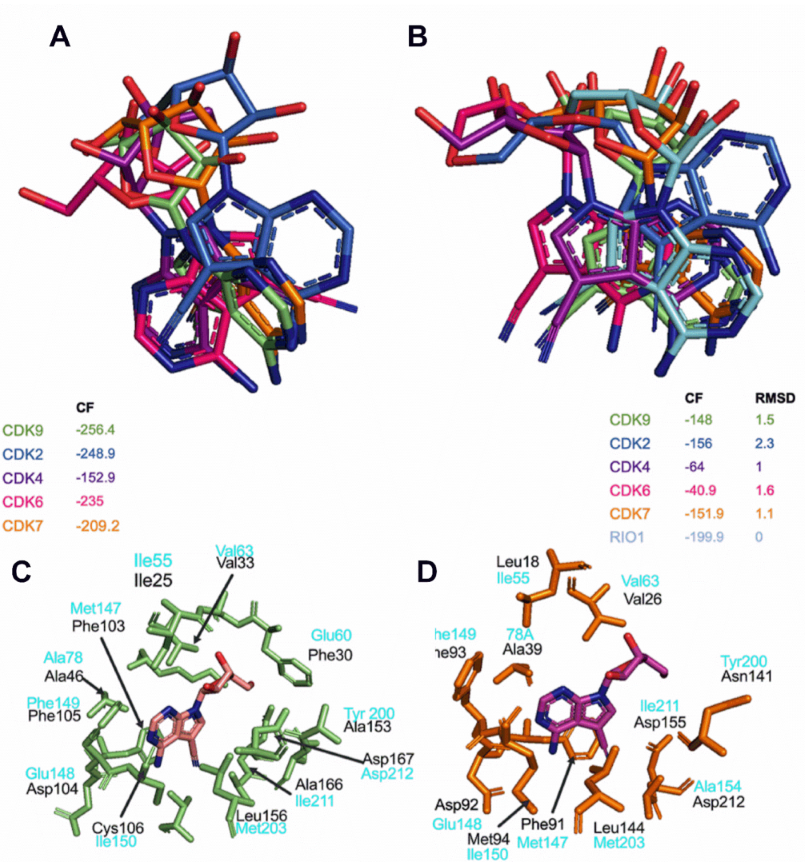
3.7. Toyocamycin Adopts a Specific Position in the CDK9 Catalytic Site Compared to Other CDKs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jones, P.A.; Issa, J.P.; Baylin, S. Targeting the cancer epigenome for therapy. Nat. Rev. Genet. 2016, 17, 630–641. [Google Scholar] [CrossRef] [PubMed]
- Baylin, S.B.; Jones, P.A. Epigenetic Determinants of Cancer. Cold Spring Harbor Perspect. Biol. 2016, 8, a019505. [Google Scholar] [CrossRef] [Green Version]
- Baylin, S.B.; Jones, P.A. A decade of exploring the cancer epigenome—Biological and translational implications. Nat. Rev. Cancer 2011, 11, 726–734. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.; Pruitt, K. Functional assessment of MeCP2 in Rett syndrome and cancers of breast, colon, and prostate. Biochem. Cell Biol. Biochim. Biol. Cell. 2017, 95, 368–378. [Google Scholar] [CrossRef] [PubMed]
- Kelly, T.K.; De Carvalho, D.D.; Jones, P.A. Epigenetic modifications as therapeutic targets. Nat. Biotechnol. 2010, 28, 1069–1078. [Google Scholar] [CrossRef] [Green Version]
- Taby, R.; Issa, J.P. Cancer epigenetics. CA A Cancer J. Clin. 2010, 60, 376–392. [Google Scholar] [CrossRef]
- Pandey, S.; Simmons, G.E.; Malyarchuk, S.; Calhoun, T.N.; Pruitt, K. A novel MeCP2 acetylation site regulates interaction with ATRX and HDAC1. Genes Cancer 2015, 6, 408–421. [Google Scholar] [CrossRef] [Green Version]
- Juergens, R.A.; Wrangle, J.; Vendetti, F.P.; Murphy, S.C.; Zhao, M.; Coleman, B.; Sebree, R.; Rodgers, K.; Hooker, C.M.; Franco, N.; et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov. 2011, 1, 598–607. [Google Scholar] [CrossRef] [Green Version]
- Mohammad, H.P.; Barbash, O.; Creasy, C.L. Targeting epigenetic modifications in cancer therapy: Erasing the roadmap to cancer. Nat. Med. 2019, 25, 403–418. [Google Scholar] [CrossRef]
- Julia, E.; Salles, G. EZH2 inhibition by tazemetostat: Mechanisms of action, safety and efficacy in relapsed/refractory follicular lymphoma. Futur. Oncol. 2021, 17, 2127–2140. [Google Scholar] [CrossRef]
- Liu, X.; Gong, Y. Isocitrate dehydrogenase inhibitors in acute myeloid leukemia. Biomark. Res. 2019; 7, 22. [Google Scholar]
- Arrowsmith, C.H.; Bountra, C.; Fish, P.V.; Lee, K.; Schapira, M. Epigenetic protein families: A new frontier for drug discovery. Nat. Rev. Drug Discov. 2012, 11, 384–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raynal, N.J.-M.; Si, J.; Taby, R.F.; Gharibyan, V.; Ahmed, S.; Jelinek, J.; Estécio, M.R.; Issa, J.-P.J. DNA Methylation Does Not Stably Lock Gene Expression but Instead Serves as a Molecular Mark for Gene Silencing Memory. Cancer Res. 2012, 72, 1170–1181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Si, J.; Boumber, Y.A.; Shu, J.; Qin, T.; Ahmed, S.; He, R.; Jelinek, J.; Issa, J.-P.J. Chromatin remodeling is required for gene reactivation after decitabine-mediated DNA hypomethylation. Cancer Res. 2010, 70, 6968–6977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Pandey, S.; Travers, M.; Sun, H.; Morton, G.; Madzo, J.; Chung, W.; Khowsathit, J.; Perez-Leal, O.; Barrero, C.A.; et al. Targeting CDK9 Reactivates Epigenetically Silenced Genes in Cancer. Cell 2018, 175, 1244–1258 e.26. [Google Scholar] [CrossRef] [Green Version]
- Raynal, N.J.-M.; Lee, J.T.; Wang, Y.; Beaudry, A.; Madireddi, P.; Garriga, J.; Malouf, G.; Dumont, S.N.; Dettman, E.J.; Gharibyan, V.; et al. Targeting Calcium Signaling Induces Epigenetic Reactivation of Tumor Suppressor Genes in Cancer. Cancer Res. 2016, 76, 1494–1505. [Google Scholar] [CrossRef] [Green Version]
- Raynal, N.J.-M.; Da Costa, E.M.; Lee, J.T.; Gharibyan, V.; Ahmed, S.; Zhang, H.; Sato, T.; Malouf, G.G.; Issa, J.-P.J. Repositioning FDA-Approved Drugs in Combination with Epigenetic Drugs to Reprogram Colon Cancer Epigenome. Mol. Cancer Ther. 2017, 16, 397–407. [Google Scholar] [CrossRef] [Green Version]
- Huot, M.; Caron, M.; Richer, C.; Djibo, R.; Najmanovich, R.; St-Onge, P.; Sinnett, D.; Raynal, N.J.M. Repurposing proscillaridin A in combination with decitabine against embryonal rhabdomyosarcoma RD cells. Cancer Chemother. Pharmacol. 2021, 88, 845–856. [Google Scholar] [CrossRef]
- Da Costa, E.M.; Armaos, G.; McInnes, G.; Beaudry, A.; Moquin-Beaudry, G.; Bertrand-Lehouillier, V.; Caron, M.; Richer, C.; St-Onge, P.; Johnson, J.R.; et al. Heart failure drug proscillaridin A targets MYC overexpressing leukemia through global loss of lysine acetylation. J. Exp. Clin. Cancer Res. 2019, 38, 251. [Google Scholar] [CrossRef]
- Lahlou, M. The Success of Natural Products in Drug Discovery. Pharmacol. Pharm. 2013, 4, 17–31. [Google Scholar] [CrossRef] [Green Version]
- Spannhoff, A.; Kim, Y.K.; Raynal, N.J.M.; Gharibyan, V.; Su, M.; Zhou, Y.; Li, J.; Castellano, S.; Sbardella, G.; Issa, J.-P.; et al. Histone deacetylase inhibitor activity in royal jelly might facilitate caste switching in bees. EMBO Rep. 2011, 12, 238–243. [Google Scholar] [CrossRef] [Green Version]
- Ri, M.; Tashiro, E.; Oikawa, D.; Shinjo, S.K.; Tokuda, M.; Yokouchi, Y.; Narita, T.; Masaki, A.; Ito, A.; Ding, J.; et al. Identification of Toyocamycin, an agent cytotoxic for multiple myeloma cells, as a potent inhibitor of ER stress-induced XBP1 mRNA splicing. Blood Cancer J. 2012, 2, e79. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Hausheer, F.; Beaty, R.; Zahnow, C.; Issa, J.P.; Bunz, F.; Baylin, S.B. A recombinant reporter system for monitoring reactivation of an endogenously DNA hypermethylated gene. Cancer Res. 2014, 74, 3834–3843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henry, R.A.; Kuo, Y.M.; Andrews, A.J. Differences in Specificity and Selectivity Between CBP and p300 acetylation of histone H3 and H3/H4. Biochemistry 2013, 52, 5746–5759. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Shafiq, M.I.; Steinbrecher, T.; Schmid, R. Fascaplysin as a specific inhibitor for CDK4: Insights from molecular modelling. PLoS ONE 2012, 7, e42612. [Google Scholar] [CrossRef]
- Gaudreault, F.; Najmanovich, R.J. FlexAID: Revisiting Docking on Non-Native-Complex Structures. J. Chem. Inf. Model. 2015, 55, 1323–1336. [Google Scholar] [CrossRef]
- Raynal, N.J.; Momparler, L.F.; Rivard, G.E.; Momparler, R.L. 3-Deazauridine enhances the antileukemic action of 5-aza-2’-deoxycytidine and targets drug-resistance due to deficiency in deoxycytidine kinase. Leuk. Res. 2011, 35, 110–118. [Google Scholar] [CrossRef]
- Cohen, M.B.; Glazer, R.I. Comparison of the cellular and RNA-dependent effects of sangivamycin and toyocamycin in human colon carcinoma cells. Mol. Pharmacol. 1985, 27, 349–355. [Google Scholar]
- Nishimura, H.; Katagiri, K.; Sato, K.; Mayama, M.; Shimaoka, N. Toyocamycin, a new anti-candida antibiotics. J. Antibiot. 1956, 9, 60–62. [Google Scholar]
- Lamb, J.; Crawford, E.D.; Peck, D.; Modell, J.W.; Blat, I.C.; Wrobel, M.J.; Lerner, J.; Brunet, J.-P.; Subramanian, A.; Ross, K.N.; et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science 2006, 313, 1929–1935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schultz, C.; Link, A.; Leost, M.; Zaharevitz, D.W.; Gussio, R.; Sausville, E.A.; Meijer, A.L.; Kunick, C. Paullones, a series of cyclin-dependent kinase inhibitors: Synthesis, evaluation of CDK1/cyclin B inhibition, and in vitro antitumor activity. J. Med. Chem. 1999, 42, 2909–2919. [Google Scholar] [CrossRef]
- Ren, P.X.; Shang, W.J.; Yin, W.C.; Ge, H.; Wang, L.; Zhang, X.L.; Li, B.Q.; Li, H.L.; Xu, Y.C.; Xu, E.H.; et al. A multi-targeting drug design strategy for identifying potent anti-SARS-CoV-2 inhibitors. Acta Pharmacol. Sin. 2021, 43, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.; Liu, L.; Majdzadeh, N.; Chavez, C.; Chin, P.; Morrison, B.; Wang, L.; Park, J.; Chugh, P.; Chen, H.; et al. Inhibition of neuronal apoptosis by the cyclin-dependent kinase inhibitor GW8510: Identification of 3’ substituted indolones as a scaffold for the development of neuroprotective drugs. J. Neurochem. 2005, 93, 538–548. [Google Scholar] [CrossRef] [PubMed]
- Chung, F.H.; Jin, Z.H.; Hsu, T.T.; Hsu, C.L.; Liu, H.C.; Lee, H.C. Gene-Set Local Hierarchical Clustering (GSLHC)--A Gene Set-Based Approach for Characterizing Bioactive Compounds in Terms of Biological Functional Groups. PLoS ONE 2015, 10, e0139889. [Google Scholar] [CrossRef]
- Felipo, V.; Minana, M.D.; Cabedo, H.; Perez-Minguez, F.; Llombart-Bosch, A.; Grisolia, S. H7, an inhibitor of protein kinase C, inhibits tumour cell division in mice bearing ascitic Ehrlich’s carcinoma. Eur. J. Cancer 1994, 30, 525–527. [Google Scholar] [CrossRef]
- Pommier, Y. Topoisomerase I inhibitors: Camptothecins and beyond. Nat. Rev. Cancer 2006, 6, 789–802. [Google Scholar] [CrossRef]
- Da Costa, E.M.; McInnes, G.; Beaudry, A.; Raynal, N.J. DNA Methylation-Targeted Drugs. Cancer J. 2017, 23, 270–276. [Google Scholar] [CrossRef]
- Bailly, C. Irinotecan: 25 years of cancer treatment. Pharmacol. Res. 2019, 148, 104398. [Google Scholar] [CrossRef]
- Coulthard, S.A.; McGarrity, S.; Sahota, K.; Berry, P.; Redfern, C.P.F. Three Faces of Mercaptopurine Cytotoxicity In Vitro: Methylation, Nucleotide Homeostasis, and Deoxythioguanosine in DNA. Drug Metab. Dispos. 2018, 46, 1191–1199. [Google Scholar] [CrossRef] [PubMed]
- Anshabo, A.T.; Milne, R.; Wang, S.; Albrecht, H. CDK9: A Comprehensive Review of Its Biology, and Its Role as a Potential Target for Anti-Cancer Agents. Front. Oncol. 2021, 11, 678559. [Google Scholar] [CrossRef] [PubMed]
- Mayer, A.; di Iulio, J.; Maleri, S.; Eser, U.; Vierstra, J.; Reynolds, A.; Sandstrom, R.; Stamatoyannopoulos, J.A.; Churchman, L.S. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell 2015, 161, 541–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pack, L.R.; Daigh, L.H.; Chung, M.; Meyer, T. Clinical CDK4/6 inhibitors induce selective and immediate dissociation of p21 from cyclin D-CDK4 to inhibit CDK2. Nat. Commun. 2021, 12, 3356. [Google Scholar] [CrossRef] [PubMed]
- McGovern, S.L.; Shoichet, B.K. Information decay in molecular docking screens against holo, apo, and modeled conformations of enzymes. J. Med. Chem. 2003, 46, 2895–2907. [Google Scholar] [CrossRef]
- Kiburu, I.N.; LaRonde-LeBlanc, N. Interaction of Rio1 kinase with toyocamycin reveals a conformational switch that controls oligomeric state and catalytic activity. PLoS ONE 2012, 7, e37371. [Google Scholar] [CrossRef] [Green Version]
- Kurihara, Y.; Egawa, K.; Kunimoto, S.; Takeuchi, T.; Nose, K. Induction of p16/INK4a gene expression and cellular senescence by toyocamycin. Biol. Pharm. Bull. 2002, 25, 1272–1276. [Google Scholar] [CrossRef] [Green Version]
- Weiss, J.W.; Pitot, H.C. Inhibition of ribosomal RNA maturation in Novikoff hepatoma cells by toyocamycin, tubercidin, and 6-thioguanosine. Cancer Res. 1974, 34, 581–587. [Google Scholar]
- Olson, C.M.; Jiang, B.; Erb, M.A.; Liang, Y.; Doctor, Z.M.; Zhang, Z.; Zhang, T.; Kwiatkowski, N.; Boukhali, M.; Green, J.L.; et al. Pharmacological perturbation of CDK9 using selective CDK9 inhibition or degradation. Nat. Chem. Biol. 2018, 14, 163–170. [Google Scholar] [CrossRef]
- Mandal, R.; Becker, S.; Strebhardt, K. Targeting CDK9 for Anti-Cancer Therapeutics. Cancers 2021, 13, 2181. [Google Scholar] [CrossRef]
- Morales, F.; Giordano, A. Overview of CDK9 as a target in cancer research. Cell Cycle 2016, 15, 519–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dolloff, N.G.; Allen, J.E.; Dicker, D.T.; Aqui, N.; Vogl, D.; Malysz, J.; Talamo, G.; El-Deiry, W.S. Sangivamycin-like molecule 6 exhibits potent anti-multiple myeloma activity through inhibition of cyclin-dependent kinase-9. Mol. Cancer Ther. 2012, 11, 2321–2330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graña, X.; De Luca, A.; Sang, N.; Fu, Y.; Claudio, P.P.; Rosenblatt, J.; O Morgan, D.; Giordano, A. PITALRE, a nuclear CDC2-related protein kinase that phosphorylates the retinoblastoma protein in vitro. Proc. Natl. Acad. Sci. USA 1994, 91, 3834–3838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boffo, S.; Damato, A.; Alfano, L.; Giordano, A. CDK9 inhibitors in acute myeloid leukemia. J. Exp. Clin. Cancer Res. 2018, 37, 36. [Google Scholar] [CrossRef] [Green Version]
- Vervoort, S.J.; Devlin, J.R.; Kwiatkowski, N.; Teng, M.; Gray, N.S.; Johnstone, R.W. Targeting transcription cycles in cancer. Nat. Rev. Cancer 2022, 22, 5–24. [Google Scholar] [CrossRef]
- Poon, E.; Liang, T.; Jamin, Y.; Walz, S.; Kwok, C.; Hakkert, A.; Barker, K.; Urban, Z.; Thway, K.; Zeid, R.; et al. Orally bioavailable CDK9/2 inhibitor shows mechanism-based therapeutic potential in MYCN-driven neuroblastoma. J. Clin. Investig. 2020, 130, 5875–5892. [Google Scholar] [CrossRef]
- Barlaam, B.; Casella, R.; Cidado, J.; Cook, C.; De Savi, C.; Dishington, A.; Donald, C.S.; Drew, L.; Ferguson, A.D.; Ferguson, D.; et al. Discovery of AZD4573, a Potent and Selective Inhibitor of CDK9 That Enables Short Duration of Target Engagement for the Treatment of Hematological Malignancies. J. Med. Chem. 2020, 63, 15564–15590. [Google Scholar] [CrossRef]
- Cidado, J.; Boiko, S.; Proia, T.; Ferguson, D.; Criscione, S.W.; Martin, M.S.; Pop-Damkov, P.; Su, N.; Franklin, V.N.R.; Chilamakuri, C.S.R.; et al. AZD4573 Is a Highly Selective CDK9 Inhibitor That Suppresses MCL-1 and Induces Apoptosis in Hematologic Cancer Cells. Clin. Cancer Res. 2020, 26, 922–934. [Google Scholar] [CrossRef] [Green Version]
- Diamond, J.R.; Boni, V.; Lim, E.; Nowakowski, G.; Cordoba, R.; Morillo, D.; Valencia, R.; Genvresse, I.; Merz, C.; Boix, O.; et al. First-in-Human Dose-Escalation Study of Cyclin-Dependent Kinase 9 Inhibitor VIP152 in Patients with Advanced Malignancies Shows Early Signs of Clinical Efficacy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2022, 28, 1285–1293. [Google Scholar] [CrossRef]
- Wilson, W.L. Phase I study with toyocamycin (NSC-63701). Cancer Chemother Rep. 1968, 52, 301–303. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandey, S.; Djibo, R.; Darracq, A.; Calendo, G.; Zhang, H.; Henry, R.A.; Andrews, A.J.; Baylin, S.B.; Madzo, J.; Najmanovich, R.; et al. Selective CDK9 Inhibition by Natural Compound Toyocamycin in Cancer Cells. Cancers 2022, 14, 3340. https://doi.org/10.3390/cancers14143340
Pandey S, Djibo R, Darracq A, Calendo G, Zhang H, Henry RA, Andrews AJ, Baylin SB, Madzo J, Najmanovich R, et al. Selective CDK9 Inhibition by Natural Compound Toyocamycin in Cancer Cells. Cancers. 2022; 14(14):3340. https://doi.org/10.3390/cancers14143340
Chicago/Turabian StylePandey, Somnath, Rahinatou Djibo, Anaïs Darracq, Gennaro Calendo, Hanghang Zhang, Ryan A. Henry, Andrew J. Andrews, Stephen B. Baylin, Jozef Madzo, Rafael Najmanovich, and et al. 2022. "Selective CDK9 Inhibition by Natural Compound Toyocamycin in Cancer Cells" Cancers 14, no. 14: 3340. https://doi.org/10.3390/cancers14143340
APA StylePandey, S., Djibo, R., Darracq, A., Calendo, G., Zhang, H., Henry, R. A., Andrews, A. J., Baylin, S. B., Madzo, J., Najmanovich, R., Issa, J.-P. J., & Raynal, N. J.-M. (2022). Selective CDK9 Inhibition by Natural Compound Toyocamycin in Cancer Cells. Cancers, 14(14), 3340. https://doi.org/10.3390/cancers14143340