Prospective Proteomic Study Identifies Potential Circulating Protein Biomarkers for Colorectal Cancer Risk
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods and Materials
2.1. Study Design, Population, and Data
2.2. Laboratory Methods
2.3. Statistical Analysis
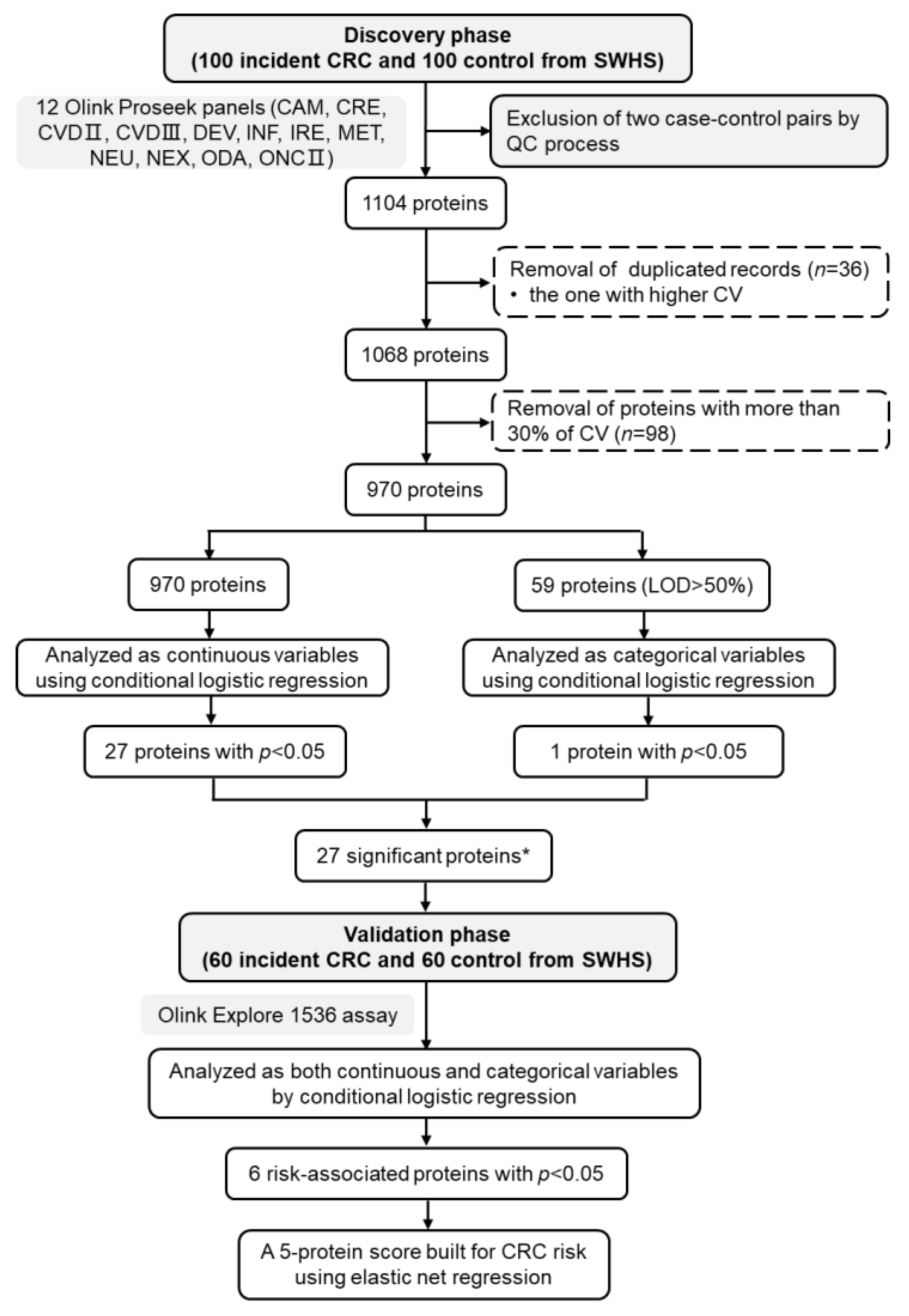
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of colorectal cancer: Incidence, mortality, survival, and risk factors. Przegląd Gastroenterol. 2019, 14, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.T.; Giovannucci, E.L. Primary prevention of colorectal cancer. Gastroenterology 2010, 138, 2029–2043.e10. [Google Scholar] [CrossRef] [PubMed]
- Keum, N.; Giovannucci, E. Global burden of colorectal cancer: Emerging trends, risk factors and prevention strategies. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 713–732. [Google Scholar] [CrossRef]
- Alberts, B.; Johnson, A.; Lewis, J.; Morgan, D.; Raff, M.; Roberts, K.; Walter, P.; Wilson, J.; Hunt, T. Molecular Biology of the Cell; WW Norton & Company: New York, NY, USA, 2017. [Google Scholar]
- Huang, W.; Luo, S.; Burgess, R.; Yi, Y.H.; Huang, G.F.; Huang, R.P. New insights into the tumor microenvironment utilizing protein array technology. Int. J. Mol. Sci. 2018, 19, 559. [Google Scholar] [CrossRef]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, inflammation, and cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef]
- Galdiero, M.R.; Marone, G.; Mantovani, A. Cancer inflammation and cytokines. Cold Spring Harb. Perspect Biol. 2018, 10, a028662. [Google Scholar] [CrossRef]
- Tsilidis, K.K.; Branchini, C.; Guallar, E.; Helzlsouer, K.J.; Erlinger, T.P.; Platz, E.A. C-reactive protein and colorectal cancer risk: A systematic review of prospective studies. Int. J. Cancer 2008, 123, 1133–1140. [Google Scholar] [CrossRef]
- Prizment, A.E.; Anderson, K.E.; Visvanathan, K.; Folsom, A.R. Association of inflammatory markers with colorectal cancer incidence in the atherosclerosis risk in communities study. Cancer Epidemiol. Biomarkers Prev. 2011, 20, 297–307. [Google Scholar] [CrossRef]
- Mehta, R.S.; Song, M.; Bezawada, N.; Wu, K.; Garcia-Albeniz, X.; Morikawa, T.; Fuchs, C.S.; Ogino, S.; Giovannucci, E.L.; Chan, A.T. A prospective study of macrophage inhibitory cytokine-1 (MIC-1/GDF15) and risk of colorectal cancer. J. Natl. Cancer Inst. 2014, 106, dju016. [Google Scholar] [CrossRef]
- Kakourou, A.; Koutsioumpa, C.; Lopez, D.S.; Hoffman-Bolton, J.; Bradwin, G.; Rifai, N.; Helzlsouer, K.J.; Platz, E.A.; Tsilidis, K.K. Interleukin-6 and risk of colorectal cancer: Results from the CLUE II cohort and a meta-analysis of prospective studies. Cancer Causes Control 2015, 26, 1449–1460. [Google Scholar] [CrossRef] [PubMed]
- Murphy, N.; Carreras-Torres, R.; Song, M.; Chan, A.T.; Martin, R.M.; Papadimitriou, N.; Dimou, N.; Tsilidis, K.K.; Banbury, B.; Bradbury, K.E.; et al. Circulating levels of insulin-like growth factor 1 and insulin-like growth factor binding protein 3 associate with risk of colorectal cancer based on serologic and mendelian randomization analyses. Gastroenterology 2020, 158, 1300–1312.e20. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zucknick, M.; Werner, S.; Knebel, P.; Brenner, H. Head-to-head comparison and evaluation of 92 plasma protein biomarkers for early detection of colorectal cancer in a true screening setting. Clin. Cancer Res. 2015, 21, 3318–3326. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, K.; Naito, Y.; Yagi, N.; Mizushima, K.; Higashimura, Y.; Hirai, Y.; Dohi, O.; Okayama, T.; Yoshida, N.; Katada, K.; et al. Selected reaction monitoring for colorectal cancer diagnosis using a set of five serum peptides identified by BLOTCHIP((R))-MS analysis. J. Gastroenterol. 2018, 53, 1179–1185. [Google Scholar] [CrossRef]
- Harlid, S.; Harbs, J.; Myte, R.; Brunius, C.; Gunter, M.J.; Palmqvist, R.; Liu, X.; Van Guelpen, B. A two-tiered targeted proteomics approach to identify pre-diagnostic biomarkers of colorectal cancer risk. Sci. Rep. 2021, 11, 5151. [Google Scholar] [CrossRef]
- Zheng, W.; Chow, W.H.; Yang, G.; Jin, F.; Rothman, N.; Blair, A.; Li, H.L.; Wen, W.; Ji, B.T.; Li, Q.; et al. The Shanghai Women’s Health Study: Rationale, study design, and baseline characteristics. Am. J. Epidemiol. 2005, 162, 1123–1131. [Google Scholar] [CrossRef]
- Harlid, S.; Gunter, M.J.; Van Guelpen, B. Risk-predictive and diagnostic biomarkers for colorectal cancer; A systematic review of studies using pre-diagnostic blood samples collected in prospective cohorts and screening settings. Cancers 2021, 13, 4406. [Google Scholar] [CrossRef]
- Nguyen, M.T.; Weinberg, D.S. Biomarkers in colorectal cancer screening. J. Natl. Compr. Canc. Netw. 2016, 14, 1033–1040. [Google Scholar] [CrossRef][Green Version]
- Suppiah, A.; Greenman, J. Clinical utility of anti-p53 auto-antibody: Systematic review and focus on colorectal cancer. World J. Gastroenterol. 2013, 19, 4651–4670. [Google Scholar] [CrossRef]
- Chen, X.; Sun, J.; Wang, X.; Yuan, Y.; Cai, L.; Xie, Y.; Fan, Z.; Liu, K.; Jiao, X. A meta-analysis of proteomic blood markers of colorectal cancer. Curr. Med. Chem. 2021, 28, 1176–1196. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Qian, J.; Werner, S.; Cuk, K.; Knebel, P.; Brenner, H. Development and validation of a panel of five proteins as blood biomarkers for early detection of colorectal cancer. Clin. Epidemiol. 2017, 9, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Rho, J.H.; Ladd, J.J.; Li, C.I.; Potter, J.D.; Zhang, Y.; Shelley, D.; Shibata, D.; Coppola, D.; Yamada, H.; Toyoda, H.; et al. Protein and glycomic plasma markers for early detection of adenoma and colon cancer. Gut 2018, 67, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Shao, S.; Neely, B.A.; Kao, T.C.; Eckhaus, J.; Bourgeois, J.; Brooks, J.; Jones, E.E.; Drake, R.R.; Zhu, K. Proteomic profiling of serial prediagnostic serum samples for early detection of colon cancer in the U.S. Military. Cancer Epidemiol. Biomarkers Prev. 2017, 26, 711–718. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Sasazuki, S.; Camargo, M.C.; Shimazu, T.; Charvat, H.; Yamaji, T.; Sawada, N.; Kemp, T.J.; Pfeiffer, R.M.; Hildesheim, A.; et al. Circulating inflammatory markers and colorectal cancer risk: A prospective case-cohort study in Japan. Int. J. Cancer 2018, 143, 2767–2776. [Google Scholar] [CrossRef]
- El-Naggar, M.H.; Mira, A.; Abdel Bar, F.M.; Shimizu, K.; Amer, M.M.; Badria, F.A. Synthesis, docking, cytotoxicity, and LTA4H inhibitory activity of new gingerol derivatives as potential colorectal cancer therapy. Bioorg. Med. Chem. 2017, 25, 1277–1285. [Google Scholar] [CrossRef]
- Young, R.E.; Voisin, M.B.; Wang, S.; Dangerfield, J.; Nourshargh, S. Role of neutrophil elastase in LTB4-induced neutrophil transmigration in vivo assessed with a specific inhibitor and neutrophil elastase deficient mice. Br. J. Pharmacol. 2007, 151, 628–637. [Google Scholar] [CrossRef]
- Haeggstrom, J.Z. Leukotriene A4 hydrolase/aminopeptidase, the gatekeeper of chemotactic leukotriene B4 biosynthesis. J. Biol. Chem. 2004, 279, 50639–50642. [Google Scholar] [CrossRef]
- Chen, X.; Li, N.; Wang, S.; Wu, N.; Hong, J.; Jiao, X.; Krasna, M.J.; Beer, D.G.; Yang, C.S. Leukotriene A4 hydrolase in rat and human esophageal adenocarcinomas and inhibitory effects of bestatin. J. Natl. Cancer Inst. 2003, 95, 1053–1061. [Google Scholar] [CrossRef]
- Zhao, S.; Yao, K.; Li, D.; Liu, K.; Jin, G.; Yan, M.; Wu, Q.; Chen, H.; Shin, S.H.; Bai, R.; et al. Inhibition of LTA4H by bestatin in human and mouse colorectal cancer. EBioMedicine 2019, 44, 361–374. [Google Scholar] [CrossRef]
- Jeong, C.H.; Bode, A.M.; Pugliese, A.; Cho, Y.Y.; Kim, H.G.; Shim, J.H.; Jeon, Y.J.; Li, H.; Jiang, H.; Dong, Z. [6]-Gingerol suppresses colon cancer growth by targeting leukotriene A4 hydrolase. Cancer Res. 2009, 69, 5584–5591. [Google Scholar] [CrossRef] [PubMed]
- Hafner, C.; Schmitz, G.; Meyer, S.; Bataille, F.; Hau, P.; Langmann, T.; Dietmaier, W.; Landthaler, M.; Vogt, T. Differential gene expression of Eph receptors and ephrins in benign human tissues and cancers. Clin. Chem. 2004, 50, 490–499. [Google Scholar] [CrossRef] [PubMed]
- Arvanitis, D.N.; Davy, A. Regulation and misregulation of Eph/ephrin expression. Cell Adh. Migr. 2012, 6, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Saintigny, P.; Peng, S.; Zhang, L.; Sen, B.; Wistuba, I.I.; Lippman, S.M.; Girard, L.; Minna, J.D.; Heymach, J.V.; Johnson, F.M. Global evaluation of Eph receptors and ephrins in lung adenocarcinomas identifies EphA4 as an inhibitor of cell migration and invasion. Mol. Cancer Ther. 2012, 11, 2021–2032. [Google Scholar] [CrossRef]
- Bernhard, O.K.; Greening, D.W.; Barnes, T.W.; Ji, H.; Simpson, R.J. Detection of cadherin-17 in human colon cancer LIM1215 cell secretome and tumour xenograft-derived interstitial fluid and plasma. Biochim. Biophys. Acta 2013, 1834, 2372–2379. [Google Scholar] [CrossRef]
- Leitinger, B. Discoidin domain receptor functions in physiological and pathological conditions. Int. Rev. Cell Mol. Biol. 2014, 310, 39–87. [Google Scholar] [CrossRef]
- Hu, Y.; Liu, J.; Jiang, B.; Chen, J.; Fu, Z.; Bai, F.; Jiang, J.; Tang, Z. MiR-199a-5p loss up-regulated DDR1 aggravated colorectal cancer by activating epithelial-to-mesenchymal transition related signaling. Dig. Dis. Sci. 2014, 59, 2163–2172. [Google Scholar] [CrossRef]
- Romayor, I.; Marquez, J.; Benedicto, A.; Herrero, A.; Arteta, B.; Olaso, E. Tumor DDR1 deficiency reduces liver metastasis by colon carcinoma and impairs stromal reaction. Am. J. Physiol. Gastrointest. Liver Physiol. 2021, 320, G1002–G1013. [Google Scholar] [CrossRef]
- Jeitany, M.; Leroy, C.; Tosti, P.; Lafitte, M.; Le Guet, J.; Simon, V.; Bonenfant, D.; Robert, B.; Grillet, F.; Mollevi, C.; et al. Inhibition of DDR1-BCR signalling by nilotinib as a new therapeutic strategy for metastatic colorectal cancer. EMBO Mol. Med. 2018, 10, e7918. [Google Scholar] [CrossRef]
- Gazit, R.; Gruda, R.; Elboim, M.; Arnon, T.I.; Katz, G.; Achdout, H.; Hanna, J.; Qimron, U.; Landau, G.; Greenbaum, E.; et al. Lethal influenza infection in the absence of the natural killer cell receptor gene Ncr1. Nat. Immunol. 2006, 7, 517–523. [Google Scholar] [CrossRef]
- Elboim, M.; Gazit, R.; Gur, C.; Ghadially, H.; Betser-Cohen, G.; Mandelboim, O. Tumor immunoediting by NKp46. J. Immunol. 2010, 184, 5637–5644. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, S.; Chiorazzi, N.; Gregersen, P.K. Alternative splicing of CD79a (Ig-alpha/mb-1) and CD79b (Ig-beta/B29) RNA transcripts in human B cells. Mol. Immunol. 1995, 32, 651–659. [Google Scholar] [CrossRef]
- Singh, M.P.; Rai, S.; Singh, N.K.; Srivastava, S. Transcriptomic landscape of early age onset of colorectal cancer identifies novel genes and pathways in Indian CRC patients. Sci. Rep. 2021, 11, 11765. [Google Scholar] [CrossRef] [PubMed]
- Flintoff, K.A.; Arudchelvan, Y.; Gong, S.G. FLRT2 interacts with fibronectin in the ATDC5 chondroprogenitor cells. J. Cell. Physiol. 2014, 229, 1538–1547. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Song, C.; Fang, L.; Li, M.; Yue, L.; Sun, Q. FLRT2 functions as Tumor Suppressor gene inactivated by promoter methylation in Colorectal Cancer. J. Cancer 2020, 11, 7329–7338. [Google Scholar] [CrossRef]
- Peng, Y.; Liu, C.; Li, M.; Li, W.; Zhang, M.; Jiang, X.; Chang, Y.; Liu, L.; Wang, F.; Zhao, Q. Identification of a prognostic and therapeutic immune signature associated with hepatocellular carcinoma. Cancer Cell Int. 2021, 21, 98. [Google Scholar] [CrossRef]
- Khaket, T.P.; Singh, M.P.; Khan, I.; Kang, S.C. In vitro and in vivo studies on potentiation of curcumin-induced lysosomal-dependent apoptosis upon silencing of cathepsin C in colorectal cancer cells. Pharmacol. Res. 2020, 161, 105156. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Y.; Ma, J.; Yang, C.; Wang, M.; Lv, J.; Wang, Y.; Miao, D.; Wang, Y.; Li, M.; et al. Determining the effects of Ephrin Type B Receptor 6 and Type A Receptor 3 on facilitating colorectal epithelial cell malignant transformation. Neoplasma 2021, 68, 955–964. [Google Scholar] [CrossRef]
- Bhardwaj, M.; Gies, A.; Weigl, K.; Tikk, K.; Benner, A.; Schrotz-King, P.; Borchers, C.H.; Brenner, H. Evaluation and validation of plasma proteins using two different protein detection methods for early detection of colorectal cancer. Cancers 2019, 11, 1426. [Google Scholar] [CrossRef]
| Characteristics | Discovery, No. (%) | Validation, No. (%) | ||||
|---|---|---|---|---|---|---|
| Case (n = 98) | Control (n = 98) | p | Case (n = 60) | Control (n = 60) | p | |
| Age at blood draw, mean (SD), y | 59.2 (8.8) | 58.92 (8.7) | 0.021 | 60.1 (8.7) | 60.1 (8.7) | 0.998 |
| BMI, mean (SD), kg/m2 | 24.4 (3.0) | 24.9 (3.5) | 0.295 | 25.2 (4.0) | 25.0 (3.4) | 0.806 |
| WHR, mean (SD) | 0.8 (0.1) | 0.8 (0.1) | 0.409 | 0.8 (0.1) | 0.8 (0.1) | 0.844 |
| Family income, % | ||||||
| <10,000 RMB | 23 (23.5) | 19 (19.4) | 0.922 | 18 (30.0) | 18 (30.0) | 0.765 |
| 10,000 RMB- | 40 (40.8) | 42 (42.9) | 22 (36.7) | 21 (35.0) | ||
| 20,000 RMB- | 20 (20.4) | 21 (21.4) | 17 (28.3) | 15 (25.0) | ||
| ≥30,000 RMB | 15 (15.3) | 16 (16.3) | 3 (5.0) | 6 (10.0) | ||
| Educational attainment, % | ||||||
| ≤Elementary school | 34 (34.7) | 43 (43.9) | 0.532 | 27 (45.0) | 25 (41.7) | 0.611 |
| Middle school | 29 (29.6) | 24 (24.5) | 16 (26.7) | 21 (35.0) | ||
| High school | 22(22.4) | 17 (17.3) | 10 (16.7) | 6 (10.0) | ||
| ≥College | 13 (13.3) | 14 (14.3) | 7 (11. 7) | 8 (13.3) | ||
| Physical activity, mean (SD), MET-hrs/day/yrs | 0.9 (1.4) | 1.0 (1.5) | 0.759 | 1.0 (1.4) | 1.0 (2.0) | 0.876 |
| Family history of adenomatous polyposis of colorectum, % | 1 (1.0) | 0 (0.0) | 1.000 | 0 (0.0) | 0 (0.0) | 1.000 |
| Family history of colorectal cancer, % | 3 (3.1) | 1 (1.0) | 0.613 | 2 (3.3) | 0 (0.0) | 0.476 |
| Current aspirin use, % | 4 (4.1) | 3 (3.1) | 1.000 | 3 (5.0) | 2 (3.3) | 1.000 |
| Current peptic ulcer medication use, % | 4 (4.1) | 3 (3.1) | 1.000 | 2 (3.3) | 1 (1.7) | 1.000 |
| Ulcerative colitis, % | 1 (1.0) | 0 (0.0) | 1.000 | 0 (0.0) | 1 (1.7) | 1.000 |
| Diabetes, % | 7 (7.1) | 11 (11.2) | 0.458 | 4 (6. 7) | 5 (8.3) | 1.000 |
| Colorectal polyp, % | 2 (2.0) | 1 (1.0) | 1.000 | 1 (1.7) | 1 (1.7) | 1.000 |
| Total energy, mean (SD), Kcal | 1677.1 (426.6) | 1673.2 (398.3) | 0.946 | 1613.8 (403.0) | 1642.6 (395.8) | 0.693 |
| Red meat, mean (SD), g/day/1000 Kcal | 30.0 (18.9) | 29.62 (22.1) | 0.897 | 25.2 (16.9) | 26.1 (17.6) | 0.773 |
| Fat, mean (SD), g/day/1000 Kcal | 17.6 (5.7) | 17.77 (6.5) | 0.858 | 15.7 (6.0) | 15.8 (4.9) | 0.922 |
| Fruit, mean (SD), g/day/1000 Kcal | 138.1 (95.4) | 150.0(93.5) | 0.381 | 107.7 (87.9) | 130.5 (79.4) | 0.140 |
| Vegetable, mean (SD), g/data/1000 Kcal | 171.7 (80.3) | 196.7 (106.6) | 0.066 | 165.3 (82.0) | 180.0 (86.6) | 0.341 |
| Proteins | Discovery | Validation | Meta-Analysis | |||
|---|---|---|---|---|---|---|
| OR (95% CI) a | p | OR (95% CI) a | p | OR (95% CI) a | p | |
| ADAM22 | 1.50 (1.01–2.22) | 0.046 | 1.37 (0.85–2.22) | 0.196 | 1.44 (1.06–1.96) | 0.018 |
| AGR3 | 0.72 (0.52–1.00) | 0.047 | 1.38 (0.91–2.09) | 0.132 | 0.98 (0.52–1.86) | 0.953 |
| Beta-NGF | 1.70 (1.07–2.69) | 0.024 | 0.85 (0.55–1.30) | 0.447 | 1.19 (0.60–2.36) | 0.611 |
| CANT1 | 1.63 (1.05–2.52) | 0.028 | 1.27 (0.86–1.88) | 0.225 | 1.42 (1.06–1.90) | 0.018 |
| CASP-8 | 1.47 (1.04–2.08) | 0.030 | 1.62 (0.96–2.74) | 0.072 | 1.51 (1.13–2.02) | 0.005 |
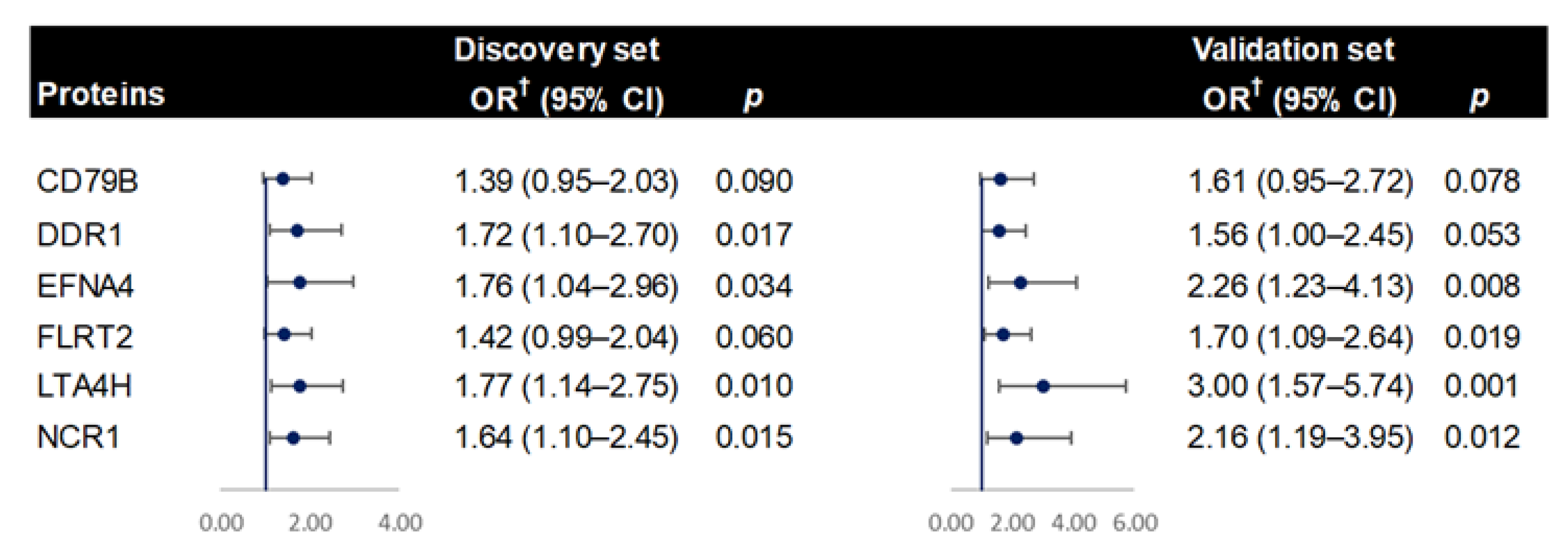
| CD79B | 1.47 (1.02–2.13) | 0.039 | 1.65 (1.03–2.66) | 0.038 | 1.54 (1.15–2.06) | 0.004 |
| CDH17 | 0.71 (0.51–0.97) | 0.034 | 1.19 (0.79–1.81) | 0.403 | 0.90 (0.54–1.51) | 0.695 |
| CLM-1 | 1.55 (1.01–2.37) | 0.044 | 1.37 (0.89–2.11) | 0.158 | 1.46 (1.08–1.98) | 0.015 |
| CRTAM | 1.48 (1.04–2.13) | 0.032 | 1.20 (0.75–1.91) | 0.449 | 1.37 (1.03–1.82) | 0.030 |
| CTSC | 1.51 (1.07–2.13) | 0.019 | 0.79 (0.11–5.87) | 0.818 | 1.48 (1.05–2.08) | 0.023 |
| DDR1 | 1.73 (1.11–2.70) | 0.015 | 1.68 (1.07–2.64) | 0.026 | 1.71 (1.24–2.34) | 0.001 |
| EFNA4 | 1.86 (1.11–3.14) | 0.019 | 2.29 (1.28–4.09) | 0.005 | 2.04 (1.39–3.01) | 3.11 × 10−4 |
| EPHB6 | 1.85 (1.20–2.85) | 0.005 | 1.33 (0.87–2.05) | 0.190 | 1.57 (1.16–2.13) | 0.004 |
| FABP9 | 0.68 (0.47–0.98) | 0.041 | 1.35 (0.88–2.07) | 0.167 | 0.95 (0.48–1.87) | 0.879 |
| FLRT2 | 1.44 (1.00–2.08) | 0.049 | 1.67 (1.09–2.54) | 0.018 | 1.54 (1.16–2.02) | 0.002 |
| HSP-27 | 0.69 (0.50–0.95) | 0.025 | 1.52 (0.89–2.58) | 0.127 | 0.99 (0.46–2.15) | 0.983 |
| HSP90B1 | 1.71 (1.18–2.48) | 0.005 | 0.77 (0.48–1.23) | 0.274 | 1.16 (0.53–2.54) | 0.707 |
| IL-6RA | 1.50 (1.04–2.17) | 0.028 | 1.27 (0.85–1.91) | 0.246 | 1.40 (1.06–1.83) | 0.016 |
| LTA4H | 1.78 (1.16–2.74) | 0.008 | 2.93 (1.57–5.46) | 0.001 | 2.09 (1.47–2.98) | 4.44 × 10−5 |
| MATN3 | 1.58 (1.09–2.29) | 0.017 | 1.09 (0.73–1.65) | 0.669 | 1.34 (1.01–1.76) | 0.039 |
| NCR1 | 1.70 (1.14–2.54) | 0.009 | 2.34 (1.29–4.23) | 0.005 | 1.88 (1.35–2.62) | 1.90 × 10−4 |
| SLAMF8 | 1.43 (1.00–2.02) | 0.047 | 1.35 (0.80–2.28) | 0.267 | 1.40 (1.05–1.88) | 0.024 |
| SPINK5 | 1.55 (1.08–2.23) | 0.018 | 1.54 (0.98–2.42) | 0.064 | 1.55 (1.16–2.05) | 0.003 |
| TR | 1.52 (1.04–2.23) | 0.031 | 1.35 (0.88–2.06) | 0.167 | 1.44 (1.09–1.91) | 0.011 |
| TRANCE | 1.52 (1.06–2.17) | 0.022 | 1.11 (0.76–1.63) | 0.580 | 1.31 (1.01–1.70) | 0.040 |
| UNC5C b | 1.91 (1.18–3.08) | 0.008 | - | - | - | - |
| WAS | 0.71 (0.52–0.97) | 0.034 | 0.94 (0.64–1.37) | 0.731 | 0.79 (0.62–1.01) | 0.065 |
| 5-Protein Score | Discovery | Validation | ||
|---|---|---|---|---|
| OR (95% CI) | p | OR (95% CI) | p | |
| Continuous score a | 2.46 (1.53–3.95) | 1.97 × 10−4 | 4.16 (1.92–8.99) | 2.97 × 10−4 |
| Categorical score b | ||||
| Low level | Ref | Ref | ||
| High level | 2.87 (1.38–5.95) | 0.005 | 4.88 (1.76–13.50) | 2.27 × 10−4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, X.; Shu, X.-O.; Lan, Q.; Laszkowska, M.; Cai, Q.; Rothman, N.; Wen, W.; Zheng, W.; Shu, X. Prospective Proteomic Study Identifies Potential Circulating Protein Biomarkers for Colorectal Cancer Risk. Cancers 2022, 14, 3261. https://doi.org/10.3390/cancers14133261
Sun X, Shu X-O, Lan Q, Laszkowska M, Cai Q, Rothman N, Wen W, Zheng W, Shu X. Prospective Proteomic Study Identifies Potential Circulating Protein Biomarkers for Colorectal Cancer Risk. Cancers. 2022; 14(13):3261. https://doi.org/10.3390/cancers14133261
Chicago/Turabian StyleSun, Xiaohui, Xiao-Ou Shu, Qing Lan, Monika Laszkowska, Qiuyin Cai, Nathaniel Rothman, Wanqing Wen, Wei Zheng, and Xiang Shu. 2022. "Prospective Proteomic Study Identifies Potential Circulating Protein Biomarkers for Colorectal Cancer Risk" Cancers 14, no. 13: 3261. https://doi.org/10.3390/cancers14133261
APA StyleSun, X., Shu, X.-O., Lan, Q., Laszkowska, M., Cai, Q., Rothman, N., Wen, W., Zheng, W., & Shu, X. (2022). Prospective Proteomic Study Identifies Potential Circulating Protein Biomarkers for Colorectal Cancer Risk. Cancers, 14(13), 3261. https://doi.org/10.3390/cancers14133261






