Implications of Gut Microbiota in Epithelial–Mesenchymal Transition and Cancer Progression: A Concise Review
Abstract
:Simple Summary
Abstract
1. Introduction
2. Microbiota
3. Microbiota-Induced Epithelial–Mesenchymal Transition
4. Microbiota-Enhanced Carcinogenesis via Epithelial–Mesenchymal Transition
4.1. Respiratory Tract Microbiota
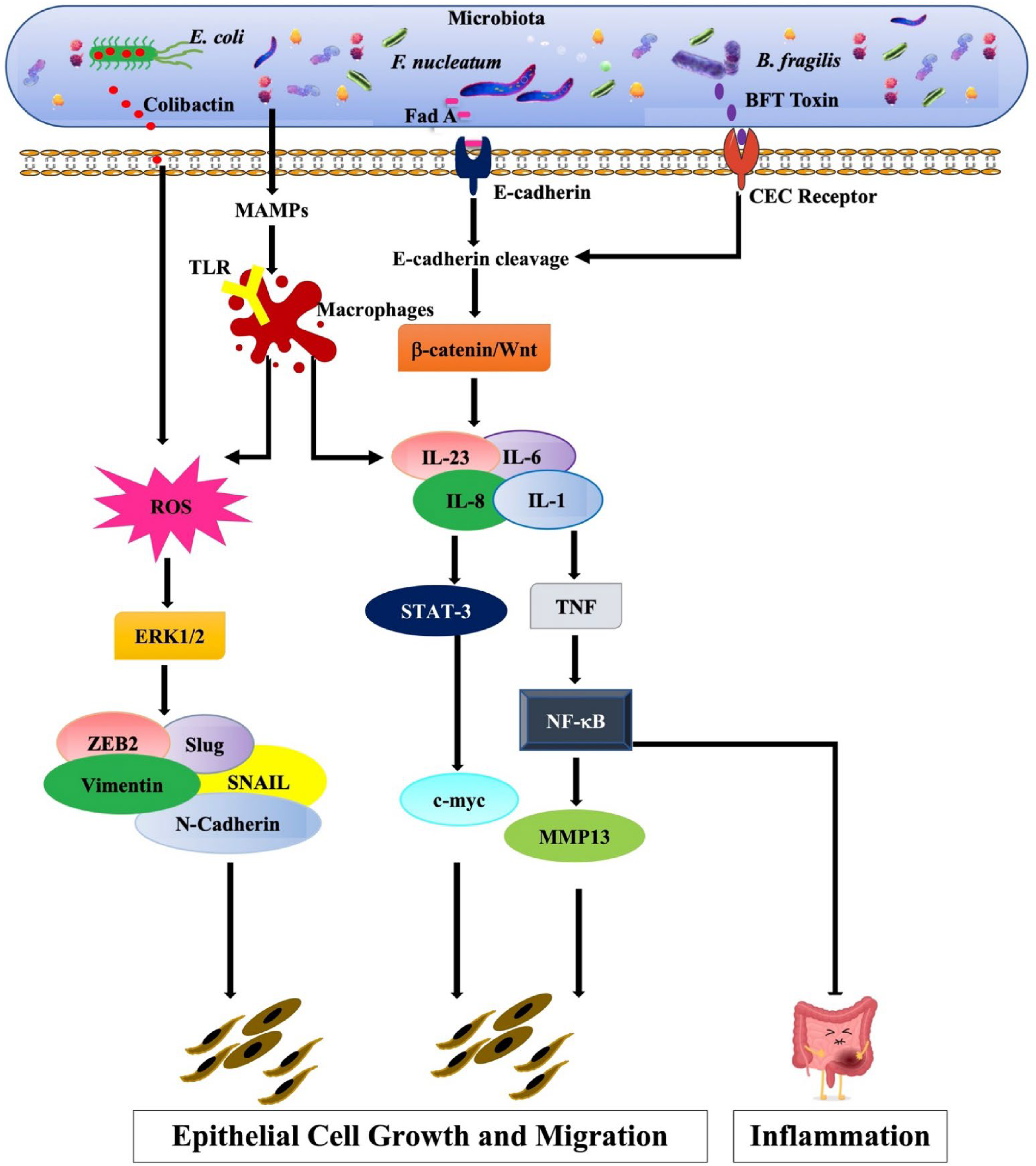
4.2. Gastrointestinal (GI) Tract Microbiota
4.3. Female Reproductive Tract Microbiota
5. Microbiome-Based Therapies (Biotherapy)
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| α-SMA | Alpha smooth muscle actin |
| β-catenin | Beta-catenin |
| γδ T cells | Gamma delta T cells |
| CagA | Cytotoxin-associated gene A |
| CIN | Cervical intraepithelial neoplasia |
| c-MYC | Cellular myelocytomatosis |
| CRC | Colorectal cancer |
| CVM | Cervicovaginal microbiome |
| DAEC | Diffusely adherent |
| EAC | Esophageal adenocarcinoma |
| E-cadherin | Epithelial cadherin |
| ECM | Extracellular matrix |
| E. coli | Escherichia coli |
| EMT | Epithelial mesenchymal transition |
| ERK | Extracellular-signal-regulated kinase |
| ESCC | Esophageal squamous cell carcinoma |
| F. nucleatum | Fusobacterium nucleatum |
| Flt-3L | FMS-like tyrosine kinase 3 ligand |
| GI | Gastrointestinal |
| HIF-1α | Hypoxia-inducible factor 1-alpha |
| H. pylori | Helicobacter pylori |
| HMP | Human Microbiome Project |
| HPV | Human papillomavirus |
| IL | Interleukin |
| IP-10 | Interferon gamma-induced protein 10 |
| Kras | Kirsten rat sarcoma viral oncogene homolog |
| LPS | Lipopolysaccharide |
| MAPK | Mitogen-activated protein kinase |
| MDP | Muramyl dipeptides |
| MIP | Macrophage Inflammatory Proteins |
| MMPs | Matrix metalloproteases |
| N-cadherin | Neural cadherin |
| NF-κB | Nuclear factor kappa light chain enhancer of activated B cells |
| NSCLC | Non-small-cell lung cancer |
| OSCC | Oral squamous cell carcinoma |
| PCR | Polymerase chain reaction |
| P. gingivalis | Porphyromonas gingivalis |
| p53 | Tumor protein 53 |
| PI3K | Phosphatidylinositol 3-kinase |
| Ptger4 | Prostaglandin E Receptor 4 |
| Ras | Rat sarcoma virus |
| ROS | Reactive oxygen species |
| rRNA | Ribosomal RNA |
| S100A | S100 Calcium Binding Protein A1 |
| S. aureus | Staphylococcus aureus |
| SCFA | Short chain fatty acids |
| SMAD | Suppressor of Mothers against Decapentaplegic |
| STAT | Signal transducer and activator of transcription |
| STDs | Sexually transmitted diseases |
| TAMs | Tumor-associated macrophages |
| TGFβ | Transforming growth factor β |
| TILs | Tumor-infiltrating lymphocytes |
| TLR | Toll-like receptor |
| TNF | Tumor necrosis factor |
| VacA | Vacuolating toxin A |
| Wnt | Wingless |
| YAP1 | Yes-associated protein 1 |
| Zeb1 | Zinc Finger E-Box Binding Homeobox 1. |
References
- Farquhar, M.G.; Palade, G.E. Junctional complexes in various epithelia. J. Cell Biol. 1963, 17, 375–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.L.; Streuli, C.H. Integrins and epithelial cell polarity. J. Cell Sci. 2014, 127, 3217–3225. [Google Scholar] [CrossRef] [Green Version]
- Grünert, S.; Jechlinger, M.; Beug, H. Diverse cellular and molecular mechanisms contribute to epithelial plasticity and metastasis. Nat. Rev. Mol. Cell Biol. 2003, 4, 657–665. [Google Scholar] [CrossRef]
- Nieto, M.A. The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu. Rev. Cell Dev. Biol. 2011, 27, 347–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef]
- Son, H.; Moon, A. Epithelial-mesenchymal Transition and Cell Invasion. Toxicol. Res. 2010, 26, 245–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofman, P.; Vouret-Craviari, V. Microbes-induced EMT at the crossroad of inflammation and cancer. Gut Microbes 2012, 3, 176–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caven, L.T.; Brinkworth, A.J.; Carabeo, R.A. Pathogen-driven induction of a host transcriptome facilitating epithelial-to-mesenchymal transition. bioRxiv 2022, in press. [Google Scholar] [CrossRef]
- Champsi, J.; Young, L.S.; Bermudez, L.E. Production of TNF-alpha, IL-6 and TGF-beta, and expression of receptors for TNF-alpha and IL-6, during murine Mycobacterium avium infection. Immunology 1995, 84, 549–554. [Google Scholar] [PubMed]
- Reed, S.G. TGF-beta in infections and infectious diseases. Microbes Infect. 1999, 1, 1313–1325. [Google Scholar] [CrossRef]
- Silva, J.S.; Twardzik, D.R.; Reed, S.G. Regulation of Trypanosoma cruzi infections in vitro and in vivo by transforming growth factor beta (TGF-beta). J. Exp. Med. 1991, 174, 539–545. [Google Scholar] [CrossRef] [Green Version]
- Chapnick, D.A.; Warner, L.; Bernet, J.; Rao, T.; Liu, X. Partners in crime: The TGFβ and MAPK pathways in cancer progression. Cell Biosci. 2011, 1, 42. [Google Scholar] [CrossRef] [Green Version]
- Zhao, M.; Mishra, L.; Deng, C.-X. The role of TGF-β/SMAD4 signaling in cancer. Int. J. Biol. Sci. 2018, 14, 111–123. [Google Scholar] [CrossRef] [Green Version]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ley, R.E.; Peterson, D.A.; Gordon, J.I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 2006, 124, 837–848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inagaki, H.; Suzuki, T.; Nomoto, K.; Yoshikai, Y. Increased susceptibility to primary infection with Listeria monocytogenes in germfree mice may be due to lack of accumulation of L-selectin+ CD44+ T cells in sites of inflammation. Infect. Immun. 1996, 64, 3280–3287. [Google Scholar] [CrossRef] [Green Version]
- Round, J.L.; Mazmanian, S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009, 9, 313–323. [Google Scholar] [CrossRef]
- Wiertsema, S.P.; van Bergenhenegouwen, J.; Garssen, J.; Knippels, L.M.J. The Interplay between the Gut Microbiome and the Immune System in the Context of Infectious Diseases throughout Life and the Role of Nutrition in Optimizing Treatment Strategies. Nutrients 2021, 13, 886. [Google Scholar] [CrossRef]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Integrative HMP (iHMP) Research Network Consortium. The Integrative Human Microbiome Project: Dynamic analysis of microbiome-host omics profiles during periods of human health and disease. Cell Host Microbe 2014, 16, 276–289. [Google Scholar] [CrossRef] [Green Version]
- Morgillo, F.; Dallio, M.; Della Corte, C.M.; Gravina, A.G.; Viscardi, G.; Loguercio, C.; Ciardiello, F.; Federico, A. Carcinogenesis as a Result of Multiple Inflammatory and Oxidative Hits: A Comprehensive Review from Tumor Microenvironment to Gut Microbiota. Neoplasia 2018, 20, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, A.P.; Redinbo, M.R.; Bultman, S.J. The role of the microbiome in cancer development and therapy. CA Cancer J. Clin. 2017, 67, 326–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coker, O.O.; Wu, W.K.K.; Wong, S.H.; Sung, J.J.Y.; Yu, J. Altered Gut Archaea Composition and Interaction with Bacteria Are Associated With Colorectal Cancer. Gastroenterology 2020, 159, 1459–1470.e1455. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Liang, S.; Jia, H.; Stadlmayr, A.; Tang, L.; Lan, Z.; Zhang, D.; Xia, H.; Xu, X.; Jie, Z.; et al. Gut microbiome development along the colorectal adenoma–carcinoma sequence. Nat. Commun. 2015, 6, 6528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Zhang, X.; Xu, H.; Li, S.; Lau, H.C.-H.; Chen, Q.; Zhang, B.; Zhao, L.; Chen, H.; Sung, J.J.-Y.; et al. Microbial Community Heterogeneity Within Colorectal Neoplasia and its Correlation With Colorectal Carcinogenesis. Gastroenterology 2021, 160, 2395–2408. [Google Scholar] [CrossRef]
- Nakatsu, G.; Zhou, H.; Wu, W.K.K.; Wong, S.H.; Coker, O.O.; Dai, Z.; Li, X.; Szeto, C.-H.; Sugimura, N.; Lam, T.Y.-T.; et al. Alterations in Enteric Virome Are Associated with Colorectal Cancer and Survival Outcomes. Gastroenterology 2018, 155, 529–541.e525. [Google Scholar] [CrossRef] [Green Version]
- Thomas, A.M.; Manghi, P.; Asnicar, F.; Pasolli, E.; Armanini, F.; Zolfo, M.; Beghini, F.; Manara, S.; Karcher, N.; Pozzi, C.; et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 2019, 25, 667–678. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wan, X.; Wu, X.; Zhang, C.; Liu, J.; Hou, S. Eubacterium rectale contributes to colorectal cancer initiation via promoting colitis. Gut Pathog. 2021, 13, 2. [Google Scholar] [CrossRef]
- Wirbel, J.; Pyl, P.T.; Kartal, E.; Zych, K.; Kashani, A.; Milanese, A.; Fleck, J.S.; Voigt, A.Y.; Palleja, A.; Ponnudurai, R.; et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. 2019, 25, 679–689. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.-W.; Lee, W.-H.; Tu, S.-J.; Huang, W.-C.; Chen, H.-M.; Sun, T.-H.; Tsai, M.-C.; Wang, C.-C.; Chen, H.-Y.; Huang, C.-C.; et al. Enterotype-based Analysis of Gut Microbiota along the Conventional Adenoma-Carcinoma Colorectal Cancer Pathway. Sci. Rep. 2019, 9, 10923. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Misra, B.B.; Liang, L.; Bi, D.; Weng, W.; Wu, W.; Cai, S.; Qin, H.; Goel, A.; Li, X.; et al. Integrated microbiome and metabolome analysis reveals a novel interplay between commensal bacteria and metabolites in colorectal cancer. Theranostics 2019, 9, 4101–4114. [Google Scholar] [CrossRef] [PubMed]
- Zackular, J.P.; Rogers, M.A.M.; Ruffin, M.T.t.; Schloss, P.D. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev. Res. 2014, 7, 1112–1121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández, M.F.; Reina-Pérez, I.; Astorga, J.M.; Rodríguez-Carrillo, A.; Plaza-Díaz, J.; Fontana, L. Breast Cancer and Its Relationship with the Microbiota. Int. J. Environ. Res. Public Health 2018, 15, 1747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gui, Q.F.; Lu, H.F.; Zhang, C.X.; Xu, Z.R.; Yang, Y.H. Well-balanced commensal microbiota contributes to anti-cancer response in a lung cancer mouse model. Genet. Mol. Res. 2015, 14, 5642–5651. [Google Scholar] [CrossRef] [PubMed]
- Raza, M.H.; Gul, K.; Arshad, A.; Riaz, N.; Waheed, U.; Rauf, A.; Aldakheel, F.; Alduraywish, S.; Rehman, M.U.; Abdullah, M.; et al. Microbiota in cancer development and treatment. J. Cancer Res. Clin. Oncol. 2019, 145, 49–63. [Google Scholar] [CrossRef]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.Q.; Zhao, S.K.; Luo, J.W.; Dong, X.P.; Hao, Y.T.; Li, H.; Shan, L.; Zhou, Y.; Shi, H.B.; Zhang, Z.Y.; et al. Alterations of fecal bacterial communities in patients with lung cancer. Am. J. Transl. Res. 2018, 10, 3171–3185. [Google Scholar]
- Banerjee, S.; Tian, T.; Wei, Z.; Shih, N.; Feldman, M.D.; Alwine, J.C.; Coukos, G.; Robertson, E.S. The ovarian cancer oncobiome. Oncotarget 2017, 8, 36225–36245. [Google Scholar] [CrossRef] [Green Version]
- Łaniewski, P.; Barnes, D.; Goulder, A.; Cui, H.; Roe, D.J.; Chase, D.M.; Herbst-Kralovetz, M.M. Linking cervicovaginal immune signatures, HPV and microbiota composition in cervical carcinogenesis in non-Hispanic and Hispanic women. Sci. Rep. 2018, 8, 7593. [Google Scholar] [CrossRef]
- Łaniewski, P.; Ilhan, Z.E.; Herbst-Kralovetz, M.M. The microbiome and gynaecological cancer development, prevention and therapy. Nat. Rev. Urol. 2020, 17, 232–250. [Google Scholar] [CrossRef]
- Zhou, B.; Sun, C.; Huang, J.; Xia, M.; Guo, E.; Li, N.; Lu, H.; Shan, W.; Wu, Y.; Li, Y.; et al. The biodiversity Composition of Microbiome in Ovarian Carcinoma Patients. Sci. Rep. 2019, 9, 1691. [Google Scholar] [CrossRef] [PubMed]
- Rubinstein, M.R.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y.W. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 2013, 14, 195–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Gibbons, D.L.; Goswami, S.; Cortez, M.A.; Ahn, Y.H.; Byers, L.A.; Zhang, X.; Yi, X.; Dwyer, D.; Lin, W.; et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat. Commun. 2014, 5, 5241. [Google Scholar] [CrossRef] [PubMed]
- Sacdalan, D.B.; Lucero, J.A. The Association Between Inflammation and Immunosuppression: Implications for ICI Biomarker Development. OncoTargets Ther. 2021, 14, 2053–2064. [Google Scholar] [CrossRef]
- Kostic, A.D.; Chun, E.; Robertson, L.; Glickman, J.N.; Gallini, C.A.; Michaud, M.; Clancy, T.E.; Chung, D.C.; Lochhead, P.; Hold, G.L.; et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013, 14, 207–215. [Google Scholar] [CrossRef] [Green Version]
- Hooi, J.K.Y.; Lai, W.Y.; Ng, W.K.; Suen, M.M.Y.; Underwood, F.E.; Tanyingoh, D.; Malfertheiner, P.; Graham, D.Y.; Wong, V.W.S.; Wu, J.C.Y.; et al. Global Prevalence of Helicobacter pylori Infection: Systematic Review and Meta-Analysis. Gastroenterology 2017, 153, 420–429. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, H.M. The epidemiology of Helicobacter pylori. Curr. Top. Microbiol. Immunol. 1999, 241, 11–30. [Google Scholar] [CrossRef]
- Smith, M.F., Jr.; Mitchell, A.; Li, G.; Ding, S.; Fitzmaurice, A.M.; Ryan, K.; Crowe, S.; Goldberg, J.B. Toll-like receptor (TLR) 2 and TLR5, but not TLR4, are required for Helicobacter pylori-induced NF-kappa B activation and chemokine expression by epithelial cells. J. Biol. Chem. 2003, 278, 32552–32560. [Google Scholar] [CrossRef] [Green Version]
- Papini, E.; Satin, B.; Norais, N.; de Bernard, M.; Telford, J.L.; Rappuoli, R.; Montecucco, C. Selective increase of the permeability of polarized epithelial cell monolayers by Helicobacter pylori vacuolating toxin. J. Clin. Investig. 1998, 102, 813–820. [Google Scholar] [CrossRef] [Green Version]
- Amieva, M.R.; Vogelmann, R.; Covacci, A.; Tompkins, L.S.; Nelson, W.J.; Falkow, S. Disruption of the epithelial apical-junctional complex by Helicobacter pylori CagA. Science 2003, 300, 1430–1434. [Google Scholar] [CrossRef] [Green Version]
- Murata-Kamiya, N.; Kurashima, Y.; Teishikata, Y.; Yamahashi, Y.; Saito, Y.; Higashi, H.; Aburatani, H.; Akiyama, T.; Peek, R.M., Jr.; Azuma, T.; et al. Helicobacter pylori CagA interacts with E-cadherin and deregulates the beta-catenin signal that promotes intestinal transdifferentiation in gastric epithelial cells. Oncogene 2007, 26, 4617–4626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brandt, S.; Kwok, T.; Hartig, R.; König, W.; Backert, S. NF-kappaB activation and potentiation of proinflammatory responses by the Helicobacter pylori CagA protein. Proc. Natl. Acad. Sci. USA 2005, 102, 9300–9305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, Y.; Grabowska, A.M.; Clarke, P.A.; Whelband, E.; Robinson, K.; Argent, R.H.; Tobias, A.; Kumari, R.; Atherton, J.C.; Watson, S.A. Helicobacter pylori potentiates epithelial: Mesenchymal transition in gastric cancer: Links to soluble HB-EGF, gastrin and matrix metalloproteinase-7. Gut 2010, 59, 1037–1045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, R.; Palm, M.; Mustonen, V.; Warringer, J.; Farewell, A.; Parts, L.; Moradigaravand, D. Genomic Epidemiology and Evolution of Escherichia coli in Wild Animals in Mexico. mSphere 2021, 6, e00738-20. [Google Scholar] [CrossRef] [PubMed]
- König, J.; Wells, J.; Cani, P.D.; García-Ródenas, C.L.; MacDonald, T.; Mercenier, A.; Whyte, J.; Troost, F.; Brummer, R.-J. Human Intestinal Barrier Function in Health and Disease. Clin. Transl. Gastroenterol. 2016, 7, e196. [Google Scholar] [CrossRef]
- Bétis, F.; Brest, P.; Hofman, V.; Guignot, J.; Bernet-Camard, M.F.; Rossi, B.; Servin, A.; Hofman, P. The Afa/Dr adhesins of diffusely adhering Escherichia coli stimulate interleukin-8 secretion, activate mitogen-activated protein kinases, and promote polymorphonuclear transepithelial migration in T84 polarized epithelial cells. Infect. Immun. 2003, 71, 1068–1074. [Google Scholar] [CrossRef] [Green Version]
- Bétis, F.; Brest, P.; Hofman, V.; Guignot, J.; Kansau, I.; Rossi, B.; Servin, A.; Hofman, P. Afa/Dr diffusely adhering Escherichia coli infection in T84 cell monolayers induces increased neutrophil transepithelial migration, which in turn promotes cytokine-dependent upregulation of decay-accelerating factor (CD55), the receptor for Afa/Dr adhesins. Infect. Immun. 2003, 71, 1774–1783. [Google Scholar] [CrossRef] [Green Version]
- Cane, G.; Moal, V.L.; Pagès, G.; Servin, A.L.; Hofman, P.; Vouret-Craviari, V. Up-regulation of intestinal vascular endothelial growth factor by Afa/Dr diffusely adhering Escherichia coli. PLoS ONE 2007, 2, e1359. [Google Scholar] [CrossRef] [Green Version]
- Cane, G.; Ginouvès, A.; Marchetti, S.; Buscà, R.; Pouysségur, J.; Berra, E.; Hofman, P.; Vouret-Craviari, V. HIF-1alpha mediates the induction of IL-8 and VEGF expression on infection with Afa/Dr diffusely adhering E. coli and promotes EMT-like behaviour. Cell. Microbiol. 2010, 12, 640–653. [Google Scholar] [CrossRef]
- Park, B.S.; Lee, J.-O. Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp. Mol. Med. 2013, 45, e66. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Yang, R.; Cheng, L.; Wang, M.; Jiang, Y.; Wang, S. LPS-induced epithelial-mesenchymal transition of intrahepatic biliary epithelial cells. J. Surg. Res. 2011, 171, 819–825. [Google Scholar] [CrossRef] [PubMed]
- Miyazono, K. Transforming growth factor-beta signaling in epithelial-mesenchymal transition and progression of cancer. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009, 85, 314–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honko, A.N.; Mizel, S.B. Effects of flagellin on innate and adaptive immunity. Immunol. Res. 2005, 33, 83–101. [Google Scholar] [CrossRef]
- Franchi, L.; Park, J.H.; Shaw, M.H.; Marina-Garcia, N.; Chen, G.; Kim, Y.G.; Núñez, G. Intracellular NOD-like receptors in innate immunity, infection and disease. Cell. Microbiol. 2008, 10, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.J.; Wang, D.D.; Sun, T.Y. Flagellin of Pseudomonas aeruginosa induces transforming growth factor beta 1 expression in normal bronchial epithelial cells through mitogen activated protein kinase cascades. Chin. Med. J. 2011, 124, 599–605. [Google Scholar] [PubMed]
- Ferrand, A.; Al Nabhani, Z.; Tapias, N.S.; Mas, E.; Hugot, J.-P.; Barreau, F. NOD2 Expression in Intestinal Epithelial Cells Protects Toward the Development of Inflammation and Associated Carcinogenesis. Cell. Mol. Gastroenterol. Hepatol. 2019, 7, 357–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dominguez, C.; David, J.M.; Palena, C. Epithelial-mesenchymal transition and inflammation at the site of the primary tumor. Semin. Cancer Biol. 2017, 47, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Suo, C.; Li, S.T.; Zhang, H.; Gao, P. Metabolic reprogramming for cancer cells and their microenvironment: Beyond the Warburg Effect. Biochim. Biophys. Acta Rev. Cancer 2018, 1870, 51–66. [Google Scholar] [CrossRef] [PubMed]
- Kovács, T.; Mikó, E.; Vida, A.; Sebő, É.; Toth, J.; Csonka, T.; Boratkó, A.; Ujlaki, G.; Lente, G.; Kovács, P.; et al. Cadaverine, a metabolite of the microbiome, reduces breast cancer aggressiveness through trace amino acid receptors. Sci. Rep. 2019, 9, 1300. [Google Scholar] [CrossRef] [Green Version]
- Mikó, E.; Vida, A.; Kovács, T.; Ujlaki, G.; Trencsényi, G.; Márton, J.; Sári, Z.; Kovács, P.; Boratkó, A.; Hujber, Z.; et al. Lithocholic acid, a bacterial metabolite reduces breast cancer cell proliferation and aggressiveness. Biochim. Biophys. Acta Bioenerg. 2018, 1859, 958–974. [Google Scholar] [CrossRef]
- François, A.; Grebert, D.; Rhimi, M.; Mariadassou, M.; Naudon, L.; Rabot, S.; Meunier, N. Olfactory epithelium changes in germfree mice. Sci. Rep. 2016, 6, 24687. [Google Scholar] [CrossRef] [PubMed]
- Rawls, M.; Ellis, A.K. The microbiome of the nose. Ann. Allergy Asthma Immunol. 2019, 122, 17–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salzano, F.A.; Marino, L.; Salzano, G.; Botta, R.M.; Cascone, G.; D’Agostino Fiorenza, U.; Selleri, C.; Casolaro, V. Microbiota Composition and the Integration of Exogenous and Endogenous Signals in Reactive Nasal Inflammation. J. Immunol. Res. 2018, 2018, 2724951. [Google Scholar] [CrossRef]
- Evans, S.E.; Xu, Y.; Tuvim, M.J.; Dickey, B.F. Inducible innate resistance of lung epithelium to infection. Annu. Rev. Physiol. 2010, 72, 413–435. [Google Scholar] [CrossRef] [Green Version]
- Ziesemer, S.; Eiffler, I.; Schönberg, A.; Müller, C.; Hochgräfe, F.; Beule, A.G.; Hildebrandt, J.P. Staphylococcus aureus α-Toxin Induces Actin Filament Remodeling in Human Airway Epithelial Model Cells. Am. J. Respir. Cell Mol. Biol. 2018, 58, 482–491. [Google Scholar] [CrossRef]
- Patou, J.; Gevaert, P.; Van Zele, T.; Holtappels, G.; van Cauwenberge, P.; Bachert, C. Staphylococcus aureus enterotoxin B, protein A, and lipoteichoic acid stimulations in nasal polyps. J. Allergy Clin. Immunol. 2008, 121, 110–115. [Google Scholar] [CrossRef]
- Meng, J.; Zhou, P.; Liu, Y.; Liu, F.; Yi, X.; Liu, S.; Holtappels, G.; Bachert, C.; Zhang, N. The development of nasal polyp disease involves early nasal mucosal inflammation and remodelling. PLoS ONE 2013, 8, e82373. [Google Scholar] [CrossRef] [Green Version]
- Dickson, R.P.; Erb-Downward, J.R.; Martinez, F.J.; Huffnagle, G.B. The Microbiome and the Respiratory Tract. Annu. Rev. Physiol. 2016, 78, 481–504. [Google Scholar] [CrossRef] [Green Version]
- Mur, L.A.; Huws, S.A.; Cameron, S.J.; Lewis, P.D.; Lewis, K.E. Lung cancer: A new frontier for microbiome research and clinical translation. Ecancermedicalscience 2018, 12, 866. [Google Scholar] [CrossRef]
- Mao, Q.; Jiang, F.; Yin, R.; Wang, J.; Xia, W.; Dong, G.; Ma, W.; Yang, Y.; Xu, L.; Hu, J. Interplay between the lung microbiome and lung cancer. Cancer Lett. 2018, 415, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Lagoudas, G.K.; Zhao, C.; Bullman, S.; Bhutkar, A.; Hu, B.; Ameh, S.; Sandel, D.; Liang, X.S.; Mazzilli, S.; et al. Commensal Microbiota Promote Lung Cancer Development via γδ T Cells. Cell 2019, 176, 998–1013.e16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhuang, H.; Cheng, L.; Wang, Y.; Zhang, Y.K.; Zhao, M.F.; Liang, G.D.; Zhang, M.C.; Li, Y.G.; Zhao, J.B.; Gao, Y.N.; et al. Dysbiosis of the Gut Microbiome in Lung Cancer. Front. Cell. Infect. Microbiol. 2019, 9, 112. [Google Scholar] [CrossRef]
- Botticelli, A.; Putignani, L.; Zizzari, I.; Chierico, F.D.; Reddel, S.; Pietro, F.D.; Quagliarello, A.; Onesti, C.E.; Raffaele, G.; Mazzuca, F.; et al. Changes of microbiome profile during nivolumab treatment in NSCLC patients. J. Clin. Oncol. 2018, 36, e15020. [Google Scholar] [CrossRef]
- Gui, Q.; Li, H.; Wang, A.; Zhao, X.; Tan, Z.; Chen, L.; Xu, K.; Xiao, C. The association between gut butyrate-producing bacteria and non-small-cell lung cancer. J. Clin. Lab. Anal. 2020, 34, e23318. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Li, J.; Guan, Y.; Lou, Y.; Chen, H.; Xu, M.; Deng, D.; Chen, J.; Ni, B.; Zhao, L.; et al. Dysbiosis of the Gut Microbiome is associated with Tumor Biomarkers in Lung Cancer. Int. J. Biol. Sci. 2019, 15, 2381–2392. [Google Scholar] [CrossRef]
- Zheng, Y.; Fang, Z.; Xue, Y.; Zhang, J.; Zhu, J.; Gao, R.; Yao, S.; Ye, Y.; Wang, S.; Lin, C.; et al. Specific gut microbiome signature predicts the early-stage lung cancer. Gut Microbes 2020, 11, 1030–1042. [Google Scholar] [CrossRef]
- Yang, J.J.; Yu, D.; Xiang, Y.B.; Blot, W.; White, E.; Robien, K.; Sinha, R.; Park, Y.; Takata, Y.; Lazovich, D.; et al. Association of Dietary Fiber and Yogurt Consumption With Lung Cancer Risk: A Pooled Analysis. JAMA Oncol. 2020, 6, e194107. [Google Scholar] [CrossRef]
- Bai, Y.; Shen, W.; Zhu, M.; Zhang, L.; Wei, Y.; Tang, H.; Zhao, J. Combined detection of estrogen and tumor markers is an important reference factor in the diagnosis and prognosis of lung cancer. J. Cell. Biochem. 2019, 120, 105–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gill, N.; Wlodarska, M.; Finlay, B.B. The future of mucosal immunology: Studying an integrated system-wide organ. Nat. Immunol. 2010, 11, 558–560. [Google Scholar] [CrossRef] [PubMed]
- Renz, H.; Brandtzaeg, P.; Hornef, M. The impact of perinatal immune development on mucosal homeostasis and chronic inflammation. Nat. Rev. Immunol. 2011, 12, 9–23. [Google Scholar] [CrossRef]
- Bingula, R.; Filaire, M.; Radosevic-Robin, N.; Bey, M.; Berthon, J.Y.; Bernalier-Donadille, A.; Vasson, M.P.; Filaire, E. Desired Turbulence? Gut-Lung Axis, Immunity, and Lung Cancer. J. Oncol. 2017, 2017, 5035371. [Google Scholar] [CrossRef] [PubMed]
- Hooper, L.V.; Littman, D.R.; Macpherson, A.J. Interactions between the microbiota and the immune system. Science 2012, 336, 1268–1273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazmanian, S.K.; Liu, C.H.; Tzianabos, A.O.; Kasper, D.L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 2005, 122, 107–118. [Google Scholar] [CrossRef] [Green Version]
- Samuelson, D.R.; Welsh, D.A.; Shellito, J.E. Regulation of lung immunity and host defense by the intestinal microbiota. Front. Microbiol. 2015, 6, 1085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dumas, A.; Bernard, L.; Poquet, Y.; Lugo-Villarino, G.; Neyrolles, O. The role of the lung microbiota and the gut-lung axis in respiratory infectious diseases. Cell. Microbiol. 2018, 20, e12966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuijt, T.J.; Lankelma, J.M.; Scicluna, B.P.; de Sousa e Melo, F.; Roelofs, J.J.; de Boer, J.D.; Hoogendijk, A.J.; de Beer, R.; de Vos, A.; Belzer, C.; et al. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut 2016, 65, 575–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.; Gu, B.; Madison, M.C.; Song, H.W.; Norwood, K.; Hill, A.A.; Wu, W.J.; Corry, D.; Kheradmand, F.; Diehl, G.E. Cigarette Smoke Induces Intestinal Inflammation via a Th17 Cell-Neutrophil Axis. Front. Immunol. 2019, 10, 75. [Google Scholar] [CrossRef] [Green Version]
- Boursi, B.; Mamtani, R.; Haynes, K.; Yang, Y.X. Recurrent antibiotic exposure may promote cancer formation--Another step in understanding the role of the human microbiota? Eur. J. Cancer 2015, 51, 2655–2664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Druzhinin, V.G.; Matskova, L.V.; Fucic, A. Induction and modulation of genotoxicity by the bacteriome in mammals. Mutat. Res./Rev. Mutat. Res. 2018, 776, 70–77. [Google Scholar] [CrossRef]
- Louis, P.; Hold, G.L.; Flint, H.J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014, 12, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Van Raay, T.; Allen-Vercoe, E. Microbial Interactions and Interventions in Colorectal Cancer. Microbiol. Spectr. 2017, 5, 99–130. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Chu, M.; Huang, Z.; Yang, X.; Ran, S.; Hu, B.; Zhang, C.; Liang, J. Variations in oral microbiota associated with oral cancer. Sci. Rep. 2017, 7, 11773. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.H.; Chen, H.M.; Yang, S.F.; Liang, C.; Peng, C.Y.; Lin, F.M.; Tsai, L.L.; Wu, B.C.; Hsin, C.H.; Chuang, C.Y.; et al. Bacterial alterations in salivary microbiota and their association in oral cancer. Sci. Rep. 2017, 7, 16540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitmore, S.E.; Lamont, R.J. Oral bacteria and cancer. PLoS Pathog. 2014, 10, e1003933. [Google Scholar] [CrossRef]
- Nagy, K.N.; Sonkodi, I.; Szöke, I.; Nagy, E.; Newman, H.N. The microflora associated with human oral carcinomas. Oral Oncol. 1998, 34, 304–308. [Google Scholar] [CrossRef]
- Karpiński, T.M. Role of oral microbiota in cancer development. Microorganisms 2019, 7, 20. [Google Scholar] [CrossRef] [Green Version]
- Ha, N.H.; Woo, B.H.; Kim, D.J.; Ha, E.S.; Choi, J.I.; Kim, S.J.; Park, B.S.; Lee, J.H.; Park, H.R. Prolonged and repetitive exposure to Porphyromonas gingivalis increases aggressiveness of oral cancer cells by promoting acquisition of cancer stem cell properties. Tumour Biol. 2015, 36, 9947–9960. [Google Scholar] [CrossRef]
- Sztukowska, M.N.; Ojo, A.; Ahmed, S.; Carenbauer, A.L.; Wang, Q.; Shumway, B.; Jenkinson, H.F.; Wang, H.; Darling, D.S.; Lamont, R.J. Porphyromonas gingivalis initiates a mesenchymal-like transition through ZEB1 in gingival epithelial cells. Cell. Microbiol. 2016, 18, 844–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snider, E.J.; Compres, G.; Freedberg, D.E.; Khiabanian, H.; Nobel, Y.R.; Stump, S.; Uhlemann, A.-C.; Lightdale, C.J.; Abrams, J.A. Alterations to the esophageal microbiome associated with progression from Barrett’s esophagus to esophageal adenocarcinoma. Cancer Epidemiol. Prev. Biomark. 2019, 28, 1687–1693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Rao, Y.; Guo, X.; Liu, N.; Liu, S.; Wen, P.; Li, S.; Li, Y. Oral microbiome in patients with oesophageal squamous cell carcinoma. Sci. Rep. 2019, 9, 19055. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Winckler, B.; Lu, M.; Cheng, H.; Yuan, Z.; Yang, Y.; Jin, L.; Ye, W. Oral microbiota and risk for esophageal squamous cell carcinoma in a high-risk area of China. PLoS ONE 2015, 10, e0143603. [Google Scholar] [CrossRef] [PubMed]
- Blackett, K.; Siddhi, S.; Cleary, S.; Steed, H.; Miller, M.; Macfarlane, S.; Macfarlane, G.; Dillon, J. Oesophageal bacterial biofilm changes in gastro-oesophageal reflux disease, Barrett’s and oesophageal carcinoma: Association or causality? Aliment. Pharmacol. Ther. 2013, 37, 1084–1092. [Google Scholar] [CrossRef] [PubMed]
- Cass, S.; Hamilton, C.; Miller, A.; Jupiter, D.; Khanipov, K.; Booth, A.; Pyles, R.; Krill, T.; Reep, G.; Okereke, I. Novel ex vivo model to examine the mechanism and relationship of esophageal microbiota and disease. Biomedicines 2021, 9, 142. [Google Scholar] [CrossRef] [PubMed]
- Elliott, D.R.F.; Walker, A.W.; O’Donovan, M.; Parkhill, J.; Fitzgerald, R.C. A non-endoscopic device to sample the oesophageal microbiota: A case-control study. Lancet Gastroenterol. Hepatol. 2017, 2, 32–42. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; He, R.; Hou, G.; Ming, W.; Fan, T.; Chen, L.; Zhang, L.; Jiang, W.; Wang, W.; Lu, Z. Characterization of the esophageal microbiota and prediction of the metabolic pathways involved in esophageal cancer. Front. Cell. Infect. Microbiol. 2020, 10, 268. [Google Scholar] [CrossRef]
- Shao, D.; Vogtmann, E.; Liu, A.; Qin, J.; Chen, W.; Abnet, C.C.; Wei, W. Microbial characterization of esophageal squamous cell carcinoma and gastric cardia adenocarcinoma from a high-risk region of China. Cancer 2019, 125, 3993–4002. [Google Scholar] [CrossRef]
- Peters, B.A.; Wu, J.; Pei, Z.; Yang, L.; Purdue, M.P.; Freedman, N.D.; Jacobs, E.J.; Gapstur, S.M.; Hayes, R.B.; Ahn, J. Oral microbiome composition reflects prospective risk for esophageal cancers. Cancer Res. 2017, 77, 6777–6787. [Google Scholar] [CrossRef] [Green Version]
- Narikiyo, M.; Tanabe, C.; Yamada, Y.; Igaki, H.; Tachimori, Y.; Kato, H.; Muto, M.; Montesano, R.; Sakamoto, H.; Nakajima, Y. Frequent and preferential infection of Treponema denticola, Streptococcus mitis, and Streptococcus anginosus in esophageal cancers. Cancer Sci. 2004, 95, 569–574. [Google Scholar] [CrossRef]
- Lopetuso, L.R.; Severgnini, M.; Pecere, S.; Ponziani, F.R.; Boskoski, I.; Larghi, A.; Quaranta, G.; Masucci, L.; Ianiro, G.; Camboni, T. Esophageal microbiome signature in patients with Barrett’s esophagus and esophageal adenocarcinoma. PLoS ONE 2020, 15, e0231789. [Google Scholar] [CrossRef]
- Liu, Y.; Lin, Z.; Lin, Y.; Chen, Y.; Peng, X.-E.; He, F.; Liu, S.; Yan, S.; Huang, L.; Lu, W. Streptococcus and Prevotella are associated with the prognosis of oesophageal squamous cell carcinoma. J. Med. Microbiol. 2018, 67, 1058–1068. [Google Scholar] [CrossRef]
- Chen, M.-F.; Lu, M.-S.; Hsieh, C.-C.; Chen, W.-C. Porphyromonas gingivalis promotes tumor progression in esophageal squamous cell carcinoma. Cell. Oncol. 2021, 44, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Kaakoush, N.O.; Lecomte, V.; Maloney, C.A.; Morris, M.J. Cross-talk among metabolic parameters, esophageal microbiota, and host gene expression following chronic exposure to an obesogenic diet. Sci. Rep. 2017, 7, 45753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Münch, N.S.; Fang, H.-Y.; Ingermann, J.; Maurer, H.C.; Anand, A.; Kellner, V.; Sahm, V.; Wiethaler, M.; Baumeister, T.; Wein, F. High-fat diet accelerates carcinogenesis in a mouse model of Barrett’s esophagus via interleukin 8 and alterations to the gut microbiome. Gastroenterology 2019, 157, 492–506.e2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pham, V.T.; Fehlbaum, S.; Seifert, N.; Richard, N.; Bruins, M.J.; Sybesma, W.; Rehman, A.; Steinert, R.E. Effects of colon-targeted vitamins on the composition and metabolic activity of the human gut microbiome—A pilot study. Gut Microbes 2021, 13, 1875774. [Google Scholar] [CrossRef]
- Pan, F.; Xu, X.; Zhang, L.-L.; Luo, H.-J.; Chen, Y.; Long, L.; Wang, X.; Zhuang, P.-T.; Li, E.-M.; Xu, L.-Y. Correction: Dietary riboflavin deficiency induces genomic instability of esophageal squamous cells that is associated with gut microbiota dysbiosis in rats. Food Funct. 2020, 11, 10979. [Google Scholar] [CrossRef]
- Pan, F.; Zhang, L.-L.; Luo, H.-J.; Chen, Y.; Long, L.; Wang, X.; Zhuang, P.-T.; Li, E.-M.; Xu, L.-Y. Dietary riboflavin deficiency induces ariboflavinosis and esophageal epithelial atrophy in association with modification of gut microbiota in rats. Eur. J. Nutr. 2021, 60, 807–820. [Google Scholar] [CrossRef]
- Chow, W.-H.; Blaser, M.J.; Blot, W.J.; Gammon, M.D.; Vaughan, T.L.; Risch, H.A.; Perez-Perez, G.I.; Schoenberg, J.B.; Stanford, J.L.; Rotterdam, H. An inverse relation between cagA+ strains of Helicobacter pylori infection and risk of esophageal and gastric cardia adenocarcinoma. Cancer Res. 1998, 58, 588–590. [Google Scholar]
- Nie, S.; Chen, T.; Yang, X.; Huai, P.; Lu, M. Association of h elicobacter pylori infection with esophageal adenocarcinoma and squamous cell carcinoma: A meta-analysis. Dis. Esophagus 2014, 27, 645–653. [Google Scholar] [CrossRef]
- Peek, R.M.; Blaser, M.J. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat. Rev. Cancer 2002, 2, 28–37. [Google Scholar] [CrossRef]
- Kumar, S.; Metz, D.C.; Ginsberg, G.G.; Kaplan, D.E.; Goldberg, D.S. Oesophageal and proximal gastric adenocarcinomas are rare after detection of Helicobacter pylori infection. Aliment. Pharmacol. Ther. 2020, 51, 781–788. [Google Scholar] [CrossRef]
- Doorakkers, E.; Lagergren, J.; Santoni, G.; Engstrand, L.; Brusselaers, N. Helicobacter pylori eradication treatment and the risk of Barrett’s esophagus and esophageal adenocarcinoma. Helicobacter 2020, 25, e12688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira, R.M.; Pereira-Marques, J.; Pinto-Ribeiro, I.; Costa, J.L.; Carneiro, F.; Machado, J.C.; Figueiredo, C. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut 2018, 67, 226–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amieva, M.; Peek Jr, R.M. Pathobiology of Helicobacter pylori–induced gastric cancer. Gastroenterology 2016, 150, 64–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, A.; Johannesen, T.B.; Spiegelhauer, M.; Kupcinskas, J.; Urba, M.; Skieceviciene, J.; Jonaitis, L.; Frandsen, T.; Kupcinskas, L.; Fuursted, K. Distinct composition and distribution of the gastric mycobiota observed between dyspeptic and gastric cancer patients evaluated from gastric biopsies. Microb. Health Dis. 2020, 2, e340. [Google Scholar]
- Spiegelhauer, M.R.; Kupcinskas, J.; Johannesen, T.B.; Urba, M.; Skieceviciene, J.; Jonaitis, L.; Frandsen, T.H.; Kupcinskas, L.; Fuursted, K.; Andersen, L.P. Transient and persistent gastric microbiome: Adherence of bacteria in gastric cancer and dyspeptic patient biopsies after washing. J. Clin. Med. 2020, 9, 1882. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Torres, J.; Hu, N.; Medrano-Guzman, R.; Herrera-Goepfert, R.; Humphrys, M.S.; Wang, L.; Wang, C.; Ding, T.; Ravel, J. Molecular characterization of the human stomach microbiota in gastric cancer patients. Front. Cell. Infect. Microbiol. 2017, 7, 302. [Google Scholar] [CrossRef]
- Liu, X.; Shao, L.; Liu, X.; Ji, F.; Mei, Y.; Cheng, Y.; Liu, F.; Yan, C.; Li, L.; Ling, Z. Alterations of gastric mucosal microbiota across different stomach microhabitats in a cohort of 276 patients with gastric cancer. EBioMedicine 2019, 40, 336–348. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.-L.; Pang, W.; Huang, Y.; Zhang, Y.; Zhang, C.-J. The gastric microbiome is perturbed in advanced gastric adenocarcinoma identified through shotgun metagenomics. Front. Cell. Infect. Microbiol. 2018, 8, 433. [Google Scholar] [CrossRef] [Green Version]
- Aviles-Jimenez, F.; Vazquez-Jimenez, F.; Medrano-Guzman, R.; Mantilla, A.; Torres, J. Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Sci. Rep. 2014, 4, 4202. [Google Scholar] [CrossRef] [Green Version]
- Sohn, S.-H.; Kim, N.; Jo, H.J.; Kim, J.; Park, J.H.; Nam, R.H.; Seok, Y.-J.; Kim, Y.-R.; Lee, D.H. Analysis of Gastric Body Microbiota by Pyrosequencing: Possible Role of Bacteria Other Than Helicobacter pylori in the Gastric Carcinogenesis. J. Cancer Prev. 2017, 22, 115–125. [Google Scholar] [CrossRef] [Green Version]
- Gantuya, B.; El-Serag, H.B.; Matsumoto, T.; Ajami, N.J.; Oyuntsetseg, K.; Azzaya, D.; Uchida, T.; Yamaoka, Y. Gastric Microbiota in Helicobacter pylori-Negative and -Positive Gastritis Among High Incidence of Gastric Cancer Area. Cancers 2019, 11, 504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Yu, H. The role of non-H. pylori bacteria in the development of gastric cancer. Am. J. Cancer Res. 2020, 10, 2271–2281. [Google Scholar] [PubMed]
- Thorell, K.; Bengtsson-Palme, J.; Liu, O.H.-F.; Gonzales, R.V.P.; Nookaew, I.; Rabeneck, L.; Paszat, L.; Graham, D.Y.; Nielsen, J.; Lundin, S.B.; et al. In Vivo Analysis of the Viable Microbiota and Helicobacter pylori Transcriptome in Gastric Infection and Early Stages of Carcinogenesis. Infect. Immun. 2017, 85, e00031-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molendijk, J.; Nguyen, T.-M.-T.; Brown, I.; Mohamed, A.; Lim, Y.; Barclay, J.; Hodson, M.P.; Hennessy, T.P.; Krause, L.; Morrison, M.; et al. Chronic High-Fat Diet Induces Early Barrett’s Esophagus in Mice through Lipidome Remodeling. Biomolecules 2020, 10, 776. [Google Scholar] [CrossRef]
- Hsieh, Y.-Y.; Tung, S.-Y.; Pan, H.-Y.; Yen, C.-W.; Xu, H.-W.; Lin, Y.-J.; Deng, Y.-F.; Hsu, W.-T.; Wu, C.-S.; Li, C. Increased Abundance of Clostridium and Fusobacterium in Gastric Microbiota of Patients with Gastric Cancer in Taiwan. Sci. Rep. 2018, 8, 158. [Google Scholar] [CrossRef]
- Boehm, E.T.; Thon, C.; Kupcinskas, J.; Steponaitiene, R.; Skieceviciene, J.; Canbay, A.; Malfertheiner, P.; Link, A. Fusobacterium nucleatum is associated with worse prognosis in Lauren’s diffuse type gastric cancer patients. Sci. Rep. 2020, 10, 16240. [Google Scholar] [CrossRef]
- Arita, S.; Ogawa, T.; Murakami, Y.; Kinoshita, Y.; Okazaki, M.; Inagaki-Ohara, K. Dietary Fat-Accelerating Leptin Signaling Promotes Protumorigenic Gastric Environment in Mice. Nutrients 2019, 11, 2127. [Google Scholar] [CrossRef] [Green Version]
- Arita, S.; Inagaki-Ohara, K. High-fat-diet–induced modulations of leptin signaling and gastric microbiota drive precancerous lesions in the stomach. Nutrition 2019, 67–68, 110556. [Google Scholar] [CrossRef]
- Sears, C.L.; Pardoll, D.M. Perspective: Alpha-bugs, their microbial partners, and the link to colon cancer. J. Infect. Dis. 2011, 203, 306–311. [Google Scholar] [CrossRef] [Green Version]
- Tjalsma, H.; Boleij, A.; Marchesi, J.R.; Dutilh, B.E. A bacterial driver-passenger model for colorectal cancer: Beyond the usual suspects. Nat. Rev. Microbiol. 2012, 10, 575–582. [Google Scholar] [CrossRef]
- Mori, G.; Rampelli, S.; Orena, B.S.; Rengucci, C.; De Maio, G.; Barbieri, G.; Passardi, A.; Casadei Gardini, A.; Frassineti, G.L.; Gaiarsa, S.; et al. Shifts of Faecal Microbiota During Sporadic Colorectal Carcinogenesis. Sci. Rep. 2018, 8, 10329. [Google Scholar] [CrossRef] [PubMed]
- Castellarin, M.; Warren, R.L.; Freeman, J.D.; Dreolini, L.; Krzywinski, M.; Strauss, J.; Barnes, R.; Watson, P.; Allen-Vercoe, E.; Moore, R.A.; et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012, 22, 299–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, T.; Guo, F.; Yu, Y.; Sun, T.; Ma, D.; Han, J.; Qian, Y.; Kryczek, I.; Sun, D.; Nagarsheth, N.; et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell 2017, 170, 548–563.e16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abed, J.; Emgård, J.E.; Zamir, G.; Faroja, M.; Almogy, G.; Grenov, A.; Sol, A.; Naor, R.; Pikarsky, E.; Atlan, K.A.; et al. Fap2 Mediates Fusobacterium nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host Microbe 2016, 20, 215–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonnet, M.; Buc, E.; Sauvanet, P.; Darcha, C.; Dubois, D.; Pereira, B.; Déchelotte, P.; Bonnet, R.; Pezet, D.; Darfeuille-Michaud, A. Colonization of the Human Gut by E. coli and Colorectal Cancer Risk. Clin. Cancer Res. 2014, 20, 859–867. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Hobert, M.E.; Duan, Y.; Rao, A.S.; He, T.-C.; Chang, E.B.; Madara, J.L. Crosstalk between NF-κB and β-catenin pathways in bacterial-colonized intestinal epithelial cells. Am. J. Physiol.-Gastrointest. Liver Physiol. 2005, 289, G129–G137. [Google Scholar] [CrossRef]
- Moncrief, J.S.; Duncan, A.J.; Wright, R.L.; Barroso, L.A.; Wilkins, T.D. Molecular characterization of the fragilysin pathogenicity islet of enterotoxigenic Bacteroides fragilis. Infect. Immun. 1998, 66, 1735–1739. [Google Scholar] [CrossRef] [Green Version]
- Cheng, W.T.; Kantilal, H.K.; Davamani, F. The Mechanism of Bacteroides fragilis Toxin Contributes to Colon Cancer Formation. Malays. J. Med. Sci. 2020, 27, 9–21. [Google Scholar] [CrossRef]
- Sears, C.L.; Geis, A.L.; Housseau, F. Bacteroides fragilis subverts mucosal biology: From symbiont to colon carcinogenesis. J. Clin. Investig. 2014, 124, 4166–4172. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Rhee, K.-J.; Zhang, M.; Franco, A.; Sears, C.L. Bacteroides fragilis toxin stimulates intestinal epithelial cell shedding and γ-secretase-dependent E-cadherin cleavage. J. Cell Sci. 2007, 120, 1944–1952. [Google Scholar] [CrossRef] [Green Version]
- Ertz-Archambault, N.; Keim, P.; Von Hoff, D. Microbiome and pancreatic cancer: A comprehensive topic review of literature. World J. Gastroenterol. 2017, 23, 1899–1908. [Google Scholar] [CrossRef]
- Michaud, D.S.; Izard, J.; Wilhelm-Benartzi, C.S.; You, D.H.; Grote, V.A.; Tjønneland, A.; Dahm, C.C.; Overvad, K.; Jenab, M.; Fedirko, V.; et al. Plasma antibodies to oral bacteria and risk of pancreatic cancer in a large European prospective cohort study. Gut 2013, 62, 1764–1770. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Alekseyenko, A.V.; Wu, J.; Peters, B.A.; Jacobs, E.J.; Gapstur, S.M.; Purdue, M.P.; Abnet, C.C.; Stolzenberg-Solomon, R.; Miller, G.; et al. Human oral microbiome and prospective risk for pancreatic cancer: A population-based nested case-control study. Gut 2018, 67, 120–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, M.Y.; Shi, S.; Liang, C.; Meng, Q.C.; Hua, J.; Zhang, Y.Y.; Liu, J.; Zhang, B.; Xu, J.; Yu, X.J. The microbiota and microbiome in pancreatic cancer: More influential than expected. Mol. Cancer 2019, 18, 97. [Google Scholar] [CrossRef] [PubMed]
- Jesnowski, R.; Isaksson, B.; Möhrcke, C.; Bertsch, C.; Bulajic, M.; Schneider-Brachert, W.; Klöppel, G.; Lowenfels, A.B.; Maisonneuve, P.; Löhr, J.M. Helicobacter pylori in autoimmune pancreatitis and pancreatic carcinoma. Pancreatology 2010, 10, 462–466. [Google Scholar] [CrossRef]
- Knorr, J.; Ricci, V.; Hatakeyama, M.; Backert, S. Classification of Helicobacter pylori Virulence Factors: Is CagA a Toxin or Not? Trends Microbiol. 2019, 27, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Pushalkar, S.; Hundeyin, M.; Daley, D.; Zambirinis, C.P.; Kurz, E.; Mishra, A.; Mohan, N.; Aykut, B.; Usyk, M.; Torres, L.E.; et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov. 2018, 8, 403–416. [Google Scholar] [CrossRef] [Green Version]
- Thomas, R.M.; Gharaibeh, R.Z.; Gauthier, J.; Beveridge, M.; Pope, J.L.; Guijarro, M.V.; Yu, Q.; He, Z.; Ohland, C.; Newsome, R.; et al. Intestinal microbiota enhances pancreatic carcinogenesis in preclinical models. Carcinogenesis 2018, 39, 1068–1078. [Google Scholar] [CrossRef]
- Benner, M.; Ferwerda, G.; Joosten, I.; van der Molen, R.G. How uterine microbiota might be responsible for a receptive, fertile endometrium. Hum. Reprod. Update 2018, 24, 393–415. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Song, X.; Wei, W.; Zhong, H.; Dai, J.; Lan, Z.; Li, F.; Yu, X.; Feng, Q.; Wang, Z.; et al. The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases. Nat. Commun. 2017, 8, 875. [Google Scholar] [CrossRef] [Green Version]
- Ravel, J.; Gajer, P.; Abdo, Z.; Schneider, G.M.; Koenig, S.S.; McCulle, S.L.; Karlebach, S.; Gorle, R.; Russell, J.; Tacket, C.O.; et al. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4680–4687. [Google Scholar] [CrossRef] [Green Version]
- Smith, B.C.; Zolnik, C.P.; Usyk, M.; Chen, Z.; Kaiser, K.; Nucci-Sack, A.; Peake, K.; Diaz, A.; Viswanathan, S.; Strickler, H.D.; et al. Distinct Ecological Niche of Anal, Oral, and Cervical Mucosal Microbiomes in Adolescent Women. Yale J. Biol. Med. 2016, 89, 277–284. [Google Scholar]
- Dethlefsen, L.; McFall-Ngai, M.; Relman, D.A. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature 2007, 449, 811–818. [Google Scholar] [CrossRef] [PubMed]
- Valenti, P.; Rosa, L.; Capobianco, D.; Lepanto, M.S.; Schiavi, E.; Cutone, A.; Paesano, R.; Mastromarino, P. Role of Lactobacilli and Lactoferrin in the Mucosal Cervicovaginal Defense. Front. Immunol. 2018, 9, 376. [Google Scholar] [CrossRef] [PubMed]
- Petrova, M.I.; Lievens, E.; Malik, S.; Imholz, N.; Lebeer, S. Lactobacillus species as biomarkers and agents that can promote various aspects of vaginal health. Front. Physiol. 2015, 6, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, B.; Forney, L.J.; Ravel, J. Vaginal microbiome: Rethinking health and disease. Annu. Rev. Microbiol. 2012, 66, 371–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, S.B.; Ravel, J. The vaginal microbiota, host defence and reproductive physiology. J. Physiol. 2017, 595, 451–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bik, E.M.; Bird, S.W.; Bustamante, J.P.; Leon, L.E.; Nieto, P.A.; Addae, K.; Alegría-Mera, V.; Bravo, C.; Bravo, D.; Cardenas, J.P.; et al. A novel sequencing-based vaginal health assay combining self-sampling, HPV detection and genotyping, STI detection, and vaginal microbiome analysis. PLoS ONE 2019, 14, e0215945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freitas, A.C.; Bocking, A.; Hill, J.E.; Money, D.M. Increased richness and diversity of the vaginal microbiota and spontaneous preterm birth. Microbiome 2018, 6, 117. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Fettweis, J.M.; Brooks, J.P.; Jefferson, K.K.; Buck, G.A. The changing landscape of the vaginal microbiome. Clin. Lab. Med. 2014, 34, 747–761. [Google Scholar] [CrossRef] [Green Version]
- Torcia, M.G. Interplay among Vaginal Microbiome, Immune Response and Sexually Transmitted Viral Infections. Int. J. Mol. Sci. 2019, 20, 266. [Google Scholar] [CrossRef] [Green Version]
- Garrett, W.S. Cancer and the microbiota. Science 2015, 348, 80–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwabe, R.F.; Jobin, C. The microbiome and cancer. Nat. Rev. Cancer 2013, 13, 800–812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walther-António, M.R.; Chen, J.; Multinu, F.; Hokenstad, A.; Distad, T.J.; Cheek, E.H.; Keeney, G.L.; Creedon, D.J.; Nelson, H.; Mariani, A.; et al. Potential contribution of the uterine microbiome in the development of endometrial cancer. Genome Med. 2016, 8, 122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitra, A.; MacIntyre, D.A.; Lee, Y.S.; Smith, A.; Marchesi, J.R.; Lehne, B.; Bhatia, R.; Lyons, D.; Paraskevaidis, E.; Li, J.V.; et al. Cervical intraepithelial neoplasia disease progression is associated with increased vaginal microbiome diversity. Sci. Rep. 2015, 5, 16865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Paola, M.; Sani, C.; Clemente, A.M.; Iossa, A.; Perissi, E.; Castronovo, G.; Tanturli, M.; Rivero, D.; Cozzolino, F.; Cavalieri, D.; et al. Characterization of cervico-vaginal microbiota in women developing persistent high-risk Human Papillomavirus infection. Sci. Rep. 2017, 7, 10200. [Google Scholar] [CrossRef]
- Lee, J.E.; Lee, S.; Lee, H.; Song, Y.M.; Lee, K.; Han, M.J.; Sung, J.; Ko, G. Association of the vaginal microbiota with human papillomavirus infection in a Korean twin cohort. PLoS ONE 2013, 8, e63514. [Google Scholar] [CrossRef] [PubMed]
- Norenhag, J.; Du, J.; Olovsson, M.; Verstraelen, H.; Engstrand, L.; Brusselaers, N. The vaginal microbiota, human papillomavirus and cervical dysplasia: A systematic review and network meta-analysis. BJOG 2020, 127, 171–180. [Google Scholar] [CrossRef]
- Kwasniewski, W.; Wolun-Cholewa, M.; Kotarski, J.; Warchol, W.; Kuzma, D.; Kwasniewska, A.; Gozdzicka-Jozefiak, A. Microbiota dysbiosis is associated with HPV-induced cervical carcinogenesis. Oncol. Lett. 2018, 16, 7035–7047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brusselaers, N.; Shrestha, S.; van de Wijgert, J.; Verstraelen, H. Vaginal dysbiosis and the risk of human papillomavirus and cervical cancer: Systematic review and meta-analysis. Am. J. Obstet. Gynecol. 2019, 221, 9–18.e18. [Google Scholar] [CrossRef]
- Kyrgiou, M.; Moscicki, A.B. Vaginal microbiome and cervical cancer. Semin. Cancer Biol. 2022; in press. [Google Scholar] [CrossRef] [PubMed]
- Baker, J.M.; Al-Nakkash, L.; Herbst-Kralovetz, M.M. Estrogen–gut microbiome axis: Physiological and clinical implications. Maturitas 2017, 103, 45–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, L.; Liu, C.; Zhou, Q.; Feng, M.; Wang, J. Association of estradiol and HPV/HPV16 infection with the occurrence of cervical squamous cell carcinoma. Oncol. Lett. 2019, 17, 3548–3554. [Google Scholar] [CrossRef] [Green Version]
- James, C.D.; Morgan, I.M.; Bristol, M.L. The relationship between estrogen-related signaling and human papillomavirus positive cancers. Pathogens 2020, 9, 403. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.-Y.; Liu, H.-Z.; Liu, J.-F.; Sun, Y.; Song, Y. Pathogenic mechanism, detection methods and clinical significance of group B Streptococcus. Future Microbiol. 2021, 16, 671–685. [Google Scholar] [CrossRef] [PubMed]
- Antonio, M.A.; Rabe, L.K.; Hillier, S.L. Colonization of the rectum by Lactobacillus species and decreased risk of bacterial vaginosis. J. Infect. Dis. 2005, 192, 394–398. [Google Scholar] [CrossRef] [Green Version]
- Homayouni, A.; Bastani, P.; Ziyadi, S.; Mohammad-Alizadeh-Charandabi, S.; Ghalibaf, M.; Mortazavian, A.M.; Mehrabany, E.V. Effects of probiotics on the recurrence of bacterial vaginosis: A review. J. Low. Genit. Tract Dis. 2014, 18, 79–86. [Google Scholar] [CrossRef]
- Blaak, E.; Canfora, E.; Theis, S.; Frost, G.; Groen, A.; Mithieux, G.; Nauta, A.; Scott, K.; Stahl, B.; van Harsselaar, J. Short chain fatty acids in human gut and metabolic health. Benef. Microbes 2020, 11, 411–455. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Diaz, D.J.; Tyssen, D.; Hayward, J.A.; Gugasyan, R.; Hearps, A.C.; Tachedjian, G. Distinct immune responses elicited from cervicovaginal epithelial cells by lactic acid and short chain fatty acids associated with optimal and non-optimal vaginal microbiota. Front. Cell. Infect. Microbiol. 2020, 9, 446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, S.; Liu, Z.; Lv, M.; Chen, Y.; Liu, Y. Intestinal dysbiosis promotes epithelial-mesenchymal transition by activating tumor-associated macrophages in ovarian cancer. Pathog. Dis 2019, 77, ftz019. [Google Scholar] [CrossRef]
- Magnus, M.C.; Håberg, S.E.; Stigum, H.; Nafstad, P.; London, S.J.; Vangen, S.; Nystad, W. Delivery by Cesarean section and early childhood respiratory symptoms and disorders: The Norwegian mother and child cohort study. Am. J. Epidemiol. 2011, 174, 1275–1285. [Google Scholar] [CrossRef]
- Almqvist, C.; Cnattingius, S.; Lichtenstein, P.; Lundholm, C. The impact of birth mode of delivery on childhood asthma and allergic diseases—A sibling study. Clin. Exp. Allergy 2012, 42, 1369–1376. [Google Scholar] [CrossRef]
- Gondwe, T.; Betha, K.; Kusneniwar, G.; Bunker, C.H.; Tang, G.; Simhan, H.; Reddy, P.; Haggerty, C.L. Mode of delivery and short-term infant health outcomes: A prospective cohort study in a peri-urban Indian population. BMC Pediatr. 2018, 18, 346. [Google Scholar] [CrossRef] [Green Version]
- Mima, K.; Nishihara, R.; Qian, Z.R.; Cao, Y.; Sukawa, Y.; Nowak, J.A.; Yang, J.; Dou, R.; Masugi, Y.; Song, M.; et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 2016, 65, 1973–1980. [Google Scholar] [CrossRef] [Green Version]
- Mira-Pascual, L.; Cabrera-Rubio, R.; Ocon, S.; Costales, P.; Parra, A.; Suarez, A.; Moris, F.; Rodrigo, L.; Mira, A.; Collado, M.C. Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. J. Gastroenterol. 2015, 50, 167–179. [Google Scholar] [CrossRef]
- Tahara, T.; Yamamoto, E.; Suzuki, H.; Maruyama, R.; Chung, W.; Garriga, J.; Jelinek, J.; Yamano, H.-O.; Sugai, T.; An, B.; et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014, 74, 1311–1318. [Google Scholar] [CrossRef] [Green Version]
- Nugent, J.L.; McCoy, A.N.; Addamo, C.J.; Jia, W.; Sandler, R.S.; Keku, T.O. Altered tissue metabolites correlate with microbial dysbiosis in colorectal adenomas. J. Proteome Res. 2014, 13, 1921–1929. [Google Scholar] [CrossRef]
- Wu, N.; Yang, X.; Zhang, R.; Li, J.; Xiao, X.; Hu, Y.; Chen, Y.; Yang, F.; Lu, N.; Wang, Z.; et al. Dysbiosis signature of fecal microbiota in colorectal.l cancer patients. Microb. Ecol. 2013, 66, 462–470. [Google Scholar] [CrossRef]
- Warren, R.L.; Freeman, D.J.; Pleasance, S.; Watson, P.; Moore, R.A.; Cochrane, K.; Allen-Vercoe, E.; Holt, R.A. Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome 2013, 1, 16. [Google Scholar] [CrossRef] [Green Version]
- McCoy, A.N.; Araújo-Pérez, F.; Azcárate-Peril, A.; Yeh, J.J.; Sandler, R.S.; Keku, T.O. Fusobacterium is associated with colorectal adenomas. PLoS ONE 2013, 8, e53653. [Google Scholar] [CrossRef] [PubMed]
- Brim, H.; Yooseph, S.; Zoetendal, E.G.; Lee, E.; Torralbo, M.; Laiyemo, A.O.; Shokrani, B.; Nelson, K.; Ashktorab, H. Microbiome analysis of stool samples from African Americans with colon polyps. PLoS ONE 2013, 8, e81352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanapareddy, N.; Legge, R.M.; Jovov, B.; McCoy, A.; Burcal, L.; Araujo-Perez, F.; Randall, T.A.; Galanko, J.; Benson, A.; Sandler, R.S.; et al. Increased rectal microbial richness is associated with the presence of colorectal adenomas in humans. ISME J. 2012, 6, 1858–1868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchesi, J.R.; Dutilh, B.E.; Hall, N.; Peters, W.H.M.; Roelofs, R.; Boleij, A.; Tjalsma, H. Towards the Human Colorectal Cancer Microbiome. PLoS ONE 2011, 6, e20447. [Google Scholar] [CrossRef] [Green Version]
- Shen, X.J.; Rawls, J.F.; Randall, T.; Burcal, L.; Mpande, C.N.; Jenkins, N.; Jovov, B.; Abdo, Z.; Sandler, R.S.; Keku, T.O. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes 2010, 1, 138–147. [Google Scholar] [CrossRef] [Green Version]
- Dicksved, J.; Lindberg, M.; Rosenquist, M.; Enroth, H.; Jansson, J.K.; Engstrand, L. Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. J. Med. Microbiol. 2009, 58, 509–516. [Google Scholar] [CrossRef]
- Sobhani, I.; Tap, J.; Roudot-Thoraval, F.; Roperch, J.P.; Letulle, S.; Langella, P.; Corthier, G.; Tran Van Nhieu, J.; Furet, J.P. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS ONE 2011, 6, e16393. [Google Scholar] [CrossRef]
- Apostolou, P.; Tsantsaridou, A.; Papasotiriou, I.; Toloudi, M.; Chatziioannou, M.; Giamouzis, G. Bacterial and fungal microflora in surgically removed lung cancer samples. J. Cardiothorac. Surg. 2011, 6, 137. [Google Scholar] [CrossRef] [Green Version]
- Chan, P.J.; Seraj, I.M.; Kalugdan, T.H.; King, A. Prevalence of Mycoplasma Conserved DNA in Malignant Ovarian Cancer Detected Using Sensitive PCR–ELISA. Gynecol. Oncol. 1996, 63, 258–260. [Google Scholar] [CrossRef]
- van Vliet, M.J.; Tissing, W.J.; Dun, C.A.; Meessen, N.E.; Kamps, W.A.; de Bont, E.S.; Harmsen, H.J. Chemotherapy treatment in pediatric patients with acute myeloid leukemia receiving antimicrobial prophylaxis leads to a relative increase of colonization with potentially pathogenic bacteria in the gut. Clin. Infect. Dis. 2009, 49, 262–270. [Google Scholar] [CrossRef] [Green Version]
- Jenq, R.R.; Ubeda, C.; Taur, Y.; Menezes, C.C.; Khanin, R.; Dudakov, J.A.; Liu, C.; West, M.L.; Singer, N.V.; Equinda, M.J.; et al. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. J. Exp. Med. 2012, 209, 903–911. [Google Scholar] [CrossRef]
- Viaud, S.; Saccheri, F.; Mignot, G.; Yamazaki, T.; Daillère, R.; Hannani, D.; Enot, D.P.; Pfirschke, C.; Engblom, C.; Pittet, M.J.; et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 2013, 342, 971–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iida, N.; Dzutsev, A.; Stewart, C.A.; Smith, L.; Bouladoux, N.; Weingarten, R.A.; Molina, D.A.; Salcedo, R.; Back, T.; Cramer, S.; et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013, 342, 967–970. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.L.; Wilson, I.D.; Teare, J.; Marchesi, J.R.; Nicholson, J.K.; Kinross, J.M. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 356–365. [Google Scholar] [CrossRef]
- Montassier, E.; Gastinne, T.; Vangay, P.; Al-Ghalith, G.A.; Bruley des Varannes, S.; Massart, S.; Moreau, P.; Potel, G.; de La Cochetière, M.F.; Batard, E.; et al. Chemotherapy-driven dysbiosis in the intestinal microbiome. Aliment. Pharmacol. Ther. 2015, 42, 515–528. [Google Scholar] [CrossRef]
- Gerassy-Vainberg, S.; Blatt, A.; Danin-Poleg, Y.; Gershovich, K.; Sabo, E.; Nevelsky, A.; Daniel, S.; Dahan, A.; Ziv, O.; Dheer, R.; et al. Radiation induces proinflammatory dysbiosis: Transmission of inflammatory susceptibility by host cytokine induction. Gut 2018, 67, 97–107. [Google Scholar] [CrossRef] [Green Version]
- Schwan, A.; Sjölin, S.; Trottestam, U.; Aronsson, B. Relapsing clostridium difficile enterocolitis cured by rectal infusion of homologous faeces. Lancet 1983, 2, 845. [Google Scholar] [CrossRef]
- Gough, E.; Shaikh, H.; Manges, A.R. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin. Infect. Dis. 2011, 53, 994–1002. [Google Scholar] [CrossRef] [Green Version]
- Brandt, L.J. Fecal Microbiota Transplant: Respice, Adspice, Prospice. J. Clin. Gastroenterol. 2015, 49 (Suppl. S1), S65–S68. [Google Scholar] [CrossRef]
- Kao, D.; Roach, B.; Silva, M.; Beck, P.; Rioux, K.; Kaplan, G.G.; Chang, H.-J.; Coward, S.; Goodman, K.J.; Xu, H.; et al. Effect of Oral Capsule– vs Colonoscopy-Delivered Fecal Microbiota Transplantation on Recurrent Clostridium difficile Infection: A Randomized Clinical Trial. JAMA 2017, 318, 1985–1993. [Google Scholar] [CrossRef] [Green Version]
- Khanna, S.; Raffals, L.E. The Microbiome in Crohn’s Disease: Role in Pathogenesis and Role of Microbiome Replacement Therapies. Gastroenterol. Clin. N. Am. 2017, 46, 481–492. [Google Scholar] [CrossRef]
- Khanna, S. Microbiota Replacement Therapies: Innovation in Gastrointestinal Care. Clin. Pharmacol. Ther. 2018, 103, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Vindigni, S.M.; Surawicz, C.M. Fecal Microbiota Transplantation. Gastroenterol. Clin. N. Am. 2017, 46, 171–185. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Saha, S.; Khanna, S. Therapies to modulate gut microbiota: Past, present and future. World J. Gastroenterol. 2020, 26, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Davar, D.; Dzutsev, A.K.; McCulloch, J.A.; Rodrigues, R.R.; Chauvin, J.M.; Morrison, R.M.; Deblasio, R.N.; Menna, C.; Ding, Q.; Pagliano, O.; et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science 2021, 371, 595–602. [Google Scholar] [CrossRef]
- Mohty, M.; Malard, F.; D’Incan, E.; Thomas, X.; Recher, C.; Michallet, A.-S.; Peterlin, P.; Vekhoff, A.; Vey, N.; Plantamura, E. Prevention of dysbiosis complications with autologous fecal microbiota transplantation (auto-FMT) in acute myeloid leukemia (AML) patients undergoing intensive treatment (ODYSSEE study): First results of a prospective multicenter trial. Blood 2017, 130, 2624. [Google Scholar]
- Taur, Y.; Jenq, R.R.; Ubeda, C.; van den Brink, M.; Pamer, E.G. Role of intestinal microbiota in transplantation outcomes. Best Pract. Res. Clin. Haematol. 2015, 28, 155–161. [Google Scholar] [CrossRef] [Green Version]
- Guarner, F.; Schaafsma, G.J. Probiotics. Int. J. Food Microbiol. 1998, 39, 237–238. [Google Scholar] [CrossRef]
- Zhu, Y.; Michelle Luo, T.; Jobin, C.; Young, H.A. Gut microbiota and probiotics in colon tumorigenesis. Cancer Lett. 2011, 309, 119–127. [Google Scholar] [CrossRef] [Green Version]
- Appleyard, C.B.; Cruz, M.L.; Isidro, A.A.; Arthur, J.C.; Jobin, C.; De Simone, C. Pretreatment with the probiotic VSL#3 delays transition from inflammation to dysplasia in a rat model of colitis-associated cancer. Am. J. Physiol.-Gastrointest. Liver Physiol. 2011, 301, G1004–G1013. [Google Scholar] [CrossRef] [Green Version]
- Arthur, J.C.; Gharaibeh, R.Z.; Uronis, J.M.; Perez-Chanona, E.; Sha, W.; Tomkovich, S.; Mühlbauer, M.; Fodor, A.A.; Jobin, C. VSL#3 probiotic modifies mucosal microbial composition but does not reduce colitis-associated colorectal cancer. Sci. Rep. 2013, 3, 2868. [Google Scholar] [CrossRef] [Green Version]
- Hibberd, A.A.; Lyra, A.; Ouwehand, A.C.; Rolny, P.; Lindegren, H.; Cedgård, L.; Wettergren, Y. Intestinal microbiota is altered in patients with colon cancer and modified by probiotic intervention. BMJ Open Gastroenterol. 2017, 4, e000145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gianotti, L.; Morelli, L.; Galbiati, F.; Rocchetti, S.; Coppola, S.; Beneduce, A.; Gilardini, C.; Zonenschain, D.; Nespoli, A.; Braga, M. A randomized double-blind trial on perioperative administration of probiotics in colorectal cancer patients. World J. Gastroenterol. 2010, 16, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Riehl, T.E.; Alvarado, D.; Ee, X.; Zuckerman, A.; Foster, L.; Kapoor, V.; Thotala, D.; Ciorba, M.A.; Stenson, W.F. Lactobacillus rhamnosus GG protects the intestinal epithelium from radiation injury through release of lipoteichoic acid, macrophage activation and the migration of mesenchymal stem cells. Gut 2019, 68, 1003–1013. [Google Scholar] [CrossRef] [PubMed]
- Dizman, N.; Hsu, J.; Bergerot, P.G.; Gillece, J.D.; Folkerts, M.; Reining, L.; Trent, J.; Highlander, S.K.; Pal, S.K. Randomized trial assessing impact of probiotic supplementation on gut microbiome and clinical outcome from targeted therapy in metastatic renal cell carcinoma. Cancer Med. 2021, 10, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Li, M.; Song, W.; Jiang, R.; Li, Y.Q. Effects of probiotics on chemotherapy in patients with lung cancer. Oncol. Lett. 2019, 17, 2836–2848. [Google Scholar] [CrossRef] [Green Version]
- Carmody, R.N.; Gerber, G.K.; Luevano, J.M., Jr.; Gatti, D.M.; Somes, L.; Svenson, K.L.; Turnbaugh, P.J. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 2015, 17, 72–84. [Google Scholar] [CrossRef] [Green Version]
- Le Chatelier, E.; Nielsen, T.; Qin, J.; Prifti, E.; Hildebrand, F.; Falony, G.; Almeida, M.; Arumugam, M.; Batto, J.-M.; Kennedy, S.; et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013, 500, 541–546. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [Green Version]
- Cotillard, A.; Kennedy, S.P.; Kong, L.C.; Prifti, E.; Pons, N.; Le Chatelier, E.; Almeida, M.; Quinquis, B.; Levenez, F.; Galleron, N.; et al. Dietary intervention impact on gut microbial gene richness. Nature 2013, 500, 585–588. [Google Scholar] [CrossRef]
- Taper, H.S.; Roberfroid, M.B. Possible adjuvant cancer therapy by two prebiotics--inulin or oligofructose. In Vivo 2005, 19, 201–204. [Google Scholar]
- O’Keefe, S.J.D. Diet, microorganisms and their metabolites, and colon cancer. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 691–706. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, H.E.; Jernberg, C.; Andersson, A.F.; Sjölund-Karlsson, M.; Jansson, J.K.; Engstrand, L. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS ONE 2010, 5, e9836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pflug, N.; Kluth, S.; Vehreschild, J.J.; Bahlo, J.; Tacke, D.; Biehl, L.; Eichhorst, B.; Fischer, K.; Cramer, P.; Fink, A.M.; et al. Efficacy of antineoplastic treatment is associated with the use of antibiotics that modulate intestinal microbiota. Oncoimmunology 2016, 5, e1150399. [Google Scholar] [CrossRef] [PubMed]
- Cieplak, T.; Soffer, N.; Sulakvelidze, A.; Nielsen, D.S. A bacteriophage cocktail targeting Escherichia coli reduces E. coli in simulated gut conditions, while preserving a non-targeted representative commensal normal microbiota. Gut Microbes 2018, 9, 391–399. [Google Scholar] [CrossRef] [Green Version]
- Zuo, T.; Wong, S.H.; Lam, K.; Lui, R.; Cheung, K.; Tang, W.; Ching, J.Y.L.; Chan, P.K.S.; Chan, M.C.W.; Wu, J.C.Y.; et al. Bacteriophage transfer during faecal microbiota transplantation in Clostridium difficile infection is associated with treatment outcome. Gut 2018, 67, 634–643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Study | Detection Method | Bacterium Species | Expression Levels |
|---|---|---|---|
| Colorectal Cancer | |||
| Boehm et al. (2020) [146] | Probe-based quantitative PCR | Fusobacterium nucleatum | Upregulated |
| Mori et al. (2018) [151] | 16S rRNA gene sequencing | Sutterella and Escherichia/Shigella | Upregulated |
| Yu et al. (2017) [153] | Quantitative PCR | Fusobacterium nucleatum | Upregulated |
| Mima et al. (2015) [204] | Molecular pathological epidemiology database | Fusobacterium nucleatum | Upregulated |
| Mira-Pascual et al. (2015) [205] | 16S rRNA gene pyrosequencing and quantitative PCR | Methanobacteriales, Methanobrevibacterium, Fusobacterium nucleatum, Enterobacteriaceae, Akkermansia muciniphila, and Blautia coccoides | Upregulated |
| Bifidobacterium, Faecalibacterium prausnitzii, and Lactobacillus | Downregulated | ||
| Tahara et al. (2014) [206] | Quantitative real-time PCR | Fusobacterium nucleatum and pan-fusobacterium | Upregulated |
| Zackular et al. (2014) [32] | 16S rRNA gene sequencing | Ruminococcaceae, Clostridium, Pseudomonas, and Porphyromonadaceae | Upregulated |
| Bonnet et al. (2014) [155] | PCR | Escherichia coli | Upregulated |
| Nugent et al. (2014) [207] | Quantitative real-time PCR | Bifidobacterium, Eubacteria, Escherichia coli, Clostridium, and Bacteroides | Upregulated |
| Wu et al. (2013) [208] | Pyrosequencing of the 16S rRNA gene V3 region | Bacteroids, Fusobacterium, and Campylobacter | Upregulated |
| Faecalibacterium and Roseburia | Downregulated | ||
| Warren et al. (2013) [209] | Metatranscriptomic analysis | Fusobacterium, Leptotrichia, and Campylobacter | Upregulated |
| McCoy et al. (2013) [210] | 16S rRNA quantitative PCR and pyrosequencing | Fusobacterium | Upregulated |
| Brim et al. (2013) [211] | Human intestinal Tract Chip (HITChip) and 16S rRNA gene barcoded 454 pyrosequencing | Bacteroidetes and Firmicutes | Upregulated |
| Castellarin et al. (2012) [152] | Quantitative PCR | Fusobacterium nucleatum | Upregulated |
| Sanapareddy et al. (2012) [212] | 454 titanium pyrosequencing of the V1–V2 region of the 16S rRNA gene | Firmicutes, Bacteroidetes, Pseudomonas, Helicobacter, Actinobacteria, Lactobacillus, Acinetobacter, and Proteobacteria | Upregulated |
| Marchesi et al. (2011) [213] | Deep rRNA sequencing | Roseburia, Fusobacterium, and Faecalibacterium | Upregulated |
| Citrobacter, Shigella, Cronobacter, Kluyvera, Serratia, and Salmonella spp. | Downregulated | ||
| Shen et al. (2010) [214] | Terminal restriction fragment length polymorphism, clone sequencing and fluorescent in situ hybridization analysis of the 16S rRNA genes | Dorea spp. and Faecalibacterium spp. | Upregulated |
| Esophageal Cancer | |||
| Nie et al. (2014) [128] | Meta-analysis | Helicobacter pylori | Downregulated |
| Chow et al. (1998) [127] | Antigen-specific ELISA | Helicobacter pylori | Downregulated |
| Gastric Cancer | |||
| Boehm et al. (2020) [146] | Probe-based quantitative PCR | Fusobacterium nucleatum | Upregulated |
| Hansen et al. (2020) [134] | 18S rDNA sequencing | Malassezia | Upregulated |
| Hsieh et al. (2018) [145] | 16S ribosomal DNA analysis | Fusobacterium and Clostridium | Upregulated |
| Helicobacter pylori | Downregulated | ||
| Ferriera et al. (2018) [132] | 16S rRNA next-generation sequencing | Helicobacter pylori | Downregulated |
| Yu et al. (2017) [136] | 16S rRNA gene sequencing | Helicobacter pylori | Upregulated |
| Sohn et al. (2017) [140] | Bar-coded 454 pyrosequencing of the 16S rRNA gene | Streptococcus pseudopneumoniae, S. parasanguinis, and S. oralis | Upregulated |
| Aviles-Jimenez et al. (2014) [139] | Microarray G3 PhyloChip analysis | Pseudomonas, Lactobacillus coleohominis, and Lachnospiraceae | Upregulated |
| Porphyromonas, TM7, Neisseria, and Streptococcus sinensis | Downregulated | ||
| Dicksved et al. (2009) [215] | Terminal restriction fragment length polymorphism analysis in combination with 16S rRNA gene cloning and sequencing | Streptococcus, Lactobacillus, Veillonella, and Prevotella | Upregulated |
| Chow et al. (1998) [127] | Antigen-specific ELISA | Helicobacter pylori | Downregulated |
| Lung Cancer | |||
| Sobhani et al. (2011) [216] | Quantitative PCR and pyrosequencing | Helicobacter pylori | Downregulated |
| Bifidobacterium, Faecalibacterium, Streptococcus, and Veillonella | Downregulated | ||
| Gui et al. (2020) [84] | Quantitative PCR | Faecalibacterium prausnitzii, Clostridium leptum, Ruminococcus spp., Clostridial cluster I, Clostridial cluster XIVa, and Roseburia spp. | Downregulated |
| Zhuang at el. (2019) [82] | 16S rRNA next-generation sequencing | Enterococcus | Upregulated |
| Bifidobacterium | Downregulated | ||
| Liu et al. (2019) [137] | 16S rRNA gene amplicon sequencing | Fusobacteria, Prevotella Proteobacteria, Streptococcus, Verrucomicrobia, and Veillonella | Upregulated |
| Bacteroidetes, Firmicutes, and Actinobacteria | Downregulated | ||
| Zhang et al. (2018) [37] | 16S rRNA gene sequencing | Bacteroides, Veillonella, and Fusobacterium | Upregulated |
| Escherichia-Shigella, Kluyvera, Fecalibacterium, Enterobacter, and Dialister | Downregulated | ||
| Apostolou et al. (2011) [217] | Reverse-transcription polymerase chain reaction | Staphylococcus epidermidis, Streptococcus mitis, and Bacillus strains | Upregulated |
| Pancreatic Ductal Adenocarcinoma | |||
| Jesnowski et al. (2010) [165] | Nested PCR | Helicobacter pylori | No expression |
| Ovarian Cancer | |||
| Chan et al. (1996) [218] | Combined PCR-ELISA Assay | Mycoplasma | Upregulated |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gupta, I.; Pedersen, S.; Vranic, S.; Al Moustafa, A.-E. Implications of Gut Microbiota in Epithelial–Mesenchymal Transition and Cancer Progression: A Concise Review. Cancers 2022, 14, 2964. https://doi.org/10.3390/cancers14122964
Gupta I, Pedersen S, Vranic S, Al Moustafa A-E. Implications of Gut Microbiota in Epithelial–Mesenchymal Transition and Cancer Progression: A Concise Review. Cancers. 2022; 14(12):2964. https://doi.org/10.3390/cancers14122964
Chicago/Turabian StyleGupta, Ishita, Shona Pedersen, Semir Vranic, and Ala-Eddin Al Moustafa. 2022. "Implications of Gut Microbiota in Epithelial–Mesenchymal Transition and Cancer Progression: A Concise Review" Cancers 14, no. 12: 2964. https://doi.org/10.3390/cancers14122964
APA StyleGupta, I., Pedersen, S., Vranic, S., & Al Moustafa, A.-E. (2022). Implications of Gut Microbiota in Epithelial–Mesenchymal Transition and Cancer Progression: A Concise Review. Cancers, 14(12), 2964. https://doi.org/10.3390/cancers14122964









