Combined High-Throughput Approaches Reveal the Signals Driven by Skin and Blood Environments and Define the Tumor Heterogeneity in Sézary Syndrome
Abstract
Simple Summary
Abstract
1. Introduction
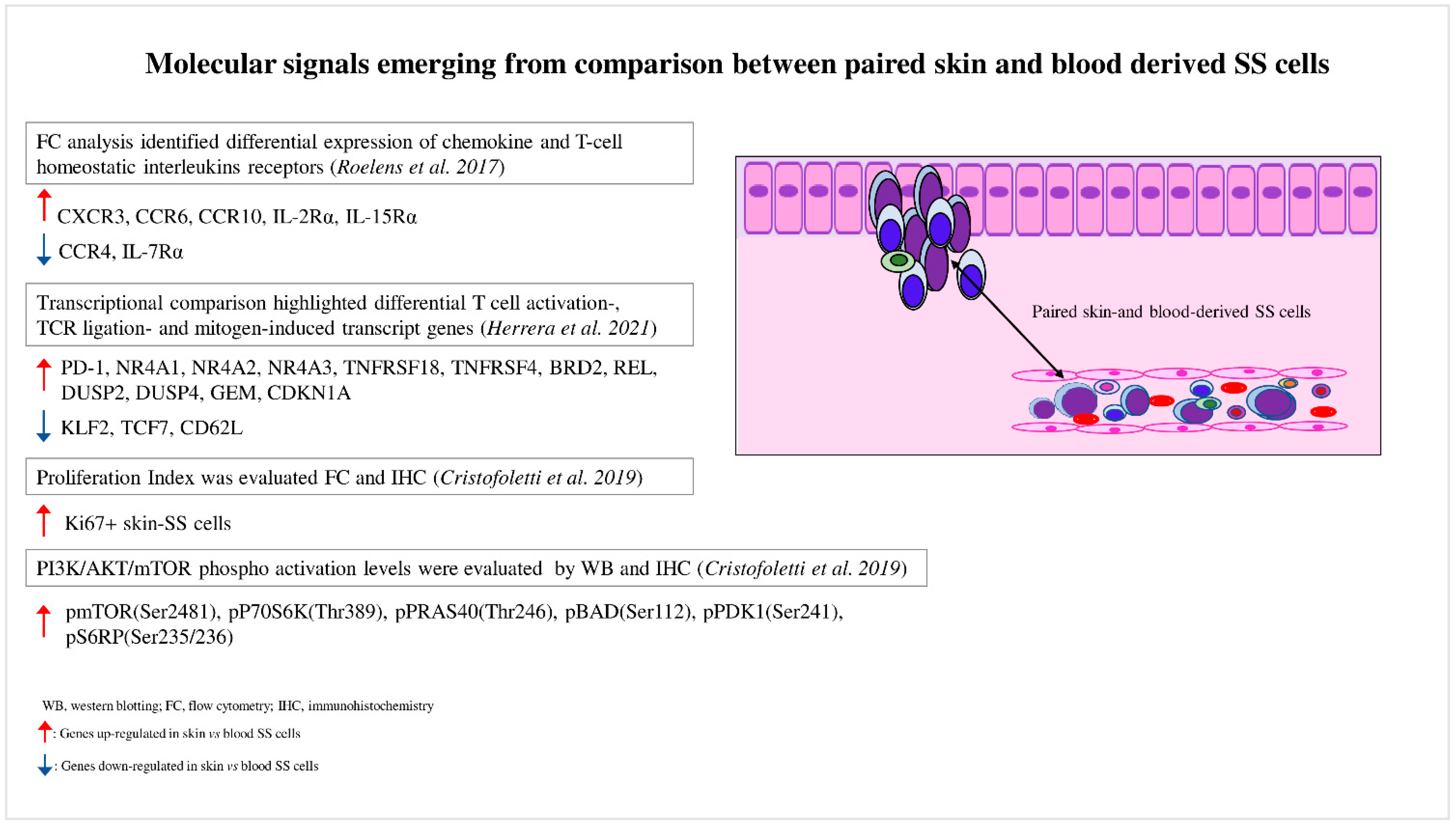
2. Blood and Skin Microenvironments Drive the Transcriptional Program and Signaling of SS Cells
3. Skin SS Cells Exhibit a Hyperactivated PI3K/AKT/mTOR Signaling Compared to the Matched Blood Counterpart: Spotlight on Skin Microenvironment
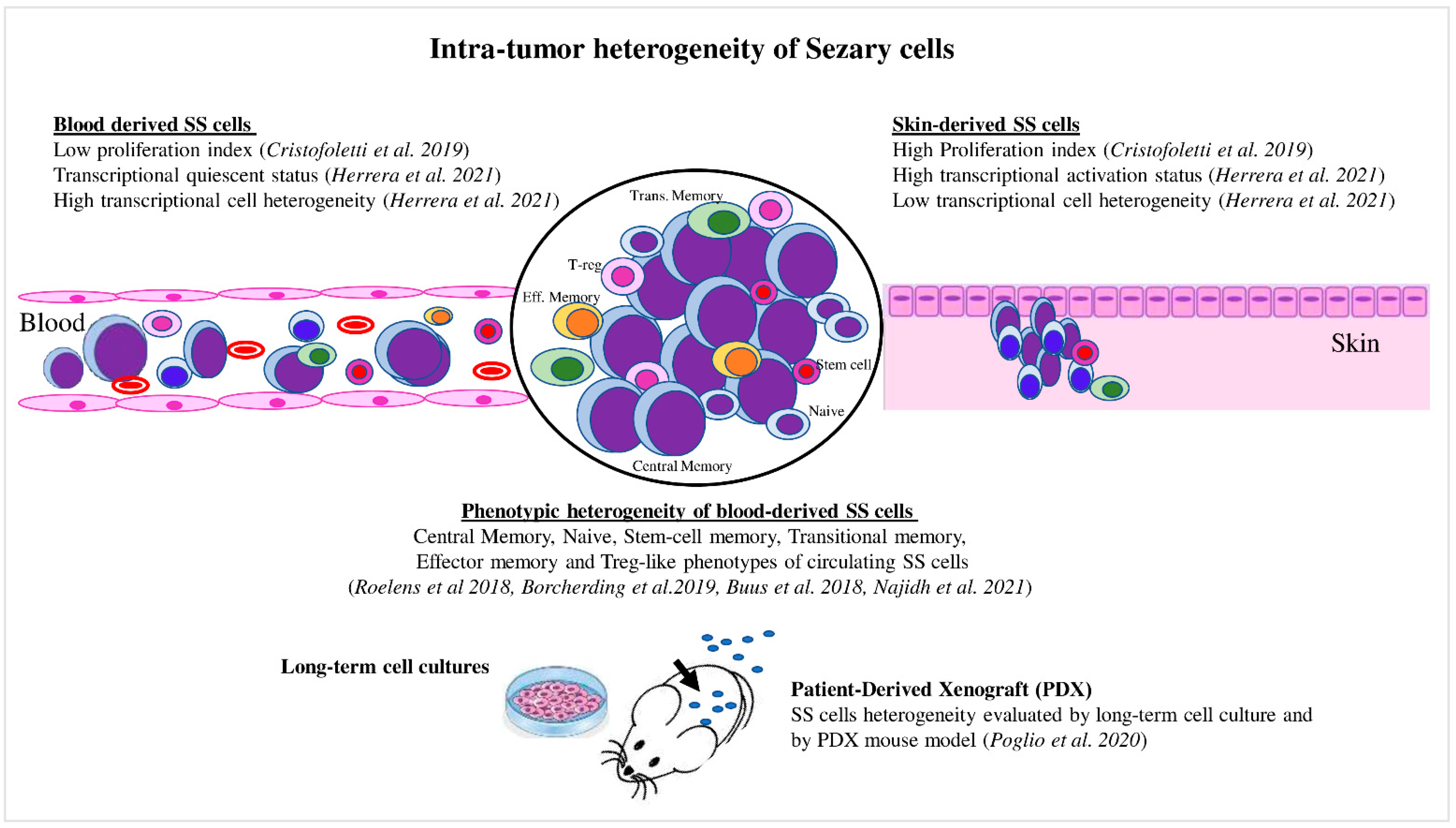
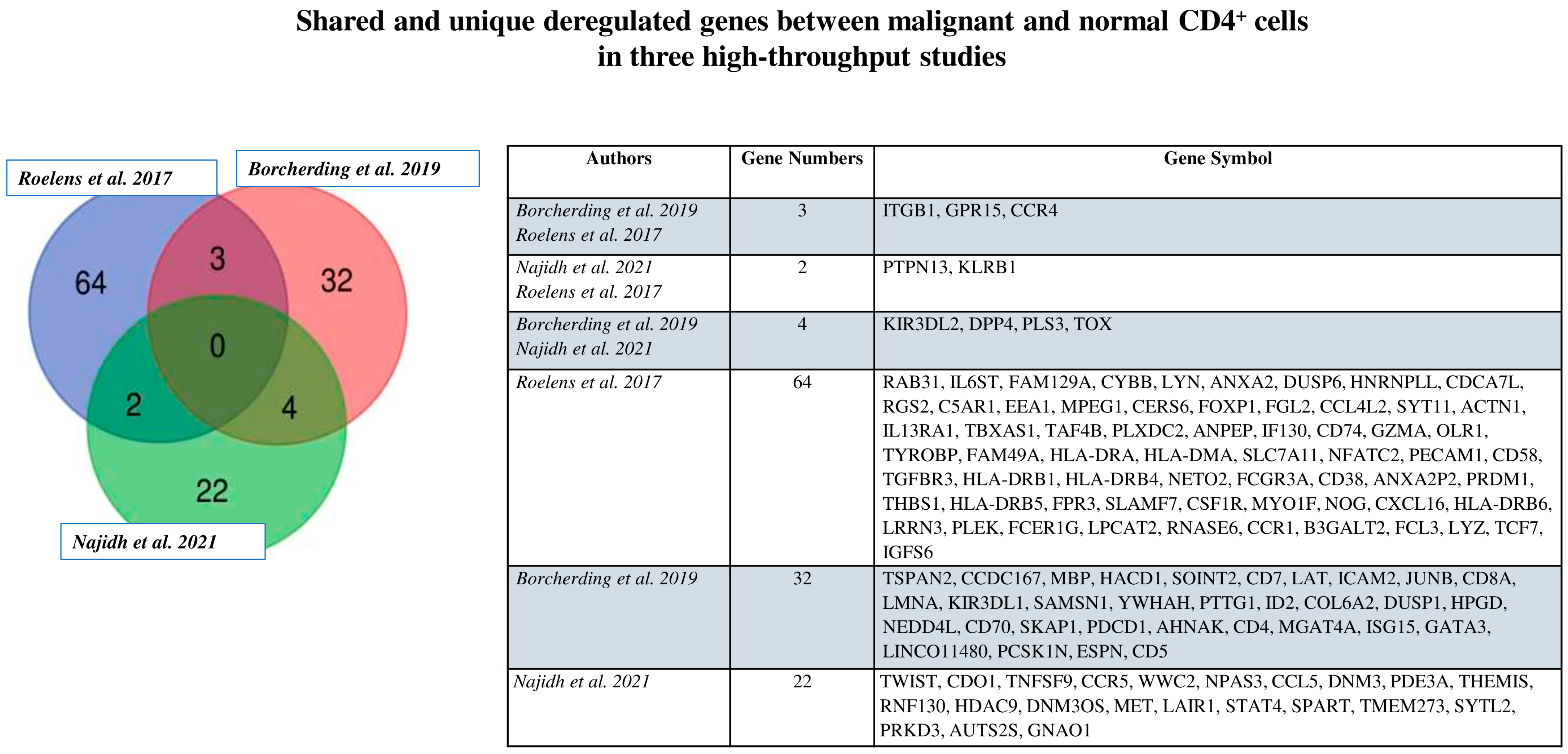
4. Recent Advances in Intratumor Heterogeneity in Circulating SS Cells
5. Intratumor Heterogeneity Influences the Drug Resistance in SS: A Possible Tool to Orient the Therapies In Vivo
6. Beyond Genetic Interpatient Heterogeneity: What Is Next?
7. Conclusions
8. Summary
Author Contributions
Funding
Conflicts of Interest
References
- Scarisbrick, J.J.; Quaglino, P.; Prince, H.M.; Papadavid, E.; Hodak, E.; Bagot, M.; Servitje, O.; Berti, E.; Ortiz-Romero, P.; Stadler, R.; et al. The PROCLIPI International Registry of Early-Stage Mycosis Fungoides Identifies Substantial Diagnostic Delay in Most Patients. Br. J. Dermatol. 2019, 181, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Scarisbrick, J.J.; Prince, H.M.; Vermeer, M.H.; Quaglino, P.; Horwitz, S.; Porcu, P.; Stadler, R.; Wood, G.S.; Beylot-Barry, M.; Pham-Ledard, A.; et al. Cutaneous Lymphoma International Consortium Study of Outcome in Advanced Stages of Mycosis Fungoides and Sézary Syndrome: Effect of Specific Prognostic Markers on Survival and Development of a Prognostic Model. J. Clin. Oncol. 2015, 33, 3766–3773. [Google Scholar] [CrossRef] [PubMed]
- Quaglino, P.; Pimpinelli, N.; Berti, E.; Calzavara-Pinton, P.; Alfonso Lombardo, G.; Rupoli, S.; Alaibac, M.; Bottoni, U.; Carbone, A.; Fava, P.; et al. Time Course, Clinical Pathways, and Long-Term Hazards Risk Trends of Disease Progression in Patients with Classic Mycosis Fungoides: A Multicenter, Retrospective Follow-up Study from the Italian Group of Cutaneous Lymphomas. Cancer 2012, 118, 5830–5839. [Google Scholar] [CrossRef] [PubMed]
- Jonak, C.; Tittes, J.; Brunner, P.M.; Guenova, E. Mycosis Fungoides Und Sézary-Syndrom. JDDG J. der Dtsch. Dermatologischen Gesellschaft 2021, 19, 1307–1335. [Google Scholar] [CrossRef]
- Campbell, J.J.; Clark, R.A.; Watanabe, R.; Kupper, T.S. Sezary Syndrome and Mycosis Fungoides Arise from Distinct T-Cell Subsets: A Biologic Rationale for Their Distinct Clinical Behaviors. Blood 2010, 116, 767–771. [Google Scholar] [CrossRef]
- Olsen, E.; Vonderheid, E.; Pimpinelli, N.; Willemze, R.; Kim, Y.; Knobler, R.; Zackheim, H.; Duvic, M.; Estrach, T.; Lamberg, S.; et al. Revisions to the Staging and Classification of Mycosis Fungoides and Sézary Syndrome: A Proposal of the International Society for Cutaneous Lymphomas (ISCL) and the Cutaneous Lymphoma Task Force of the European Organization of Research and Treatment of Cancer (EORTC). Blood 2007, 110, 1713–1722. [Google Scholar] [CrossRef]
- Wang, L.; Ni, X.; Covington, K.R.; Yang, B.Y.; Shiu, J.; Zhang, X.; Xi, L.; Meng, Q.; Langridge, T.; Drummond, J.; et al. Genomic Profiling of Sézary Syndrome Identifies Alterations of Key T Cell Signaling and Differentiation Genes. Nat. Genet. 2015, 47, 1426–1434. [Google Scholar] [CrossRef]
- Vermeer, M.H.; Van Doorn, R.; Dijkman, R.; Mao, X.; Whittaker, S.; Van Voorst Vader, P.C.; Gerritsen, M.J.P.; Geerts, M.L.; Gellrich, S.; Söderberg, O.; et al. Novel and Highly Recurrent Chromosomal Alterations in Sézary Syndrome. Cancer Res. 2008, 68, 2689–2698. [Google Scholar] [CrossRef]
- Da Silva Almeida, A.C.; Abate, F.; Khiabanian, H.; Martinez-Escala, E.; Guitart, J.; Tensen, C.P.; Vermeer, M.H.; Rabadan, R.; Ferrando, A.; Palomero, T. The Mutational Landscape of Cutaneous T Cell Lymphoma and Sézary Syndrome. Nat. Genet. 2015, 47, 1465–1470. [Google Scholar] [CrossRef]
- Tensen, C.P.; Quint, K.D.; Vermeer, M.H. Genetic and Epigenetic Insights into Cutaneous T-Cell Lymphoma. Blood 2022, 139, 15–33. [Google Scholar] [CrossRef]
- Mirza, A.S.; Horna, P.; Teer, J.K.; Song, J.; Akabari, R.; Hussaini, M.; Sokol, L. New Insights Into the Complex Mutational Landscape of Sézary Syndrome. Front. Oncol. 2020, 10, 514. [Google Scholar] [CrossRef]
- Arneth, B. Tumor Microenvironment. Medicina 2019, 56, 15. [Google Scholar] [CrossRef] [PubMed]
- Miyagaki, T.; Sugaya, M. Immunological Milieu in Mycosis Fungoides and Sézary Syndrome. J. Dermatol. 2014, 41, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Krejsgaard, T.; Lindahl, L.M.; Mongan, N.P.; Wasik, M.A.; Litvinov, I.V.; Iversen, L.; Langhoff, E.; Woetmann, A.; Odum, N. Malignant Inflammation in Cutaneous T-cell Lymphoma—A Hostile Takeover. Semin. Immunopathol. 2017, 39, 269–282. [Google Scholar] [CrossRef] [PubMed]
- Pileri, A.; Guglielmo, A.; Fuligni, F.; Lastrucci, I.; Patrizi, A.; Pimpinelli, N. Second Neoplasm in Cutaneous T-Cell Lymphoma Patients: A Marker of Worse Prognosis? Ital. J. Dermatology Venereol. 2021, 156, 484–488. [Google Scholar] [CrossRef] [PubMed]
- Quaglino, P.; Fava, P.; Pileri, A.; Grandi, V.; Sanlorenzo, M.; Panasiti, V.; Guglielmo, A.; Alberti-Violetti, S.; Novelli, M.; Astrua, C.; et al. Phenotypical Markers, Molecular Mutations, and Immune Microenvironment as Targets for New Treatments in Patients with Mycosis Fungoides and/or Sézary Syndrome. J. Investig. Dermatol. 2021, 141, 484–495. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, B.R.; Zain, J.; Rosen, S.T.; Querfeld, C. Tumor Microenvironment in Mycosis Fungoides and Sézary Syndrome. Curr. Opin. Oncol. 2016, 28, 88–96. [Google Scholar] [CrossRef]
- Pals, S.T.; De Gorter, D.J.J.; Spaargaren, M. Lymphoma Dissemination: The Other Face of Lymphocyte Homing. Blood 2007, 110, 3102–3111. [Google Scholar] [CrossRef]
- Yamanaka, K.; Clark, R.; Rich, B.; Dowgiert, R.; Hirahara, K.; Hurwitz, D.; Shibata, M.; Mirchandani, N.; Jones, D.A.; Goddard, D.S.; et al. Skin-Derived Interleukin-7 Contributes to the Proliferation of Lymphocytes in Cutaneous T-Cell Lymphoma. Blood 2006, 107, 2440–2445. [Google Scholar] [CrossRef]
- Berger, C.L.; Hanlon, D.; Kanada, D.; Dhodapkar, M.; Lombillo, V.; Wang, N.; Christensen, I.; Howe, G.; Crouch, J.; El-Fishawy, P.; et al. The Growth of Cutaneous T-Cell Lymphoma Is Stimulated by Immature Dendritic Cells. Blood 2002, 99, 2929–2939. [Google Scholar] [CrossRef]
- Wilcox, R.A.; Wada, D.A.; Ziesmer, S.C.; Elsawa, S.F.; Comfere, N.I.; Dietz, A.B.; Novak, A.J.; Witzig, T.E.; Feldman, A.L.; Pittelkow, M.R.; et al. Monocytes Promote Tumor Cell Survival in T-Cell Lymphoproliferative Disorders and Are Impaired in Their Ability to Differentiate into Mature Dendritic Cells. Blood 2009, 114, 2936–2944. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Schulte, B.C.; Zhou, Y.; Haribhai, D.; Mackinnon, A.C.; Plaza, J.A.; Williams, C.B.; Hwang, S.T. Depletion of M2-like Tumor-Associated Macrophages Delays Cutaneous T-Cell Lymphoma Development in Vivo. J. Investig. Dermatol. 2014, 134, 2814–2822. [Google Scholar] [CrossRef] [PubMed]
- Rabenhorst, A.; Schlaak, M.; Heukamp, L.C.; Förster, A.; Theurich, S.; von Bergwelt-Baildon, M.; Büttner, R.; Kurschat, P.; Mauch, C.; Roers, A.; et al. Mast Cells Play a Protumorigenic Role in Primary Cutaneous Lymphoma. Blood 2012, 120, 2042–2054. [Google Scholar] [CrossRef] [PubMed]
- Dagogo-Jack, I.; Shaw, A.T. Tumour Heterogeneity and Resistance to Cancer Therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Stolearenco, V.; Namini, M.R.J.; Hasselager, S.S.; Gluud, M.; Buus, T.B.; Willerslev-Olsen, A.; Ødum, N.; Krejsgaard, T. Cellular Interactions and Inflammation in the Pathogenesis of Cutaneous T-Cell Lymphoma. Front. Cell Dev. Biol. 2020, 8, 851. [Google Scholar] [CrossRef]
- Rassek, K.; Iżykowska, K. Single-Cell Heterogeneity of Cutaneous T-Cell Lymphomas Revealed Using RNA-Seq Technologies. Cancers 2020, 12, 2129. [Google Scholar] [CrossRef]
- García-Díaz, N.; Piris, M.Á.; Ortiz-Romero, P.L.; Vaqué, J.P. Mycosis Fungoides and Sézary Syndrome: An Integrative Review of the Pathophysiology, Molecular Drivers, and Targeted Therapy. Cancers 2021, 13, 1931. [Google Scholar] [CrossRef]
- Roelens, M.; Delord, M.; Ram-Wolff, C.; Marie-Cardine, A.; Alberdi, A.; Maki, G.; Homyrda, L.; Bensussan, A.; Bagot, M.; Toubert, A.; et al. Circulating and Skin-Derived Sézary Cells: Clonal but with Phenotypic Plasticity. Blood 2017, 130, 1468–1471. [Google Scholar] [CrossRef]
- Kim, Y.H.; Bagot, M.; Pinter-Brown, L.; Rook, A.H.; Porcu, P.; Horwitz, S.M.; Whittaker, S.; Tokura, Y.; Vermeer, M.; Zinzani, P.L.; et al. Mogamulizumab versus Vorinostat in Previously Treated Cutaneous T-Cell Lymphoma (MAVORIC): An International, Open-Label, Randomised, Controlled Phase 3 Trial. Lancet. Oncol. 2018, 19, 1192–1204. [Google Scholar] [CrossRef]
- Herrera, A.; Cheng, A.; Mimitou, E.P.; Seffens, A.; George, D.; Bar-Natan, M.; Heguy, A.; Ruggles, K.V.; Scher, J.U.; Hymes, K.; et al. Multimodal Single-Cell Analysis of Cutaneous T-Cell Lymphoma Reveals Distinct Subclonal Tissue-Dependent Signatures. Blood 2021, 138, 1456–1464. [Google Scholar] [CrossRef]
- Cristofoletti, C.; Bresin, A.; Picozza, M.; Picchio, M.C.M.C.; Monzo, F.; Helmer Citterich, M.; Passarelli, F.; Frezzolini, A.; Scala, E.; Monopoli, A.; et al. Blood and Skin-Derived Sezary Cells: Differences in Proliferation-Index, Activation of PI3K/AKT/MTORC1 Pathway and Its Prognostic Relevance. Leukemia 2019, 33, 1231–1242. [Google Scholar] [CrossRef] [PubMed]
- Preston, G.C.; Feijoo-Carnero, C.; Schurch, N.; Cowling, V.H.; Cantrell, D.A. The Impact of KLF2 Modulation on the Transcriptional Program and Function of CD8 T Cells. PLoS ONE 2013, 8, e77537. [Google Scholar] [CrossRef] [PubMed]
- Willinger, T.; Freeman, T.; Herbert, M.; Hasegawa, H.; McMichael, A.J.; Callan, M.F.C. Human Naive CD8 T Cells Down-Regulate Expression of the WNT Pathway Transcription Factors Lymphoid Enhancer Binding Factor 1 and Transcription Factor 7 (T Cell Factor-1) Following Antigen Encounter in Vitro and in Vivo. J. Immunol. 2006, 176, 1439–1446. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Liu, F.; Wang, Q.J.; Rosenberg, S.A.; Morgan, R.A. The Shedding of CD62L (L-Selectin) Regulates the Acquisition of Lytic Activity in Human Tumor Reactive T Lymphocytes. PLoS ONE 2011, 6, e22560. [Google Scholar] [CrossRef] [PubMed]
- Kulpa, D.A.; Lawani, M.; Cooper, A.; Peretz, Y.; Ahlers, J.; Sékaly, R.P. PD-1 Coinhibitory Signals: The Link Between Pathogenesis and Protection. Semin. Immunol. 2013, 25, 219–227. [Google Scholar] [CrossRef]
- Howden, A.J.M.; Hukelmann, J.L.; Brenes, A.; Spinelli, L.; Sinclair, L.V.; Lamond, A.I.; Cantrell, D.A. Quantitative Analysis of T Cell Proteomes and Environmental Sensors during T Cell Differentiation. Nat. Immunol. 2019, 20, 1542–1554. [Google Scholar] [CrossRef]
- Choi, J.; Goh, G.; Walradt, T.; Hong, B.S.; Bunick, C.G.; Chen, K.; Bjornson, R.D.; Maman, Y.; Wang, T.; Tordoff, J.; et al. Genomic Landscape of Cutaneous T Cell Lymphoma. Nat. Genet. 2015, 47, 1011–1019. [Google Scholar] [CrossRef]
- Park, J.; Yang, J.; Wenzel, A.T.; Ramachandran, A.; Lee, W.J.; Daniels, J.C.; Kim, J.; Martinez-Escala, E.; Amankulor, N.; Pro, B.; et al. Genomic Analysis of 220 CTCLs Identifies a Novel Recurrent Gain-of-Function Alteration in RLTPR (p.Q575E). Blood 2017, 130, 1430–1440. [Google Scholar] [CrossRef]
- Ungewickell, A.; Bhaduri, A.; Rios, E.; Reuter, J.; Lee, C.S.; Mah, A.; Zehnder, A.; Ohgami, R.; Kulkarni, S.; Armstrong, R.; et al. Genomic Analysis of Mycosis Fungoides and Sézary Syndrome Identifies Recurrent Alterations in TNFR2. Nat. Genet. 2015, 47, 1056–1060. [Google Scholar] [CrossRef]
- Vaqué, J.P.; Gómez-López, G.; Monsálvez, V.; Varela, I.; Martínez, N.; Pérez, C.; Domínguez, O.; Graña, O.; Rodríguez-Peralto, J.L.; Rodríguez-Pinilla, S.M.; et al. PLCG1 Mutations in Cutaneous T-Cell Lymphomas. Blood 2014, 123, 2034–2043. [Google Scholar] [CrossRef]
- Okkenhaug, K.; Fruman, D.A. PI3Ks in Lymphocyte Signaling and Development. Curr. Top. Microbiol. Immunol. 2010, 346, 57–85. [Google Scholar] [CrossRef] [PubMed]
- Laharanne, E.; Oumouhou, N.; Bonnet, F.; Carlotti, M.; Gentil, C.; Chevret, E.; Jouary, T.; Longy, M.; Vergier, B.; Beylot-Barry, M.; et al. Genome-Wide Analysis of Cutaneous T-Cell Lymphomas Identifies Three Clinically Relevant Classes. J. Investig. Dermatol. 2010, 130, 1707–1718. [Google Scholar] [CrossRef]
- Scarisbrick, J.J.; Woolford, A.J.; Russell-Jones, R.; Whittaker, S.J. Loss of Heterozygosity on 10q and Microsatellite Instability in Advanced Stages of Primary Cutaneous T-Cell Lymphoma and Possible Association with Homozygous Deletion of PTEN. Blood 2000, 95, 2937–2942. [Google Scholar] [CrossRef] [PubMed]
- Cristofoletti, C.; Picchio, M.C.; Lazzeri, C.; Tocco, V.; Pagani, E.; Bresin, A.; Mancini, B.; Passarelli, F.; Facchiano, A.; Scala, E.; et al. Comprehensive Analysis of PTEN Status in Sézary Syndrome. Blood J. Am. Soc. Hematol. 2013, 122, 3511–3520. [Google Scholar] [CrossRef]
- Bresin, A.; Cristofoletti, C.; Caprini, E.; Cantonetti, M.; Monopoli, A.; Russo, G.; Narducci, M.G.M.G.M.G. Preclinical Evidence for Targeting PI3K/MTOR Signaling with Dual-Inhibitors as a Therapeutic Strategy against Cutaneous T-Cell Lymphoma. J. Investig. Dermatol. 2020, 140, 1045–1053.e6. [Google Scholar] [CrossRef] [PubMed]
- Marzec, M.; Liu, X.; Kasprzycka, M.; Witkiewicz, A.; Raghunath, P.N.; El-Salem, M.; Robertson, E.; Odum, N.; Wasik, M.A. IL-2- and IL-15-Induced Activation of the Rapamycin-Sensitive MTORC1 Pathway in Malignant CD4+ T Lymphocytes. Blood 2008, 111, 2181–2189. [Google Scholar] [CrossRef] [PubMed]
- Marzec, M.; Halasa, K.; Kasprzycka, M.; Wysocka, M.; Liu, X.; Tobias, J.W.; Baldwin, D.; Zhang,, Q.; Odum, N.; Rook, A.H.; et al. Differential effects of interleukin-2 and interleukin-15 versus interleukin-21 on CD4+ cutaneous T-cell lymphoma cells. Cancer Res. 2008, 68, 1083–1091. [Google Scholar] [CrossRef]
- Kittipongdaja, W.; Wu, X.; Garner, J.; Liu, X.; Komas, S.M.; Hwang, S.T.; Schieke, S.M. Rapamycin Suppresses Tumor Growth and Alters the Metabolic Phenotype in T-Cell Lymphoma. J. Investig. Dermatol. 2015, 135, 2301–2308. [Google Scholar] [CrossRef]
- Murga-Zamalloa, C.; Rolland, D.C.M.; Polk, A.; Wolfe, A.; Dewar, H.; Chowdhury, P.; Onder, O.; Dewar, R.; Brown, N.A.; Bailey, N.G.; et al. Colony-Stimulating Factor 1 Receptor (CSF1R) Activates Akt/MTOR Signaling and Promotes T-Cell Lymphoma Viability. Clin. Cancer Res. 2020, 26, 690–703. [Google Scholar] [CrossRef]
- Abreu, M.; Miranda, M.; Castro, M.; Fernandes, I.; Cabral, R.; Santos, A.H.; Fonseca, S.; Rodrigues, J.; Leander, M.; Lau, C.; et al. IL-31 and IL-8 in Cutaneous T-Cell Lymphoma: Looking for Their Role in Itch. Adv. Hematol. 2021, 2021, 5582581. [Google Scholar] [CrossRef]
- Ferretti, E.; Corcione, A.; Pistoia, V. The IL-31/IL-31 Receptor Axis: General Features and Role in Tumor Microenvironment. J. Leukoc. Biol. 2017, 102, 711–717. [Google Scholar] [CrossRef] [PubMed]
- Levidou, G.; Siakantaris, M.; Papadaki, T.; Papadavid, E.; Vassilakopoulos, T.P.; Angelopoulou, M.K.; Marinos, L.; Nikolaou, V.; Economidi, A.; Antoniou, C.; et al. A Comprehensive Immunohistochemical Approach of AKT/MTOR Pathway and p-STAT3 in Mycosis Fungoides. J. Am. Acad. Dermatol. 2013, 69, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Aronovich, A.; Moyal, L.; Gorovitz, B.; Amitay-Laish, I.; Naveh, H.P.; Forer, Y.; Maron, L.; Knaneh, J.; Ad-El, D.; Yaacobi, D.; et al. Cancer-Associated Fibroblasts in Mycosis Fungoides Promote Tumor Cell Migration and Drug Resistance through CXCL12/CXCR4. J. Investig. Dermatol. 2021, 141, 619–627.e2. [Google Scholar] [CrossRef] [PubMed]
- Hong, C.H.; Lin, S.H.; Lee, C.H. CCL21 Induces MTOR-Dependent MALAT1 Expression, Leading to Cell Migration in Cutaneous T-Cell Lymphoma. In Vivo 2019, 33, 793–800. [Google Scholar] [CrossRef] [PubMed]
- Narducci, M.G.; Scala, E.; Bresin, A.; Caprini, E.; Picchio, M.C.; Remotti, D.; Ragone, G.; Nasorri, F.; Frontani, M.; Arcelli, D.; et al. Skin Homing of Sézary Cells Involves SDF-1-CXCR4 Signaling and down-Regulation of CD26/Dipeptidylpeptidase IV. Blood 2006, 107, 1108–1115. [Google Scholar] [CrossRef]
- Picchio, M.C.; Scala, E.; Pomponi, D.; Caprini, E.; Frontani, M.; Angelucci, I.; Mangoni, A.; Lazzeri, C.; Perez, M.; Remotti, D.; et al. CXCL13 Is Highly Produced by Sézary Cells and Enhances Their Migratory Ability via a Synergistic Mechanism Involving CCL19 and CCL21 Chemokines. Cancer Res. 2008, 68, 7137–7146. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and Quantifying Mammalian Transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Buus, T.B.; Willerslev-Olsen, A.; Fredholm, S.; Blümel, E.; Nastasi, C.; Gluud, M.; Hu, T.; Lindahl, L.M.; Iversen, L.; Fogh, H.; et al. Single-Cell Heterogeneity in Sézary Syndrome. Blood Adv. 2018, 2, 2115–2126. [Google Scholar] [CrossRef]
- Najidh, S.; Tensen, C.P.; Van Der Sluijs-Gelling, A.J.; Teodosio, C.; Cats, D.; Mei, H.; Kuipers, T.B.; Out-Luijting, J.J.; Zoutman, W.H.; Van Hall, T.; et al. Improved S Ezary Cell Detection and Novel Insights into Immunophenotypic and Molecular Heterogeneity in S Ezary Syndrome. Blood J. Am. Soc. Hematol. 2021, 138, 2539–2554. [Google Scholar]
- Borcherding, N.; Voigt, A.P.; Liu, V.; Link, B.K.; Zhang, W.; Jabbari, A. Single-Cell Profiling of Cutaneous T-Cell Lymphoma Reveals Underlying Heterogeneity Associated with Disease Progression. Clin. Cancer Res. 2019, 25, 2996–3005. [Google Scholar] [CrossRef]
- Park, J.; Daniels, J.; Wartewig, T.; Ringbloom, K.G.; Martinez-Escala, M.E.; Choi, S.; Thomas, J.J.; Doukas, P.G.; Yang, J.; Snowden, C.; et al. Integrated Genomic Analyses of Cutaneous T-Cell Lymphomas Reveal the Molecular Bases for Disease Heterogeneity. Blood 2021, 138, 1225–1236. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Park, S.M.; Tumanov, A.V.; Hau, A.; Sawada, K.; Feig, C.; Turner, J.R.; Fu, Y.X.; Romero, I.L.; Lengyel, E.; et al. CD95 Promotes Tumour Growth. Nature 2010, 465, 492–496. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.L.; Wain, E.M.; Chu, C.-C.; Tosi, I.; Foster, R.; McKenzie, R.C.T.; Whittaker, S.J.; Mitchell, T.J. Downregulation of Fas Gene Expression in Sézary Syndrome Is Associated with Promoter Hypermethylation. J. Investig. Dermatol. 2010, 130, 1116–1125. [Google Scholar] [CrossRef] [PubMed]
- Nagai, Y.; Kawahara, M.; Hishizawa, M.; Shimazu, Y.; Sugino, N.; Fujii, S.; Kadowaki, N.; Takaori-Kondo, A. T Memory Stem Cells Are the Hierarchical Apex of Adult T-Cell Leukemia. Blood 2015, 125, 3527–3535. [Google Scholar] [CrossRef]
- Pulitzer, M.P.; Horna, P.; Almeida, J. Sézary Syndrome and Mycosis Fungoides: An Overview, Including the Role of Immunophenotyping. Cytom. Part B Clin. Cytom. 2021, 100, 132–138. [Google Scholar] [CrossRef]
- Li, J.Y.; Horwitz, S.; Moskowitz, A.; Myskowski, P.L.; Pulitzer, M.; Querfeld, C. Management of Cutaneous T Cell Lymphoma: New and Emerging Targets and Treatment Options. Cancer Manag. Res. 2012, 4, 75–89. [Google Scholar] [CrossRef][Green Version]
- Tracey, L.; Villuendas, R.; Ortiz, P.; Dopazo, A.; Spiteri, I.; Lombardia, L.; Rodríguez-Peralto, J.L.; Fernández-Herrera, J.; Hernández, A.; Fraga, J.; et al. Identification of Genes Involved in Resistance to Interferon-Alpha in Cutaneous T-Cell Lymphoma. Am. J. Pathol. 2002, 161, 1825–1837. [Google Scholar] [CrossRef]
- Otsubo, C.; Otomo, R.; Miyazaki, M.; Matsushima-Hibiya, Y.; Kohno, T.; Iwakawa, R.; Takeshita, F.; Okayama, H.; Ichikawa, H.; Saya, H.; et al. TSPAN2 Is Involved in Cell Invasion and Motility during Lung Cancer Progression. Cell Rep. 2014, 7, 527–538. [Google Scholar] [CrossRef]
- Willerslev-Olsen, A.; Buus, T.B.; Nastasi, C.; Blümel, E.; Gluud, M.; Bonefeld, C.M.; Geisler, C.; Lindahl, L.M.; Vermeer, M.; Wasik, M.A.; et al. Staphylococcus Aureus Enterotoxins Induce FOXP3 in Neoplastic T Cells in Sézary Syndrome. Blood Cancer J. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- RA, C. Regulation Gone Wrong: A Subset of Sézary Patients Have Malignant Regulatory T Cells. J. Investig. Dermatol. 2009, 129, 2747–2750. [Google Scholar] [CrossRef]
- Krejsgaard, T.; Odum, N.; Geisler, C.; Wasik, M.A.; Woetmann, A. REVIEW Regulatory T Cells and Immunodeficiency in Mycosis Fungoides and Sézary Syndrome. Leukemia 2012, 26, 424–432. [Google Scholar] [CrossRef] [PubMed]
- Heid, J.B.; Schmidt, A.; Oberle, N.; Goerdt, S.; Krammer, P.H.; Suri-Payer, E.; Klemke, C.D. FOXP3+CD25- Tumor Cells with Regulatory Function in Sézary Syndrome. J. Investig. Dermatol. 2009, 129, 2875–2885. [Google Scholar] [CrossRef] [PubMed]
- Boonk, S.E.; Zoutman, W.H.; Marie-Cardine, A.; van der Fits, L.; Out-Luiting, J.J.; Mitchell, T.J.; Tosi, I.; Morris, S.L.; Moriarty, B.; Booken, N.; et al. Evaluation of Immunophenotypic and Molecular Biomarkers for Sézary Syndrome Using Standard Operating Procedures: A Multicenter Study of 59 Patients. J. Investig. Dermatol. 2016, 136, 1364–1372. [Google Scholar] [CrossRef]
- Walia, R.; Yeung, C.C.S. An Update on Molecular Biology of Cutaneous T Cell Lymphoma. Front. Oncol. 2020, 9, 1558. [Google Scholar] [CrossRef] [PubMed]
- Jegodzinski, L.; Sezin, T.; Loser, K.; Mousavi, S.; Zillikens, D.; Sadik, C.D. The G Protein-Coupled Receptor (GPR) 15 Counteracts Antibody-Mediated Skin Inflammation. Front. Immunol. 2020, 11, 1858. [Google Scholar] [CrossRef]
- Bauer, M. The Role of GPR15 Function in Blood and Vasculature. Int. J. Mol. Sci. 2021, 22, 10824. [Google Scholar] [CrossRef]
- McHeik, S.; Aptecar, L.; Coopman, P.; D’hondt, V.; Freiss, G. Dual Role of the PTPN13 Tyrosine Phosphatase in Cancer. Biomolecules 2020, 10, 1659. [Google Scholar] [CrossRef]
- Ye, W.; Luo, C.; Li, C.; Liu, Z.; Liu, F. CD161, a Promising Immune Checkpoint, Correlates with Patient Prognosis: A Pan-Cancer Analysis. J. Cancer 2021, 12, 6588–6599. [Google Scholar] [CrossRef]
- Nurzat, Y.; Su, W.; Min, P.; Li, K.; Xu, H.; Zhang, Y. Identification of Therapeutic Targets and Prognostic Biomarkers Among Integrin Subunits in the Skin Cutaneous Melanoma Microenvironment. Front. Oncol. 2021, 11, 3907. [Google Scholar] [CrossRef]
- Nwagwu, C.D.; Immidisetti, A.V.; Bukanowska, G.; Vogelbaum, M.A.; Carbonell, A.M. Convection-Enhanced Delivery of a First-in-Class Anti-Β1 Integrin Antibody for the Treatment of High-Grade Glioma Utilizing Real-Time Imaging. Pharmaceutics 2020, 13, 40. [Google Scholar] [CrossRef]
- Duvic, M.; Talpur, R.; Ni, X.; Zhang, C.; Hazarika, P.; Kelly, C.; Chiao, J.H.; Reilly, J.F.; Ricker, J.L.; Richon, V.M.; et al. Phase 2 Trial of Oral Vorinostat (Suberoylanilide Hydroxamic Acid, SAHA) for Refractory Cutaneous T-Cell Lymphoma (CTCL). Blood 2007, 109, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Piekarz, R.L.; Frye, R.; Turner, M.; Wright, J.J.; Allen, S.L.; Kirschbaum, M.H.; Zain, J.; Prince, H.M.; Leonard, J.P.; Geskin, L.J.; et al. Phase II Multi-Institutional Trial of the Histone Deacetylase Inhibitor Romidepsin as Monotherapy for Patients with Cutaneous T-Cell Lymphoma. J. Clin. Oncol. 2009, 27, 5410–5417. [Google Scholar] [CrossRef] [PubMed]
- Poglio, S.; Prochazkova-Carlotti, M.; Cherrier, F.; Gros, A.; Laharanne, E.; Pham-Ledard, A.; Beylot-Barry, M.; Merlio, J.-P.P. Xenograft and cell culture models of Sézary syndrome reveal cell of origin diversity and subclonal heterogeneity. Leukemia 2021, 35, 1696–1709. [Google Scholar] [CrossRef] [PubMed]
- Bresin, A.; Caprini, E.; Russo, G.; Narducci, M.G. Challenging Cutaneous T-Cell Lymphoma: What Animal Models Tell Us So Far. J. Investig. Dermatol. 2022, 142, 1533–1540. [Google Scholar] [CrossRef]
- Izykowska, K.; Przybylski, G.K. Genetic Alterations in Sezary Syndrome. Leuk. Lymphoma 2011, 52, 745–753. [Google Scholar] [CrossRef]
- Iżykowska, K.; Przybylski, G.K.; Gand, C.; Braun, F.C.; Grabarczyk, P.; Kuss, A.W.; Olek-Hrab, K.; Bastidas Torres, A.N.; Vermeer, M.H.; Zoutman, W.H.; et al. Genetic Rearrangements Result in Altered Gene Expression and Novel Fusion Transcripts in Sézary Syndrome. Oncotarget 2017, 8, 39627–39639. [Google Scholar] [CrossRef]
- Chevret, E.; Merlio, J.P. Sézary Syndrome: Translating Genetic Diversity into Personalized Medicine. J. Investig. Dermatol. 2016, 136, 1319–1324. [Google Scholar] [CrossRef][Green Version]
- McLane, L.M.; Abdel-Hakeem, M.S.; Wherry, E.J. CD8 T Cell Exhaustion During Chronic Viral Infection and Cancer. Annu. Rev. Immunol. 2019, 37, 457–495. [Google Scholar] [CrossRef]
- Gupta, P.K.; Godec, J.; Wolski, D.; Adland, E.; Yates, K.; Pauken, K.E.; Cosgrove, C.; Ledderose, C.; Junger, W.G.; Robson, S.C.; et al. CD39 Expression Identifies Terminally Exhausted CD8+ T Cells. PLoS Pathog. 2015, 11, e1005177. [Google Scholar] [CrossRef]
- Sánchez-Beato, M. PD-1 Loss and T-Cell Exhaustion in CTCL Tumoral T Cells. Blood 2021, 138, 1201–1203. [Google Scholar] [CrossRef]
- Khodadoust, M.S.; Rook, A.H.; Porcu, P.; Foss, F.; Moskowitz, A.J.; Shustov, A.; Shanbhag, S.; Sokol, L.; Fling, S.P.; Ramchurren, N.; et al. Pembrolizumab in Relapsed and Refractory Mycosis Fungoides and Sezary Syndrome: A Multicenter Phase II Study. J Clin Oncol 2020, 38, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Takenaka, M.C.; Robson, S.; Quintana, F.J. Regulation of the T Cell Response by CD39. Trends Immunol. 2016, 37, 427–439. [Google Scholar] [CrossRef] [PubMed]
- Friedman, D.J.; Künzli, B.M.; A-Rahim, Y.I.; Sevigny, J.; Berberat, P.O.; Enjyoji, K.; Csizmadia, E.; Friess, H.; Robson, S.C. From the Cover: CD39 Deletion Exacerbates Experimental Murine Colitis and Human Polymorphisms Increase Susceptibility to Inflammatory Bowel Disease. Proc. Natl. Acad. Sci. USA 2009, 106, 16788–16793. [Google Scholar] [CrossRef] [PubMed]
- Rissiek, A.; Baumann, I.; Cuapio, A.; Mautner, A.; Kolster, M.; Arck, P.C.; Dodge-Khatami, A.; Mittrücker, H.-W.; Koch-Nolte, F.; Haag, F.; et al. The Expression of CD39 on Regulatory T Cells Is Genetically Driven and Further Upregulated at Sites of Inflammation. J. Autoimmun. 2015, 58, 12–20. [Google Scholar] [CrossRef]
- Fang, F.; Yu, M.; Cavanagh, M.M.; Hutter Saunders, J.; Qi, Q.; Ye, Z.; Le Saux, S.; Sultan, W.; Turgano, E.; Dekker, C.L.; et al. Expression of CD39 on Activated T Cells Impairs Their Survival in Older Individuals. Cell Rep. 2016, 14, 1218–1231. [Google Scholar] [CrossRef]
- Gallerano, D.; Ciminati, S.; Grimaldi, A.; Piconese, S.; Cammarata, I.; Focaccetti, C.; Pacella, I.; Accapezzato, D.; Lancellotti, F.; Sacco, L.; et al. Genetically Driven CD39 Expression Shapes Human Tumor-Infiltrating CD8+ T-Cell Functions. Int. J. Cancer 2020, 147, 2597–2610. [Google Scholar] [CrossRef]
- Picozza, M.; Cristofoletti, C.; Bresin, A.; Fioretti, M.; Sambucci, M.; Scala, E.; Monopoli, A.; Cantonetti, M.; Pilla, M.A.; Accetturi, M.P.; et al. Genetically Driven CD39 Expression Affects Sezary Cell Viability, IL-2 Production and Detects Two Patient Subsets with Distinct Prognosis. J. Investig. Dermatol. 2022. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cristofoletti, C.; Bresin, A.; Fioretti, M.; Russo, G.; Narducci, M.G. Combined High-Throughput Approaches Reveal the Signals Driven by Skin and Blood Environments and Define the Tumor Heterogeneity in Sézary Syndrome. Cancers 2022, 14, 2847. https://doi.org/10.3390/cancers14122847
Cristofoletti C, Bresin A, Fioretti M, Russo G, Narducci MG. Combined High-Throughput Approaches Reveal the Signals Driven by Skin and Blood Environments and Define the Tumor Heterogeneity in Sézary Syndrome. Cancers. 2022; 14(12):2847. https://doi.org/10.3390/cancers14122847
Chicago/Turabian StyleCristofoletti, Cristina, Antonella Bresin, Martina Fioretti, Giandomenico Russo, and Maria Grazia Narducci. 2022. "Combined High-Throughput Approaches Reveal the Signals Driven by Skin and Blood Environments and Define the Tumor Heterogeneity in Sézary Syndrome" Cancers 14, no. 12: 2847. https://doi.org/10.3390/cancers14122847
APA StyleCristofoletti, C., Bresin, A., Fioretti, M., Russo, G., & Narducci, M. G. (2022). Combined High-Throughput Approaches Reveal the Signals Driven by Skin and Blood Environments and Define the Tumor Heterogeneity in Sézary Syndrome. Cancers, 14(12), 2847. https://doi.org/10.3390/cancers14122847





