Somatic Mutations in Exon 7 of the TP53 Gene in Index Colorectal Lesions Are Associated with the Early Occurrence of Metachronous Adenoma
Abstract
:Simple Summary
Abstract
1. Introduction
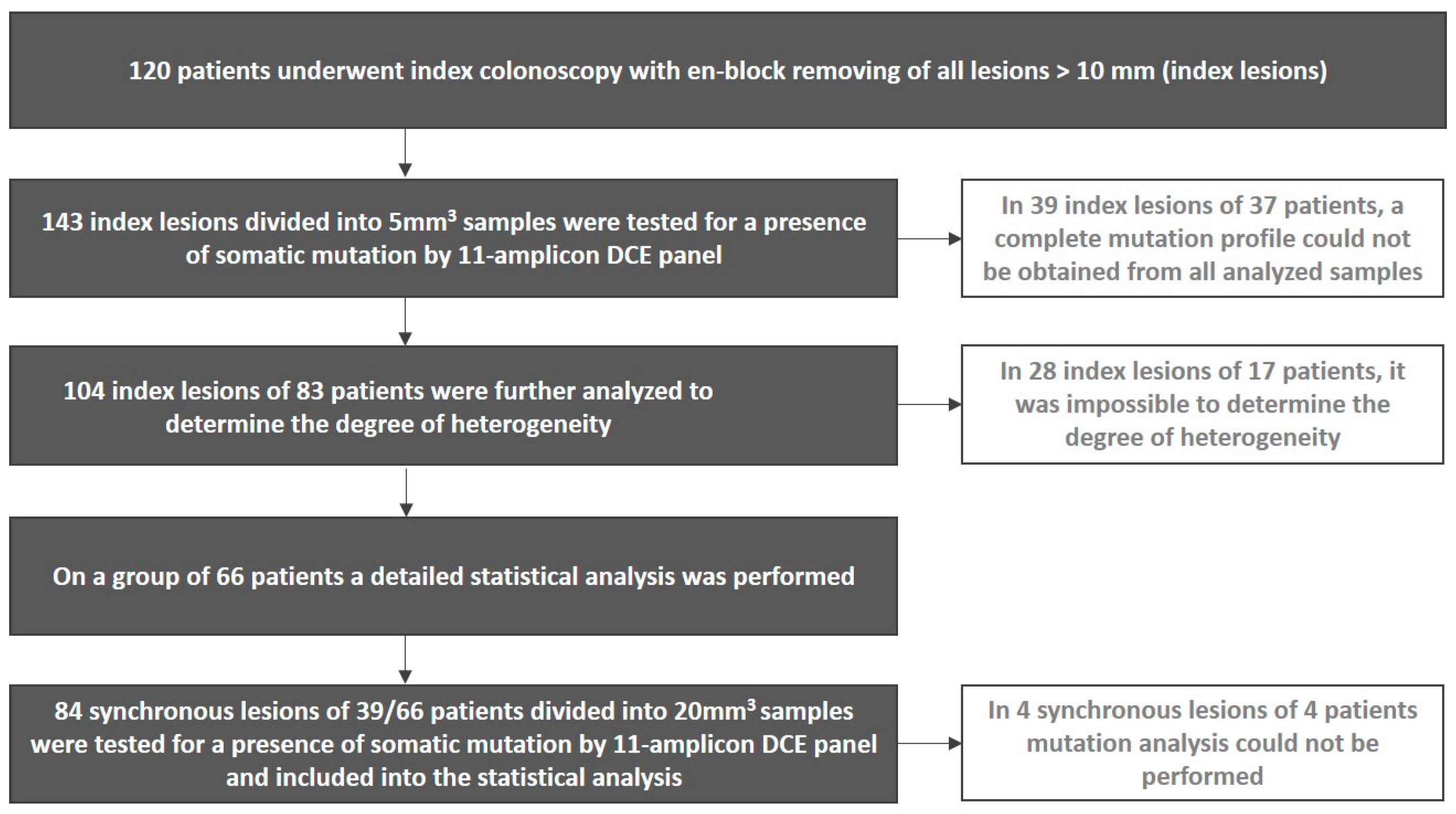
2. Materials and Methods
2.1. Patients
2.2. Index and Surveillance Colonoscopy
2.3. Samples
2.4. Molecular-Genetic Analysis
2.5. Detection of TP53 Mutations in Metachronous Adenomas
2.6. Determination of the Degree of Heterogeneity
2.7. Statistical Analysis
3. Results
3.1. Surveillance Colonoscopy Results
3.2. Mutation Analyses of Index and Synchronous Lesions
3.3. Statistical Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferlay, J.; Laversanne, M.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Tomorrow; International Agency for Research on Cancer: Lyon, France, 2020. Available online: https://gco.iarc.fr/tomorrow (accessed on 13 April 2022).
- Brenner, H.; Chang-Claude, J.; Seiler, C.M.; Rickert, A.; Hoffmeister, M. Protection from colorectal cancer after colonoscopy: A population-based, case-control study. Ann. Intern. Med. 2011, 154, 22–30. [Google Scholar] [CrossRef]
- Hassan, C.; Antonelli, G.; Dumonceau, J.M.; Regula, J.; Bretthauer, M.; Chaussade, S.; Dekker, E.; Ferlitsch, M.; Gimeno-Garcia, A.; Jover, R.; et al. Post-polypectomy colonoscopy surveillance: European Society of Gastrointestinal Endoscopy (ESGE) Guideline-Update 2020. Endoscopy 2020, 52, 687–700. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaqué, J.P.; Martínez, N.; Varela, I.; Fernández, F.; Mayorga, M.; Derdak, S.; Beltrán, S.; Moreno, T.; Almaraz, C.; De Las Heras, G.; et al. Colorectal adenomas contain multiple somatic mutations that do not coincide with synchronous adenocarcinoma specimens. PLoS ONE 2015, 10, e0119946. [Google Scholar]
- Lee, S.H.; Jung, S.H.; Kim, T.M.; Rhee, J.K.; Park, H.C.; Kim, M.S.; Kim, S.S.; An, C.H.; Lee, S.H.; Chung, Y.J. Whole-exome sequencing identified mutational profiles of high-grade colon adenomas. Oncotarget 2017, 8, 6579–6588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S.H.; Raju, G.S.; Huff, C.; Ye, Y.; Gu, J.; Chen, J.S.; Hildebrandt, M.A.T.; Liang, H.; Menter, D.G.; Morris, J.; et al. The somatic mutation landscape of premalignant colorectal adenoma. Gut 2018, 67, 1299–1305. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.H.; Yoo, J.; Song, Y.S.; Lim, C.H.; Kim, T.M. Mutation analysis of colorectal and gastric carcinomas originating from adenomas: Insights into genomic evolution associated with malignant progression. Cancers 2020, 12, 325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diaz, L.A., Jr.; Williams, R.T.; Wu, J.; Kinde, I.; Hecht, J.R.; Berlin, J.; Allen, B.; Bozic, I.; Reiter, J.G.; Nowak, M.A.; et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 2012, 486, 537–540. [Google Scholar] [CrossRef] [Green Version]
- Misale, S.; Yaeger, R.; Hobor, S.; Scala, E.; Janakiraman, M.; Liska, D.; Valtorta, E.; Schiavo, R.; Buscarino, M.; Siravegna, G.; et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 2012, 486, 532–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yaeger, R.; Cowell, E.; Chou, J.F.; Gewirtz, A.N.; Borsu, L.; Vakiani, E.; Solit, D.B.; Rosen, N.; Capanu, M.; Ladanyi, M.; et al. RAS mutations affect pattern of metastatic spread and increase propensity for brain metastasis in colorectal cancer. Cancer 2015, 121, 1195–1203. [Google Scholar] [CrossRef] [Green Version]
- Schweiger, T.; Liebmann-Reindl, S.; Glueck, O.; Starlinger, P.; Laengle, J.; Birner, P.; Klepetko, W.; Pils, D.; Streubel, B.; Hoetzenecker, K. Mutational profile of colorectal cancer lung metastases and paired primary tumors by targeted next generation sequencing: Implications on clinical outcome after surgery. J. Thorac. Dis. 2018, 10, 6147–6157. [Google Scholar] [CrossRef]
- COSMIC Database. Available online: https://cancer.sanger.ac.uk (accessed on 13 April 2022).
- Benešová, L.; Hálková, T.; Ptáčková, R.; Semyakina, A.; Menclová, K.; Pudil, J.; Ryska, M.; Levý, M.; Šimša, J.; Pazdírek, F.; et al. Significance of postoperative follow-up of patients with metastatic colorectal cancer using circulating tumor DNA. World J. Gastroenterol. 2019, 25, 6939–6948. [Google Scholar] [CrossRef]
- Benesova, L.; Belsanova, B.; Kramar, F.; Halkova, T.; Benes, V.; Minarik, M. Application of denaturing capillary electrophoresis for the detection of prognostic mutations in isocitrate dehydrogenase 1 and isocitrate dehydrogenase 2 genes in brain tumors. J. Sep. Sci. 2018, 41, 2819–2827. [Google Scholar] [CrossRef]
- Losi, L.; Baisse, B.; Bouzourene, H.; Benhattar, J. Evolution of intratumoral genetic heterogeneity during colorectal cancer progression. Carcinogenesis 2005, 26, 916–922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hershkovitz, D.; Simon, E.; Bick, T.; Prinz, E.; Noy, S.; Sabo, E.; Ben-Izhak, O.; Vieth, M. Adenoma and carcinoma components in colonic tumors show discordance for KRAS mutation. Hum. Pathol. 2014, 45, 1866–1871. [Google Scholar] [CrossRef]
- Cappellesso, R.; Lo Mele, M.; Munari, G.; Rosa-Rizzotto, E.; Guido, E.; De Lazzari, F.; Pilati, P.; Tonello, M.; Farinati, F.; Realdon, S. Molecular characterization of “sessile serrated” adenoma to carcinoma transition in six early colorectal cancers. Pathol. Res. Pract. 2019, 215, 957–962. [Google Scholar] [CrossRef] [PubMed]
- Harada, T.; Yamamoto, E.; Yamano, H.O.; Aoki, H.; Matsushita, H.O.; Yoshikawa, K.; Takagi, R.; Harada, E.; Tanaka, Y.; Yoshida, Y.; et al. Surface microstructures are associated with mutational intratumoral heterogeneity in colorectal tumors. J. Gastroenterol. 2018, 53, 1241–1252. [Google Scholar] [CrossRef] [PubMed]
- Xi, X.; Fu, Z.; Liu, T.; Lin, Y.; Wu, W.; Li, J.; Luo, M.; Zhang, B. Establishment and Verification of Scoring System for Colorectal Adenoma Recurrence. Risk Manag. Healthc. Policy 2021, 14, 4545–4552. [Google Scholar] [CrossRef] [PubMed]
- Kandoth, C.; McLellan, M.D.; Vandin, F.; Ye, K.; Niu, B.; Lu, C.; Xie, M.; Zhang, Q.; McMichael, J.F.; Wyczalkowski, M.A.; et al. Mutational landscape and significance across 12 major cancer types. Nature 2013, 502, 333–339. [Google Scholar] [CrossRef] [Green Version]
- Robles, A.I.; Jen, J.; Harris, C.C. Clinical Outcomes of TP53 Mutations in Cancers. Cold Spring Harb. Perspect. Med. 2016, 6, a026294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rivlin, N.; Brosh, R.; Oren, M.; Rotter, V. Mutations in the p53 Tumor Suppressor Gene: Important Milestones at the Various Steps of Tumorigenesis. Genes Cancer 2011, 2, 466–474. [Google Scholar] [CrossRef] [Green Version]
- Nakayama, M.; Oshima, M. Mutant p53 in colon cancer. J. Mol. Cell Biol. 2019, 11, 267–276. [Google Scholar] [CrossRef] [Green Version]
- Gafà, R.; Lanza, G. Espressione della proteina p53 nella sequenza adenoma-carcinoma del colon-retto [Expression of protein p53 in the adenoma-colorectal carcinoma sequence]. Pathologica 1998, 90, 351–356. (In Italian) [Google Scholar]
- Saleh, H.A.; Aburashed, A.; Bober, P.; Tabaczka, P. P53 protein immunohistochemical expression in colonic adenomas with and without associated carcinoma. Am. J. Gastroenterol. 1998, 93, 980–984. [Google Scholar] [CrossRef]
- Chang, P.Y.; Chen, J.S.; Chang, S.C.; Wang, M.C.; Chang, N.C.; Wen, Y.H.; Tsai, W.S.; Liu, W.H.; Liu, H.L.; Lu, J.J. Acquired somatic TP53 or PIK3CA mutations are potential predictors of when polyps evolve into colorectal cancer. Oncotarget 2017, 8, 72352–72362. [Google Scholar] [CrossRef] [Green Version]
- Minarikova, P.; Benesova, L.; Halkova, T.; Belsanova, B.; Suchanek, S.; Cyrany, J.; Tuckova, I.; Bures, J.; Zavoral, M.; Minarik, M. Longitudinal molecular characterization of endoscopic specimens from colorectal lesions. World J. Gastroenterol. 2016, 22, 4936–4945. [Google Scholar] [CrossRef]
- Iniesta, P.; Vega, F.J.; Caldés, T.; Massa, M.; de Juan, C.; Cerdán, F.J.; Sánchez, A.; López, J.A.; Torres, A.J.; Balibrea, J.L.; et al. p53 exon 7 mutations as a predictor of poor prognosis in patients with colorectal cancer. Cancer Lett. 1998, 130, 153–160. [Google Scholar] [CrossRef]
- Vidaurreta, M.; Maestro, M.L.; Sanz-Casla, M.T.; Rafael, S.; Veganzones, S.; de la Orden, V.; Cerdán, J.; Arroyo, M.; Torres, A. Colorectal carcinoma prognosis can be predicted by alterations in gene p53 exons 5 and 8. Int. J. Colorectal. Dis. 2008, 23, 581–586. [Google Scholar] [CrossRef]
- Juárez, M.; Egoavil, C.; Rodríguez-Soler, M.; Hernández-Illán, E.; Guarinos, C.; García-Martínez, A.; Alenda, C.; Giner-Calabuig, M.; Murcia, O.; Mangas, C.; et al. KRAS and BRAF somatic mutations in colonic polyps and the risk of metachronous neoplasia. PLoS ONE 2017, 12, e0184937. [Google Scholar] [CrossRef] [Green Version]
- Speroni, A.H.; Vanzulli, S.I.; Meiss, R.P. Adenomas of the colon: Overexpression of p53 protein and risk factors. Endoscopy 1998, 30, 623–626. [Google Scholar] [CrossRef] [PubMed]
- Remvikos, Y.; Laurent-Puig, P.; Salmon, R.J.; Frelat, G.; Dutrillaux, B.; Thomas, G. Simultaneous monitoring of p53 protein and DNA content of colorectal adenocarcinomas by flow cytometry. Int. J. Cancer 1990, 45, 450–456. [Google Scholar] [CrossRef]
- Rodrigues, N.R.; Rowan, A.; Smith, M.E.; Kerr, I.B.; Bodmer, W.F.; Gannon, J.V.; Lane, D.P. p53 mutations in colorectal cancer. Proc. Natl. Acad. Sci. USA 1990, 87, 7555–7559. [Google Scholar] [CrossRef] [Green Version]
- Brazda, V.; Muller, P.; Brozkova, K.; Vojtesek, B. Restoring wild-type conformation and DNA-binding activity of mutant p53 is insufficient for restoration of transcriptional activity. Biochem. Biophys. Res. Commun. 2006, 351, 499–506. [Google Scholar] [CrossRef]
- Cripps, K.J.; Purdie, C.A.; Carder, P.J.; White, S.; Komine, K.; Bird, C.C.; Wyllie, A.H. A study of stabilisation of p53 protein versus point mutation in colorectal carcinoma. Oncogene 1994, 9, 2739–2743. [Google Scholar]
- Dix, B.; Robbins, P.; Carrello, S.; House, A.; Iacopetta, B. Comparison of p53 gene mutation and protein overexpression in colorectal carcinomas. Br. J. Cancer 1994, 70, 585–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Degree of Heterogeneity | Number of Mutation Clones | Percentage Difference in Mutated Fractions |
|---|---|---|
| I | 0 or 1 | low |
| II | 1 | middle |
| III | 1 | high |
| 2 | low | |
| IV | 2 | middle/high |
| 3 | low | |
| V | 3 | middle/high |
| ˃3 | low/middle/high |
| Number of Mutation Clones | Number of Index Lesions (%) | Number of Synchronous Lesions (%) |
|---|---|---|
| 0 | 27 (26%) | 44 (55%) |
| 1 | 44 (42%) | 30 (37.5%) |
| 2 | 18 (17%) | 6 (7.5%) |
| 3 | 10 (10%) | 0 (0%) |
| 4 | 3 (3%) | 0 (0%) |
| 5 | 2 (2%) | 0 (0%) |
| Patient ID/Gender | Number of Index Lesions | Mutations Found in Index Lesion(s) | Degree of Heterogeneity | Number of Synchronous Lesions | Mutations Found in Synchronous Lesion(s) | Interval to Surveillance Colonoscopy (Months) | Result of Surveillance Colonoscopy | TP53 Mutations Found in Metachronous Adenoma(s) |
|---|---|---|---|---|---|---|---|---|
| 11/M | 1x hyperplastic polyp | BRAF | 1 | 2 | BRAF | 18 | 1x sessile serrated lesion, no dysplasia 1x tubular adenoma, LGD | none |
| 1x hyperplastic polyp | BRAF | 1 | ||||||
| 1x tubulovillous adenoma, predominantly LGD, in one section HGD | TP53-ex7 | 3 | ||||||
| 1x tubulovillous adenoma, LGD | APC | 2 | ||||||
| 39/M | 1x sessile serrated lesion, no dysplasia | TP53-ex7, TP53-ex8, TP53-ex5, BRAF, APC | 5 | 2 | No mutation | 12 | 1x tubular adenoma, LGD, 2x hyperplastic polyp | none |
| 52/M | 1x hyperplastic polyp, no dysplasia | TP53-ex7, BRAF | 4 | 5 | APC | 15 | 2x tubular adenoma, LGD | none |
| 58/M | 1x tubular adenoma, LGD | none | 1 | 2 | No mutation | 11 | 3x tubular adenoma, LGD, 1x tubular adenoma, predominantly LGD, sometimes HGD | none |
| 1x tubular adenoma, LGD | TP53-ex7 | 1 | ||||||
| 74/M | 1x sessile serrated lesion, no dysplasia | TP53-ex7, TP53-ex8, TP53-ex5, BRAF | 5 | 3 | BRAF, APC | 12 | 3x tubular adenoma, LGD | none |
| 86/M | 1x tubulovillous adenoma, LGD | TP53-ex7, TP53-ex8, TP53-ex5, KRAS | 5 | 3 | APC | 12 | 1x tubulovillous adenoma, LGD | none |
| 126/F | 1x tubulovillous adenoma, LGD | TP53-ex7, TP53-ex8, APC | 5 | 3 | KRAS, BRAF | 12 | 2x tubular adenoma, LGD, 1x hyperplastic polyp | none |
| 134/M | 1x tubulovillous adenoma, LGD | TP53-ex7 | not determined | 3 | No mutation | 15 | 1x tubular adenoma, LGD | none |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hálková, T.; Ptáčková, R.; Semyakina, A.; Suchánek, Š.; Traboulsi, E.; Ngo, O.; Hejcmanová, K.; Májek, O.; Bureš, J.; Zavoral, M.; et al. Somatic Mutations in Exon 7 of the TP53 Gene in Index Colorectal Lesions Are Associated with the Early Occurrence of Metachronous Adenoma. Cancers 2022, 14, 2823. https://doi.org/10.3390/cancers14122823
Hálková T, Ptáčková R, Semyakina A, Suchánek Š, Traboulsi E, Ngo O, Hejcmanová K, Májek O, Bureš J, Zavoral M, et al. Somatic Mutations in Exon 7 of the TP53 Gene in Index Colorectal Lesions Are Associated with the Early Occurrence of Metachronous Adenoma. Cancers. 2022; 14(12):2823. https://doi.org/10.3390/cancers14122823
Chicago/Turabian StyleHálková, Tereza, Renata Ptáčková, Anastasiya Semyakina, Štěpán Suchánek, Eva Traboulsi, Ondřej Ngo, Kateřina Hejcmanová, Ondřej Májek, Jan Bureš, Miroslav Zavoral, and et al. 2022. "Somatic Mutations in Exon 7 of the TP53 Gene in Index Colorectal Lesions Are Associated with the Early Occurrence of Metachronous Adenoma" Cancers 14, no. 12: 2823. https://doi.org/10.3390/cancers14122823
APA StyleHálková, T., Ptáčková, R., Semyakina, A., Suchánek, Š., Traboulsi, E., Ngo, O., Hejcmanová, K., Májek, O., Bureš, J., Zavoral, M., Minárik, M., & Benešová, L. (2022). Somatic Mutations in Exon 7 of the TP53 Gene in Index Colorectal Lesions Are Associated with the Early Occurrence of Metachronous Adenoma. Cancers, 14(12), 2823. https://doi.org/10.3390/cancers14122823






