RP-Rs-fMRIomics as a Novel Imaging Analysis Strategy to Empower Diagnosis of Brain Gliomas
Abstract
Simple Summary
Abstract
1. Introduction
2. Material and Methods
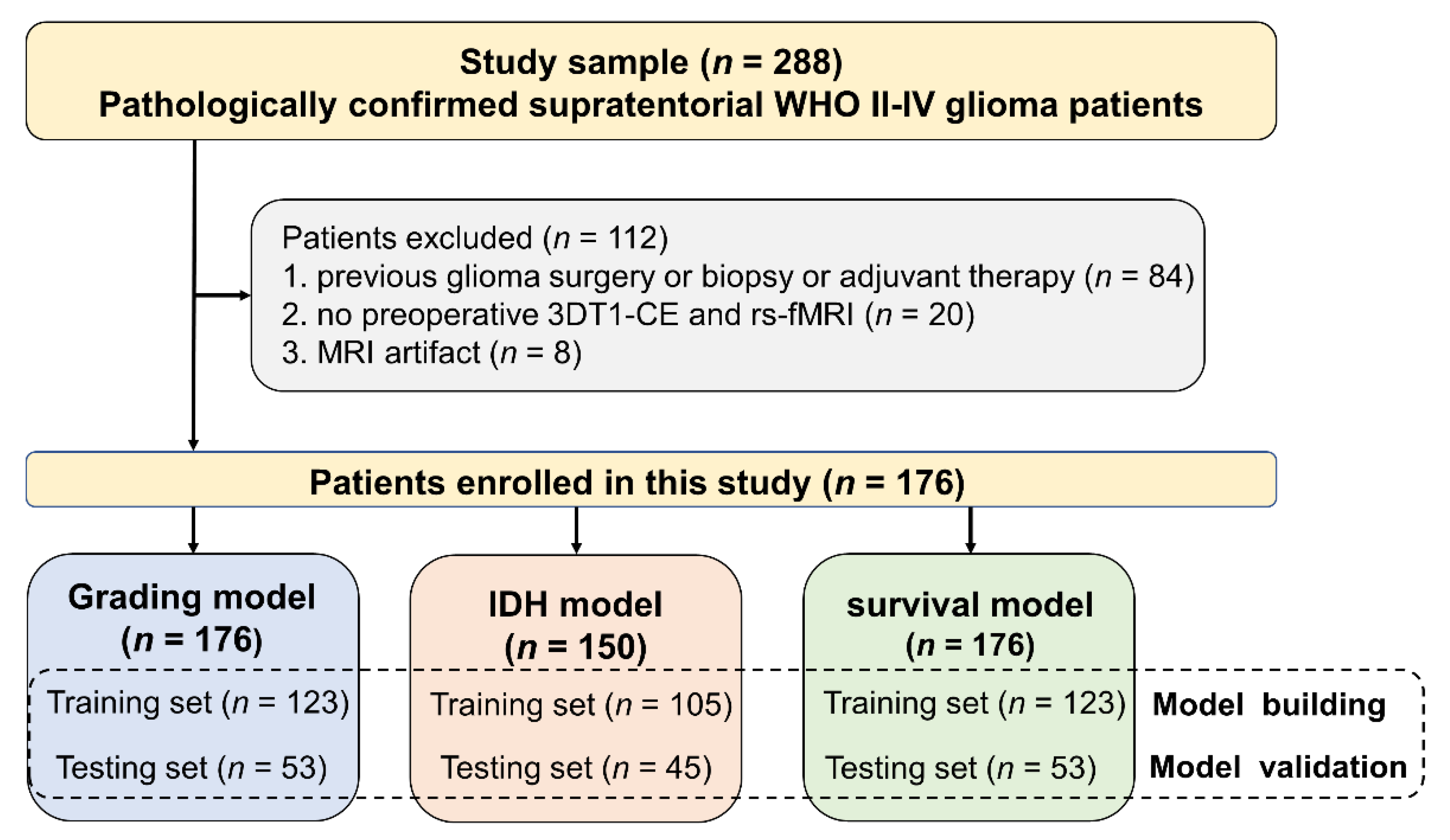
2.1. Patients Enrollment
2.2. Clinical Data
2.3. MRI Data Acquisition
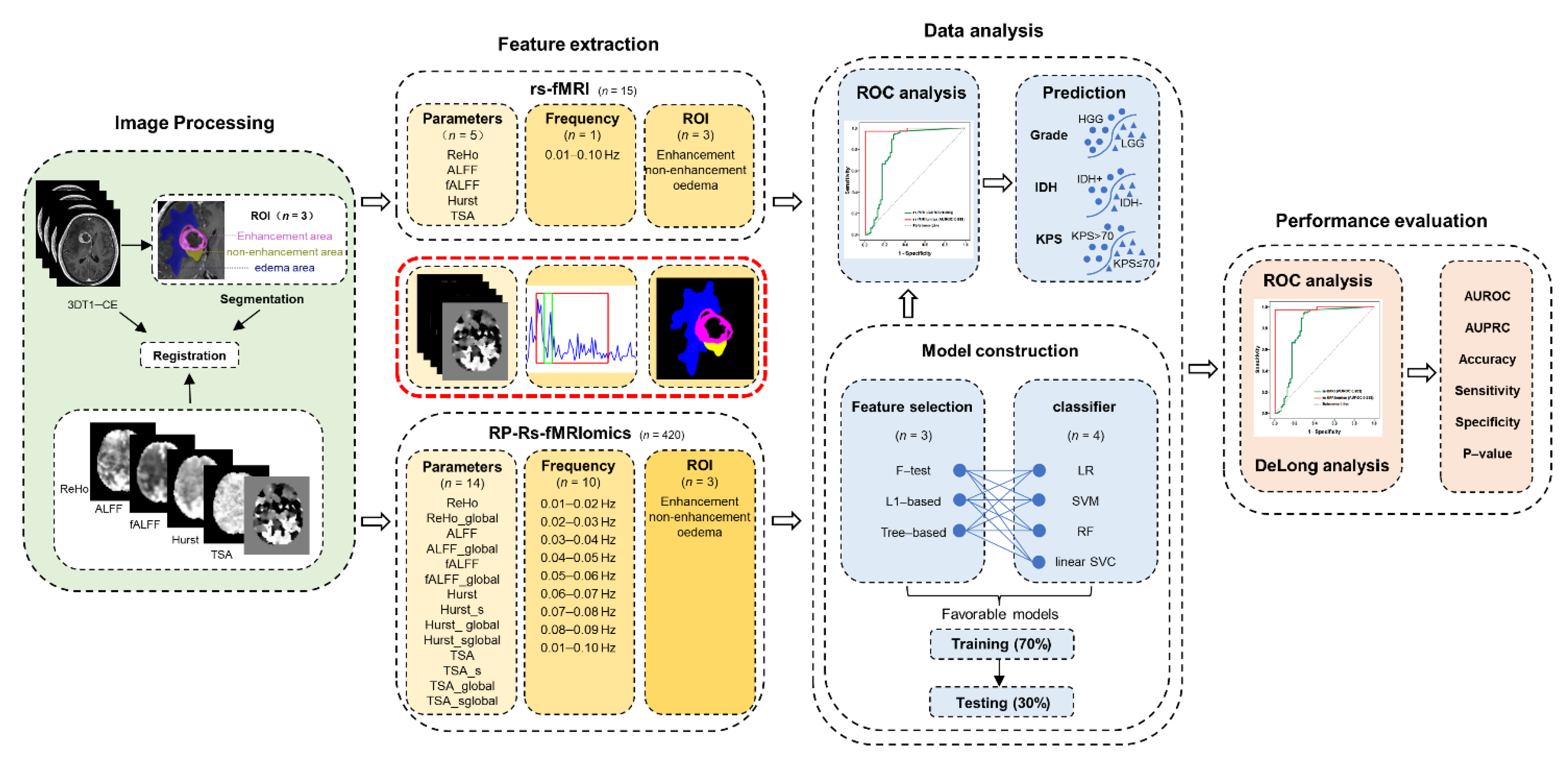
2.4. Imaging Processing
2.4.1. Data Preprocessing
2.4.2. Tumor Segmentation and Feature Extraction
2.5. Statistical Analysis
2.5.1. Conventional rs-fMRI Analysis
2.5.2. Feature Selection, RP-Rs-fMRIomics Model Construction and Validation
3. Results
3.1. Patient Characteristics
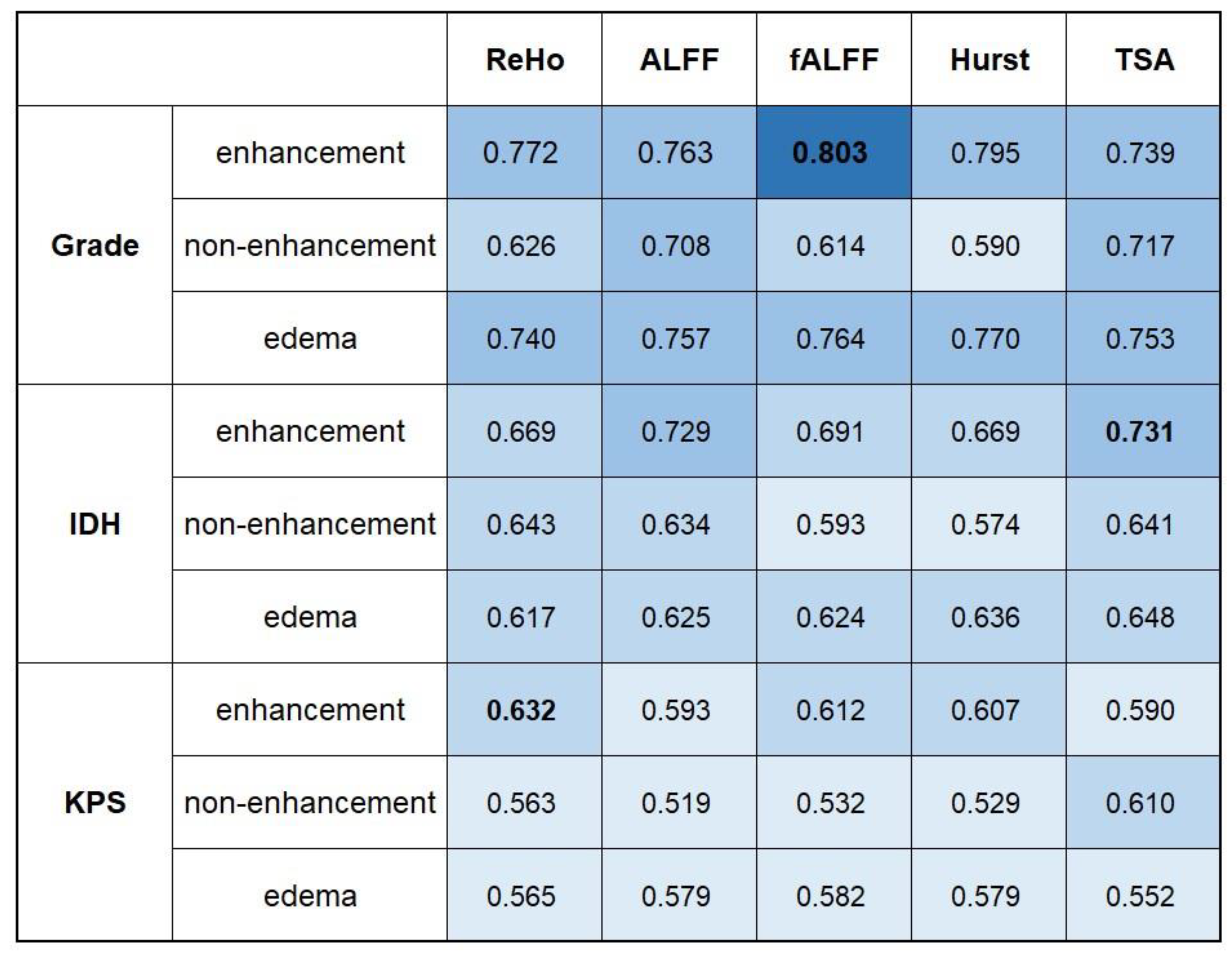
3.2. Performance of Conventional rs-fMRI Analysis
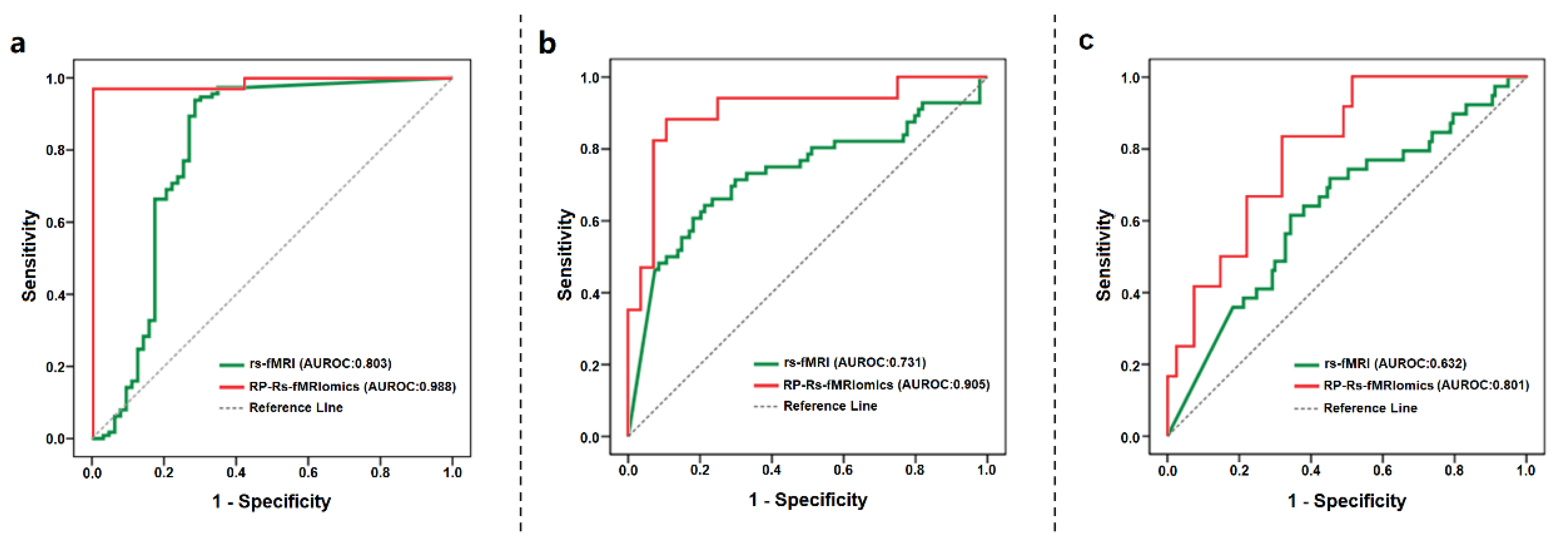
3.3. Performance of RP-Rs-fMRIomics Models
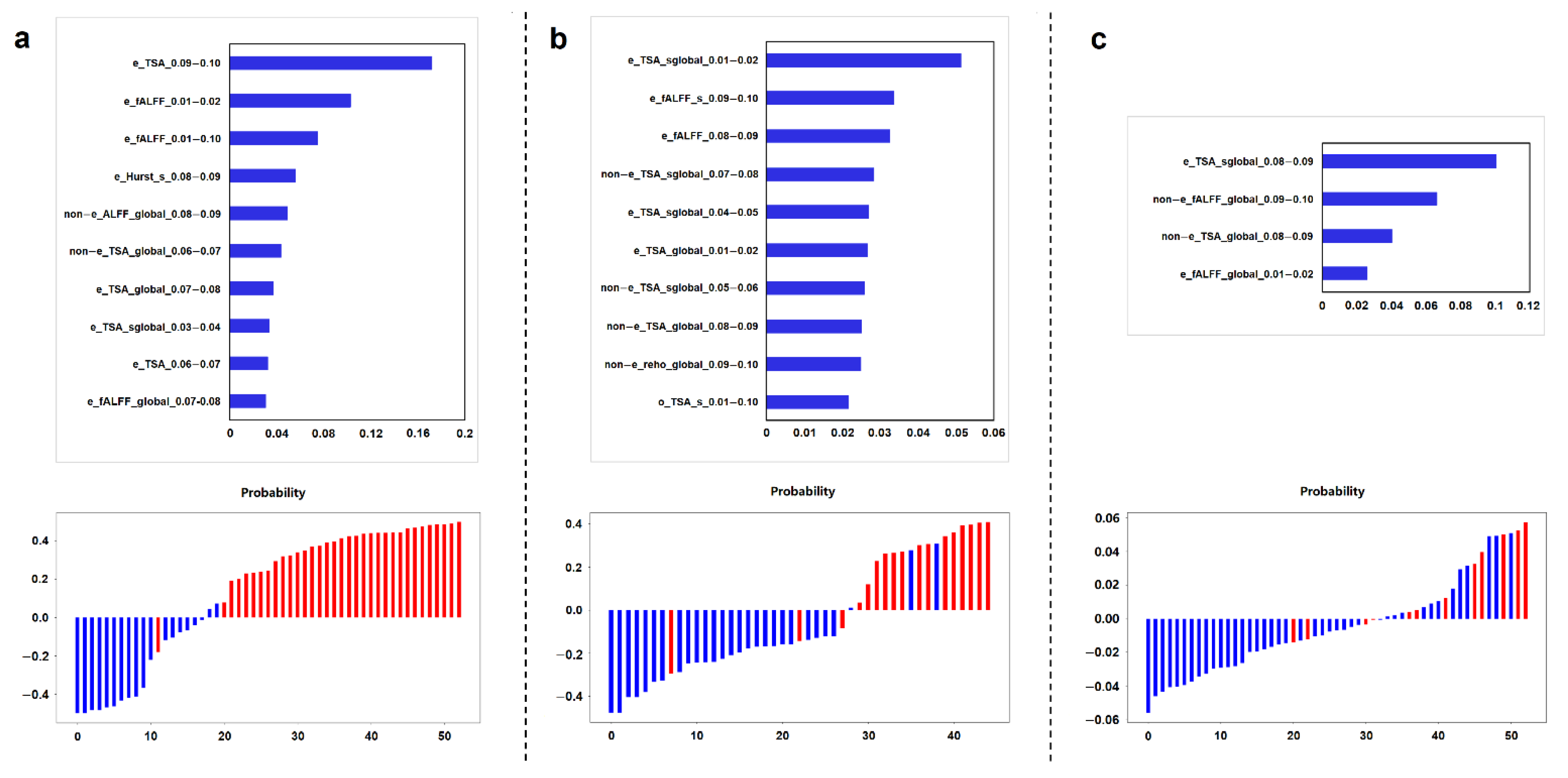
3.4. Key Imaging Features in RP-Rs-fMRIomics Models
3.5. Comparisons of Prediction Performance between Conventional rs-fMRI and RP-Rs-fMRIomics Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lee, M.H.; Smyser, C.D.; Shimony, J.S. Resting-State fMRI: A Review of Methods and Clinical Applications. Am. J. Neuroradiol. 2013, 34, 1866–1872. [Google Scholar] [CrossRef] [PubMed]
- Biswal, B.B. Resting state fMRI: A personal history. Neuroimage 2012, 62, 938–944. [Google Scholar] [CrossRef] [PubMed]
- Fox, M.D.; Raichle, M.E. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat. Rev. Neurosci. 2007, 8, 700–711. [Google Scholar] [CrossRef]
- Zhang, D.; Raichle, M.E. Disease and the brain’s dark energy. Nat. Rev. Neurol. 2010, 6, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Daniel, A.G.S.; Park, K.Y.; Roland, J.L.; Dierker, D.; Gross, J.; Humphries, J.B.; Hacker, C.D.; Snyder, A.Z.; Shimony, J.S.; Leuthardt, E.C. Functional connectivity within glioblastoma impacts overall survival. Neuro-Oncology 2021, 23, 412–421. [Google Scholar] [CrossRef]
- Yuan, B.K.; Zhang, N.; Yan, J.; Cheng, J.L.; Lu, J.F.; Wu, J.S. Tumor grade-related language and control network reorganization in patients with left cerebral glioma. Cortex 2020, 129, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Zou, Q.H.; Zhu, C.Z.; Yang, Y.H.; Zuo, X.N.; Long, X.Y.; Cao, Q.J.; Wang, Y.F.; Zang, Y.F. An improved approach to detection of amplitude of low-frequency fluctuation (ALFF) for resting-state fMRI: Fractional ALFF. J. Neurosci. Methods 2008, 172, 137–141. [Google Scholar] [CrossRef]
- Zang, Y.F.; Jiang, T.Z.; Lu, Y.L.; He, Y.; Tian, L.X. Regional homogeneity approach to fMRI data analysis. Neuroimage 2004, 22, 394–400. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.L.; Zuo, X.N. Regional Homogeneity: A Multimodal, Multiscale Neuroimaging Marker of the Human Connectome. Neuroscientist 2016, 22, 486–505. [Google Scholar] [CrossRef]
- Maxim, V.; Sendur, L.; Fadili, J.; Suckling, J.; Gould, R.; Howard, R.; Bullmore, E.T. Fractional Gaussian noise, functional MRI and Alzheimer’s disease. Neuroimage 2005, 25, 141–158. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zhou, J.S.; Zhang, H.; Chen, Y.C.; Mao, C.N.; Chen, X.L.; Ni, L.; Zhuo, Z.Z.; Zhang, Y.D.; Geng, W.; et al. One-step analysis of brain perfusion and function for acute stroke patients after reperfusion: A resting-state fMRI study. J. Magn. Reson. Imaging 2019, 50, 221–229. [Google Scholar] [CrossRef]
- Stoecklein, V.M.; Stoecklein, S.; Galie, F.; Ren, J.X.; Schmutzer, M.; Unterrainer, M.; Albert, N.L.; Kreth, F.W.; Thon, N.; Liebig, T.; et al. Resting-state fMRI detects alterations in whole brain connectivity related to tumor biology in glioma patients. Neuro-Oncology 2020, 22, 1388–1398. [Google Scholar] [CrossRef]
- Haberg, A.; Kvistad, K.A.; Unsgard, G.; Haraldseth, O. Preoperative blood oxygen level-dependent functional magnetic resonance imaging in patients with primary brain tumors: Clinical application and outcome. Neurosurgery 2004, 54, 902–914. [Google Scholar] [CrossRef]
- Metwali, H.; Raemaekers, M.; Ibrahim, T.; Samii, A. The Fluctuations of Blood Oxygen Level-Dependent Signals as a Method of Brain Tumor Characterization: A Preliminary Report. World Neurosurg. 2020, 142, E10–E17. [Google Scholar] [CrossRef]
- Wu, J.F.; Qian, Z.Y.; Tao, L.; Yin, J.H.; Ding, S.W.; Zhang, Y.M.; Yu, Z. Resting state fMRI feature-based cerebral glioma grading by support vector machine. Int. J. Comput. Assist. Radiol. Surg. 2015, 10, 1167–1174. [Google Scholar] [CrossRef] [PubMed]
- Gupta, L.; Gupta, R.K.; Postma, A.A.; Sahoo, P.; Gupta, P.K.; Patir, R.; Ahlawat, S.; Saha, I.; Backes, W.H. Advanced and Amplified BOLD Fluctuations in High-Grade Gliomas. J. Magn. Reson. Imaging 2018, 47, 1616–1625. [Google Scholar] [CrossRef]
- Holodny, A.I.; Schulder, M.; Liu, W.C.; Wolko, J.; Maldjian, J.A.; Kalnin, A.J. The effect of brain tumors on BOLD functional MR imaging activation in the adjacent motor cortex: Implications for image-guided neurosurgery. Am. J. Neuroradiol. 2000, 21, 1415–1422. [Google Scholar] [PubMed]
- Petridis, P.D.; Horenstein, C.; Pereira, B.; Wu, P.; Samanamud, J.; Marie, T.; Boyett, D.; Sudhakar, T.; Sheth, S.A.; McKhann, G.M.; et al. BOLD Asynchrony Elucidates Tumor Burden in IDH-Mutated Gliomas. Neuro-Oncology 2022, 24, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Chow, D.S.; Horenstein, C.I.; Canoll, P.; Lignelli, A.; Hillman, E.M.C.; Filippi, C.G.; Grinband, J. Glioblastoma Induces Vascular Dysregulation in Nonenhancing Peritumoral Regions in Humans. Am. J. Roentgenol. 2016, 206, 1073–1081. [Google Scholar] [CrossRef][Green Version]
- Maralani, P.J.; Das, S.; Mainprize, T.; Phan, N.; Bharatha, A.; Keith, J.; Munoz, D.G.; Sahgal, A.; Symons, S.; Ironside, S.; et al. Hypoxia Detection in Infiltrative Astrocytoma: Ferumoxytol-based Quantitative BOLD MRI with Intraoperative and Histologic Validation. Radiology 2018, 288, 821–829. [Google Scholar] [CrossRef]
- Agarwal, S.; Sair, H.I.; Yahyavi-Firouz-Abadi, N.; Airan, R.; Pillai, J.J. Neurovascular Uncoupling in Resting State fMRI Demonstrated in Patients With Primary Brain Gliomas. J. Magn. Reson. Imaging 2016, 43, 620–626. [Google Scholar] [CrossRef]
- Logothetis, N.K. What we can do and what we cannot do with fMRI. Nature 2008, 453, 869–878. [Google Scholar] [CrossRef] [PubMed]
- Olivier, M.; Asmis, R.; Hawkins, G.A.; Howard, T.D.; Cox, L.A. The Need for Multi-Omics Biomarker Signatures in Precision Medicine. Int. J. Mol. Sci. 2019, 20, 4781. [Google Scholar] [CrossRef] [PubMed]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Aerts, H.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Cavalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 8. [Google Scholar] [CrossRef] [PubMed]
- Tomaszewski, M.R.; Gillies, R.J. The Biological Meaning of Radiomic Features. Radiology 2021, 299, E256. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.Y.; Zhang, H.; Wu, J.S.; Yu, Z.D.; Chen, X.B.; Rekik, I.; Wang, Q.; Lu, J.F.; Shen, D.G. Overall survival time prediction for high-grade glioma patients based on large-scale brain functional networks. Brain Imaging Behav. 2019, 13, 1333–1351. [Google Scholar] [CrossRef]
- Sun, K.; Liu, Z.; Chen, G.; Zhou, Z.; Zhong, S.; Tang, Z.; Wang, S.; Zhou, G.; Zhou, X.; Shao, L.; et al. A two-center radiomic analysis for differentiating major depressive disorder using multi-modality MRI data under different parcellation methods. J. Affect. Disord. 2022, 300, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, K.; Liu, Z.; Chen, G.; Jia, Y.; Zhong, S.; Pan, J.; Huang, L.; Tian, J. Classification of Unmedicated Bipolar Disorder Using Whole-Brain Functional Activity and Connectivity: A Radiomics Analysis. Cereb. Cortex 2020, 30, 1117–1128. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, S.; Liu, X.; Chen, S.; Ke, Y.; Qi, S.; Wei, X.; Ming, D. Discriminating subclinical depression from major depression using multi-scale brain functional features: A radiomics analysis. J. Affect. Disord. 2022, 297, 542–552. [Google Scholar] [CrossRef]
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K.; Burger, P.C.; Jouvet, A.; Scheithauer, B.W.; Kleihues, P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007, 114, 97–109. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Park, J.E.; Kim, H.S.; Jo, Y.; Yoo, R.E.; Choi, S.H.; Nam, S.J.; Kim, J.H. Radiomics prognostication model in glioblastoma using diffusion- and perfusion-weighted MRI. Sci. Rep. 2020, 10, 9. [Google Scholar] [CrossRef]
- Pan, Z.Q.; Zhang, S.J.; Wang, X.L.; Jiao, Y.X.; Qiu, J.J. Machine Learning Based on a Multiparametric and Multiregional Radiomics Signature Predicts Radiotherapeutic Response in Patients with Glioblastoma. Behav. Neurol. 2020, 2020, 1712604. [Google Scholar] [CrossRef]
- Chao-Gan, Y.; Yu-Feng, Z. DPARSF: A MATLAB Toolbox for “Pipeline” Data Analysis of Resting-State fMRI. Front. Syst. Neurosci. 2010, 4, 13. [Google Scholar] [CrossRef]
- He, B.Y.J. Scale-Free Properties of the Functional Magnetic Resonance Imaging Signal during Rest and Task. J. Neurosci. 2011, 31, 13786–13795. [Google Scholar] [CrossRef] [PubMed]
- He, B.Y.J.; Zempel, J.M.; Snyder, A.Z.; Raichle, M.E. The Temporal Structures and Functional Significance of Scale-free Brain Activity. Neuron 2010, 66, 353–369. [Google Scholar] [CrossRef] [PubMed]
- Dehghani, N.; Hatsopoulos, N.G.; Haga, Z.D.; Parker, R.A.; Greger, B.; Halgren, E.; Cash, S.S.; Destexhe, A. Avalanche analysis from multielectrode ensemble recordings in cat, monkey, and human cerebral cortex during wakefulness and sleep. Front. Physiol. 2012, 3, 18. [Google Scholar] [CrossRef]
- Lv, Y.T.; Wei, W.; Song, Y.L.; Han, Y.; Zhou, C.S.; Zhou, D.; Zhang, F.D.; Xue, Q.M.; Liu, J.L.; Zhao, L.J.; et al. Non-invasive evaluation of cerebral perfusion in patients with transient ischemic attack: An fMRI study. J. Neurol. 2019, 266, 157–164. [Google Scholar] [CrossRef]
- Wang, C.; Shen, Z.J.; Huang, P.Y.; Yu, H.L.; Qian, W.; Guan, X.J.; Gu, Q.Q.; Yang, Y.H.; Zhang, M.M. Altered spontaneous brain activity in chronic smokers revealed by fractional ramplitude of lowfrequency fluctuation analysis: A preliminary study. Sci. Rep. 2017, 7, 7. [Google Scholar] [CrossRef]
- Lei, X.; Zhao, Z.Y.; Chen, H. Extraversion is encoded by scale-free dynamics of default mode network. Neuroimage 2013, 74, 52–57. [Google Scholar] [CrossRef]
- Lv, Y.T.; Margulies, D.S.; Craddock, R.C.; Long, X.Y.; Winter, B.; Gierhake, D.; Endres, M.; Villringer, K.; Fiebach, J.; Villringer, A. Identifying the Perfusion Deficit in Acute Stroke with Resting-State Functional Magnetic Resonance Imaging. Ann. Neurol. 2013, 73, 136–139. [Google Scholar] [CrossRef] [PubMed]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans. Med. Imaging 2015, 34, 1993–2024. [Google Scholar] [CrossRef]
- Hanley, J.A.; McNeil, B.J. A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology 1983, 148, 839–843. [Google Scholar] [CrossRef]
- Saito, T.; Rehmsmeier, M. The Precision-Recall Plot Is More Informative than the ROC Plot When Evaluating Binary Classifiers on Imbalanced Datasets. PLoS ONE 2015, 10, e0118432. [Google Scholar] [CrossRef]
- Delong, E.R.; Delong, D.M.; Clarkepearson, D.I. Comparing the areas under 2 or more correlated receiver operating characteristic curves—A nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef]
- Englander, Z.K.; Horenstein, C.I.; Bowden, S.G.; Chow, D.S.; Otten, M.L.; Lignelli, A.; Bruce, J.N.; Canoll, P.; Grinband, J. Extent of BOLD Vascular Dysregulation Is Greater in Diffuse Gliomas without Isocitrate Dehydrogenase 1 R132H Mutation. Radiology 2018, 287, 965–972. [Google Scholar] [CrossRef]
- Su, X.R.; Chen, N.; Sun, H.Q.; Liu, Y.H.; Yang, X.B.; Wang, W.N.; Zhang, S.M.; Tan, Q.Y.; Su, J.K.; Gong, Q.Y.; et al. Automated machine learning based on radiomics features predicts H3 K27M mutation in midline gliomas of the brain. Neuro-Oncolpgy 2020, 22, 393–401. [Google Scholar] [CrossRef]
- Zhuo, Z.Z.; Qu, L.Y.; Zhang, P.; Duan, Y.Y.; Cheng, D.; Xu, X.L.; Sun, T.; Ding, J.L.; Xie, C.; Liu, X.; et al. Prediction of H3K27M-mutant brainstem glioma by amide proton transfer-weighted imaging and its derived radiomics. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 4426–4436. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.F.; Hsu, F.T.; Hsieh, K.L.C.; Kao, Y.C.J.; Cheng, S.J.; Hsu, J.B.K.; Tsai, P.H.; Chen, R.J.; Huang, C.C.; Yen, Y.; et al. Machine Learning-Based Radiomics for Molecular Subtyping of Gliomas. Clin. Cancer Res. 2018, 24, 4429–4436. [Google Scholar] [CrossRef]
- Liu, X.X.; Li, J.R.; Liao, X.; Luo, Z.Q.; Xu, Q.; Pan, H.; Zhou, Q.; Tao, Y.; Shi, F.; Lu, G.M.; et al. Radiomics-based MRI for predicting Erythropoietin-producing hepatocellular receptor A2 expression and tumor grade in brain diffuse gliomas. Neuroradiology 2022, 64, 323–331. [Google Scholar] [CrossRef]
- Li, M.M.; Wang, H.F.; Shang, Z.G.; Yang, Z.L.; Zhang, Y.; Wan, H. Ependymoma and pilocytic astrocytoma: Differentiation using radiomics approach based on machine learning. J. Clin. Neurosci. 2020, 78, 175–180. [Google Scholar] [CrossRef]
- Tan, Y.; Zhang, S.T.; Wei, J.W.; Dong, D.; Wang, X.C.; Yang, G.Q.; Tian, J.; Zhang, H. A radiomics nomogram may improve the prediction of IDH genotype for astrocytoma before surgery. Eur. Radiol. 2019, 29, 3325–3337. [Google Scholar] [CrossRef]
- Wu, S.; Meng, J.; Yu, Q.; Li, P.; Fu, S. Radiomics-based machine learning methods for isocitrate dehydrogenase genotype prediction of diffuse gliomas. J. Cancer Res. Clin. Oncol. 2019, 145, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Arita, H.; Kinoshita, M.; Kawaguchi, A.; Takahashi, M.; Narita, Y.; Terakawa, Y.; Tsuyuguchi, N.; Okita, Y.; Nonaka, M.; Moriuchi, S.; et al. Lesion location implemented magnetic resonance imaging radiomics for predicting IDH and TERT promoter mutations in grade II/III gliomas. Sci. Rep. 2018, 8, 11773. [Google Scholar] [CrossRef]
- Li, Z.C.; Bai, H.M.; Sun, Q.C.; Zhao, Y.S.; Lv, Y.C.; Zhou, J.; Liang, C.F.; Chen, Y.S.; Liang, D.; Zheng, H.R. Multiregional radiomics profiling from multiparametric MRI: Identifying an imaging predictor of IDH1 mutation status in glioblastoma. Cancer Med. 2018, 7, 5999–6009. [Google Scholar] [CrossRef]
- Kim, M.; Jung, S.Y.; Park, J.E.; Jo, Y.; Park, S.Y.; Nam, S.J.; Kim, J.H.; Kim, H.S. Diffusion- and perfusion-weighted MRI radiomics model may predict isocitrate dehydrogenase (IDH) mutation and tumor aggressiveness in diffuse lower grade glioma. Eur. Radiol. 2020, 30, 2142–2151. [Google Scholar] [CrossRef]
- Hashido, T.; Saito, S.; Ishida, T. A radiomics-based comparative study on arterial spin labeling and dynamic susceptibility contrast perfusion-weighted imaging in gliomas. Sci. Rep. 2020, 10, 6121. [Google Scholar] [CrossRef]
- Cho, H.H.; Park, H. Classification of Low-grade and High-grade Glioma using Multi-modal Image Radiomics Features. In Proceedings of the 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Jeju, Korea, 11–15 July 2017; pp. 3081–3084. [Google Scholar]
- Villringer, A.; Dirnagl, U. Coupling of brain activity and cerebral blood-flow—Basis of functional neuroimaging. Cerebrovasc. Brain Metab. Rev. 1995, 7, 240–276. [Google Scholar]
- Ni, L.; Li, J.W.; Li, W.P.; Zhou, F.; Wang, F.F.; Schwarz, C.G.; Liu, R.Y.; Zhao, H.; Wu, W.B.; Zhang, X.; et al. The value of resting-state functional MRI in subacute ischemic stroke: Comparison with dynamic susceptibility contrast-enhanced perfusion MRI. Sci. Rep. 2017, 7, 41586. [Google Scholar] [CrossRef]
- Amemiya, S.; Kunimatsu, A.; Saito, N.; Ohtomo, K. Cerebral Hemodynamic Impairment: Assessment with Resting-State Functional MR Imaging. Radiology 2014, 270, 548–555. [Google Scholar] [CrossRef] [PubMed]
- Fanelli, G.N.; Grassini, D.; Ortenzi, V.; Pasqualetti, F.; Montemurro, N.; Perrini, P.; Naccarato, A.G.; Scatena, C. Decipher the Glioblastoma Microenvironment: The First Milestone for New Groundbreaking Therapeutic Strategies. Genes 2021, 12, 445. [Google Scholar] [CrossRef]
- Yekula, A.; Yekula, A.; Muralidharan, K.; Kang, K.K.; Carter, B.S.; Balaj, L. Extracellular Vesicles in Glioblastoma Tumor Microenvironment. Front. Immunol. 2020, 10, 3137. [Google Scholar] [CrossRef]
- Rooj, A.K.; Mineo, M.; Godlewski, J. MicroRNA and extracellular vesicles in glioblastoma: Small but powerful. Brain Tumor Pathol. 2016, 33, 77–88. [Google Scholar] [CrossRef]
- Li, Z.C.; Bai, H.M.; Sun, Q.C.; Li, Q.H.; Liu, L.; Zou, Y.; Chen, Y.S.; Liang, C.F.; Zheng, H.R. Multiregional radiomics features from multiparametric MRI for prediction of MGMT methylation status in glioblastoma multiforme: A multicentre study. Eur. Radiol. 2018, 28, 3640–3650. [Google Scholar] [CrossRef]
- Prasanna, P.; Patel, J.; Partovi, S.; Madabhushi, A.; Tiwari, P. Radiomic features from the peritumoral brain parenchyma on treatment-naive multi-parametric MR imaging predict long versus short-term survival in glioblastoma multiforme: Preliminary findings. Eur. Radiol. 2017, 27, 4198–4199. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Tha, K.K.; Terasaka, S.; Yamaguchi, S.; Wang, J.; Kudo, K.; Xing, L.; Shirato, H.; Li, R.J. Prognostic Imaging Biomarkers in Glioblastoma: Development and Independent Validation on the Basis of Multiregion and Quantitative Analysis of MR Images. Radiology 2016, 278, 546–553. [Google Scholar] [CrossRef]
- Kiviniemi, A.; Gardberg, M.; Ek, P.; Frantzen, J.; Bobacka, J.; Minn, H. Gadolinium retention in gliomas and adjacent normal brain tissue: Association with tumor contrast enhancement and linear/macrocyclic agents. Neuroradiology 2019, 61, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Xia, D.; Davis, R.L.; Crawford, J.A.; Abraham, J.L. Gadolinium released from MR contrast agents is deposited in brain tumors: In situ demonstration using scanning electron microscopy with energy dispersive X-ray spectroscopy. Acta Radiol. 2010, 51, 1126–1136. [Google Scholar] [CrossRef]
| Variables | Grading Model | IDH Model | Survival Model | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All Patients (n = 176) | Training Set (n = 123) | Testing Set (n = 53) | p Value | All Patients (n = 150) | Training Set (n = 105) | Testing Set (n = 45) | p Value | All Patients (n = 176) | Training Set (n = 123) | Testing Set (n = 53) | p Value | |
| Age (±SD), years | 51.11 ± 13.74 | 51.67 ± 12.24 | 49.83 ± 16.77 | 0.474 | 50.65 ± 13.37 | 52.31 ± 14.53 | 50.44 ± 12.43 | 0.503 | 51.11 ± 13.74 | 50.97 ± 13.54 | 51.45 ± 14.32 | 0.830 |
| Gender | 0.019 * | 0.390 | 0.008 * | |||||||||
| male | 96 | 60 | 36 | 82 | 55 | 27 | 96 | 59 | 37 | |||
| female | 80 | 63 | 17 | 68 | 50 | 18 | 80 | 64 | 16 | |||
| WHO grade | 0.992 | 0.682 | 0.308 | |||||||||
| II | 63 | 44 | 19 | 53 | 36 | 17 | 63 | 47 | 16 | |||
| III-IV | 113 | 79 | 34 | 97 | 69 | 28 | 113 | 76 | 37 | |||
| IDH status | 0.242 | 0.941 | 0.528 | |||||||||
| Mutant | 56 | 38 | 18 | 56 | 39 | 17 | 56 | 42 | 14 | |||
| Wild type | 94 | 72 | 22 | 94 | 66 | 28 | 94 | 66 | 28 | |||
| Extent of resection | 0.070 | 0.669 | 0.107 | |||||||||
| gross-total | 90 | 57 | 33 | 76 | 52 | 24 | 90 | 58 | 32 | |||
| partial | 86 | 66 | 20 | 74 | 53 | 21 | 86 | 65 | 21 | |||
| KPS | 0.198 | 0.001 * | 0.619 | |||||||||
| >70 | 39 | 24 | 15 | 32 | 15 | 17 | 39 | 26 | 13 | |||
| ≤70 | 137 | 99 | 38 | 118 | 90 | 28 | 137 | 97 | 40 | |||
| Optimal Model | Grading Model | IDH Model | Survival Model | |
|---|---|---|---|---|
| Classifier | Random Forest | Random Forest | Logistic Regression | |
| Feature Selection | F Test | F Test | F Test | |
| Training set | AUROC | 0.999 | 1.000 | 0.706 |
| ACC | 0.984 | 0.991 | 0.642 | |
| AUPRC | 0.987 | 1.000 | 0.667 | |
| SEN | 0.987 | 1.000 | 0.667 | |
| SPE | 0.977 | 0.985 | 0.635 | |
| F1 score | 0.987 | 0.987 | 0.450 | |
| Testing set | AUROC | 0.988 | 0.905 | 0.801 |
| ACC | 0.943 | 0.867 | 0.698 | |
| AUPRC | 0.971 | 0.824 | 0.667 | |
| SEN | 0.971 | 0.824 | 0.667 | |
| SPE | 0.895 | 0.893 | 0.707 | |
| F1 score | 0.957 | 0.824 | 0.500 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Li, J.; Xu, Q.; Zhang, Q.; Zhou, X.; Pan, H.; Wu, N.; Lu, G.; Zhang, Z. RP-Rs-fMRIomics as a Novel Imaging Analysis Strategy to Empower Diagnosis of Brain Gliomas. Cancers 2022, 14, 2818. https://doi.org/10.3390/cancers14122818
Liu X, Li J, Xu Q, Zhang Q, Zhou X, Pan H, Wu N, Lu G, Zhang Z. RP-Rs-fMRIomics as a Novel Imaging Analysis Strategy to Empower Diagnosis of Brain Gliomas. Cancers. 2022; 14(12):2818. https://doi.org/10.3390/cancers14122818
Chicago/Turabian StyleLiu, Xiaoxue, Jianrui Li, Qiang Xu, Qirui Zhang, Xian Zhou, Hao Pan, Nan Wu, Guangming Lu, and Zhiqiang Zhang. 2022. "RP-Rs-fMRIomics as a Novel Imaging Analysis Strategy to Empower Diagnosis of Brain Gliomas" Cancers 14, no. 12: 2818. https://doi.org/10.3390/cancers14122818
APA StyleLiu, X., Li, J., Xu, Q., Zhang, Q., Zhou, X., Pan, H., Wu, N., Lu, G., & Zhang, Z. (2022). RP-Rs-fMRIomics as a Novel Imaging Analysis Strategy to Empower Diagnosis of Brain Gliomas. Cancers, 14(12), 2818. https://doi.org/10.3390/cancers14122818







