Simple Summary
Homeobox (HOX) genes encode homeodomain proteins that regulate a wide range of molecular pathways. The homeodomain is highly conserved and binds to DNA. One exception is homeodomain-only protein (HOPX) that lacks DNA-binding capacity. HOPX plays a crucial role in development and its functional impairment is associated with a variety of diseases, including cancer. Loss of HOPX function occurs in a wide range of cancer types, where it functions as a tumor suppressor gene. Understanding the molecular mechanisms by which HOPX regulates carcinogenesis will likely lead to the development of new therapeutic approaches.
Abstract
Homeobox genes are master regulators of morphogenesis and differentiation by acting at the top of genetic hierarchies and their deregulation is associated with a variety of human diseases. They usually contain a highly conserved sequence that codes for the homeodomain of the protein, a specialized motif with three α helices and an N-terminal arm that aids in DNA binding. However, one homeodomain protein, HOPX, is unique among its family members in that it lacks the capacity to bind DNA and instead functions by interacting with transcriptional regulators. HOPX plays crucial roles in organogenesis and is expressed in both embryonic and adult stem cells. Loss of HOPX expression is common in cancer, where it functions primarily as a tumor suppressor gene. In this review, we describe the function of HOPX in development and discuss its role in carcinogenesis.
1. Introduction
Homeobox genes are master regulators of morphogenesis and cell differentiation. All such genes contain a “homeobox”, which is a DNA sequence of roughly 180 base pairs that encodes the homeodomain, which comprises three helical regions folded into a tight globular structure that binds a 5’-TAAT-3’ core motif. The homeodomain is highly conserved and is identical from flies to humans [1,2] and the 60-amino-acid motif is responsible for binding to target genes to activate transcription [1,3,4]. As transcription factors, homeodomain proteins are involved in embryonic patterning and cell differentiation, and many have been connected to human diseases [5]. Based on the percentage of sequence similarity, homeobox genes are classified into several families, such as HOX, NKX, PAX, GBX2, and MSX [6]. Different functional properties amongst families are determined by DNA-binding specificities, co-factors, and other mechanisms, such as additional domains or motifs found within homeoprotein families [6].
Homeobox genes are necessary for diverse biological functions as they control the expression of a wide range of genes that play crucial roles in proliferation, differentiation, survival, cell cycle, migration, and apoptosis. Consequently, there are many target genes with a variety of known functions, and these include p53, p21, VEGF, osteopontin, IGFBP1, NCAM, keratins, and integrins [1]. The different homeodomain families possess varied biological functions. For example, MSX genes regulate epithelial to mesenchymal transition during organogenesis [7], whereas members of NKX, PAX, and DLX are involved in differentiation in a tissue-specific manner [6]. Homeodomain proteins are not specific to adult differentiated cells but are also seen in adult stem cells, such as CD34+ hematopoietic stem cells [8]. In particular, the crosstalk between embryonic- and differentiation-specific homeodomain proteins provides a link between organogenesis and cancer.
Not surprisingly, the expression of homeobox genes is often altered in human diseases, with certain homeobox genes being upregulated while others are downregulated. For example, the homeobox gene Oct-4, which is associated with embryonic stem cells [9], has been found to be upregulated in prostate cancer whilst the homeobox gene NKX3.1, on the other hand, has been shown to be downregulated [10]. Collectively, homeobox genes play critical roles in inducing oncogenic transformation in vitro by modifying a range of biological processes [8].
Whilst the majority of homeodomain proteins bind DNA and function as transcription factors, one related protein, HOPX, is unusual in that it lacks DNA-binding capability and functions only as a transcriptional regulator. Here, we will review our current understanding regarding the function of HOPX in development and its role in carcinogenesis.
2. HOPX Structure
HOPX was initially identified by two independent research groups through the screening of EST databases for novel homeobox genes expressed in the developing mouse heart [11,12]. HOPX has also been termed Toto, Cameo, Ob1, SMAP31, NECC1, and LAGY in the published literature [13,14,15]. Subsequently, the HOPX gene symbol was given to homeodomain-only protein (HOP) to differentiate it from other genes such as hopscotch in Drosophila and hop-sterile in mouse. HOPX belongs to the PRD class of homeodomain proteins [16,17].
The human HOPX gene, located on chromosome 4q12, encodes a nuclear protein [11,12] and primary sequence analysis and secondary structural prediction indicates that HOPX is closely related to members of the homeodomain family and forms three ∝ helices, with a helix-turn-helix motif characteristic of the homeodomain [12] (Figure 1). HOPX contains the 60-amino-acid homeodomain but has a number of unusual characteristics that distinguish it from other known homeodomain proteins [11,12] (Figure 1). HOPX forms a classical homeodomain fold and a single amino acid insertion between helix I and II, but it lacks DNA-binding capacity due to the absence of several key residues known to mediate critical contacts with DNA that are conserved among other homeodomain proteins [11,12,18] (Figure 1). Exons 1, 5, 6, and 7 of the HOPX genes are alternatively spliced to generate five mRNA transcripts. These transcripts encode three protein isoforms: isoform a (previously γ) consists of 91 amino acids, isoform b (previously β) contains 73 amino acids, and isoform c (previously α) is longer with 112 amino acids, giving a different C-terminal sequence [19]. Only the HOPX b (β) promoter harbors CpG islands, which is relevant to the HOPX silencing observed in cancers discussed later. The precise function of the individual splice variants and protein isoforms remains to be determined.
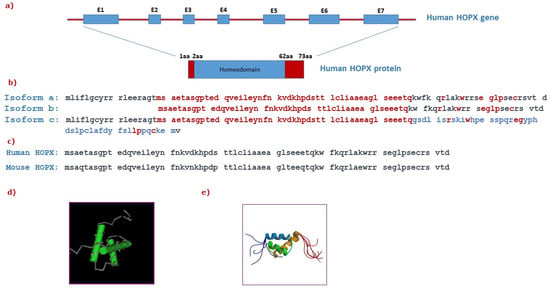
Figure 1.
Structure of the HOPX gene and protein: (a) Graphical representation of the human HOPX gene containing seven exons (E1-E7) and intronic sequences (depicted as a red thick line) encoding for a 73aa protein. (b) Comparison of the amino acid sequences between human HOPX protein isoforms a, b, and c. The common sequence in all three isoforms is highlighted in red. The amino acids that are only found in isoform c are shown in blue, indicating a distinct C-terminal sequence. (c) Comparison of the amino acid sequences between human and mouse HOPX proteins. (d) Graphical representation of the homeodomain that is conserved among homeodomain proteins and its respective sequence (adapted from NCBI). (e) The solution structure of HOPX (adapted from RCSB PDB).
3. HOPX in Development and Differentiation
Homeodomain-containing proteins act as transcription factors/regulators to regulate cell differentiation and developmental processes, and their target genes are diverse and complex [6]. In this regard, HOPX expression has been detected in both embryonic and adult stem cells in various tissues to regulate crucial biological processes.
3.1. Embryonic Stem Cells
HOPX is expressed during embryonic development of skeletal muscle, stratified epithelium (upper digestive tract and skin), epithelium of developing airways, midbrain/hindbrain junction, meninges, cartilage, chondrocytes, and lens fiber cells [13,20]. HOPX also has a role in placental development, controlling transcription factors in concert with other homeobox genes. HOPX expression has been detected in the trophoblast stem cells of E8.5–E9.5 wild-type mouse placentas [21]. The syncytiotrophoblast of humans, an epithelial layer of the highly vascular embryonic placental villi, which invades the uterine wall to create nutritional circulation between the embryo and the mother, expresses HOPX. In response to differentiation stimuli, HOPX induces the differentiation of trophoblast stem cells into a trophoblastic lineage [21]. HOPX regulates the development of endothelial cells, thereby contributing to primitive hematopoiesis by at least partially inhibiting Wnt signaling [22].
3.2. Adult Stem Cells
HOPX expression has been detected in various adult stem cells such as bone marrow, intestine, hair follicles, and lung [23,24,25]. HOPX is crucial for the proper maintenance of intestinal epithelium. There are two stem cell niches within the intestinal epithelium: one at the +4 position that contains slow-cycling and label-retaining cells and the other at the crypt base with crypt base columnar (CBC) cells. The cells at the +4 position have a unique transcriptional profile, which includes the expression of HOPX [26,27]. These +4 cells have previously been described as part of the resting stem cell population, which rebuilds Lgr5+ CBCs upon injury and has been found to be radiation resistant, implying that they can survive crypt injury and replace their damaged counterparts [28]. Furthermore, HOPX-positive intestinal stem cells in the +4 locations are resistant to anoikis, a type of apoptosis caused by aberrant cell matrix connections [29]. HOPX-positive stem cell populations have also been shown to express CD24 and CD44 [30].
Recently, the epigenetic factor, enhancer of zeste homolog 2 (EZH2), was found to interact directly with the HOPX promotor regions during bone marrow stromal cell (BMSC) differentiation to promote osteogenesis [31]. The telogen basal bulge’s resident stem cells of hair follicles have also been found to express HOPX. These HOPX+ hair follicle cells perform a function in epidermal wound repair in addition to differentiating into all lineages. In the lower hair bulb of the anagen phase, an HOPX+/Lgr5+/Shh- progenitor population was identified; when these cells differentiate into k6-positive inner bulge cells in telogen, they govern nearby hair follicle stem cells [24].
The type I cells of the mouse lung epithelium, which maintain gas exchange in alveoli, are HOPX-positive. Despite undergoing proliferation, these HOPX-positive type I cells give rise to type II cells upon partial pneumonectomy-stimulated alveolar regeneration in mice [32]. Furthermore, during the differentiation of mouse splenic marginal zone precursors (MZPs) into marginal zone B (MZB) cells caused by the Notch 2 signaling cascade, HOPX was found to be downregulated [33]. HOPX is a stem cell marker that is also associated with stem cell features. Zhou et al. identified a significant association between HOPX and hematopoietic stem/progenitor cell (HSPC) frequency by using genome-wide association studies (GWASs) to uncover genetic determinants [34]. In mice, hematopoietic cell-specific knockout of HOPX causes stemness and quiescence to be disrupted [35].
In adult pancreatic tissues, pan-islet cells are positive for HOPX, and they may play a possible role in maintaining endocrine cell identity [36]. Several lines of evidence have suggested that HOPX functions as a negative cell cycle regulator. High levels of HOPX have been found in the inner trabecular layer of the heart during mouse development, where cell proliferation is lower than in the outer compact zone, and HOPX appears to be essential for appropriate cell cycle withdrawal in the perinatal period [11,12]. A number of genes involved in cell proliferation were found to be elevated in the heart tissues of HOPX-null mice [12]. HOPX has also been demonstrated to control the balance between proliferation and differentiation in the growing heart and cells of the central nervous system [12,37]. HOPX regulates apoptosis of radial astrocytes in the dentate gyrus of the mouse hippocampus, which are required for the formation of hippocampal granular neurons throughout life [38].
4. HOPX Regulation of Molecular Signaling Networks
HOPX functions as a mediator of multiple signaling pathways in both normal and pathological conditions. HOPX has been identified as a direct downstream transcriptional target of Nkx2.5, a cardiac-specific transcription factor, in a series of in vitro and in vivo experiments in mice [11,12,39]. During cardiac development, HOPX directly binds with the serum response factor (SRF) to enhance its detachment from DNA, thereby inhibiting SRF-mediated transcription. HOPX also interferes with SRF’s cooperation with its coactivator [11,12] and can inhibit SRF-dependent transcriptional activation by recruiting histone deacetylases (HDACs) such as HDAC2 [40]. Two distinct regions on the surface of the HOPX protein are thought to be needed for its capacity to interact with other proteins, such as SRF and HDAC, to control transcription rather than binding to DNA [18]. Moreover, because it lacks intrinsic DNA-binding capability, HOPX interacts with transcription activators or repressors [11,12,40]. The association between HOPX and HDAC2 was further strengthened by the co-expression of these two proteins in developing pulmonary airway epithelium, where HOPX functions as a HDAC-dependent negative regulator of surfactant protein development. During pulmonary maturation, HOPX and HDAC act as downstream regulators of Nkx2-1 and GATA6 [41].
HOPX interacts with SRF and prevents the survival of dentate gyrus progenitor cells by inhibiting the activation of SRF target genes such as Bcl2 [38]. However, HOPX expression has been demonstrated to be spatially and temporally controlled in the developing central nervous system, even at sites where SRF was not present, suggesting that HOPX’s role in neurogenesis is distinct from that of cardiogenesis [37]. HOPX promotes cardiomyogenesis by fostering the differentiation of cardiomyoblasts into cardiomyocytes, interacting with activated Smads of the BMP signaling cascade and inhibiting Wnt signaling [42]. In addition to SRF and HDAC, HOPX interacts with enhancer of polycomb 1 (Epc 1), which is highly expressed in the embryonic heart and skeletal muscles. A physical interaction between HOPX and Epc 1 was found in yeast and mammalian cells. For example, myogenin is activated and myotube formation is stimulated in H9c2 cells following co-transfection of HOPX and Epc 1, and Epc 1-mediated skeletal muscle differentiation was disrupted in HOPX-knockout mice [43].
Lee et al. showed a significant downregulation of HOPX during the transdifferentiation of myogenic satellite cells (MSCs) into adipocyte-like cells (ALCs) [44]. HOPX has also been demonstrated to be required for keratinocyte differentiation, and it is controlled by a number of signaling pathways following cell cycle exit, including PKC, phospholipase C, AKT, and NFĸB. Interestingly, the activation of HOPX during keratinocyte differentiation is dependent on the PKC pathway rather than DNA methylation [45].
The role of HOPX in T cell lineages is distinct from that of other cell types. Higher levels of HOPX expression were seen only in terminally differentiated T-helper type I (Th 1) cells and not in naïve T-helper (Th) cells. The AP-1 complex, which includes c-Fos and c-Jun, is well known for maintaining cell proliferation. By inactivating c-Fos, which is controlled by SRF, HOPX modulates T cell proliferation and antigenic responses and enables T cell antigen unresponsiveness [46]. HOPX, in particular, suppresses Fas-induced apoptosis, and only HOPX-positive murine Th 1 cells persist in vivo [47]. Jones et al. found a negative association between HOPX and IL-2 in peripheral regulatory T cells (pTregs), which protects pTregs against autoimmune responses to self-antigens [48].
It has been demonstrated that altering normal HOPX function causes significant developmental heart abnormalities in mouse and zebrafish models [11,12,40]. HOPX expression is reduced by the SRF- and GATA4-dependent pro-hypertrophic response, and it is completely absent in severe heart failure [49]. During HOPX-induced hypertrophy in rats, inhibition of histone deacetylase (HDACi) is critical for atrial fibrosis and arrhythmic inducibility [50]. Two variants in the HOPX gene have been shown to cause syncope, which is defined as a loss of consciousness and muscle strength in patients with hypertrophic cardiomyopathy (HCM) [51].
The molecular mechanisms of HOPX during development are summarized in Figure 2.
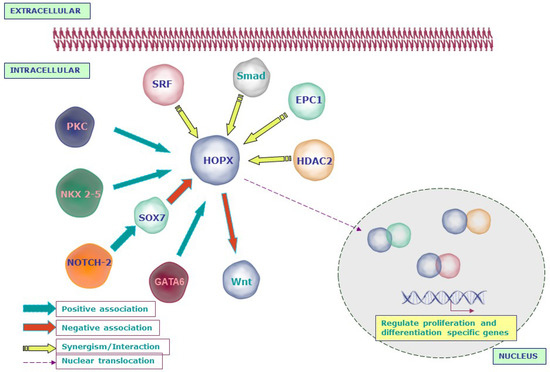
Figure 2.
Schematic representation of the molecular networks regulated by HOPX in development: HOPX is activated by the signaling molecules that are essential for normal development such as NKX2-5, PKC, and GATA6. However, NOTCH2 inhibits HOPX through SOX7. HOPX interacts with serum response factor (SRF) and inhibits SRF-mediated transcription. On the other hand, HOPX recruits histone deacetylase 2 (HDAC2) to modulate the function of SRF. Apart from SRF and HDAC, HOPX interacts with enhancer of polycomb 1 (Epc-1) to activate myogenin and induces myotube formation. During cardiomyogenesis, HOPX and Bmp signals synergistically inhibit Wnt signaling through activated Smads.
5. HOPX in Carcinogenesis
5.1. HOPX as a Tumor Suppressor Gene
There is now a wide body of evidence, which we outline below, that indicates that HOPX functions to suppress the carcinogenic process. The expression, methylation status, and function of HOPX in different cancer types is summarized in Table 1.

Table 1.
Deregulation and functional significance of HOPX in cancer.
5.1.1. HOPX Expression in Normal and Malignant Tissues
HOPX was initially isolated using a PCR-based subtracted fragmentary cDNA library between normal placental villi and a choriocarcinoma cell line [14]. HOPX expression was later detected in normal tissues of the lung, smooth muscle, brain, placenta, bladder, spleen, and kidney [14]. Subsequently, HOPX was shown to be downregulated in a wide range of human tumor types, including lung cancer [15], choriocarcinoma [14], head and neck cancers [52,54,63,64,65,66], esophageal squamous cell carcinoma [53], glioblastoma [38], human uterine endometrial cancers (HECs) [59], gastric cancer [55], colorectal cancer [60,61], and pancreatic cancer [62], suggesting a tumor suppressor role for HOPX.
More direct evidence for a tumor suppressor role for HOPX has come from in vivo studies, where ectopic expression of HOPX in choriocarcinoma [14], esophageal [53], glioblastoma [38], HEC [59], gastric [55], colorectal [61], and lung cancer [56] cells resulted in decreased tumor growth suppression in vivo. Interestingly, loss of HOPX expression has clinical relevance, at least in lung cancer, because decreased HOPX expression in lung SCC has been linked to high-grade and advanced-stage cancers [15] and miR-421 has been shown to target HOPX, which then stimulates the Wnt/catenin signaling pathway, promoting the development of non-small cell lung cancer [67].
Despite its function as a tumor suppressor, HOPX gene mutations appear to be rare in cancer [59]. The precise role of the individual splice variants is still unknown; however, all variants were detected in HNSCCs [66], and in skin cancer [68] and acute myeloid leukemia [69], the level of transcripts 1, 3, and 5 were reported to be low while variants 2 and 4 were predominantly expressed.
5.1.2. HOPX Promoter Methylation
Methylation of the cytosine residues of the CpG island within the promoter region of tumor suppressor genes is one of the mechanisms that suppresses their function in cancer [70]. HOPX promoter methylation has been detected in human uterine endometrial cancer [59], head and neck cancer [66], gastric cancer [55], colorectal cancer [60,61], pancreatic cancer [62], and lung cancer [56]. Methylation of the HOPX promoter can occur as an early event in carcinogenesis because it has been detected in the early stages of gastric cancer [55]. Interestingly, HOPX has been ranked second among the top priority genes that undergo methylation in primary esophageal squamous cell carcinoma [53]. Treatment of head and neck, esophageal, and pancreatic cancer cells with demethylating drugs such as 5-aza-2-deoxycytidine restored HOPX expression [53,62,66]. HOPX does not harbor mutations in human uterine endometrial cancer; however, its hypermethylation demonstrates epigenetic modification as a crucial event for inactivation [59]. Loss of HOPX expression as a result of promoter hypermethylation has clinical significance because in patients with gastric and colorectal cancer, the methylation status has been linked to a worse prognosis [55,61] and patients with strong HOPX expression in lung cancer have a considerably longer survival [56]. Epigenetic suppression of HOPX by promoter methylation plays a critical role in aggressive phenotypes in papillary thyroid cancer and is correlated with a poor prognosis of patients [71].
5.1.3. HOPX Affects Tumor Cell Behavior
Numerous in vitro and in vivo studies have shown that HOPX affects a variety of cellular processes and loss of HOPX promotes the malignant phenotype of tumor cells by a variety of mechanisms, as summarized below. The possible signaling networks regulated by HOPX in cancer are depicted in Figure 3.
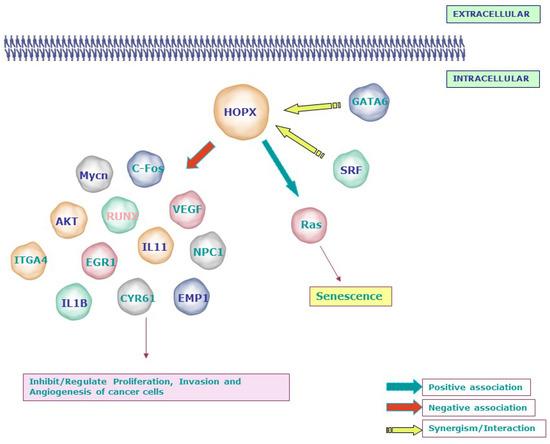
Figure 3.
Signaling molecules related to the tumor suppressor function of HOPX: Loss or gain experiments and microarray analyses revealed the negative association between HOPX and molecules involved in invasion, angiogenesis, and transformation, including Mycn, c-Fos, AKT, VEGF, EGR1, IL11, IL1B, and EMP1. In line with its mechanism during development, HOPX affects the proliferation of cancer cells by synergizing with SRF and GATA6. Notably, HOPX induces senescence through the RAS-ERK-MAPK signaling cascade in lung cancer. SLC2A3—Solute Carrier Family 2 (Facilitated Glucose Transporter), Member 3; NPC1—Niemann–Pick disease, type C1; RUNX1—Runt-related transcription factor 1; CNN1/CYR61—Cysteine-rich, angiogenic inducer, 61; WTAP—Wilms tumor 1-associated protein; PRDX2—Peroxiredoxin 2; EREG—Epiregulin; ITGA4—Integrin, alpha 4; EMP1—Epithelial membrane protein 1; EPHA2—EPH receptor A2; EGR1—Early growth response; MYCN—v-myc avian myelocytomatosis viral oncogene neuroblastoma-derived homolog; IGFII—Insulin-like growth factor 2 (somatomedin A).
HOPX was found to be involved in the regulation of the cell cycle and apoptosis. By positively regulating the subG1 and GO/G1 phases, HOPX reduces DNA synthesis and promotes cell death in pancreatic cancer [62]. Glioblastomas (GBMs) are derived from neural cancer stem cells and HOPX regulates the hippocampal stem cell niche through controlling cell survival and apoptosis, but it is downregulated in GBMs in mice; a key finding of this work is that reactivating HOPX causes apoptosis and lowers the tumorigenic ability of GBM cancer stem cells [38]. HOPX is epigenetically silenced in breast cancer. HOPX overexpression causes apoptosis and cell cycle arrest in breast cancer cells, suggesting that it could be utilized as a therapeutic target for breast cancer patients [72].
HOPX is activated by 17b-estradiol (E2) and inhibits cell proliferation in vitro and in vivo, suppressing uterine endometrial malignancies (HEC) [59]. This occurs by influencing the activity of SRF and the expression of its targets, c-Fos and Cyclin D1. HOPX activity was restored when HEC cell lines were treated with the histone deacetylase inhibitor TSA and inhibiting HOPX also stimulated immortalized human endometrial cells to proliferate [59]. In esophageal squamous cell carcinoma, however, HOPX had no effect on SRF activity, showing that the tumor suppressor functions of HOPX are cell type dependent [53]. DNA microarray analysis in colorectal cancer cells showed that the re-expression of HOPX upregulates WTAP and PRDX2 while downregulating genes associated with proliferation, angiogenesis, and invasion both in vitro and in vivo [61].
HOPX expression has been reported to be reduced at both the DNA and protein levels of oral and nasopharyngeal cancer cells, and its re-expression promoted cell proliferation and invasion and made the cells more susceptible to UV and cisplatin-induced cell death [66].
Interestingly, a recent study showed that the HOPX knockdown in squamous cell carcinoma (SCC) cells reduces their proliferative and invasive capabilities through the acceleration of apoptosis. The methylation status of two distinct HOPX promoters, promoter 1 and 2, causes the HOPX transcript expression to differ in normal keratinocytes and SCC cells. The activation of the second promoter of HOPX may contribute to the pathogenesis of SCC [68].
5.1.4. HOPX and Senescence
There are a limited number of reports suggesting a role for HOPX in cellular senescence. For example, HOPX transfection of lung cancer cells resulted in enlarged and flattened morphologies, with positive SA—Gal staining indicating senescence, and stimulated the expression of proteins associated with aging, such as SAHFs and p-H2AX. When these cells were treated with senescence-inducing drugs such as Adriamycin (ADR) and H2O2, senescence was caused in HOPX-transfected cells but not in mock transfectants [56]. In H2228 lung adenocarcinoma cells, however, HOPX did not cause senescence, showing that it is involved in oxidative stress and DNA damage-induced cellular senescence but does not cause senescence on its own [56].
It was reported that re-expressing HOPX in immortalized bronchial epithelial cells promotes senescence. The RAS-ERK-MAPK signaling cascade has been identified to be involved in HOPX-induced senescence in lung cancer cells. A significant reduction of AKT/MDM2 accompanied by an upregulation of P53/ P21 was observed in HOPX-transfected lung cancer and normal bronchial epithelial cells. This study revealed HOPX as a regulator of the expression of senescence mediators [56]. Furthermore, the HOPX gene was shown to be overexpressed in keratinocytes with shortened telomeres, which elicits senescence [73].
5.1.5. HOPX in Cancer Cell Metastasis
A critical molecular event in the development of lung cancer is the downregulation of HOPX and overexpression of Mycn [74], and re-expression of HOPX suppresses lung cancer cell proliferation migration and invasion [56]. Furthermore, the alveolar lineage transcription factors, GATA6 and HOPX, were reported to be essential in airway epithelial specification and lung adenocarcinoma subtype metastasis inhibition [57].
In a chicken metastatic cell line derived from v-src-induced sarcoma and its non-metastatic subclone, the role of HOPX in metastasis was investigated. In v-src-transformed cells, knocking down HOPX reduced metastatic activity by inactivating metastatic-associated genes such as ITGA4 [75]. The expression of EMT-related molecules, such as ZEB1/2, TWIST1, E-cadherin, and vimentin, however, was unaffected by GATA6 and HOPX knockdown. It also had no effect on Wnt or focal adhesion kinase (FAK), although the expression of src and integrin 5, two mediators of metastasis, was upregulated.
HOPX hypermethylation promotes metastasis in nasopharyngeal cancer by increasing SNAIL transcription, which facilitates epithelial to mesenchymal transition [76]. Additional metastasis-related molecules (IL1B, IL11, and EREG) and angiogenesis and ECM remodeling genes (VEGFA and PLAU) were upregulated when HOPX was knocked down [57].
Bioinformatic analysis using STRING software predicts the possible interacting partners of HOPX in mice and humans as indicated in Figure 4 [77].
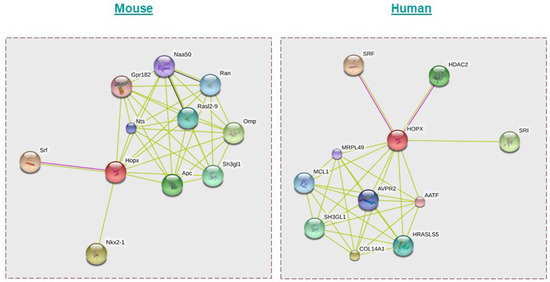
Figure 4.
Possible interaction partners of HOPX in mice and humans as predicted using the STRING database. Apart from SRF, HDAC2, and Epc1, HOPX could interact with proteins such as Apc, Nts, Ran, and Rasl2-9 in mice and COL14A1, SH3GL1, MRPL49, and HRASLS5 in humans. Mouse: Nkx2-1—NK2 homeobox 1; Ran—RAN, member RAS oncogene family; Naa50—N(alpha)-acetyltransferase 50, NatE catalytic subunit Gene; Nts—neurotensin; Srf—serum response factor; Sh3gl1—SH3-domain GRB2-like 1; Gpr182—G protein-coupled receptor 182; Rasl2-9—RAS-like, family 2, locus 9; Hopx—Homeodomain-only protein; Omp—olfactory marker protein; Apc—adenomatosis polyposis coli. Human: SRI—sorcin; HDAC2—histone deacetylase 2; COL14A1—collagen, type XIV, alpha 1; HOPX—Homeodomain-only protein; SH3GL1—SH3-domain GRB2-like 1; SRF—serum response factor (c-fos serum response element-binding transcription factor); AVPR2—arginine vasopressin receptor 2; MRPL49—mitochondrial ribosomal protein L49; MCL1—myeloid cell leukemia sequence 1 (BCL2-related); HRASLS5—HRAS-like suppressor family, member 5; AATF—apoptosis antagonizing transcription factor.
5.2. Evidence for an Oncogenic Function for HOPX
The expression level of HOPX is decreased in most cancer types and it functions as a tumor suppressor. However, HOPX expression has been reported to be elevated in a small number of esophageal carcinoma cell lines (TE2, TE5, KYSE410, and KYSE520), lung adenocarcinoma, and pancreatic cancer Langerhans islet cells [53,56,62]. Strong HOPX expression was associated with specific clinical and biochemical characteristics in de novo acute myeloid leukemia and predicted a poor prognosis [69]. HOPX is one of a group of genes, including deoxynucleotidyl transferase (DNTT) and B cell liners (BLNK), that are upregulated in patients with RUNX-1 mutant cytogenetically normal acute leukemia [78]. Interestingly, HOPX expression is increased in HPV 16- and 18-infected stage IB-IIA cervical cancer cells [79]. The expression of HOPX was also noticed in the tumor microenvironment; in particular, HOPX expression was higher in normal stromal cells than in stromal cells from colorectal malignant tissue [60,61]. There is increasing evidence that HOPX is a tumor suppressor gene in numerous cancer types. However, the function of HOPX in cancer could well be context dependent and more studies are needed to confirm a possible oncogenic role for HOPX in certain circumstances.
6. Conclusions and Future Perspectives
HOPX is a unique homeodomain protein that does not bind DNA but rather interacts with other proteins to function as a transcriptional regulator in development. During organogenesis, HOPX regulates the differentiation of various cell types, including keratinocytes, skeletal muscle, cardiomyocytes, and chondrocytes. In cardiac tissues, HOPX performs its function by directly controlling SRF and additional roles unrelated to SRF have been described. However, the entirety of the signaling networks associated with HOPX and organogenesis has yet to be uncovered and further studies are needed to fully elucidate the critical role of HOPX in organogenesis and tissue homeostasis.
HOPX is crucial to the prevention of tumor development and progression by promoting tumor suppression mechanisms. In malignancies of the head and neck, stomach, colon, pancreas, lung, placenta, breast, and esophagus, the HOPX promoter is hypermethylated and its role as a tumor suppressor gene was proven in a variety of functional studies. In the future, it is important to understand whether these findings can be utilized in a therapeutic context. Approaches to activate downstream signaling pathways that are compromised following HOPX loss could reverse some of the effects of HOPX loss in tumor cells. Similarly, the re-expression of epigenetically silenced HOPX with demethylating drugs would likely restore its tumor suppressive functions, as demonstrated in vivo in mouse models. A fuller understanding of the molecular mechanisms by which HOPX regulates carcinogenesis will likely lead to the development of new therapeutic approaches.
Author Contributions
Conceptualization, R.C.G., L.F.Y. and I.C.P.; writing—original draft preparation, R.C.G.; writing—review and editing, L.F.Y. and I.C.P. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a Fundamental Research Grant Scheme award (FRGS/1/2018/SKK08/UM/01/2) from the Ministry of Higher Education Malaysia.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Svingen, T.; Tonissen, K.F. Hox transcription factors and their elusive mammalian gene targets. Heredity 2006, 97, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Desplan, C.; Theis, J.; O’Farrell, P.H. The sequence specificity of homeodomain-DNA interaction. Cell 1988, 54, 1081–1090. [Google Scholar] [CrossRef]
- Lewis, E.B. A gene complex controlling segmentation in Drosophila. Nature 1978, 276, 565–570. [Google Scholar] [CrossRef]
- Kornberg, T.B. Understanding the homeodomain. J. Biol. Chem. 1993, 268, 26813–26816. [Google Scholar] [CrossRef]
- Boncinelli, E. Homeobox genes and disease. Curr. Opin. Genet. Dev. 1997, 7, 331–337. [Google Scholar] [CrossRef]
- Abate-Shen, C. Deregulated homeobox gene expression in cancer: Cause or consequence? Nat. Rev. Cancer 2002, 2, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Davidson, D. The function and evolution of Msx genes: Pointers and paradoxes. Trends Genet. 1995, 11, 405–411. [Google Scholar] [CrossRef]
- Del Bene, F.; Wittbrodt, J. Cell cycle control by homeobox genes in development and disease. Semin. Cell Dev. Biol. 2005, 16, 449–460. [Google Scholar] [CrossRef]
- Chiou, S.H.; Wang, M.L.; Chou, Y.T.; Chen, C.J.; Hong, C.F.; Hsieh, W.J.; Chang, H.T.; Chen, Y.S.; Lin, T.W.; Hsu, H.S.; et al. Coexpression of Oct4 and Nanog enhances malignancy in lung adenocarcinoma by inducing cancer stem cell-like properties and epithelial-mesenchymal transdifferentiation. Cancer Res. 2010, 70, 10433–10444. [Google Scholar] [CrossRef]
- Samuel, S.; Naora, H. Homeobox gene expression in cancer: Insights from developmental regulation and deregulation. Eur. J. Cancer 2005, 41, 2428–2437. [Google Scholar] [CrossRef]
- Chen, F.; Kook, H.; Milewski, R.; Gitler, A.D.; Lu, M.M.; Li, J.; Nazarian, R.; Schnepp, R.; Jen, K.; Biben, C.; et al. Hop is an unusual homeobox gene that modulates cardiac development. Cell 2002, 110, 713–723. [Google Scholar] [CrossRef]
- Shin, C.H.; Liu, Z.P.; Passier, R.; Zhang, C.L.; Wang, D.Z.; Harris, T.M.; Yamagishi, H.; Richardson, J.A.; Childs, G.; Olson, E.N. Modulation of cardiac growth and development by HOP, an unusual homeodomain protein. Cell 2002, 110, 725–735. [Google Scholar] [CrossRef]
- Adu, J.; Leong, F.T.; Smith, N.R.; Leek, J.P.; Markham, A.F.; Robinson, P.A.; Mighell, A.J. Expression of mOb1, a novel atypical 73 amino acid K50-homeodomain protein, during mouse development. Gene Expr. Patterns 2002, 2, 39–43. [Google Scholar] [CrossRef]
- Asanoma, K.; Matsuda, T.; Kondo, H.; Kato, K.; Kishino, T.; Niikawa, N.; Wake, N.; Kato, H. NECC1, a candidate choriocarcinoma suppressor gene that encodes a homeodomain consensus motif. Genomics 2003, 81, 15–25. [Google Scholar] [CrossRef]
- Chen, Y.; Petersen, S.; Pacyna-Gengelbach, M.; Pietas, A.; Petersen, I. Identification of a novel homeobox-containing gene, LAGY, which is downregulated in lung cancer. Oncology 2003, 64, 450–458. [Google Scholar] [CrossRef]
- Holland, P.W.; Booth, H.A.; Bruford, E.A. Classification and nomenclature of all human homeobox genes. BMC Biol. 2007, 5, 47. [Google Scholar] [CrossRef]
- Burglin, T.R.; Affolter, M. Homeodomain proteins: An update. Chromosoma 2016, 125, 497–521. [Google Scholar] [CrossRef]
- Kook, H.; Yung, W.W.; Simpson, R.J.; Kee, H.J.; Shin, S.; Lowry, J.A.; Loughlin, F.E.; Yin, Z.; Epstein, J.A.; Mackay, J.P. Analysis of the structure and function of the transcriptional coregulator HOP. Biochemistry 2006, 45, 10584–10590. [Google Scholar] [CrossRef]
- Mariotto, A.; Pavlova, O.; Park, H.S.; Huber, M.; Hohl, D. HOPX: The Unusual Homeodomain-Containing Protein. J. Investig. Dermatol. 2016, 136, 905–911. [Google Scholar] [CrossRef]
- Vasiliev, O.; Rhodes, S.J.; Beebe, D.C. Identification and expression of Hop, an atypical homeobox gene expressed late in lens fiber cell terminal differentiation. Mol. Vis. 2007, 13, 114–124. [Google Scholar]
- Asanoma, K.; Kato, H.; Yamaguchi, S.; Shin, C.H.; Liu, Z.P.; Kato, K.; Inoue, T.; Miyanari, Y.; Yoshikawa, K.; Sonoda, K.; et al. HOP/NECC1, a novel regulator of mouse trophoblast differentiation. J. Biol. Chem. 2007, 282, 24065–24074. [Google Scholar] [CrossRef] [PubMed]
- Palpant, N.J.; Wang, Y.; Hadland, B.; Zaunbrecher, R.J.; Redd, M.; Jones, D.; Pabon, L.; Jain, R.; Epstein, J.; Ruzzo, W.L.; et al. Chromatin and Transcriptional Analysis of Mesoderm Progenitor Cells Identifies HOPX as a Regulator of Primitive Hematopoiesis. Cell Rep. 2017, 20, 1597–1608. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Georgantas, R.W., 3rd; Tanadve, V.; Malehorn, M.; Heimfeld, S.; Chen, C.; Carr, L.; Martinez-Murillo, F.; Riggins, G.; Kowalski, J.; Civin, C.I. Microarray and serial analysis of gene expression analyses identify known and novel transcripts overexpressed in hematopoietic stem cells. Cancer Res. 2004, 64, 4434–4441. [Google Scholar] [CrossRef]
- Takeda, N.; Jain, R.; Leboeuf, M.R.; Padmanabhan, A.; Wang, Q.; Li, L.; Lu, M.M.; Millar, S.E.; Epstein, J.A. Hopx expression defines a subset of multipotent hair follicle stem cells and a progenitor population primed to give rise to K6+ niche cells. Development 2013, 140, 1655–1664. [Google Scholar] [CrossRef]
- Barkauskas, C.E.; Cronce, M.J.; Rackley, C.R.; Bowie, E.J.; Keene, D.R.; Stripp, B.R.; Randell, S.H.; Noble, P.W.; Hogan, B.L. Type 2 alveolar cells are stem cells in adult lung. J. Clin. Investig. 2013, 123, 3025–3036. [Google Scholar] [CrossRef] [PubMed]
- Takeda, N.; Jain, R.; LeBoeuf, M.R.; Wang, Q.; Lu, M.M.; Epstein, J.A. Interconversion between intestinal stem cell populations in distinct niches. Science 2011, 334, 1420–1424. [Google Scholar] [CrossRef]
- Roth, S.; Franken, P.; Sacchetti, A.; Kremer, A.; Anderson, K.; Sansom, O.; Fodde, R. Paneth cells in intestinal homeostasis and tissue injury. PLoS ONE 2012, 7, e38965. [Google Scholar] [CrossRef]
- Rees, W.D.; Tandun, R.; Yau, E.; Zachos, N.C.; Steiner, T.S. Regenerative Intestinal Stem Cells Induced by Acute and Chronic Injury: The Saving Grace of the Epithelium? Front. Cell Dev. Biol. 2020, 8, 583919. [Google Scholar] [CrossRef]
- Kawasaki, H.; Yoshida, T.; Horiguchi, K.; Ohama, T.; Sato, K. Characterization of anoikis-resistant cells in mouse colonic epithelium. J. Vet. Med. Sci. 2013, 75, 1173–1180. [Google Scholar] [CrossRef][Green Version]
- Gracz, A.D.; Fuller, M.K.; Wang, F.; Li, L.; Stelzner, M.; Dunn, J.C.; Martin, M.G.; Magness, S.T. Brief report: CD24 and CD44 mark human intestinal epithelial cell populations with characteristics of active and facultative stem cells. Stem Cells 2013, 31, 2024–2030. [Google Scholar] [CrossRef]
- Hng, C.H.; Camp, E.; Anderson, P.; Breen, J.; Zannettino, A.; Gronthos, S. HOPX regulates bone marrow-derived mesenchymal stromal cell fate determination via suppression of adipogenic gene pathways. Sci. Rep. 2020, 10, 11345. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.; Barkauskas, C.E.; Takeda, N.; Bowie, E.J.; Aghajanian, H.; Wang, Q.; Padmanabhan, A.; Manderfield, L.J.; Gupta, M.; Li, D.; et al. Plasticity of Hopx(+) type I alveolar cells to regenerate type II cells in the lung. Nat. Commun. 2015, 6, 6727. [Google Scholar] [CrossRef] [PubMed]
- Grego-Bessa, J.; Luna-Zurita, L.; del Monte, G.; Bolos, V.; Melgar, P.; Arandilla, A.; Garratt, A.N.; Zang, H.; Mukouyama, Y.S.; Chen, H.; et al. Notch signaling is essential for ventricular chamber development. Dev. Cell 2007, 12, 415–429. [Google Scholar] [CrossRef]
- Zhou, X.; Crow, A.L.; Hartiala, J.; Spindler, T.J.; Ghazalpour, A.; Barsky, L.W.; Bennett, B.J.; Parks, B.W.; Eskin, E.; Jain, R.; et al. The Genetic Landscape of Hematopoietic Stem Cell Frequency in Mice. Stem Cell Rep. 2015, 5, 125–138. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lin, C.C.; Yao, C.Y.; Hsu, Y.C.; Hou, H.A.; Yuan, C.T.; Li, Y.H.; Kao, C.J.; Chuang, P.H.; Chiu, Y.C.; Chen, Y.; et al. Knock-out of Hopx disrupts stemness and quiescence of hematopoietic stem cells in mice. Oncogene 2020, 39, 5112–5123. [Google Scholar] [CrossRef]
- Dorrell, C.; Schug, J.; Lin, C.F.; Canaday, P.S.; Fox, A.J.; Smirnova, O.; Bonnah, R.; Streeter, P.R.; Stoeckert, C.J., Jr.; Kaestner, K.H.; et al. Transcriptomes of the major human pancreatic cell types. Diabetologia 2011, 54, 2832–2844. [Google Scholar] [CrossRef]
- Muhlfriedel, S.; Kirsch, F.; Gruss, P.; Stoykova, A.; Chowdhury, K. A roof plate-dependent enhancer controls the expression of Homeodomain only protein in the developing cerebral cortex. Dev. Biol. 2005, 283, 522–534. [Google Scholar] [CrossRef]
- De Toni, A.; Zbinden, M.; Epstein, J.A.; Ruiz i Altaba, A.; Prochiantz, A.; Caille, I. Regulation of survival in adult hippocampal and glioblastoma stem cell lineages by the homeodomain-only protein HOP. Neural Dev. 2008, 3, 13. [Google Scholar] [CrossRef]
- Liu, F.; Ismat, F.A.; Patel, V.V. Role of homeodomain-only protein in the cardiac conduction system. Trends Cardiovasc. Med. 2006, 16, 193–198. [Google Scholar] [CrossRef][Green Version]
- Kook, H.; Lepore, J.J.; Gitler, A.D.; Lu, M.M.; Wing-Man Yung, W.; Mackay, J.; Zhou, R.; Ferrari, V.; Gruber, P.; Epstein, J.A. Cardiac hypertrophy and histone deacetylase-dependent transcriptional repression mediated by the atypical homeodomain protein Hop. J. Clin. Investig. 2003, 112, 863–871. [Google Scholar] [CrossRef]
- Yin, Z.; Gonzales, L.; Kolla, V.; Rath, N.; Zhang, Y.; Lu, M.M.; Kimura, S.; Ballard, P.L.; Beers, M.F.; Epstein, J.A.; et al. Hop functions downstream of Nkx2.1 and GATA6 to mediate HDAC-dependent negative regulation of pulmonary gene expression. Am. J. Physiol. Lung Cell Mol. Physiol. 2006, 291, L191–L199. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.; Li, D.; Gupta, M.; Manderfield, L.J.; Ifkovits, J.L.; Wang, Q.; Liu, F.; Liu, Y.; Poleshko, A.; Padmanabhan, A.; et al. HEART DEVELOPMENT. Integration of Bmp and Wnt signaling by Hopx specifies commitment of cardiomyoblasts. Science 2015, 348, aaa6071. [Google Scholar] [CrossRef] [PubMed]
- Kee, H.J.; Kim, J.R.; Nam, K.I.; Park, H.Y.; Shin, S.; Kim, J.C.; Shimono, Y.; Takahashi, M.; Jeong, M.H.; Kim, N.; et al. Enhancer of polycomb1, a novel homeodomain only protein-binding partner, induces skeletal muscle differentiation. J. Biol. Chem. 2007, 282, 7700–7709. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.J.; Lee, H.J.; Kamli, M.R.; Pokharel, S.; Bhat, A.R.; Lee, Y.H.; Choi, B.H.; Chun, T.; Kang, S.W.; Lee, Y.S.; et al. Depot-specific gene expression profiles during differentiation and transdifferentiation of bovine muscle satellite cells, and differentiation of preadipocytes. Genomics 2012, 100, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.M.; Sim, S.M.; Kim, H.Y.; Park, G.T. Expression of the homeobox gene, HOPX, is modulated by cell differentiation in human keratinocytes and is involved in the expression of differentiation markers. Eur. J. Cell Biol. 2010, 89, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Hawiger, D.; Wan, Y.Y.; Eynon, E.E.; Flavell, R.A. The transcription cofactor Hopx is required for regulatory T cell function in dendritic cell-mediated peripheral T cell unresponsiveness. Nat. Immunol. 2010, 11, 962–968. [Google Scholar] [CrossRef]
- Albrecht, I.; Niesner, U.; Janke, M.; Menning, A.; Loddenkemper, C.; Kuhl, A.A.; Lepenies, I.; Lexberg, M.H.; Westendorf, K.; Hradilkova, K.; et al. Persistence of effector memory Th1 cells is regulated by Hopx. Eur. J. Immunol. 2010, 40, 2993–3006. [Google Scholar] [CrossRef]
- Jones, A.; Opejin, A.; Henderson, J.G.; Gross, C.; Jain, R.; Epstein, J.A.; Flavell, R.A.; Hawiger, D. Peripherally Induced Tolerance Depends on Peripheral Regulatory T Cells That Require Hopx To Inhibit Intrinsic IL-2 Expression. J. Immunol. 2015, 195, 1489–1497. [Google Scholar] [CrossRef]
- Trivedi, C.M.; Cappola, T.P.; Margulies, K.B.; Epstein, J.A. Homeodomain only protein x is down-regulated in human heart failure. J. Mol. Cell Cardiol. 2011, 50, 1056–1058. [Google Scholar] [CrossRef]
- Liu, F.; Levin, M.D.; Petrenko, N.B.; Lu, M.M.; Wang, T.; Yuan, L.J.; Stout, A.L.; Epstein, J.A.; Patel, V.V. Histone-deacetylase inhibition reverses atrial arrhythmia inducibility and fibrosis in cardiac hypertrophy independent of angiotensin. J. Mol. Cell Cardiol. 2008, 45, 715–723. [Google Scholar] [CrossRef]
- Gulec, C.; Abaci, N.; Bayrak, F.; Komurcu Bayrak, E.; Kahveci, G.; Guven, C.; Erginel Unaltuna, N. Association between non-coding polymorphisms of HOPX gene and syncope in hypertrophic cardiomyopathy. Anadolu. Kardiyol. Derg. 2014, 14, 617–624. [Google Scholar] [CrossRef] [PubMed]
- Lemaire, F.; Millon, R.; Muller, D.; Rabouel, Y.; Bracco, L.; Abecassis, J.; Wasylyk, B. Loss of HOP tumour suppressor expression in head and neck squamous cell carcinoma. Br. J. Cancer 2004, 91, 258–261. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, K.; Kim, M.S.; Park, H.L.; Tokumaru, Y.; Osada, M.; Inoue, H.; Mori, M.; Sidransky, D. HOP/OB1/NECC1 promoter DNA is frequently hypermethylated and involved in tumorigenic ability in esophageal squamous cell carcinoma. Mol. Cancer Res. 2008, 6, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Toruner, G.A.; Ulger, C.; Alkan, M.; Galante, A.T.; Rinaggio, J.; Wilk, R.; Tian, B.; Soteropoulos, P.; Hameed, M.R.; Schwalb, M.N.; et al. Association between gene expression profile and tumor invasion in oral squamous cell carcinoma. Cancer Genet. Cytogenet 2004, 154, 27–35. [Google Scholar] [CrossRef]
- Ooki, A.; Yamashita, K.; Kikuchi, S.; Sakuramoto, S.; Katada, N.; Kokubo, K.; Kobayashi, H.; Kim, M.S.; Sidransky, D.; Watanabe, M. Potential utility of HOP homeobox gene promoter methylation as a marker of tumor aggressiveness in gastric cancer. Oncogene 2010, 29, 3263–3275. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, L.; Cui, T.; Pacyna-Gengelbach, M.; Petersen, I. HOPX is methylated and exerts tumour-suppressive function through Ras-induced senescence in human lung cancer. J. Pathol. 2015, 235, 397–407. [Google Scholar] [CrossRef]
- Cheung, W.K.; Zhao, M.; Liu, Z.; Stevens, L.E.; Cao, P.D.; Fang, J.E.; Westbrook, T.F.; Nguyen, D.X. Control of alveolar differentiation by the lineage transcription factors GATA6 and HOPX inhibits lung adenocarcinoma metastasis. Cancer Cell 2013, 23, 725–738. [Google Scholar] [CrossRef]
- Chen, Y.; Pacyna-Gengelbach, M.; Deutschmann, N.; Niesporek, S.; Petersen, I. Homeobox gene HOP has a potential tumor suppressive activity in human lung cancer. Int. J. Cancer J. Int. Cancer 2007, 121, 1021–1027. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Asanoma, K.; Takao, T.; Kato, K.; Wake, N. Homeobox gene HOPX is epigenetically silenced in human uterine endometrial cancer and suppresses estrogen-stimulated proliferation of cancer cells by inhibiting serum response factor. Int. J. Cancer 2009, 124, 2577–2588. [Google Scholar] [CrossRef]
- Harada, Y.; Kijima, K.; Shinmura, K.; Sakata, M.; Sakuraba, K.; Yokomizo, K.; Kitamura, Y.; Shirahata, A.; Goto, T.; Mizukami, H.; et al. Methylation of the homeobox gene, HOPX, is frequently detected in poorly differentiated colorectal cancer. Anticancer Res. 2011, 31, 2889–2892. [Google Scholar]
- Katoh, H.; Yamashita, K.; Waraya, M.; Margalit, O.; Ooki, A.; Tamaki, H.; Sakagami, H.; Kokubo, K.; Sidransky, D.; Watanabe, M. Epigenetic silencing of HOPX promotes cancer progression in colorectal cancer. Neoplasia 2012, 14, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Waraya, M.; Yamashita, K.; Katoh, H.; Ooki, A.; Kawamata, H.; Nishimiya, H.; Nakamura, K.; Ema, A.; Watanabe, M. Cancer specific promoter CpG Islands hypermethylation of HOP homeobox (HOPX) gene and its potential tumor suppressive role in pancreatic carcinogenesis. BMC Cancer 2012, 12, 397. [Google Scholar] [CrossRef] [PubMed]
- Cromer, A.; Carles, A.; Millon, R.; Ganguli, G.; Chalmel, F.; Lemaire, F.; Young, J.; Dembele, D.; Thibault, C.; Muller, D.; et al. Identification of genes associated with tumorigenesis and metastatic potential of hypopharyngeal cancer by microarray analysis. Oncogene 2004, 23, 2484–2498. [Google Scholar] [CrossRef] [PubMed]
- Roepman, P.; Wessels, L.F.; Kettelarij, N.; Kemmeren, P.; Miles, A.J.; Lijnzaad, P.; Tilanus, M.G.; Koole, R.; Hordijk, G.J.; van der Vliet, P.C.; et al. An expression profile for diagnosis of lymph node metastases from primary head and neck squamous cell carcinomas. Nat. Genet. 2005, 37, 182–186. [Google Scholar] [CrossRef]
- Hunter, K.D.; Thurlow, J.K.; Fleming, J.; Drake, P.J.; Vass, J.K.; Kalna, G.; Higham, D.J.; Herzyk, P.; Macdonald, D.G.; Parkinson, E.K.; et al. Divergent routes to oral cancer. Cancer Res. 2006, 66, 7405–7413. [Google Scholar] [CrossRef]
- Yap, L.F.; Lai, S.L.; Patmanathan, S.N.; Gokulan, R.; Robinson, C.M.; White, J.B.; Chai, S.J.; Rajadurai, P.; Prepageran, N.; Liew, Y.T.; et al. HOPX functions as a tumour suppressor in head and neck cancer. Sci. Rep. 2016, 6, 38758. [Google Scholar] [CrossRef]
- Liang, H.; Wang, C.; Gao, K.; Li, J.; Jia, R. MuicroRNA421 promotes the progression of nonsmall cell lung cancer by targeting HOPX and regulating the Wnt/betacatenin signaling pathway. Mol. Med. Rep. 2019, 20, 151–161. [Google Scholar] [CrossRef]
- Pavlova, O.; Lefort, K.; Mariotto, A.; Huber, M.; Hohl, D. HOPX Exhibits Oncogenic Activity during Squamous Skin Carcinogenesis. J. Investig. Dermatol. 2021, 141, 2354–2368. [Google Scholar] [CrossRef]
- Lin, C.C.; Hsu, Y.C.; Li, Y.H.; Kuo, Y.Y.; Hou, H.A.; Lan, K.H.; Chen, T.C.; Tzeng, Y.S.; Kuo, Y.Y.; Kao, C.J.; et al. Higher HOPX expression is associated with distinct clinical and biological features and predicts poor prognosis in de novo acute myeloid leukemia. Haematologica 2017, 102, 1044–1053. [Google Scholar] [CrossRef]
- Kazanets, A.; Shorstova, T.; Hilmi, K.; Marques, M.; Witcher, M. Epigenetic silencing of tumor suppressor genes: Paradigms, puzzles, and potential. Biochim. Biophys. Acta 2016, 1865, 275–288. [Google Scholar] [CrossRef]
- Ooizumi, Y.; Katoh, H.; Yokota, M.; Watanabe, M.; Yamashita, K. Epigenetic silencing of HOPX is critically involved in aggressive phenotypes and patient prognosis in papillary thyroid cancer. Oncotarget 2019, 10, 5906–5918. [Google Scholar] [CrossRef] [PubMed]
- You, Q.; Geng, Y.; Ye, H.; Zhu, G.; Gao, X.; Zhu, H. HOPX Is an Epigenetically Inactivated Tumor Suppressor and Overexpression of HOPX Induce Apoptosis and Cell Cycle Arrest in Breast Cancer. Onco. Targets Ther. 2020, 13, 5955–5965. [Google Scholar] [CrossRef] [PubMed]
- Minty, F.; Thurlow, J.K.; Harrison, P.R.; Parkinson, E.K. Telomere dysfunction in human keratinocytes elicits senescence and a novel transcription profile. Exp. Cell Res. 2008, 314, 2434–2447. [Google Scholar] [CrossRef] [PubMed]
- Pacheco-Pinedo, E.C.; Durham, A.C.; Stewart, K.M.; Goss, A.M.; Lu, M.M.; Demayo, F.J.; Morrisey, E.E. Wnt/beta-catenin signaling accelerates mouse lung tumorigenesis by imposing an embryonic distal progenitor phenotype on lung epithelium. J. Clin. Investig. 2011, 121, 1935–1945. [Google Scholar] [CrossRef] [PubMed]
- Kovarova, D.; Plachy, J.; Kosla, J.; Trejbalova, K.; Cermak, V.; Hejnar, J. Downregulation of HOPX controls metastatic behavior in sarcoma cells and identifies genes associated with metastasis. Mol. Cancer Res. 2013, 11, 1235–1247. [Google Scholar] [CrossRef]
- Ren, X.; Yang, X.; Cheng, B.; Chen, X.; Zhang, T.; He, Q.; Li, B.; Li, Y.; Tang, X.; Wen, X.; et al. HOPX hypermethylation promotes metastasis via activating SNAIL transcription in nasopharyngeal carcinoma. Nat. Commun. 2017, 8, 14053. [Google Scholar] [CrossRef]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; von Mering, C.; et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013, 41, D808–D815. [Google Scholar] [CrossRef]
- Greif, P.A.; Konstandin, N.P.; Metzeler, K.H.; Herold, T.; Pasalic, Z.; Ksienzyk, B.; Dufour, A.; Schneider, F.; Schneider, S.; Kakadia, P.M.; et al. RUNX1 mutations in cytogenetically normal acute myeloid leukemia are associated with a poor prognosis and up-regulation of lymphoid genes. Haematologica 2012, 97, 1909–1915. [Google Scholar] [CrossRef]
- Santin, A.D.; Zhan, F.; Bignotti, E.; Siegel, E.R.; Cane, S.; Bellone, S.; Palmieri, M.; Anfossi, S.; Thomas, M.; Burnett, A.; et al. Gene expression profiles of primary HPV16- and HPV18-infected early stage cervical cancers and normal cervical epithelium: Identification of novel candidate molecular markers for cervical cancer diagnosis and therapy. Virology 2005, 331, 269–291. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).