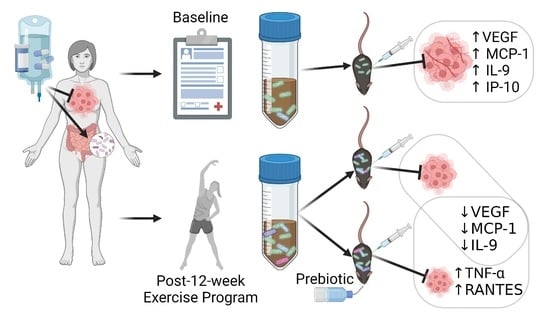
Exercise and Prebiotic Fiber Provide Gut Microbiota-Driven Benefit in a Survivor to Germ-Free Mouse Translational Model of Breast Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
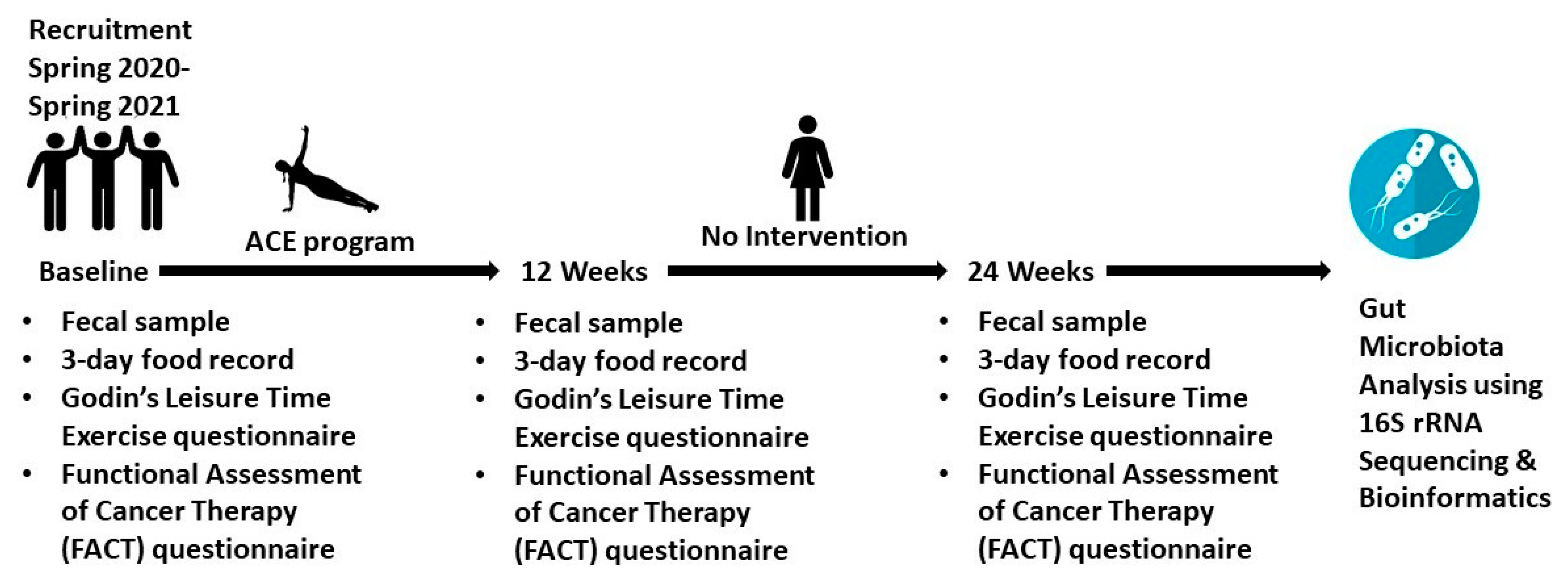
2.1. Clinical Study
2.1.1. Alberta Cancer Exercise (ACE) Program
2.1.2. Recruitment
2.1.3. Sample Size and Power
2.1.4. Demographic Information
2.1.5. Godin’s Leisure Time Exercise Questionnaire (GLTEQ)
2.1.6. Patient-Reported Psychosocial Outcomes
2.1.7. Dietary Intake
2.1.8. ACE Participant Fecal Samples and 16S rRNA Analysis
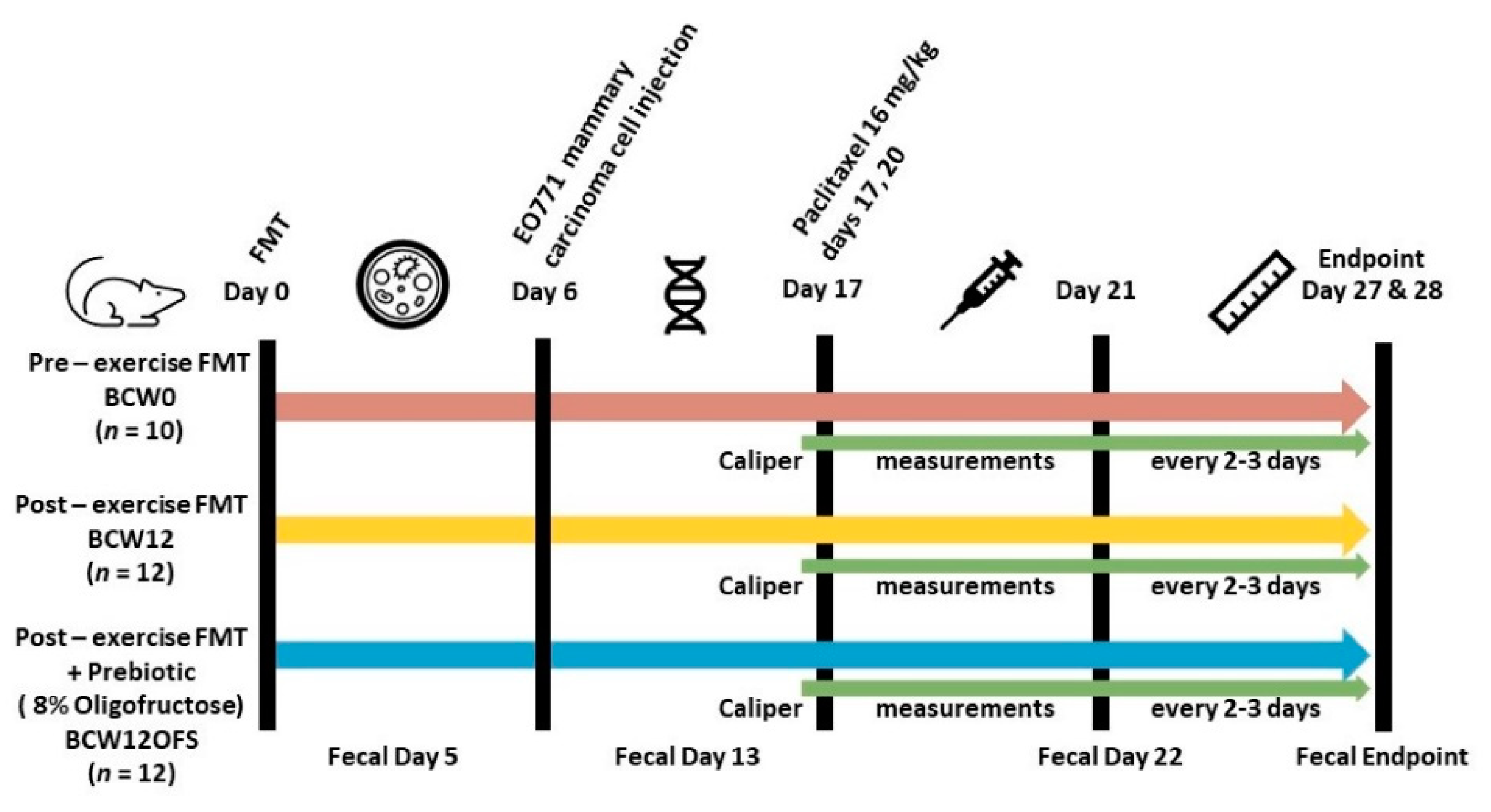
2.2. Murine FMT Study
2.2.1. Animals
2.2.2. Cell Culture
2.2.3. EO771 Cell Injections
2.2.4. Paclitaxel Injections
2.2.5. Oligofructose Supplementation
2.2.6. Mouse Fecal Samples and 16S rRNA Analysis
2.2.7. Tumor Measurements
2.2.8. Tissue Collection
2.2.9. Tissue Real-Time PCR Analysis
2.2.10. Serum and Tumor Cytokine Analysis
2.2.11. Statistical Analysis
3. Results
3.1. Clinical Study Results: ACE’s Impact on Gut Microbiota in Breast Cancer Survivors
3.1.1. Demographics
3.1.2. Participant Clinical Characteristics
3.1.3. Godin’s Leisure Time Exercise Questionnaire
3.1.4. Three-Day Food Record Dietary Analysis
3.1.5. Health-Related Quality of Life Results: FACT-G
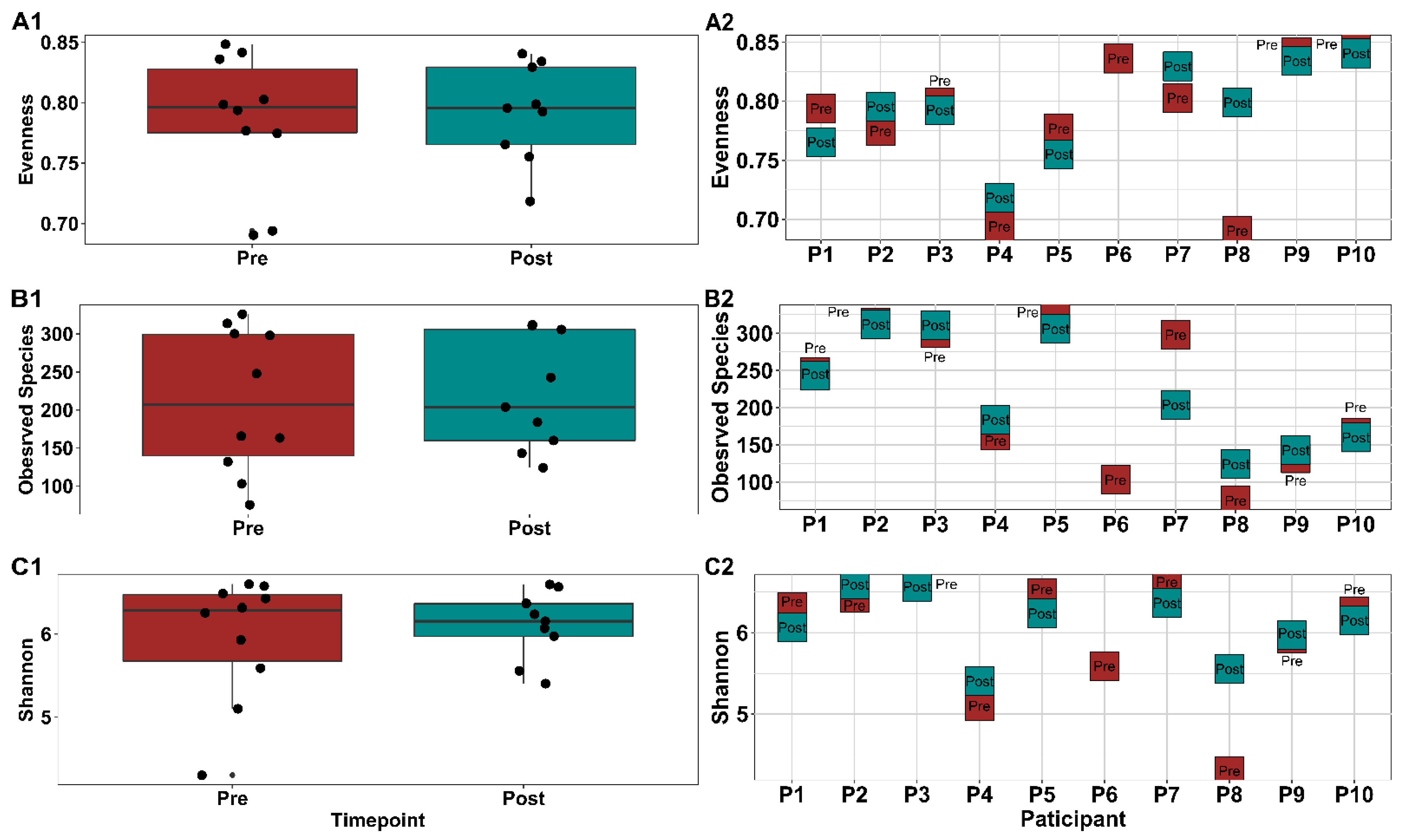
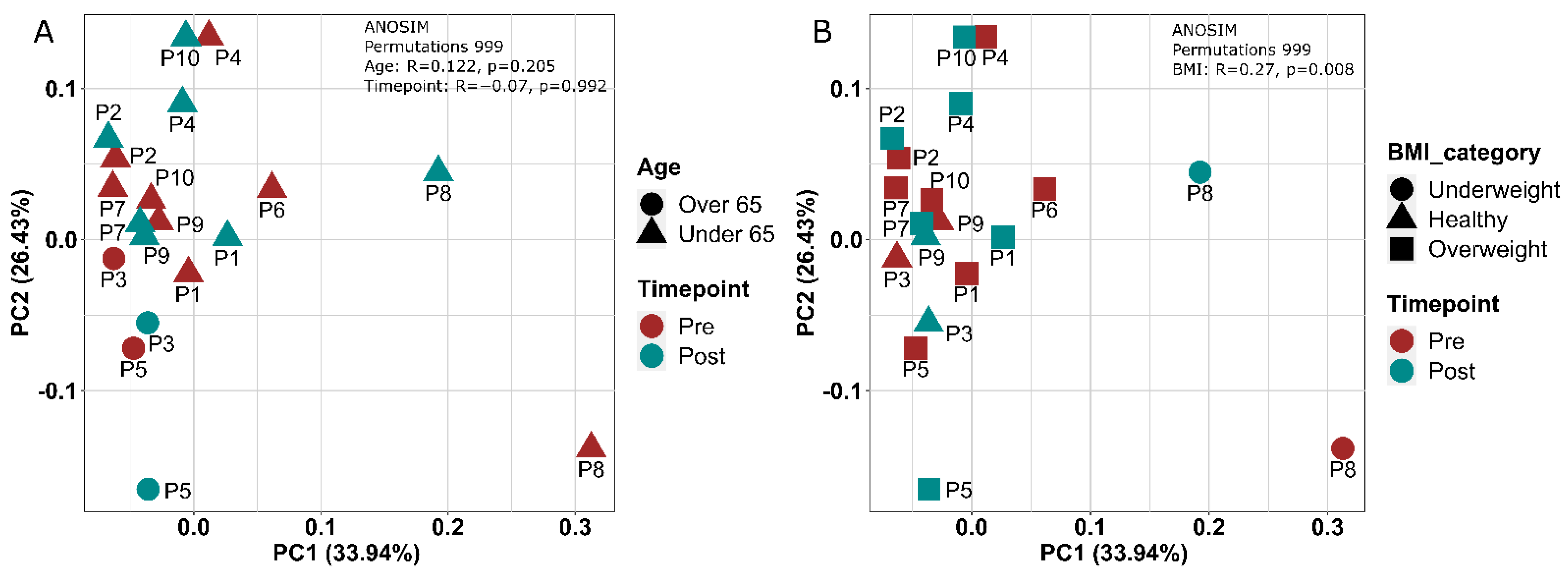
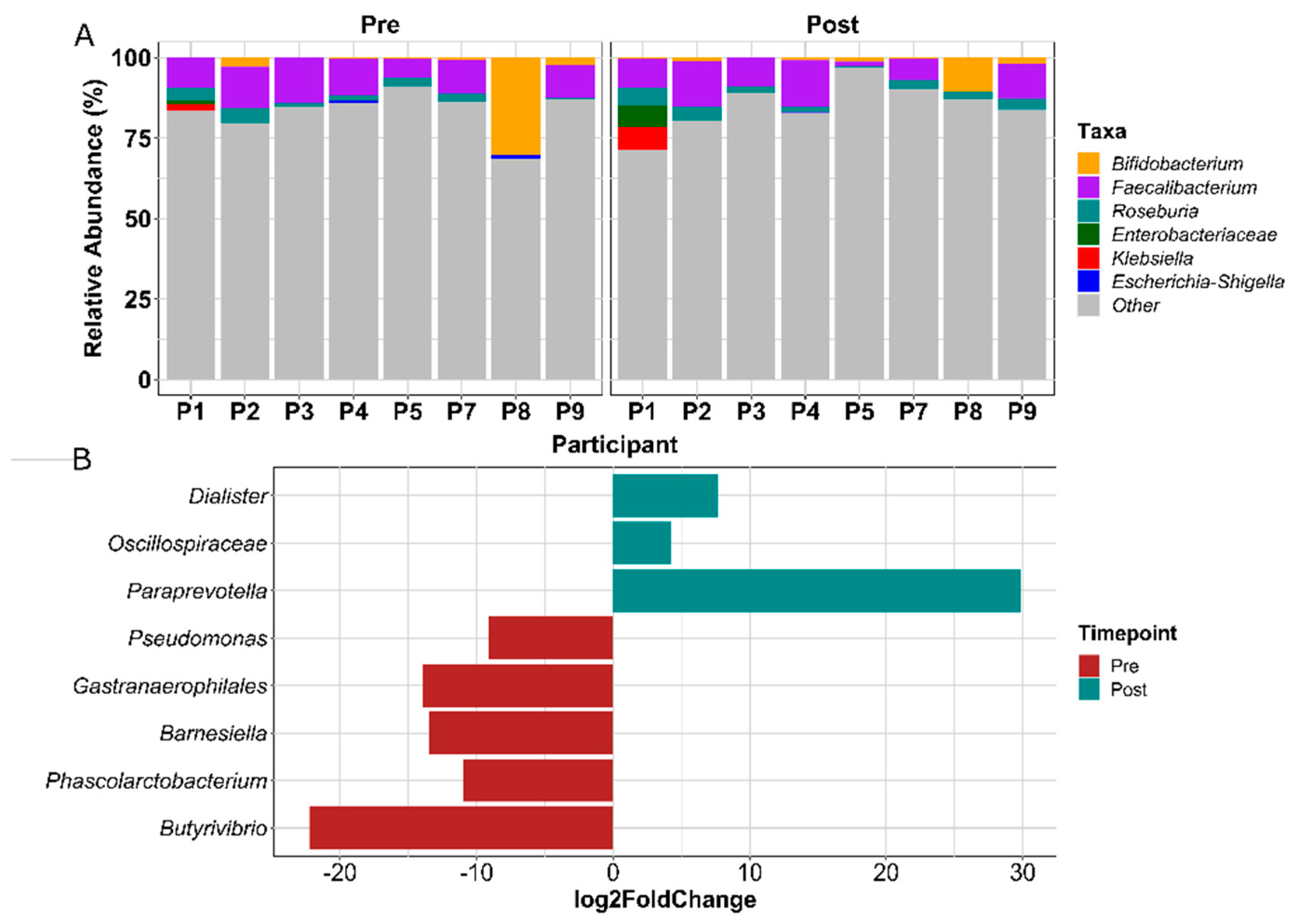
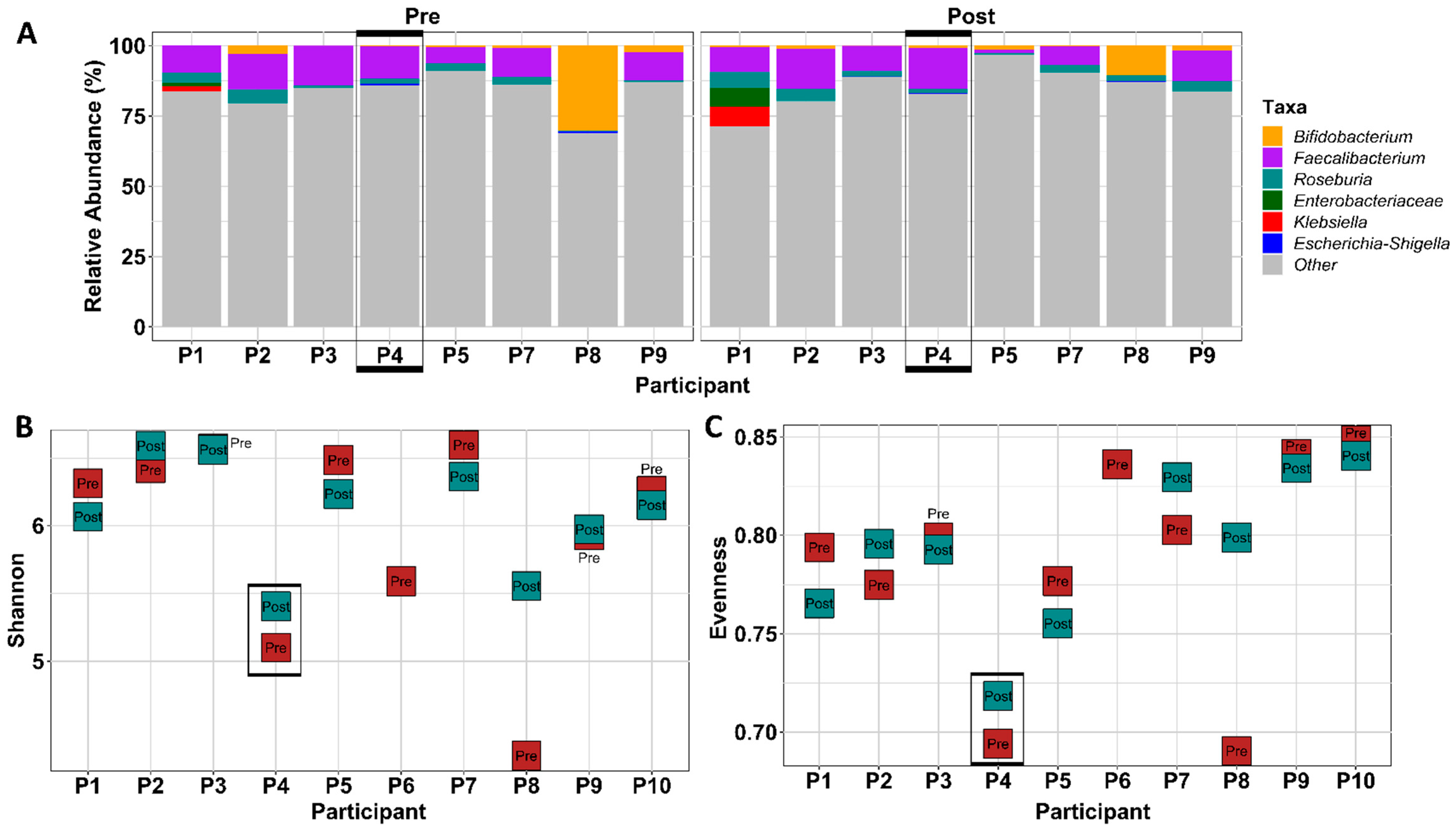
3.1.6. Gut Microbial Composition Suggests Some Response to Exercise
3.1.7. Microbial Correlations with Emotional Well-Being and Nutrient Intake
3.1.8. FMT Donor Choice for the Germ-Free Murine Study
3.2. Results of the Germ-Free Mouse Study Investigating the Impact of Exercise-Responsive Gut Microbiota in a Murine Model of Breast Cancer Treatment
3.2.1. Fluid Intake
3.2.2. Tumor Volumes Indicate Post-Exercise Microbiota-Related Benefit
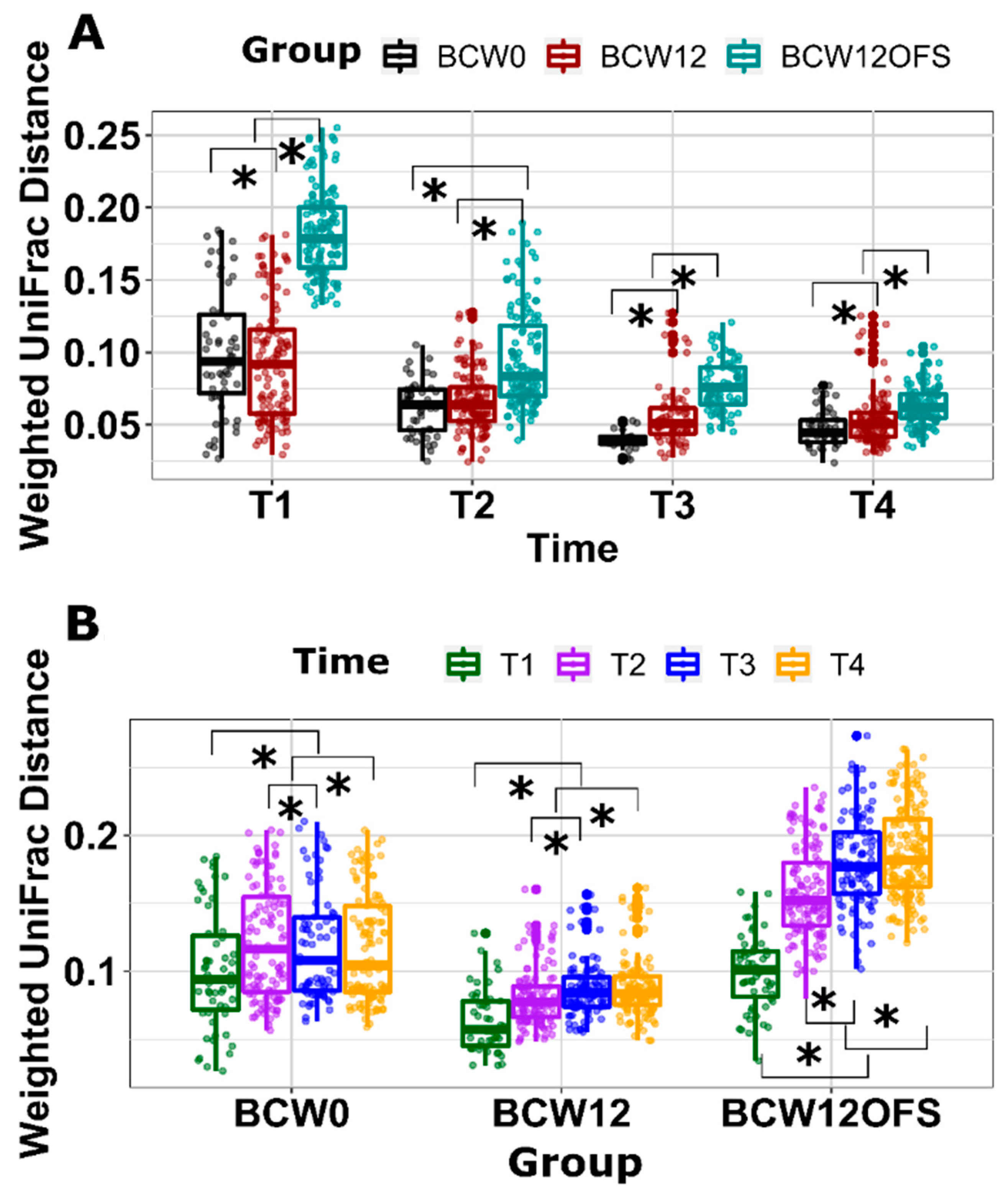
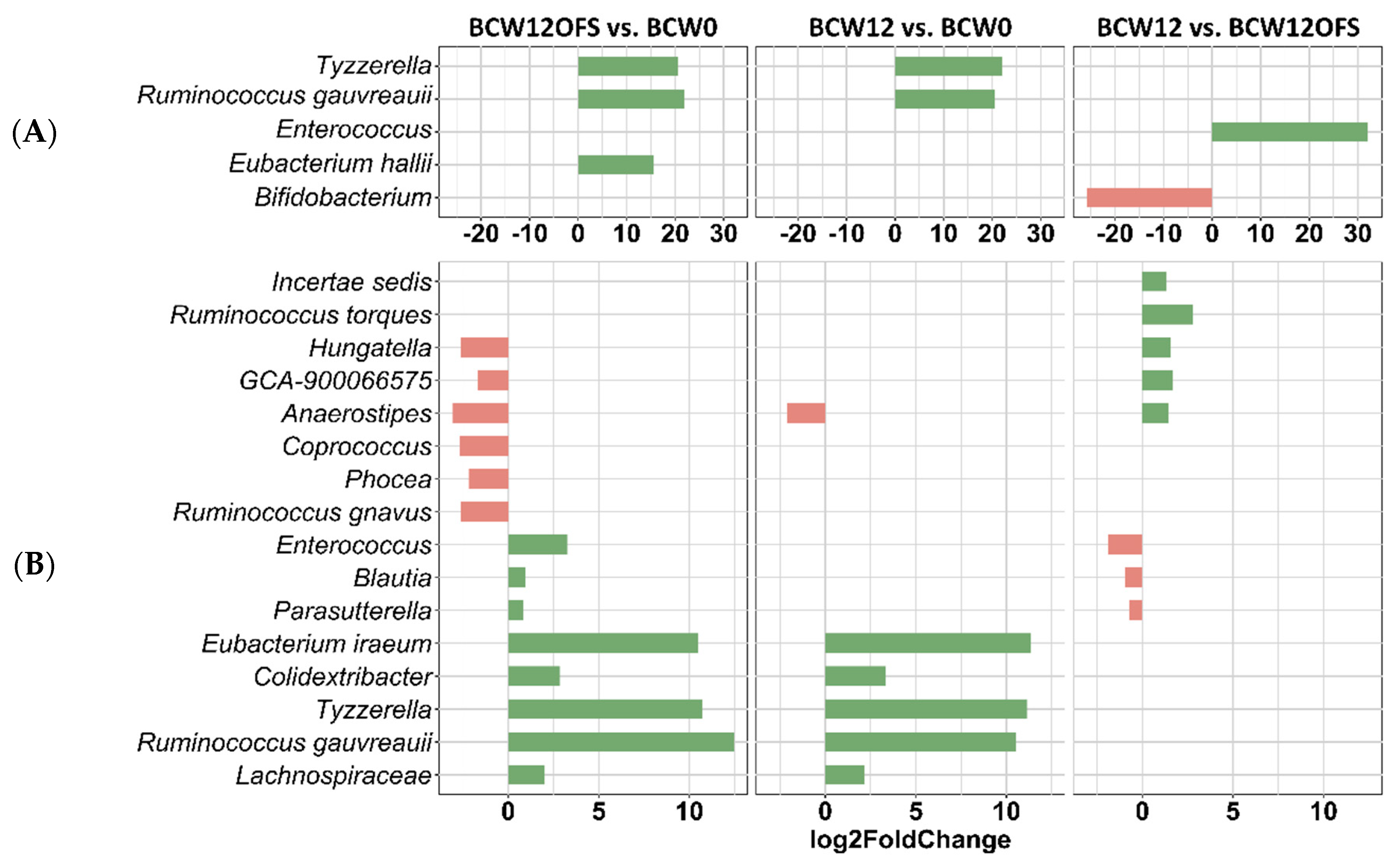
3.2.3. Mouse Gut Microbial Composition Differs Significantly across Groups
3.2.4. Ileal and Colonic Tight Junction Proteins
3.2.5. Tumor and Serum Cytokine Levels
4. Discussion
5. Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Noce, A.; Marrone, G.; Daniele, F.D.; Ottaviani, E.; Jones, G.W.; Bernini, R.; Romani, A.; Rovella, V. Impact of Gut Microbiota Composition on Onset and Progression of Chronic Non-Communicable Diseases. Nutrients 2019, 11, 1073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vivarelli, S.; Salemi, R.; Candido, S.; Falzone, L.; Santagati, M.; Stefani, S.; Torino, F.; Banna, G.L.; Tonini, G.; Libra, M. Gut microbiota and cancer: From pathogenesis to therapy. Cancers 2019, 11, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scott, A.J.; Alexander, J.L.; Merrifield, C.A.; Cunningham, D.; Jobin, C.; Brown, R.; Alverdy, J.; Keefe, S.J.O.; Gaskins, H.R.; Teare, J.; et al. International Cancer Microbiome Consortium consensus statement on the role of the human microbiome in carcinogenesis. Gut 2019, 68, 1624–1632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mager, L.F.; Burkhard, R.; Pett, N.; Cooke, N.C.A.; Brown, K.; Ramay, H.; Paik, S.; Stagg, J.; Groves, R.A.; Gallo, M.; et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science 2020, 369, 1481–1489. [Google Scholar] [CrossRef]
- Frosali, S.; Pagliari, D.; Gambassi, G.; Landolfi, R.; Pandolfi, F.; Cianci, R. How the Intricate Interaction among Toll-Like Receptors, Microbiota, and Intestinal Immunity Can Influence Gastrointestinal Pathology. J. Immunol. Res. 2015, 2015, 489821. [Google Scholar] [CrossRef]
- Soares, P.M.G.; Mota, J.M.S.C.; Souza, E.P.; Justino, P.F.C.; Franco, A.X.; Cunha, F.Q.; Ribeiro, R.A.; Souza, M.H.L.P. Inflammatory intestinal damage induced by 5-fluorouracil requires IL-4. Cytokine 2013, 61, 46–49. [Google Scholar] [CrossRef] [Green Version]
- Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; Cogdill, A.P.; Zhao, L.; Hudgens, C.W.; Hutchinson, D.S.; Manzo, T.; et al. Gut microbiome modulates response to anti—PD-1 immunotherapy in melanoma patients. Science 2018, 103, 97–103. [Google Scholar]
- Viaud, S.; Saccheri, F.; Mignot, G.; Yamazaki, T.; Daillère, R.; Hannani, D.; Enot, D.P.; Pfirschke, C.; Engblom, C.; Pittet, M.J.; et al. The Intestinal Microbiota Modulates the Anticancer Immune Effects of Cyclophosphamide. Science 2013, 342, 971–977. [Google Scholar] [CrossRef] [Green Version]
- Lam, K.C.; Araya, R.E.; Huang, A.; Chen, Q.; Di Modica, M.; Rodrigues, R.R.; Lopès, A.; Johnson, S.B.; Schwarz, B.; Bohrnsen, E.; et al. Microbiota triggers STING-type I IFN-dependent monocyte reprogramming of the tumor microenvironment. Cell 2021, 184, 5338–5356.e21. [Google Scholar] [CrossRef]
- Kumar, J.; Rani, K.; Datt, C. Molecular link between dietary fibre, gut microbiota and health. Mol. Biol. Rep. 2020, 47, 6229–6237. [Google Scholar] [CrossRef]
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zmora, N.; Suez, J.; Elinav, E. You are what you eat: Diet, health and the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 35–56. [Google Scholar] [CrossRef] [PubMed]
- Mailing, L.J.; Allen, J.M.; Buford, T.W.; Fields, C.J.; Woods, J.A. Exercise and the Gut Microbiome: A Review of the Evidence, Potential Mechanisms, and Implications for Human Health. Exerc. Sport Sci. Rev. 2019, 47, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Mohr, A.E.; Jäger, R.; Carpenter, K.C.; Kerksick, C.M.; Purpura, M.; Townsend, J.R.; West, N.P.; Black, K.; Gleeson, M.; Pyne, D.B.; et al. The athletic gut microbiota. J. Int. Soc. Sports Nutr. 2020, 17, 24. [Google Scholar] [CrossRef]
- Sheflin, A.M.; Whitney, A.K.; Weir, T.L. Cancer-Promoting Effects of Microbial Dysbiosis. Curr. Oncol. Rep. 2014, 16, 406. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Elmén, L.; Segota, I.; Xian, Y.; Tinoco, R.; Feng, Y.; Fujita, Y.; Segura Muñoz, R.R.; Schmaltz, R.; Bradley, L.M.; et al. Prebiotic-Induced Anti-tumor Immunity Attenuates Tumor Growth. Cell Rep. 2020, 30, 1753–1766.e6. [Google Scholar] [CrossRef] [Green Version]
- Betof, A.S.; Lascola, C.D.; Weitzel, D.; Landon, C.; Scarbrough, P.M.; Devi, G.R.; Palmer, G.; Jones, L.W.; Dewhirst, M.W. Modulation of Murine Breast Tumor Vascularity, Hypoxia, and Chemotherapeutic Response by Exercise. J. Natl. Cancer Inst. 2015, 107, djv040. [Google Scholar] [CrossRef] [Green Version]
- Spiliopoulou, P.; Gavriatopoulou, M.; Kastritis, E.; Dimopoulos, M.; Terzis, G. Exercise-Induced Changes in Tumor Growth via Tumor Immunity. Sports 2021, 9, 46. [Google Scholar] [CrossRef]
- Clinton, S.K.; Giovannucci, E.L.; Hursting, S.D. The World Cancer Research Fund/American Institute for Cancer Research Third Expert Report on Diet, Nutrition, Physical Activity, and Cancer: Impact and Future Directions. J. Nutr. 2020, 150, 663–671. [Google Scholar] [CrossRef]
- Ecker, B.L.; Lee, J.Y.; Sterner, C.J.; Solomon, A.C.; Pant, D.K.; Shen, F.; Peraza, J.; Vaught, L.; Mahendra, S.; Belka, G.K.; et al. Impact of obesity on breast cancer recurrence and minimal residual disease. Breast Cancer Res. 2019, 21, 41. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.; Kruper, L.; Dieli-conwright, C.M.; Mortimer, J.E. The Impact of Obesity on Breast Cancer Diagnosis and Treatment. Curr. Oncol. Rep. 2019, 21, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pierce, J.P.; Stefanick, M.L.; Flatt, S.W.; Natarajan, L.; Sternfeld, B.; Al-delaimy, W.K.; Thomson, C.A.; Kealey, S.; Hajek, R.; Parker, A.; et al. Greater Survival After Breast Cancer in Physically Active Women with High Vegetable-Fruit Intake Regardless of Obesity. J. Clin. Oncol. 2008, 25, 2345–2351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cannioto, R.A.; Hutson, A.; Dighe, S.; McCann, W.; McCann, S.E.; Zirpoli, G.R.; Barlow, W.; Kelly, K.M.; DeNysschen, C.A.; Hershman, D.L.; et al. Physical Activity Before, During, and After Chemotherapy for High-Risk Breast Cancer: Relationships with Survival. J. Natl. Cancer Inst. 2021, 113, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, K.; Sakurai, M.; Yamamoto, Y.; Suzuki, E.; Tsuda, M.; Kataoka, T.R.; Hirata, M.; Nishie, M.; Nojiri, T.; Kumazoe, M.; et al. Alteration of specific cytokine expression patterns in patients with breast cancer. Sci. Rep. 2019, 9, 2924. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Lu, P.; Beeraka, N.M.; Sukocheva, O.A.; Madhunapantula, S.V.; Liu, J.; Sinelnikov, M.Y.; Nikolenko, V.N.; Bulygin, K.V.; Mikhaleva, L.M.; et al. Mitochondrial mutations and mitoepigenetics: Focus on regulation of oxidative stress-induced responses in breast cancers. Semin. Cancer Biol. 2020, in press. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Hou, M.-F.; Ou-Yang, F.; Li, C.-L.; Chen, F.-M.; Chuang, C.-H.; Kan, J.-Y.; Wu, C.-C.; Shih, S.-L.; Shiau, J.-P.; Kao, L.-C.; et al. Comprehensive profiles and diagnostic value of menopausal-specific gut microbiota in premenopausal breast cancer. Exp. Mol. Med. 2021, 53, 1636–1646. [Google Scholar] [CrossRef]
- Byrd, D.A.; Vogtmann, E.; Wu, Z.; Han, Y.; Wan, Y.; Clegg-Lamptey, J.N.; Yarney, J.; Wiafe-Addai, B.; Wiafe, S.; Awuah, B.; et al. Associations of fecal microbial profiles with breast cancer and nonmalignant breast disease in the Ghana Breast Health Study. Int. J. Cancer 2021, 148, 2712–2723. [Google Scholar] [CrossRef]
- Goedert, J.J.; Jones, G.; Hua, X.; Xu, X.; Yu, G.; Flores, R.; Falk, R.T.; Gail, M.H.; Shi, J.; Ravel, J.; et al. Investigation of the association between the fecal microbiota and breast cancer in postmenopausal women: A population-based case-control pilot study. J. Natl. Cancer Inst. 2015, 107, djv147. [Google Scholar] [CrossRef]
- Stringer, A.M.; Gibson, R.J.; Bowen, J.M.; Keefe, D.M. Chemotherapy-induced modifications to gastrointestinal microflora: Evidence and implications of change. Curr. Drug Metab. 2009, 10, 79–83. [Google Scholar] [CrossRef]
- Montassier, E.; Gastinne, T.; Vangay, P.; Al-Ghalith, G.A.; Bruley des Varannes, S.; Massart, S.; Moreau, P.; Potel, G.; de La Cochetière, M.F.; Batard, E.; et al. Chemotherapy-driven dysbiosis in the intestinal microbiome. Aliment. Pharmacol. Ther. 2015, 42, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Loman, B.R.; Jordan, K.R.; Haynes, B.; Bailey, M.T.; Pyter, L.M. Chemotherapy-induced neuroinflammation is associated with disrupted colonic and bacterial homeostasis in female mice. Sci. Rep. 2019, 9, 16490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Bastard, Q.; Ward, T.; Sidiropoulos, D.; Hillmann, B.M.; Chun, C.L.; Sadowsky, M.J.; Knights, D.; Montassier, E. Fecal microbiota transplantation reverses antibiotic and chemotherapy-induced gut dysbiosis in mice. Sci. Rep. 2018, 8, 6219. [Google Scholar] [CrossRef] [PubMed]
- Daillère, R.; Vétizou, M.; Waldschmitt, N.; Yamazaki, T.; Isnard, C.; Poirier-Colame, V.; Duong, C.P.M.; Flament, C.; Lepage, P.; Roberti, M.P.; et al. Enterococcus hirae and Barnesiella intestinihominis Facilitate Cyclophosphamide-Induced Therapeutic Immunomodulatory Effects. Immunity 2016, 45, 931–943. [Google Scholar] [CrossRef] [Green Version]
- Heshiki, Y.; Vazquez-Uribe, R.; Li, J.; Ni, Y.; Quainoo, S.; Imamovic, L.; Sørensen, M.; Chow, B.K.C.; Weiss, G.J.; Xu, A.; et al. Predictable modulation of cancer treatment outcomes by the gut microbiota. Microbiome 2020, 8, 28. [Google Scholar] [CrossRef] [Green Version]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Vrieze, A.; Van Nood, E.; Holleman, F.; Salojärvi, J.; Kootte, R.S.; Bartelsman, J.F.W.M.; Dallinga-Thie, G.M.; Ackermans, M.T.; Serlie, M.J.; Oozeer, R.; et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 2012, 143, 913–916.e7. [Google Scholar] [CrossRef]
- Bajic, J.E.; Johnston, I.N.; Howarth, G.S.; Hutchinson, M.R. From the Bottom-Up: Chemotherapy and Gut-Brain Axis Dysregulation. Front. Behav. Neurosci. 2018, 12, 104. [Google Scholar] [CrossRef] [Green Version]
- Barandouzi, Z.A.; Starkweather, A.R.; Henderson, W.A.; Gyamfi, A.; Cong, X.S. Altered Composition of Gut Microbiota in Depression: A Systematic Review. Front. Psychiatry 2020, 11, 541. [Google Scholar] [CrossRef]
- Paulsen, J.A.; Ptacek, T.S.; Carter, S.J.; Liu, N.; Kumar, R.; Hyndman, L.K.; Lefkowitz, E.J.; Morrow, C.D.; Rogers, L.Q. Gut microbiota composition associated with alterations in cardiorespiratory fitness and psychosocial outcomes among breast cancer survivors. Support. Care Cancer 2017, 25, 1563–1570. [Google Scholar] [CrossRef]
- Deleemans, J.M.; Chleilat, F.; Reimer, R.A.; Baydoun, M.; Piedalue, K.-A.; Lowry, D.E.; Henning, J.-W.; Carlson, L.E. The Chemo-Gut Pilot Study: Associations between Gut Microbiota, Gastrointestinal Symptoms, and Psychosocial Health Outcomes in a Cross-Sectional Sample of Young Adult Cancer Survivors. Curr. Oncol. 2022, 29, 2973–2994. [Google Scholar] [CrossRef] [PubMed]
- Okubo, R.; Kinoshita, T.; Katsumata, N.; Uezono, Y.; Xiao, J.; Matsuoka, Y.J. Impact of chemotherapy on the association between fear of cancer recurrence and the gut microbiota in breast cancer survivors. Brain Behav. Immun. 2020, 85, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.D.; Nogueira, L.; Mariotto, A.B.; Rowland, J.H.; Yabroff, K.R.; Alfano, C.M.; Jemal, A.; Kramer, J.L.; Siegel, R.L. Cancer treatment and survivorship statistics, 2019. CA Cancer J. Clin. 2019, 69, 363–385. [Google Scholar] [CrossRef] [Green Version]
- Mcneely, M.L.; Sellar, C.; Williamson, T.; Shea-Budgell, M.; Joy, A.A.; Lau, H.Y.; Easaw, J.C.; Murtha, A.D.; Vallance, J.; Courneya, K.; et al. Community-based exercise for health promotion and secondary cancer prevention in Canada: Protocol for a hybrid effectiveness-implementation study. BMJ Open 2019, 9, e029975. [Google Scholar] [CrossRef]
- Carter, S.J.; Hunter, G.R.; Blackston, J.W.; Liu, N.; Lefkowitz, E.J.; Van Der Pol, W.J.; Morrow, C.D.; Paulsen, J.A.; Rogers, L.Q. Gut microbiota diversity is associated with cardiorespiratory fitness in post-primary treatment breast cancer survivors. Exp. Physiol. 2019, 104, 529–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rashidi, A.; Kaiser, T.; Shields-cutler, R.; Graiziger, C.; Holtan, S.G.; Rehman, T.U.; Wasko, J.; Weisdorf, D.J.; Dunny, G.; Khoruts, A.; et al. Dysbiosis patterns during re- induction/salvage versus induction chemotherapy for acute leukemia. Sci. Rep. 2019, 9, 6083. [Google Scholar] [CrossRef] [PubMed]
- Montassier, E.; Batard, E.; Massart, S.; Gastinne, T.; Carton, T.; Caillon, J.; Fresne, S.L.; Caroff, N.; Hardouin, J.B.; Moreau, P.; et al. 16S rRNA Gene Pyrosequencing Reveals Shift in Patient Faecal Microbiota During High-Dose Chemotherapy as Conditioning Regimen for Bone Marrow Transplantation. Microb. Ecol. 2014, 67, 690–699. [Google Scholar] [CrossRef]
- Godin, G.; Shephard, R.J. A simple method to assess exercise behavior in the community. Can. J. Appl. Sport Sci. 1985, 10, 141–146. [Google Scholar]
- Amireault, S.; Godin, G. The Godin-Shephard Leisure-Time Physical Activity Questionnaire: Validity Evidence Supporting its Use for Classifying Healthy Adults into Active and Insufficiently Active Categories. Percept. Mot. Ski. 2015, 120, 604–622. [Google Scholar] [CrossRef]
- Cella, D.F.; Tulsky, D.S.; Gray, G.; Sarafian, B.; Linn, E.; Bonomi, A.; Silberman, M.; Yellen, S.B.; Winicour, P.; Brannon, J. The Functional Assessment of Cancer Therapy scale: Development and validation of the general measure. J. Clin. Oncol. 1993, 11, 570–579. [Google Scholar] [CrossRef]
- Lambert, J.E.; Parnell, J.A.; Han, J.; Sturzenegger, T.; Paul, H.A.; Vogel, H.J.; Reimer, R.A. Evaluation of yellow pea fibre supplementation on weight loss and the gut microbiota: A randomized controlled trial. BMC Gastroenterol. 2014, 14, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicolucci, A.C.; Hume, M.P.; Martínez, I.; Mayengbam, S.; Walter, J.; Reimer, R.A. Prebiotics Reduce Body Fat and Alter Intestinal Microbiota in Children Who Are Overweight or with Obesity. Gastroenterology 2017, 153, 711–722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bomhof, M.R.; Paul, H.A.; Geuking, M.B.; Eller, L.K.; Reimer, R.A. Improvement in adiposity with oligofructose is modified by antibiotics in obese rats. FASEB J. 2016, 30, 2720–2732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Sugiura, K.; Stock, C.C. Studies in a tumor spectrum.I. Comparison of the action of methylbis(2-chloroethyl)amine and 3-bis(2-chloroethyl)aminomethyl-4-methoxymethyl-5-hydroxy-6-methylpyridine on the growth of a variety of mouse and rat tumors. Cancer 1952, 5, 382–402. [Google Scholar] [CrossRef]
- Abu Samaan, T.M.; Samec, M.; Liskova, A.; Kubatka, P.; Büsselberg, D. Paclitaxel’s Mechanistic and Clinical Effects on Breast Cancer. Biomolecules 2019, 9, 789. [Google Scholar] [CrossRef] [Green Version]
- Cividalli, A.; Arcangeli, G.; Cruciani, G.; Livdi, E.; Cordelli, E.; Danesi, D.T. Enhancement of radiation response by paclitaxel in mice according to different treatment schedules. Int. J. Radiat. Oncol. Biol. Phys. 1998, 40, 1163–1170. [Google Scholar] [CrossRef]
- Seidman, A.D.; Berry, D.; Cirrincione, C.; Harris, L.; Muss, H.; Marcom, P.K.; Gipson, G.; Burstein, H.; Lake, D.; Shapiro, C.L.; et al. Randomized phase III trial of weekly compared with every-3-weeks paclitaxel for metastatic breast cancer, with trastuzumab for all HER-2 overexpressors and random assignment to trastuzumab or not in HER-2 nonoverexpressors: Final results of Cancer and Leukemia Group B protocol 9840. J. Clin. Oncol. 2008, 26, 1642–1649. [Google Scholar] [CrossRef] [Green Version]
- Reagan-Shaw, S.; Nihal, M.; Ahmad, N. Dose translation from animal to human studies revisited. FASEB J. 2008, 22, 659–661. [Google Scholar] [CrossRef] [Green Version]
- Van Der Meulen, R.; Makras, L.; Verbrugghe, K.; Adriany, T.; De Vuyst, L. In Vitro Kinetic Analysis of Oligofructose Consumption by Bacteroides and Bifidobacterium spp. Indicates Different Degradation Mechanisms. Appl. Environ. Microbiol. 2006, 72, 1006–1012. [Google Scholar] [CrossRef] [Green Version]
- Bachmanov, A.A.; Reed, D.R.; Beauchamp, G.K.; Tordoff, M.G. Food intake, water intake, and drinking spout side preference of 28 mouse strains. Behav. Genet. 2002, 32, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Euhus, D.M.; Hudd, C.; LaRegina, M.C.; Johnson, F.E. Tumor measurement in the nude mouse. J. Surg. Oncol. 1986, 31, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Parnell, J.A.; Reimer, R.A. Differential Secretion of Satiety Hormones with Progression of Obesity in JCR:LA-corpulent Rats. Obesity 2008, 16, 736–742. [Google Scholar] [CrossRef] [PubMed]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef] [Green Version]
- O’Callaghan, A.; van Sinderen, D. Bifidobacteria and Their Role as Members of the Human Gut Microbiota. Front. Microbiol. 2016, 7, 925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira-Halder, C.V.; Faria, A.V.S.; Andrade, S.S. Action and function of Faecalibacterium prausnitzii in health and disease. Best Pract. Res. Clin. Gastroenterol. 2017, 31, 643–648. [Google Scholar] [CrossRef]
- Pryde, S.E.; Duncan, S.H.; Hold, G.L.; Stewart, C.S.; Flint, H.J. The microbiology of butyrate formation in the human colon. FEMS Microbiol. Lett. 2002, 217, 133–139. [Google Scholar] [CrossRef]
- Tamanai-Shacoori, Z.; Smida, I.; Bousarghin, L.; Loreal, O.; Meuric, V.; Fong, S.B.; Bonnaure-Mallet, M.; Jolivet-Gougeon, A. Roseburia spp.: A marker of health? Future Microbiol. 2017, 12, 157–170. [Google Scholar] [CrossRef]
- Baldelli, V.; Scaldaferri, F.; Putignani, L.; Del Chierico, F. The Role of Enterobacteriaceae in Gut Microbiota Dysbiosis in Inflammatory Bowel Diseases. Microorganisms 2021, 9, 697. [Google Scholar] [CrossRef]
- Lee, I.-A.; Kim, D.-H. Klebsiella pneumoniaeincreases the risk of inflammation and colitis in a murine model of intestinal bowel disease. Scand. J. Gastroenterol. 2011, 46, 684–693. [Google Scholar] [CrossRef]
- Mirsepasi-Lauridsen, H.C.; Vallance, B.A.; Krogfelt, K.A.; Petersen, A.M. Escherichia coli Pathobionts Associated with Inflammatory Bowel Disease. Clin. Microbiol. Rev. 2019, 32, e00060-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coussens, L.M.; Werb, Z. Inflammation and cancer. Nature 2002, 420, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Hakansson, A.; Molin, G. Gut Microbiota and Inflammation. Nutrients 2011, 3, 637–682. [Google Scholar] [CrossRef] [PubMed]
- Muscogiuri, G.; Cantone, E.; Cassarano, S.; Tuccinardi, D.; Barrea, L.; Savastano, S. Gut microbiota: A new path to treat obesity. Int. J. Obes. Suppl. 2019, 9, 10–19. [Google Scholar] [CrossRef]
- Bengmark, S. Gut microbiota, immune development and function. Pharmacol. Res. 2013, 69, 87–113. [Google Scholar] [CrossRef]
- Ortiz-Alvarez, L.; Xu, H.; Martinez-Tellez, B. Influence of Exercise on the Human Gut Microbiota of Healthy Adults: A Systematic Review. Clin. Transl. Gastroenterol. 2020, 11, e00126. [Google Scholar] [CrossRef]
- Allen, J.M.; Mailing, L.J.; Niemiro, G.M.; Moore, R.; Cook, M.D.; White, B.A.; Holscher, H.D.; Woods, J.A. Exercise Alters Gut Microbiota Composition and Function in Lean and Obese Humans. Med. Sci. Sports Exerc. 2018, 50, 747–757. [Google Scholar] [CrossRef]
- Munukka, E.; Ahtiainen, J.P.; Puigbó, P.; Jalkanen, S.; Pahkala, K.; Keskitalo, A.; Kujala, U.M.; Pietilä, S.; Hollmén, M.; Elo, L.; et al. Six-week endurance exercise alters gut metagenome that is not reflected in systemic metabolism in over-weight women. Front. Microbiol. 2018, 9, 2323. [Google Scholar] [CrossRef]
- Wan, Y.; Yuan, J.; Li, J.; Li, H.; Yin, K.; Wang, F.; Li, D. Overweight and underweight status are linked to specific gut microbiota and intestinal tricarboxylic acid cycle intermediates. Clin. Nutr. 2020, 39, 3189–3198. [Google Scholar] [CrossRef]
- Rothschild, D.; Weissbrod, O.; Barkan, E.; Kurilshikov, A.; Korem, T.; Zeevi, D.; Costea, P.I.; Godneva, A.; Kalka, I.N.; Bar, N.; et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018, 555, 210–215. [Google Scholar] [CrossRef]
- Campbell, S.C.; Wisniewski, P.J.; Noji, M.; McGuinness, L.R.; Häggblom, M.M.; Lightfoot, S.A.; Joseph, L.B.; Kerkhof, L.J. The Effect of Diet and Exercise on Intestinal Integrity and Microbial Diversity in Mice. PLoS ONE 2016, 11, e0150502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durk, R.P.; Castillo, E.; Márquez-Magaña, L.; Grosicki, G.J.; Bolter, N.D.; Matthew Lee, C.; Bagley, J.R. Gut microbiota composition is related to cardiorespiratory fitness in healthy young adults. Int. J. Sport Nutr. Exerc. Metab. 2019, 29, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Bressa, C.; Bailén-Andrino, M.; Pérez-Santiago, J.; González-Soltero, R.; Pérez, M.; Montalvo-Lominchar, M.G.; Maté-Muñoz, J.L.; Domínguez, R.; Moreno, D.; Larrosa, M. Differences in gut microbiota profile between women with active lifestyle and sedentary women. PLoS ONE 2017, 12, e0171352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garrett, W.S.; Gallini, C.A.; Yatsunenko, T.; Michaud, M.; Dubois, A.; Delaney, M.L.; Punit, S.; Karlsson, M.; Bry, L.; Glickman, J.N.; et al. Enterobacteriaceae Act in Concert with the Gut Microbiota to Induce Spontaneous and Maternally Transmitted Colitis. Cell Host Microbe 2010, 8, 292–300. [Google Scholar] [CrossRef] [Green Version]
- Pope, J.L.; Yang, Y.; Newsome, R.C.; Sun, W.; Sun, X.; Ukhanova, M.; Neu, J.; Issa, J.-P.; Mai, V.; Jobin, C. Microbial Colonization Coordinates the Pathogenesis of a Klebsiella pneumoniae Infant Isolate. Sci. Rep. 2019, 9, 3380. [Google Scholar] [CrossRef] [Green Version]
- Mckee, A.M.; Kirkup, B.M.; Madgwick, M.; Fowler, W.J.; Price, C.A.; Dreger, S.A.; Ansorge, R.; Makin, K.A.; Caim, S.; Le Gall, G.; et al. Antibiotic-induced disturbances of the gut microbiota result in accelerated breast tumor growth. iScience 2021, 24, 103012. [Google Scholar] [CrossRef]
- Markou, P.; Apidianakis, Y. Pathogenesis of intestinal Pseudomonas aeruginosa infection in patients with cancer. Front. Cell Infect Microbiol 2014, 3, 115. [Google Scholar] [CrossRef]
- Wennerberg, E.; Lhuillier, C.; Rybstein, M.D.; Dannenberg, K.; Rudqvist, N.-P.; Koelwyn, G.J.; Jones, L.W.; Demaria, S. Exercise reduces immune suppression and breast cancer progression in a preclinical model. Oncotarget 2020, 11, 452–461. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.K.; Kim, Y.; Park, S.; Kim, E.; Kim, Y.; Kim, Y.; Kim, J.-H. Effects of Steady Low-Intensity Exercise on High-Fat Diet Stimulated Breast Cancer Progression Via the Alteration of Macrophage Polarization. Integr. Cancer Ther. 2020, 19, 153473542094967. [Google Scholar] [CrossRef]
- Walsh, N.P.; Gleeson, M.; Shephard, R.J.; Woods, J.A.; Bishop, N.C.; Fleshner, M.; Green, C.; Pedersen, B.K.; Hoffman-Goetz, L.; Rogers, C.J.; et al. Position statement. Part one: Immune function and exercise. Exerc. Immunol. Rev. 2011, 17, 6–63. [Google Scholar]
- Taper, H.S.; Roberfroid, M. Influence of inulin and oligofructose on breast cancer and tumor growth. J. Nutr. 1999, 129 (Suppl. S7), 1488S–1491S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taper, H.S.; Roberfroid, M.B. Inulin/oligofructose and anticancer therapy. Br. J. Nutr. 2002, 87 (Suppl. S2), S283–S286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tandon, D.; Haque, M.M.; Gote, M.; Jain, M.; Bhaduri, A.; Dubey, A.K.; Mande, S.S. A prospective randomized, double-blind, placebo-controlled, dose-response relationship study to investigate efficacy of fructo-oligosaccharides (FOS) on human gut microflora. Sci. Rep. 2019, 9, 5473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reimer, R.A.; Willis, H.J.; Tunnicliffe, J.M.; Park, H.; Madsen, K.L.; Soto-Vaca, A. Inulin-type fructans and whey protein both modulate appetite but only fructans alter gut microbiota in adults with overweight/obesity: A randomized controlled trial. Mol. Nutr. Food Res. 2017, 61, 1700484. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Li, D.; Chen, Q.; Li, M.; Su, L.; Luo, T.; Liang, D.; Lai, G.; Shuai, O.; Jiao, C.; et al. Anti-breast Cancer Enhancement of a Polysaccharide from Spore of Ganoderma lucidum with Paclitaxel: Suppression on Tumor Metabolism with Gut Microbiota Reshaping. Front. Microbiol. 2018, 9, 3099. [Google Scholar] [CrossRef] [PubMed]
- Terrisse, S.; Derosa, L.; Iebba, V.; Ghiringhelli, F.; Vaz-Luis, I.; Kroemer, G.; Fidelle, M.; Christodoulidis, S.; Segata, N.; Thomas, A.M.; et al. Intestinal microbiota influences clinical outcome and side effects of early breast cancer treatment. Cell Death Differ. 2021, 28, 2778–2796. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishna, C.; Corleto, J.; Ruegger, P.M.; Logan, G.D.; Peacock, B.B.; Mendonca, S.; Yamaki, S.; Adamson, T.; Ermel, R.; McKemy, D.; et al. Dominant Role of the Gut Microbiota in Chemotherapy Induced Neuropathic Pain. Sci. Rep. 2019, 9, 20324. [Google Scholar] [CrossRef] [Green Version]
- Engels, C.; Ruscheweyh, H.J.; Beerenwinkel, N.; Lacroix, C.; Schwab, C. The Common Gut Microbe Eubacterium hallii also Contributes to Intestinal Propionate Formation. Front. Microbiol. 2016, 7, 713. [Google Scholar] [CrossRef] [Green Version]
- Kleessen, B.; Sykura, B.; Zunft, H.J.; Blaut, M. Effects of inulin and lactose on fecal microflora, microbial activity, and bowel habit in elderly constipated persons. Am. J. Clin. Nutr. 1997, 65, 1397–1402. [Google Scholar] [CrossRef]
- Frei, R.; Akdis, M.; O’Mahony, L. Prebiotics, probiotics, synbiotics, and the immune system. Curr. Opin. Gastroenterol. 2015, 31, 153–158. [Google Scholar] [CrossRef] [Green Version]
- Di Modica, M.; Gargari, G.; Regondi, V.; Bonizzi, A.; Arioli, S.; Belmonte, B.; De Cecco, L.; Fasano, E.; Bianchi, F.; Bertolotti, A.; et al. Gut Microbiota Condition the Therapeutic Efficacy of Trastuzumab in HER2-Positive Breast Cancer. Cancer Res. 2021, 81, 2195–2206. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, O.I.; Adel, A.M.; Diab, D.R.; Gobran, N.S. Prognostic value of serum level of interleukin-6 and interleukin-8 in metastatic breast cancer patients. Egypt. J. Immunol. 2006, 13, 61–68. [Google Scholar] [PubMed]
- Yoshimura, T. The chemokine MCP-1 (CCL2) in the host interaction with cancer: A foe or ally? Cell. Mol. Immunol. 2018, 15, 335–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehta, A.K.; Kadel, S.; Townsend, M.G.; Oliwa, M.; Guerriero, J.L. Macrophage Biology and Mechanisms of Immune Suppression in Breast Cancer. Front. Immunol. 2021, 12, 643771. [Google Scholar] [CrossRef]
- Barr, M.; Bouchier-Hayes, D.; Harmey, J. Vascular endothelial growth factor is an autocrine survival factor for breast tumour cells under hypoxia. Int. J. Oncol. 2008, 32, 41–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mercurio, A.M.; Lipscomb, E.A.; Bachelder, R.E. Non-angiogenic functions of VEGF in breast cancer. J. Mammary Gland. Biol. Neoplasia 2005, 10, 283–290. [Google Scholar] [CrossRef]
- Liang, Y.; Brekken, R.A.; Hyder, S.M. Vascular endothelial growth factor induces proliferation of breast cancer cells and inhibits the anti-proliferative activity of anti-hormones. Endocr. Relat. Cancer 2006, 13, 905–919. [Google Scholar] [CrossRef]
- Wu, X.; Sun, A.; Yu, W.; Hong, C.; Liu, Z. CXCL10 mediates breast cancer tamoxifen resistance and promotes estrogen-dependent and independent proliferation. Mol. Cell. Endocrinol. 2020, 512, 110866. [Google Scholar] [CrossRef]
- Kim, M.; Choi, H.Y.; Woo, J.W.; Chung, Y.R.; Park, S.Y. Role of CXCL10 in the progression of in situ to invasive carcinoma of the breast. Sci. Rep. 2021, 11, 18007. [Google Scholar] [CrossRef]



| Inclusion Criteria | Exclusion Criteria |
|---|---|
|
|
|
|
|
|
| |
| |
| |
|
| Gene | Forward Sequence | Reverse Sequence |
|---|---|---|
| ZO-1 (zonula occludens) | AGGGGCAGTGGTGGTTTTCTGGTTCTTTC | GCAGAGGTCAAAGTTCAAGGCTAAGAGG |
| Occludin | TCAGGGAATATCCACCTATCACTTCAG | CATCAGCAGCAGCCATGTACTCTTCAC |
| Claudin-3 | CACCGCACCATCACCACTAC | CTTCCAGCCTAGCAAGCAGAC |
| Demographic | Mean | Frequency | Percent | |
|---|---|---|---|---|
| Age | Average Age (years) | 57.9 ± 2.79 | ||
| Under 65 | 8 | 80 | ||
| Over 65 | 2 | 20 | ||
| Education | Some University | 3 | 20 | |
| Completed University | 6 | 60 | ||
| Some Graduate School | 1 | 10 | ||
| Annual Income | Between $20,000–39,999 | 2 | 20 | |
| Between $40,000–59,999 | 1 | 10 | ||
| Between $60,000–79,999 | 1 | 10 | ||
| Between $80,000–99,999 | 2 | 20 | ||
| Over $99,999 | 4 | 40 | ||
| Ethnic Background | Britain | 4 | 40 | |
| (May Report >1) | Western Europe | 2 | 20 | |
| Eastern Europe | 4 | 40 | ||
| Northern Europe | 3 | 30 | ||
| Southern Europe | 2 | 20 | ||
| Asia | 2 | 20 | ||
| Smoking Status | Never | 6 | 60 | |
| Previously | 4 | 40 | ||
| Alcohol Consumption | Never | 2 | 20 | |
| Previously | 1 | 10 | ||
| Occasionally | 5 | 50 | ||
| Socially | 1 | 10 | ||
| Regularly | 1 | 10 |
| Clinical Characteristic | Mean | Frequency | Percent | |
|---|---|---|---|---|
| Completed Treatments | Chemotherapy | 10 | 100 | |
| Surgery | 10 | 100 | ||
| Radiation Therapy | 8 | 80 | ||
| Hormone Therapy | 1 | 10 | ||
| Current Treatments | Hormone Therapy | 5 | 50 | |
| Zoledronic Acid Infusions | 1 | 10 | ||
| Body mass index | Underweight BMI (<18.5) | 1 | 10 | |
| Healthy BMI (18.5–24.9) | 2 | 20 | ||
| Overweight BMI (25–29.9) | 4 | 40 | ||
| Obese BMI (≥30) | 3 | 30 |
| Exercise Category & Week | MET Hours/Week | p Value 0 to 12 Weeks | p Value 12 to 24 Weeks | p Value 0 to 24 Weeks |
|---|---|---|---|---|
| Total 0 | 18.4 ± 4.2 | 0.002 | ||
| Total 12 | 33.6 ± 5.2 | 0.535 | ||
| Total 24 | 38.1 ± 7.7 | 0.030 | ||
| Strenuous 0 | 4.7 ± 2.6 | 0.016 | ||
| Strenuous 12 | 14.2 ± 3.8 | 0.280 | ||
| Strenuous 24 | 20.5 ± 7.0 | 0.075 | ||
| Moderate 0 | 8.8 ± 2.2 | 0.301 | ||
| Moderate 12 | 12.2 ± 3.2 | 0.452 | ||
| Moderate 24 | 10.2 ± 1.9 | 0.626 | ||
| Mild 0 | 4.8 ± 0.8 | 0.072 | ||
| Mild 12 | 7.1 ± 1.5 | 0.891 | ||
| Mild 24 | 7.3 ± 1.5 | 0.150 | ||
| Minutes/week | ||||
| Resistance 0 | 34.4 ± 20.4 | 0.113 | ||
| Resistance 12 | 69.4 ± 22.0 | 0.767 | ||
| Resistance 24 | 63.3 ± 17.1 | 0.224 | ||
| Flexibility 0 | 45.5 ± 11.5 | 0.276 | ||
| Flexibility 12 | 71.1 ± 21.3 | 0.816 | ||
| Flexibility 24 | 74.4 ± 16.1 | 0.078 |
| Nutritional Measure | Time Point | Daily Average | Consecutive Time Point p Value |
|---|---|---|---|
| Calories (kcal) | Baseline | 2069.3 ± 188.8 | |
| 12 weeks | 2260.2 ± 114.9 | 0.404 | |
| 24 weeks | 1785.2 ± 196.9 | 0.017 | |
| Protein (g) | Baseline | 79.5 ± 7.9 | |
| 12 weeks | 87.3 ± 5.0 | 0.352 | |
| 24 weeks | 73.7 ± 6.9 | 0.125 | |
| Carbohydrate (g) | Baseline | 232.3 ± 20.9 | |
| 12 weeks | 234.3 ± 10.8 | 0.936 | |
| 24 weeks | 210.4 ± 21.2 | 0.359 | |
| Total Fat (g) | Baseline | 92.6 ± 11.2 | |
| 12 weeks | 107.4 ± 9.0 | 0.338 | |
| 24 weeks | 74.1 ± 12.9 | 0.012 | |
| Polyunsaturated Fat (g) | Baseline | 17.7 ± 1.9 | |
| 12 weeks | 24.1 ± 3.2 | 0.244 | |
| 24 weeks | 13.1 ± 2.0 | 0.004 | |
| Monounsaturated Fat (g) | Baseline | 30.5 ± 3.5 | |
| 12 weeks | 38.1 ± 3.2 | 0.153 | |
| 24 weeks | 24.6 ± 3.9 | <0.001 | |
| Saturated Fat | Baseline | 29.9 ± 3.9 | |
| 12 weeks | 36.0 ± 5.3 | 0.387 | |
| 24 weeks | 22.6 ± 3.9 | 0.057 | |
| Fiber (g) | Baseline | 23.9 ± 4.4 | |
| 12 weeks | 29.7 ± 4.9 | 0.352 | |
| 24 weeks | 27.8 ± 6.2 | 0.478 | |
| Vitamin E (mg) | Baseline | 8.7 ± 1.5 | |
| 12 weeks | 10.9 ± 1.3 | 0.365 | |
| 24 weeks | 6.6 ± 1.2 | 0.029 | |
| Selenium (mcg) | Baseline | 109.9 ± 8.9 | |
| 12 weeks | 121.5 ± 8.9 | 0.466 | |
| 24 weeks | 100.9 ± 8.0 | 0.021 |
| FACT-G Total Score (108 Max) | Timepoint | Consecutive Timepoint p Value | Median Score | Sample Size | Completion% |
|---|---|---|---|---|---|
| 84.6 ± 5.2 | Baseline | 87.5 | N = 10 | 100 | |
| 84.9 ± 4.8 | 12 Weeks | 0.869 | 92 | N = 9 | 90 |
| 81.4 ± 5.3 | 24 Weeks | 0.302 | 82 | N = 9 | 90 |
| Well-being Category | Timepoint | Categorical Score | Consecutive timepoint p value | Median Score | |
| Physical | Baseline | 23.9 ± 1.3 | 25.0 | ||
| 12 weeks | 24.7 ± 0.9 | 0.560 | 24.0 | ||
| 24 weeks | 24.9 ± 0.7 | 0.816 | 25.0 | ||
| Social | Baseline | 23.7 ± 1.2 | 23.5 | ||
| 12 weeks | 23.2 ± 1.4 | 0.688 | 22.0 | ||
| 24 weeks | 22.4 ± 1.4 | 0.065 | 21.0 | ||
| Emotional | Baseline | 16.4 ± 0.9 | 17.0 | ||
| 12 weeks | 16.2 ± 1.1 | 0.327 | 21.0 | ||
| 24 weeks | 14.9 ± 1.6 | 0.291 | 14.0 | ||
| Functional | Baseline | 21.2 ± 2.0 | 20.0 | ||
| 12 weeks | 18.3 ± 1.5 | 0.091 | 18.0 | ||
| 24 weeks | 20.4 ± 1.7 | 0.082 | 21.0 | ||
| Cytokine | Spearman’s Correlation Coefficient | Significance |
|---|---|---|
| IL-10 | 0.434 | 0.030 |
| KC | 0.530 | 0.001 |
| LIF | 0.500 | 0.004 |
| MIP-2 | 0.529 | 0.002 |
| VEGF | 0.392 | 0.022 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sampsell, K.; Wang, W.; Ohland, C.; Mager, L.F.; Pett, N.; Lowry, D.E.; Sales, K.M.; McNeely, M.L.; McCoy, K.D.; Culos-Reed, S.N.; et al. Exercise and Prebiotic Fiber Provide Gut Microbiota-Driven Benefit in a Survivor to Germ-Free Mouse Translational Model of Breast Cancer. Cancers 2022, 14, 2722. https://doi.org/10.3390/cancers14112722
Sampsell K, Wang W, Ohland C, Mager LF, Pett N, Lowry DE, Sales KM, McNeely ML, McCoy KD, Culos-Reed SN, et al. Exercise and Prebiotic Fiber Provide Gut Microbiota-Driven Benefit in a Survivor to Germ-Free Mouse Translational Model of Breast Cancer. Cancers. 2022; 14(11):2722. https://doi.org/10.3390/cancers14112722
Chicago/Turabian StyleSampsell, Kara, Weilan Wang, Christina Ohland, Lukas F. Mager, Nicola Pett, Dana E. Lowry, Kate M. Sales, Margaret L. McNeely, Kathy D. McCoy, S. Nicole Culos-Reed, and et al. 2022. "Exercise and Prebiotic Fiber Provide Gut Microbiota-Driven Benefit in a Survivor to Germ-Free Mouse Translational Model of Breast Cancer" Cancers 14, no. 11: 2722. https://doi.org/10.3390/cancers14112722
APA StyleSampsell, K., Wang, W., Ohland, C., Mager, L. F., Pett, N., Lowry, D. E., Sales, K. M., McNeely, M. L., McCoy, K. D., Culos-Reed, S. N., & Reimer, R. A. (2022). Exercise and Prebiotic Fiber Provide Gut Microbiota-Driven Benefit in a Survivor to Germ-Free Mouse Translational Model of Breast Cancer. Cancers, 14(11), 2722. https://doi.org/10.3390/cancers14112722