Cytoplasmic Clusterin Suppresses Lung Cancer Metastasis by Inhibiting the ROCK1-ERK Axis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
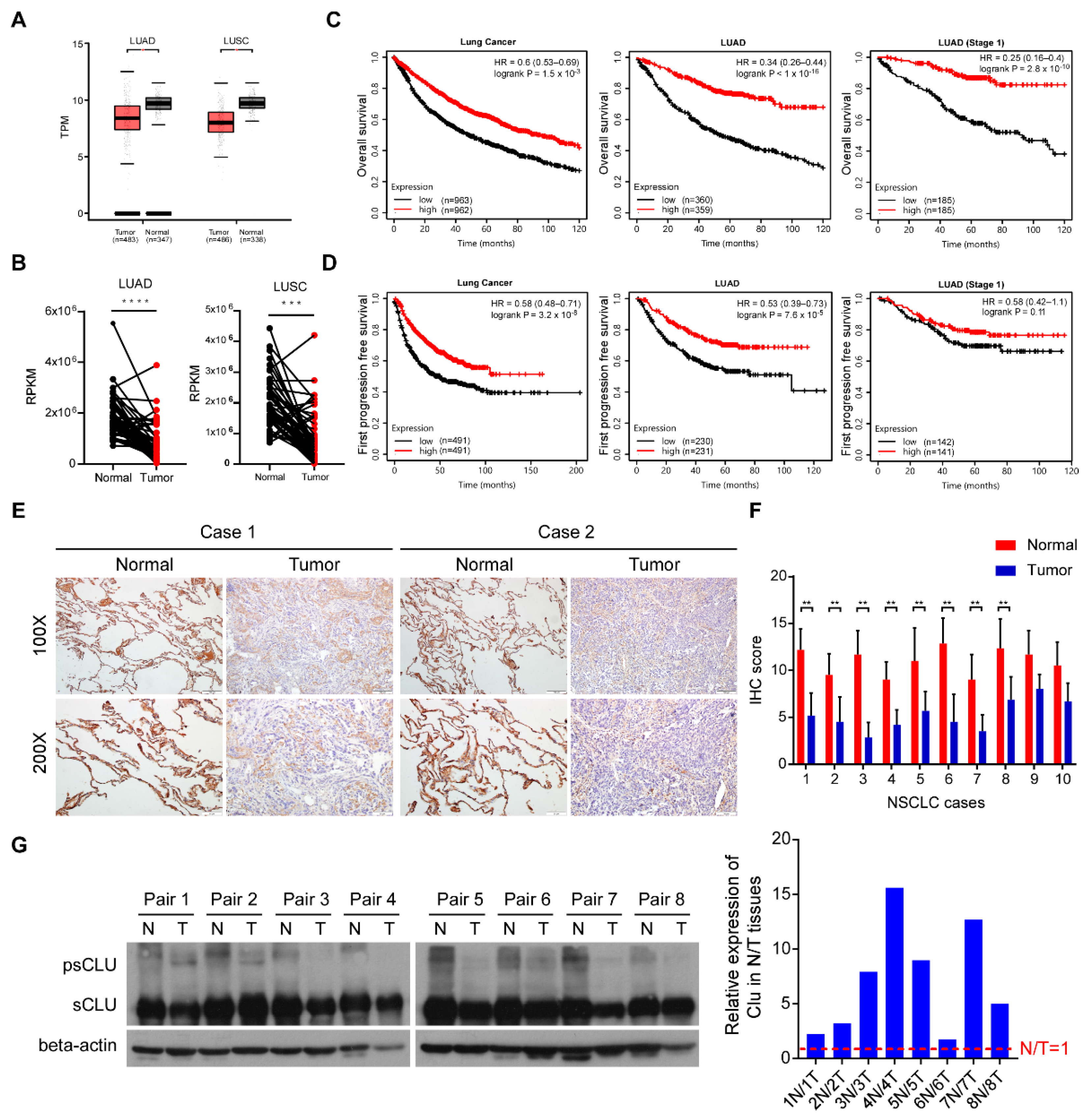
2.1. Tissue Specimens and Immunohistochemistry
2.2. Cell Culture and Recombinant Plasmid Transfection
2.3. Antibodies and Reagents
2.4. Cell Migration and Invasion and Transwell Co-Culture Assays
2.5. Wound-Healing Assay
2.6. Quantification of CLU by ELISA
2.7. Nuclear and Cytoplasmic Protein Extraction Assay
2.8. Immunofluorescence and Proximity Ligation Assay (PLA)
2.9. Co-Immunoprecipitation and Mass Spectrometry Analysis
2.10. Animal Models
2.11. Statistical Analysis
3. Results
3.1. CLU Downregulation in Lung Cancer Correlates with Poor Patient Survival
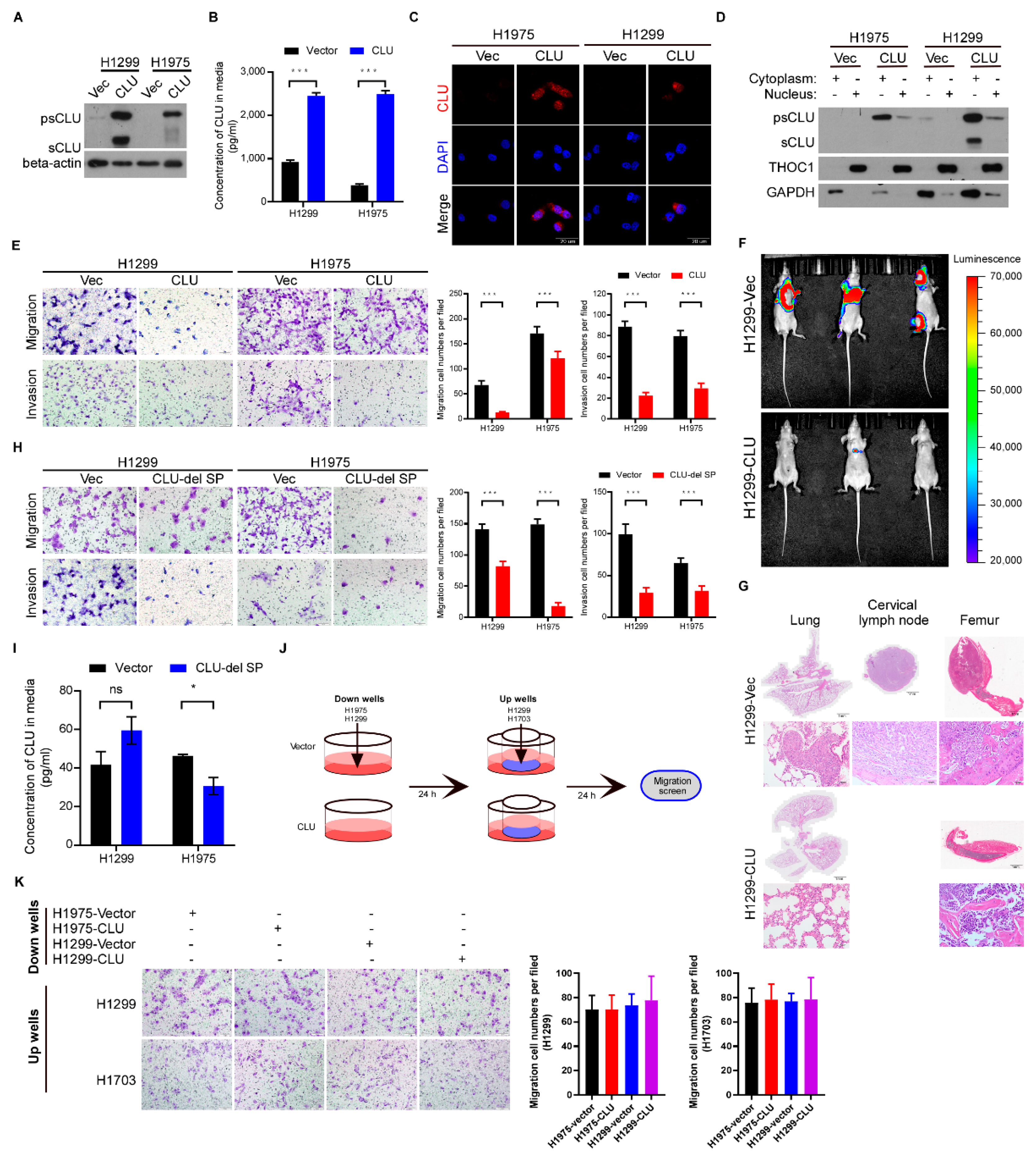
3.2. CLU Especially Cytoplasmic CLU Suppresses Invasion, Migration and Metastasis
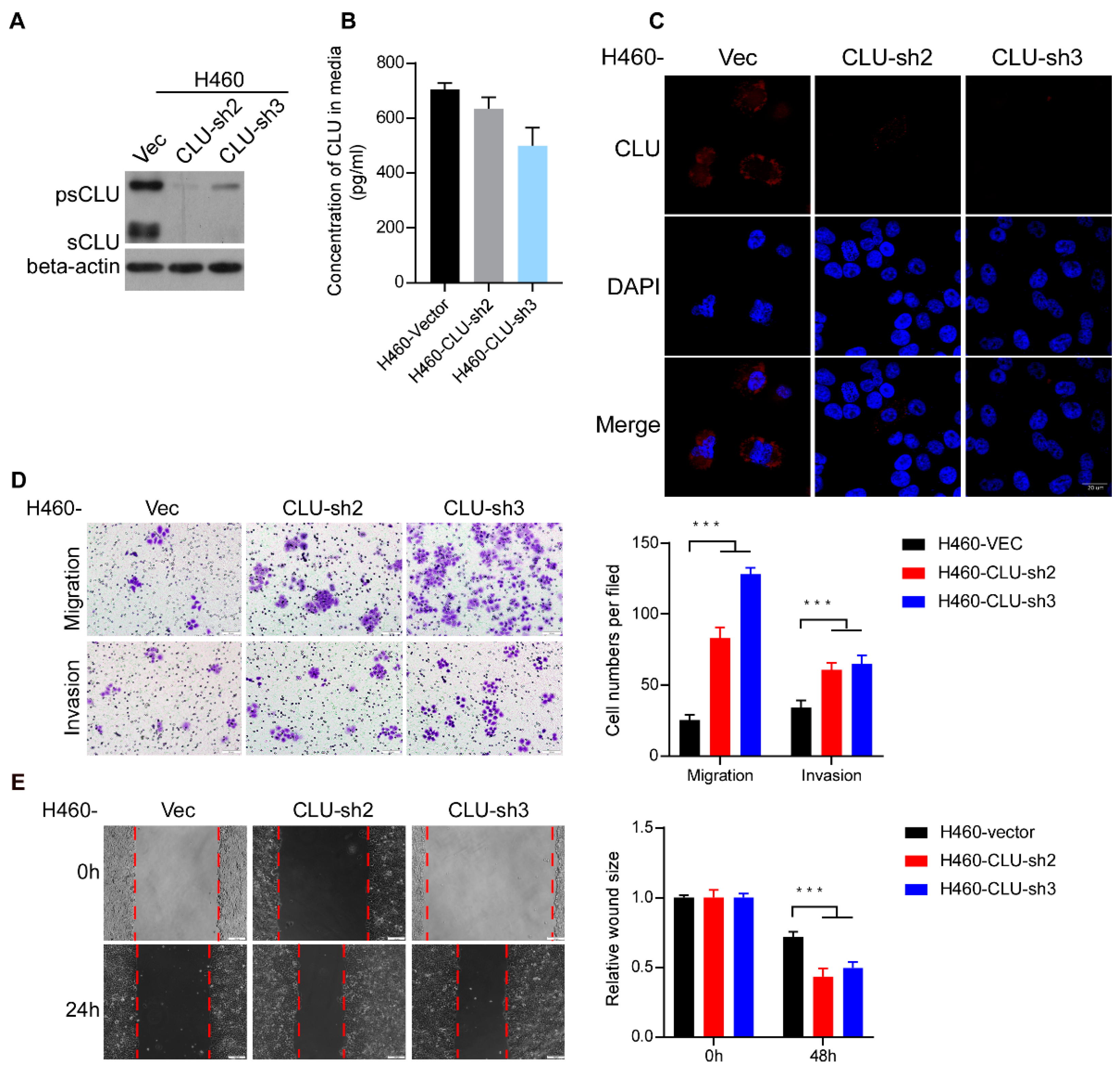
3.3. Silencing CLU Promotes Lung Cancer Invasion and Migration
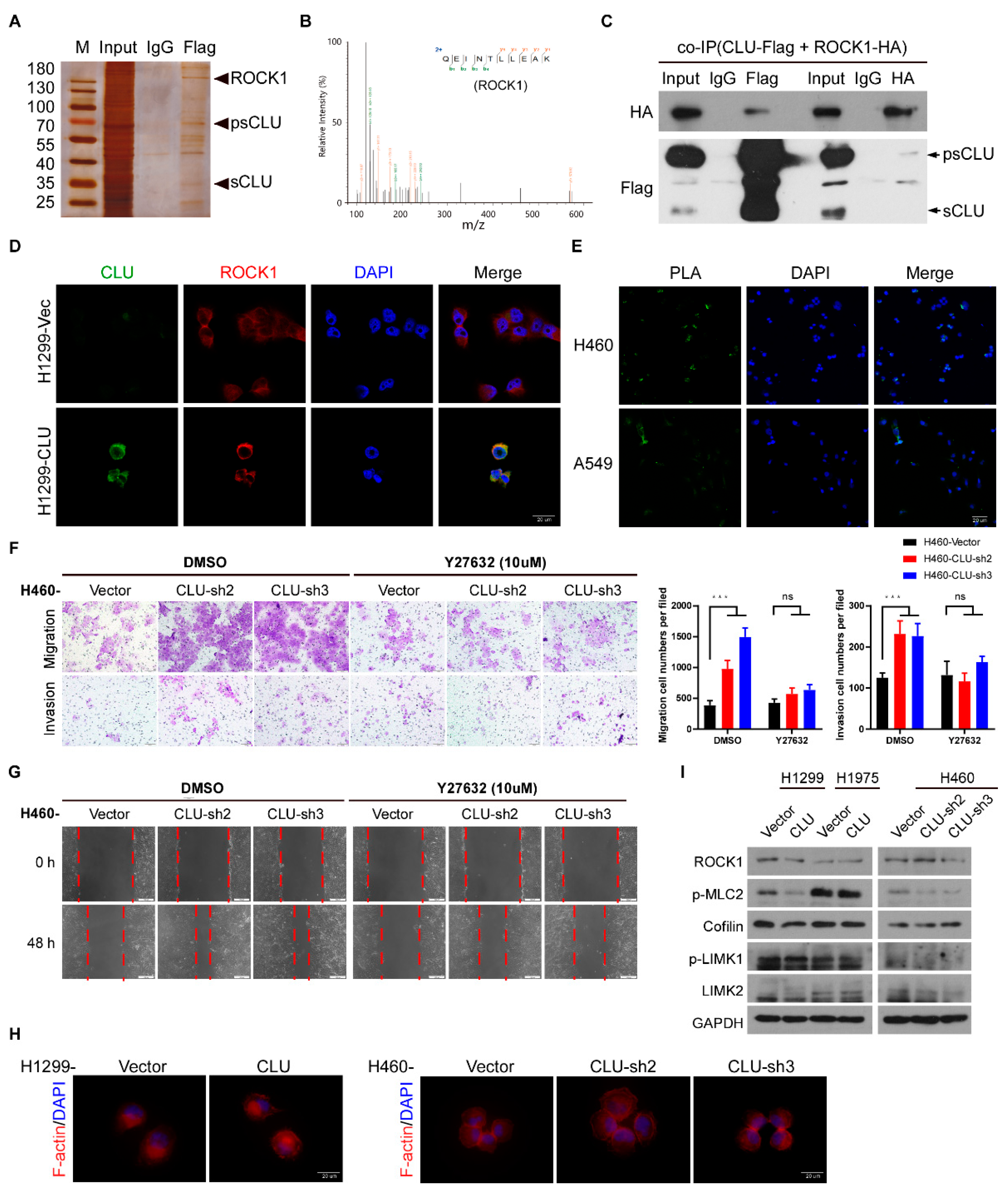
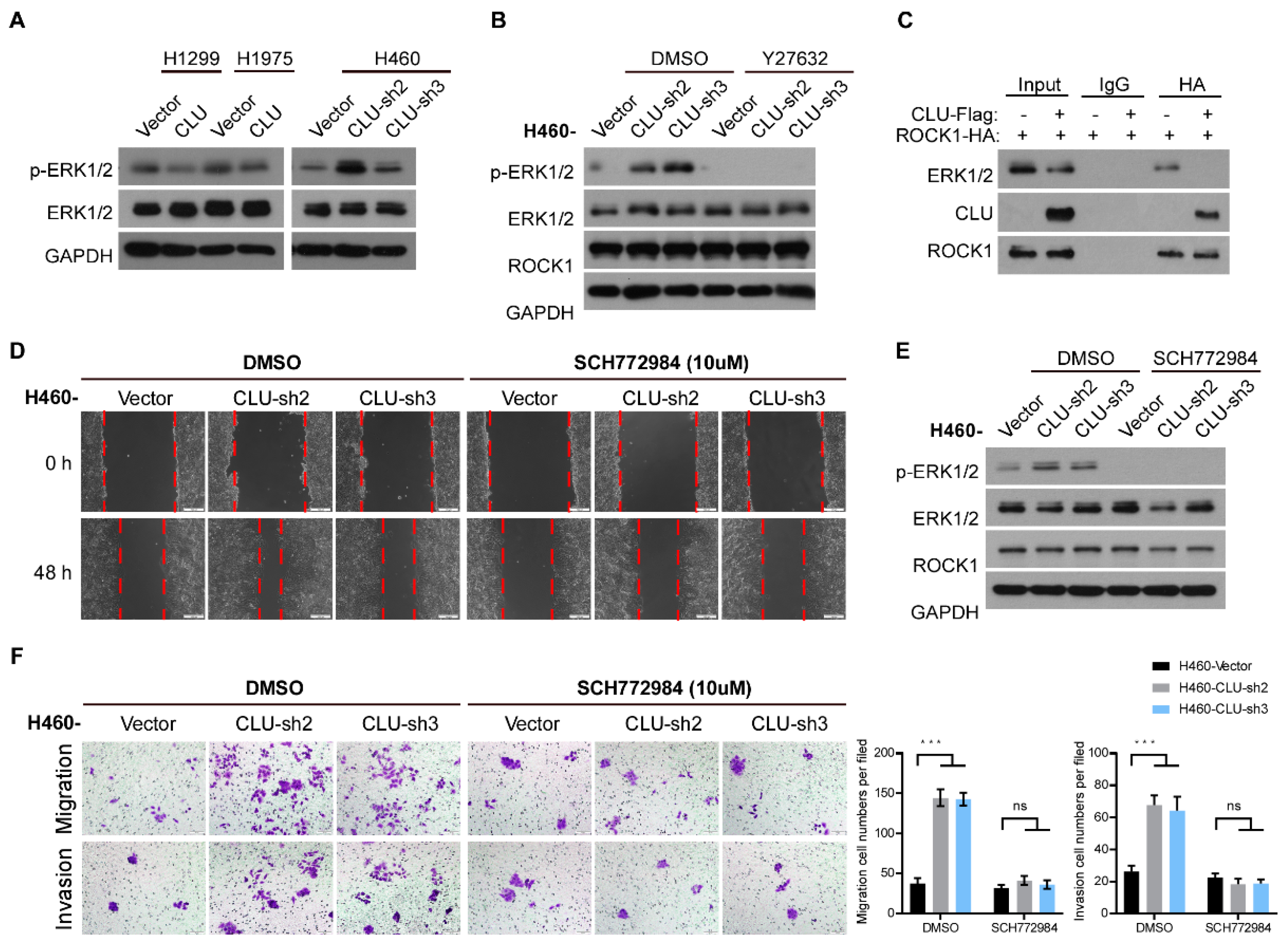
3.4. CLU Interacts with ROCK1 to Suppress Metastatic Traits
3.5. CLU Inhibits Metastasis through the ROCK1/ERK Axis
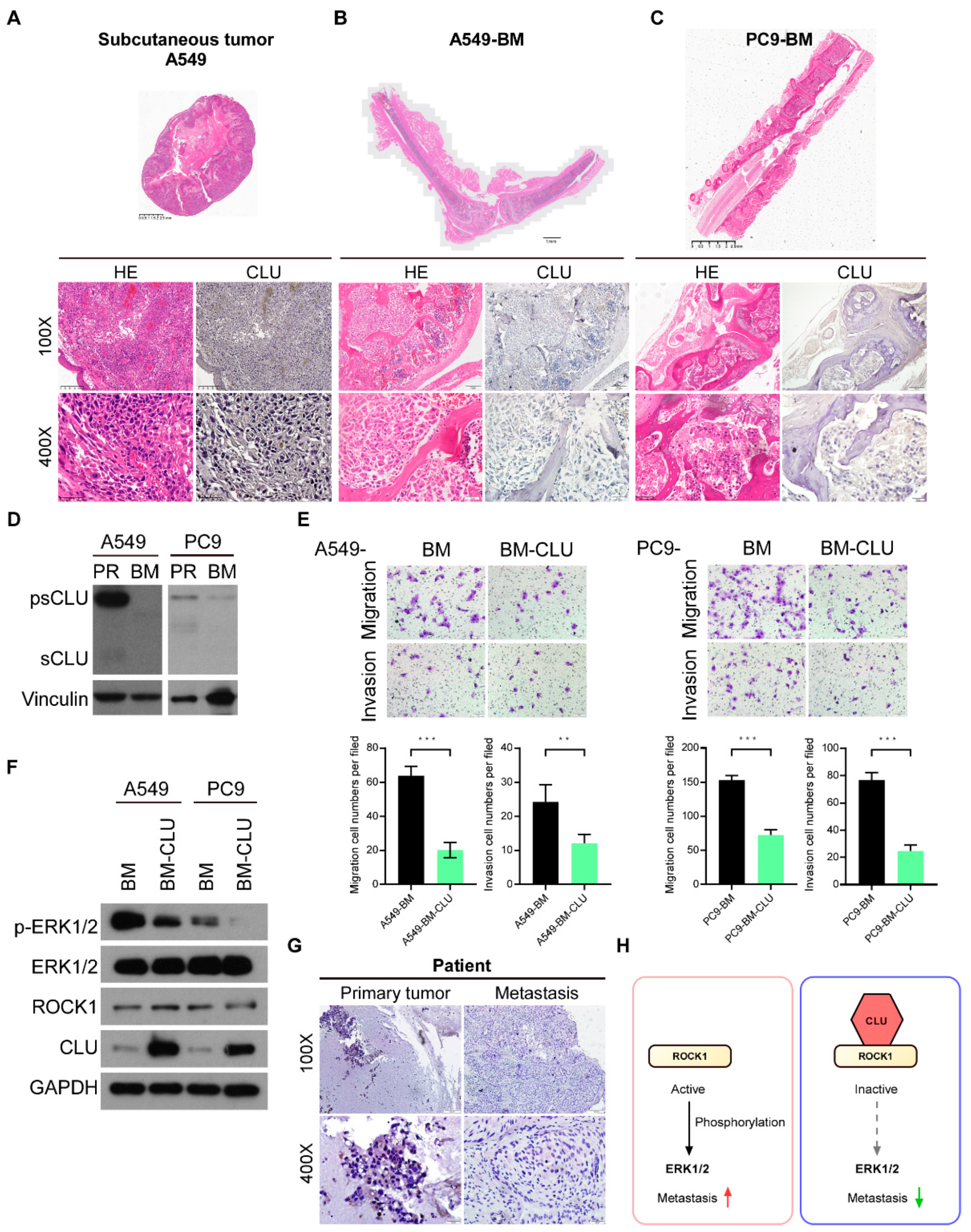
3.6. CLU Is Negatively Related to Metastasis in Models and Tissues
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Molina, J.R.; Yang, P.; Cassivi, S.D.; Schild, S.E.; Adjei, A.A. Non-small cell lung cancer: Epidemiology, risk factors, treatment, and survivorship. Mayo Clin. Proc. 2008, 83, 584–594. [Google Scholar] [CrossRef]
- Wan, L.; Pantel, K.; Kang, Y. Tumor metastasis: Moving new biological insights into the clinic. Nat. Med. 2013, 19, 1450–1464. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S.; Morgensztern, D.; Boshoff, C. The biology and management of non-small cell lung cancer. Nature 2018, 553, 446–454. [Google Scholar] [CrossRef]
- Zhang, J.; Fujimoto, J.; Zhang, J.; Wedge, D.C.; Song, X.; Zhang, J.; Seth, S.; Chow, C.W.; Cao, Y.; Gumbs, C.; et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science 2014, 346, 256–259. [Google Scholar] [CrossRef]
- Wang, X.; Huang, Y.; Guo, R.; Liu, Y.; Qian, Y.; Liu, D.; Dai, X.; Wei, Z.; Jin, F.; Liu, Y. Clinicopathological significance of ROCK1 and PIK3CA expression in nasopharyngeal carcinoma. Exp. Ther. Med. 2017, 13, 1064–1068. [Google Scholar] [CrossRef]
- Yu, G.; Wang, Z.; Zeng, S.; Liu, S.; Zhu, C.; Xu, R.; Liu, R.E. Paeoniflorin Inhibits Hepatocyte Growth Factor- (HGF-) Induced Migration and Invasion and Actin Rearrangement via Suppression of c-Met-Mediated RhoA/ROCK Signaling in Glioblastoma. Biomed Res. Int. 2019, 2019, 9053295. [Google Scholar] [CrossRef]
- Knipe, R.S.; Tager, A.M.; Liao, J.K. The Rho kinases: Critical mediators of multiple profibrotic processes and rational targets for new therapies for pulmonary fibrosis. Pharmacol. Rev. 2015, 67, 103–117. [Google Scholar] [CrossRef]
- Wu, H.; Chen, Y.; Li, B.; Li, C.; Guo, J.; You, J.; Hu, X.; Kuang, D.; Qi, S.; Liu, P.; et al. Targeting ROCK1/2 blocks cell division and induces mitotic catastrophe in hepatocellular carcinoma. Biochem. Pharmacol. 2021, 184, 114353. [Google Scholar] [CrossRef]
- Pritchard, C.A.; Hayes, L.; Wojnowski, L.; Zimmer, A.; Marais, R.M.; Norman, J.C. B-Raf acts via the ROCKII/LIMK/cofilin pathway to maintain actin stress fibers in fibroblasts. Mol. Cell Biol. 2004, 24, 5937–5952. [Google Scholar] [CrossRef]
- Luo, D.; Chen, H.; Li, X.; Lu, P.; Long, M.; Peng, X.; Lin, S.; Tan, L.; Zhu, Y.; Ouyang, N.; et al. Activation of the ROCK1/MMP-9 pathway is associated with the invasion and poor prognosis in papillary thyroid carcinoma. Int. J. Oncol. 2017, 51, 1209–1218. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhao, Y.; Lv, M.; Lin, H.; Hong, Y.; Yang, F.; Sun, Y.; Guo, Y.; Cui, Y.; Li, S.; Gao, Y. ROCK1 induces ERK nuclear translocation in PDGF-BB-stimulated migration of rat vascular smooth muscle cells. IUBMB Life 2012, 64, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.H.; Liao, S.L.; Wang, S.H.; Wang, C.C.; Yang, C.H. Simvastatin and ROCK Inhibitor Y-27632 Inhibit Myofibroblast Differentiation of Graves’ Ophthalmopathy-Derived Orbital Fibroblasts via RhoA-Mediated ERK and p38 Signaling Pathways. Front. Endocrinol. 2020, 11, 607968. [Google Scholar] [CrossRef]
- Rath, N.; Olson, M.F. Rho-associated kinases in tumorigenesis: Re-considering ROCK inhibition for cancer therapy. EMBO Rep. 2012, 13, 900–908. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Goldstein, R.H.; Scepansky, E.M.; Rosenblatt, M. Inhibition of rho-associated kinase signaling prevents breast cancer metastasis to human bone. Cancer Res. 2009, 69, 8742–8751. [Google Scholar] [CrossRef] [PubMed]
- de Toledo, M.; Anguille, C.; Roger, L.; Roux, P.; Gadea, G. Cooperative anti-invasive effect of Cdc42/Rac1 activation and ROCK inhibition in SW620 colorectal cancer cells with elevated blebbing activity. PLoS ONE. 2012, 7, e48344. [Google Scholar] [CrossRef] [PubMed]
- Mikuriya, Y.; Tashiro, H.; Kuroda, S.; Nambu, J.; Kobayashi, T.; Amano, H.; Tanaka, Y.; Ohdan, H. Fatty liver creates a pro-metastatic microenvironment for hepatocellular carcinoma through activation of hepatic stellate cells. Int. J. Cancer 2015, 136, E3–E13. [Google Scholar] [CrossRef]
- Wu, Q.; Tian, Y.; Zhang, J.; Tong, X.; Huang, H.; Li, S.; Zhao, H.; Tang, Y.; Yuan, C.; Wang, K.; et al. In vivo CRISPR screening unveils histone demethylase UTX as an important epigenetic regulator in lung tumorigenesis. Proc. Natl. Acad. Sci. USA 2018, 115, E3978–E3986. [Google Scholar] [CrossRef]
- Cheng, C.Y.; Cherng, S.H.; Wu, W.J.; Yang, T.Y.; Huang, X.Y.; Liao, F.T.; Wu, M.F.; Sheu, G.T. Regulation of chemosensitivity and migration by clusterin in non-small cell lung cancer cells. Cancer Chemother. Pharmacol. 2012, 69, 145–154. [Google Scholar] [CrossRef]
- Garcia-Aranda, M.; Serrano, A.; Redondo, M. Regulation of Clusterin Gene Expression. Curr. Protein Pept. Sci. 2018, 19, 612–622. [Google Scholar] [CrossRef]
- Koltai, T. Clusterin: A key player in cancer chemoresistance and its inhibition. Onco Targets Ther. 2014, 7, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Leskov, K.S.; Klokov, D.Y.; Li, J.; Kinsella, T.J.; Boothman, D.A. Synthesis and functional analyses of nuclear clusterin, a cell death protein. J. Biol. Chem. 2003, 278, 11590–11600. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Kim, J.K.; Edwards, C.A.; Xu, Z.; Taichman, R.; Wang, C.-Y. Clusterin inhibits apoptosis by interacting with activated Bax. Nat. Cell Biol. 2005, 7, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Trougakos, I.P.; Djeu, J.Y.; Gonos, E.S.; Boothman, D.A. Advances and challenges in basic and translational research on clusterin. Cancer Res. 2009, 69, 403–406. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wu, M.; Xu, Y.; Song, Q.; Zheng, W. Secretory Clusterin: A Promising Target for Chemoresistance of Hepatocellular Carcinoma. Mini Rev. Med. Chem. 2020, 20, 1153–1165. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Gao, W.; Gao, J. sCLU as prognostic biomarker and therapeutic target in osteosarcoma. Bioengineered 2019, 10, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Jiang, Y.; Huang, Z.; Li, D.; Chen, X.; Cao, M.; Meng, Q.; Pang, H.; Sun, L.; Zhao, Y.; et al. miRNA-378 reverses chemoresistance to cisplatin in lung adenocarcinoma cells by targeting secreted clusterin. Sci. Rep. 2016, 6, 19455. [Google Scholar] [CrossRef]
- Chen, Q.F.; Chang, L.; Su, Q.; Zhao, Y.; Kong, B. Clinical importance of serum secreted clusterin in predicting invasive breast cancer and treatment responses. Bioengineered 2021, 12, 278–285. [Google Scholar] [CrossRef]
- Jiang, L.; Lin, C.; Song, L.; Wu, J.; Chen, B.; Ying, Z.; Fang, L.; Yan, X.; He, M.; Li, J.; et al. MicroRNA-30e*promotes human glioma cell invasiveness in an orthotopic xenotransplantation model by disrupting the NF-kappa B/I kappa B alpha negative feedback loop. J. Clin. Investig. 2012, 122, 33–47. [Google Scholar] [CrossRef]
- Liang, W.; Guan, H.; He, X.; Ke, W.; Xu, L.; Liu, L.; Xiao, H.; Li, Y. Down-regulation of SOSTDC1 promotes thyroid cancer cell proliferation via regulating cyclin A2 and cyclin E2. Oncotarget 2015, 6, 31780–31791. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, L.; Cai, J.; Wu, J.; Guan, H.; Zhu, X.; Yuan, J.; Li, M. DEPDC1B enhances migration and invasion of non-small cell lung cancer cells via activating Wnt/beta-catenin signaling. Biochem. Biophys. Res. Commun. 2014, 450, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Guan, H.; Fang, L.; Yang, Y.; Zhu, X.; Yuan, J.; Wu, J.; Li, M. MicroRNA-374a activates Wnt/beta-catenin signaling to promote breast cancer metastasis. J. Clin. Investig. 2013, 123, 566–579. [Google Scholar] [PubMed]
- Senger, D.R.; Perruzzi, C.A.; Streit, M.; Koteliansky, V.E.; de Fougerolles, A.R.; Detmar, M. The alpha (1) beta (1) and alpha (2) beta (1) Integrins provide critical support for vascular endothelial growth factor signaling, endothelial cell migration, and tumor angiogenesis. Am. J. Pathol. 2002, 160, 195–204. [Google Scholar] [CrossRef]
- Maddika, S.; Chen, J. Protein kinase DYRK2 is a scaffold that facilitates assembly of an E3 ligase. Nat. Cell Biol. 2009, 11, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Shannan, B.; Seifert, M.; Leskov, K.; Willis, J.; Boothman, D.; Tilgen, W.; Reichrath, J. Challenge and promise: Roles for clusterin in pathogenesis, progression and therapy of cancer. Cell Death Differ. 2006, 13, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Fan, Z.; Dou, X.; Zhou, Q.; Zeng, G.; Liu, L.; Chen, W.; Lan, R.; Liu, W.; Ru, G.; et al. Inactivation of tumor suppressor gene Clusterin leads to hyperactivation of TAK1-NF-κB signaling axis in lung cancer cells and denotes a therapeutic opportunity. Theranostics 2020, 10, 11520–11534. [Google Scholar] [CrossRef]
- Chayka, O.; Corvetta, D.; Dews, M.; Caccamo, A.E.; Piotrowska, I.; Santilli, G.; Gibson, S.; Sebire, N.J.; Himoudi, N.; Hogarty, M.D.; et al. Clusterin, a haploinsufficient tumor suppressor gene in neuroblastomas. J. Natl. Cancer Inst. 2009, 101, 663–677. [Google Scholar] [CrossRef]
- Jones, S.E.; Jomary, C. Clusterin. Int. J. Biochem. Cell Biol. 2002, 34, 427–431. [Google Scholar] [CrossRef]
- Scaltriti, M.; Santamaria, A.; Paciucci, R.; Bettuzzi, S. Intracellular clusterin induces G2-M phase arrest and cell death in PC-3 prostate cancer cells1. Cancer Res. 2004, 64, 6174–6182. [Google Scholar] [CrossRef][Green Version]
- Moretti, R.M.; Marelli, M.M.; Mai, S.; Cariboni, A.; Scaltriti, M.; Bettuzzi, S.; Limonta, P. Clusterin isoforms differentially affect growth and motility of prostate cells: Possible implications in prostate tumorigenesis. Cancer Res. 2007, 67, 10325–10333. [Google Scholar] [CrossRef]
- Chen, W.; Mao, K.; Liu, Z.; Dinh-Xuan, A.T. The role of the RhoA/Rho kinase pathway in angiogenesis and its potential value in prostate cancer (Review). Oncol. Lett. 2014, 8, 1907–1911. [Google Scholar] [CrossRef] [PubMed]
- Kale, V.P.; Hengst, J.A.; Desai, D.H.; Amin, S.G.; Yun, J.K. The regulatory roles of ROCK and MRCK kinases in the plasticity of cancer cell migration. Cancer Lett. 2015, 361, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Mali, R.S.; Kapur, S.; Kapur, R. Role of Rho kinases in abnormal and normal hematopoiesis. Curr. Opin. Hematol. 2014, 21, 271–275. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, F.; Jiang, Q.; Shi, K.J.; Luo, H.; Yang, Y.; Xu, C.M. RhoA modulates functional and physical interaction between ROCK1 and Erk1/2 in selenite-induced apoptosis of leukaemia cells. Cell Death Dis. 2013, 4, e708. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, S.; Li, X.; Gu, W.; Li, X.; Zhao, J.; Wu, J.; Cai, J.; Feng, X.; Tao, T. Cytoplasmic Clusterin Suppresses Lung Cancer Metastasis by Inhibiting the ROCK1-ERK Axis. Cancers 2022, 14, 2463. https://doi.org/10.3390/cancers14102463
Huang S, Li X, Gu W, Li X, Zhao J, Wu J, Cai J, Feng X, Tao T. Cytoplasmic Clusterin Suppresses Lung Cancer Metastasis by Inhibiting the ROCK1-ERK Axis. Cancers. 2022; 14(10):2463. https://doi.org/10.3390/cancers14102463
Chicago/Turabian StyleHuang, Shaobo, Xu Li, Weiqi Gu, Xiaoyi Li, Jingjing Zhao, Jueheng Wu, Junchao Cai, Xianming Feng, and Tianyu Tao. 2022. "Cytoplasmic Clusterin Suppresses Lung Cancer Metastasis by Inhibiting the ROCK1-ERK Axis" Cancers 14, no. 10: 2463. https://doi.org/10.3390/cancers14102463
APA StyleHuang, S., Li, X., Gu, W., Li, X., Zhao, J., Wu, J., Cai, J., Feng, X., & Tao, T. (2022). Cytoplasmic Clusterin Suppresses Lung Cancer Metastasis by Inhibiting the ROCK1-ERK Axis. Cancers, 14(10), 2463. https://doi.org/10.3390/cancers14102463





