Microfluidic-Assisted Preparation of Targeted pH-Responsive Polymeric Micelles Improves Gemcitabine Effectiveness in PDAC: In Vitro Insights
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
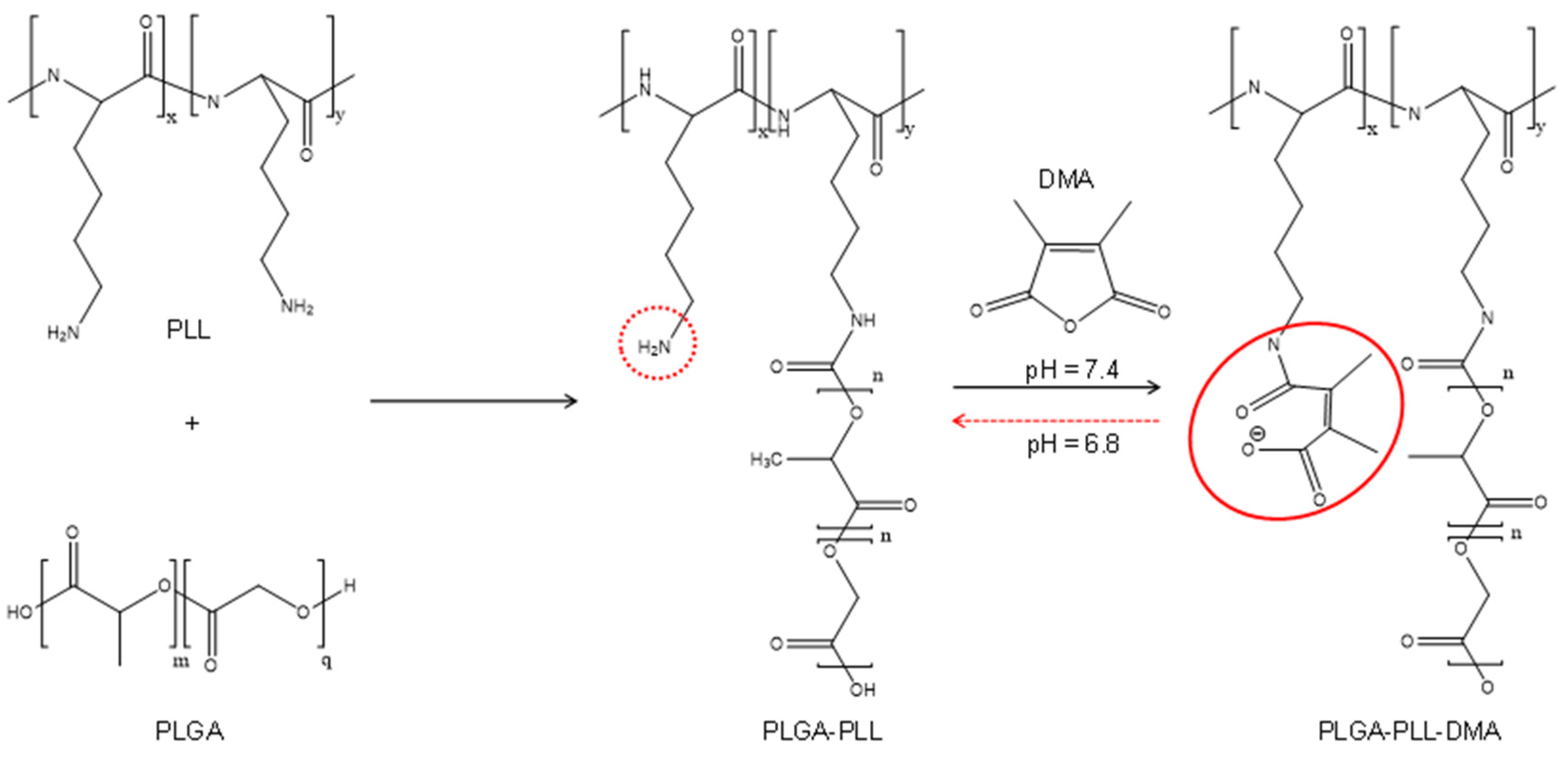
2.1. Synthesis of Copolymers
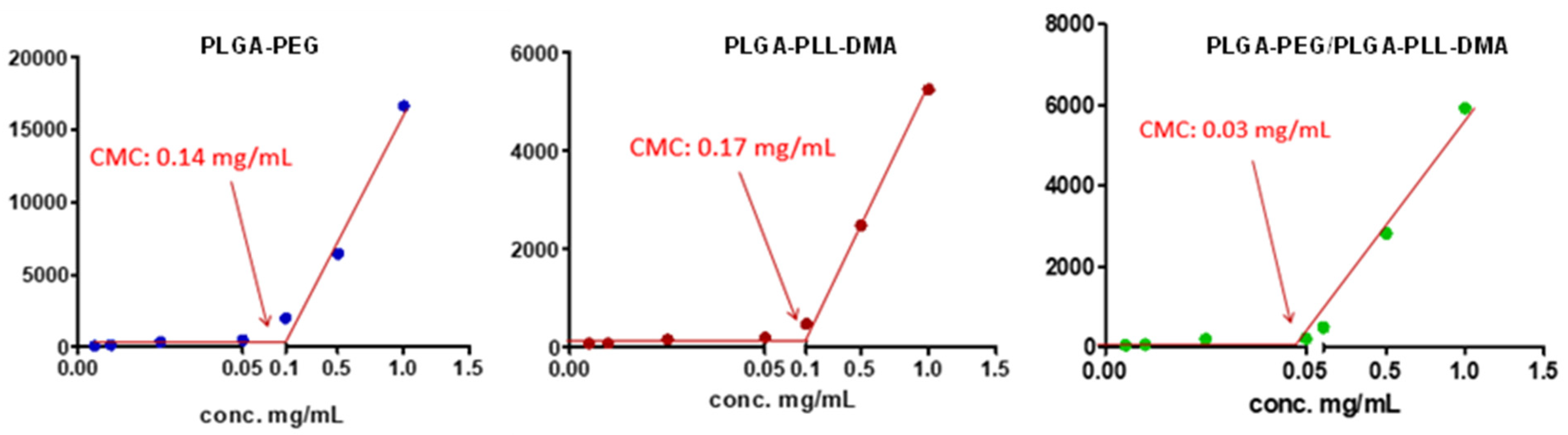
2.2. Determination of Critical Micelle Concentration (CMC)
2.3. Preparation and Characterization of the Micelles
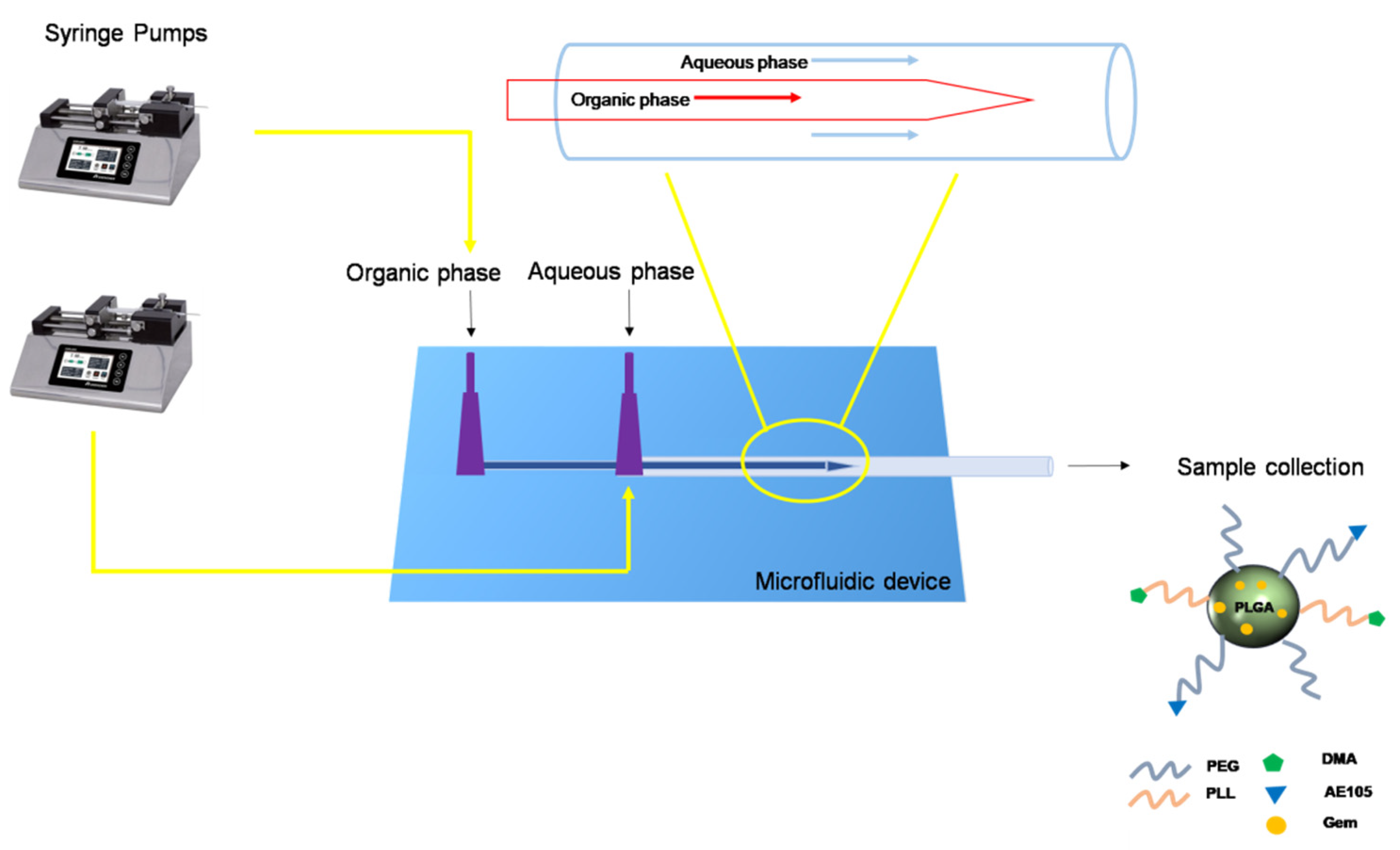
2.3.1. Preparation of Glass Capillary Microfluidic Chip
2.3.2. Preparation of Micelles by Microfluidic Technique
2.3.3. Particle Size, Size Distribution, and Surface Charge
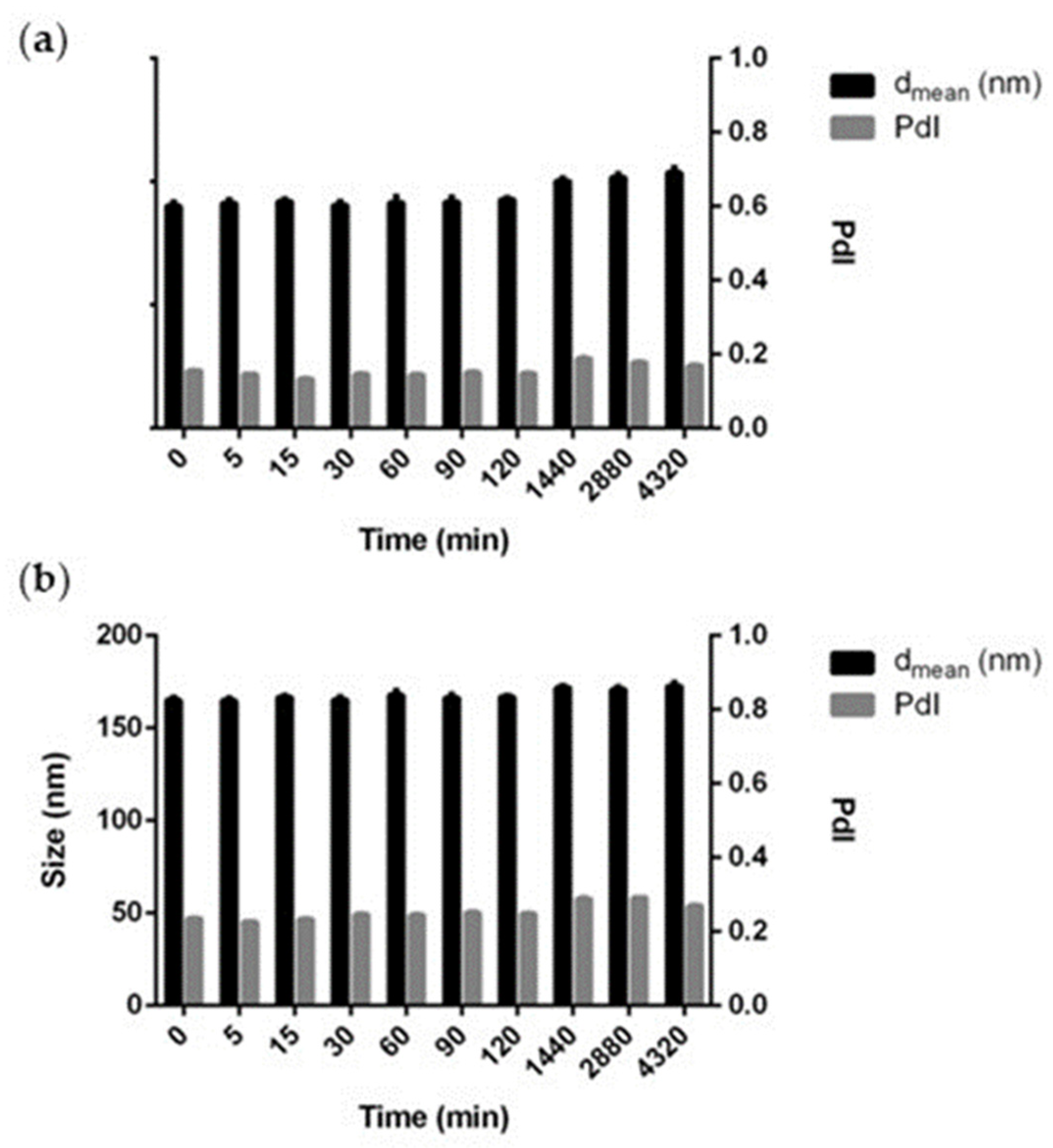
2.4. Stability Studies
2.5. Evaluation of Drug Encapsulation Efficiency
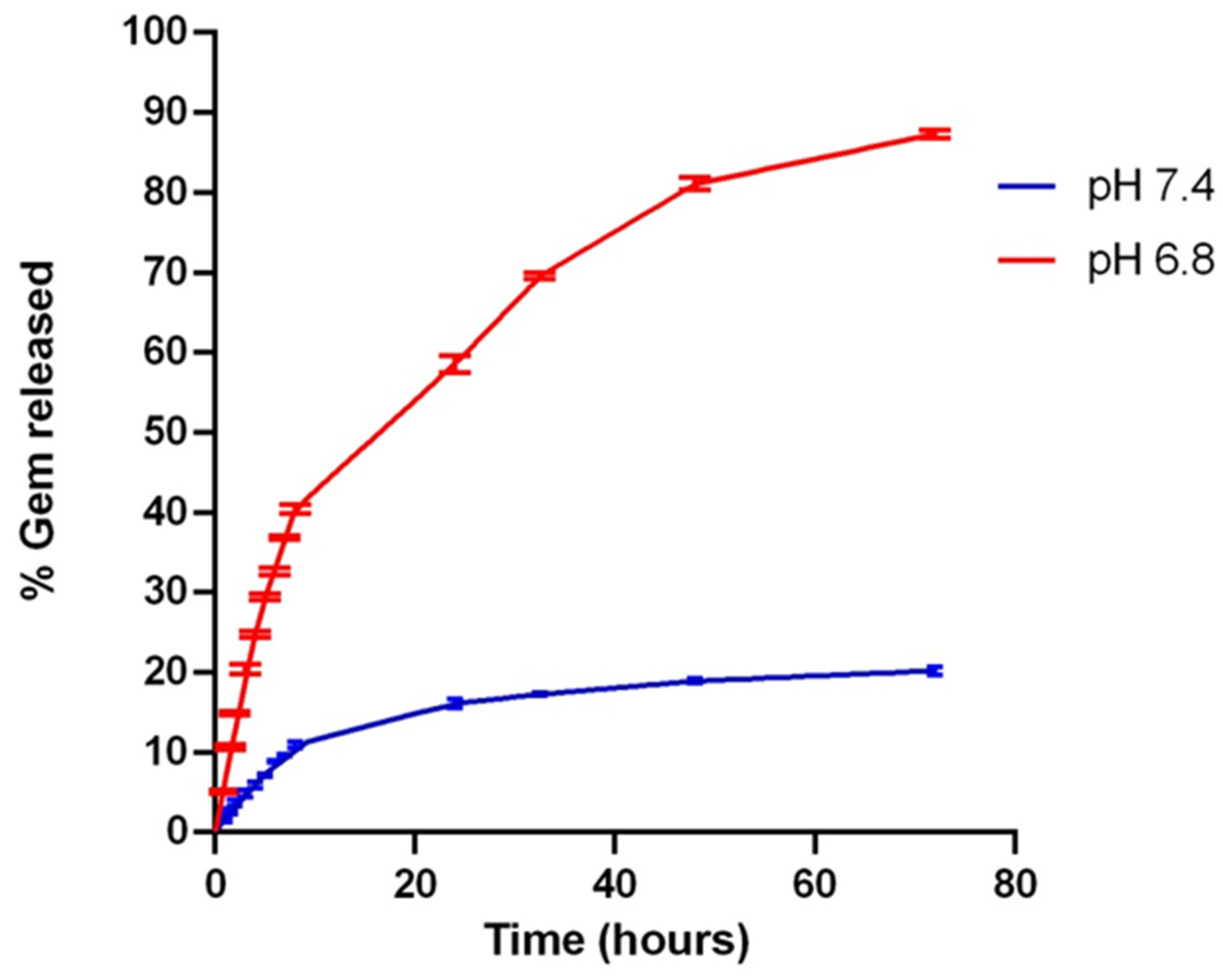
2.6. In Vitro Drug Release Study
2.7. Cell Culture
2.8. uPAR Expression Evaluation by Flow Cytometry (FCM) Analysis
2.9. In Vitro Uptake Study on 2D and 3D Model of PDAC Cells
2.10. In Vitro Proliferation Study
2.11. Evaluation of the Cell Cycle Progression, Apoptosis Induction and γH2AX Analysis
2.11.1. Cell Cycle Analysis
2.11.2. Apoptosis Assay
2.11.3. γ. H2AX Analysis
2.12. Statistical Analysis
3. Results and Discussion
3.1. Synthesis and Characterization of Copolymers
3.2. Determination of Critical Micelle Concentration
3.3. Preparation and Characterization of Micelles
3.4. In Vitro Studies
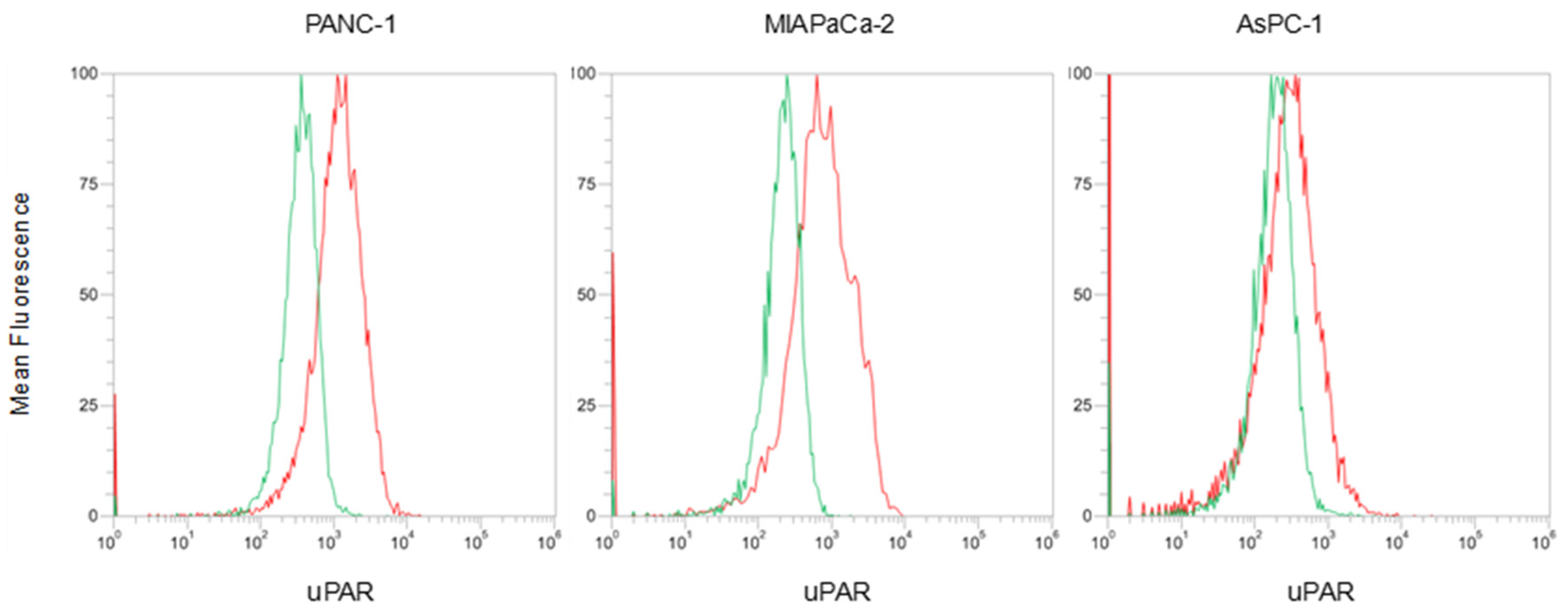
3.4.1. uPAR Expression Evaluation on Plasma Membrane of Pancreatic Cancer Cell Lines
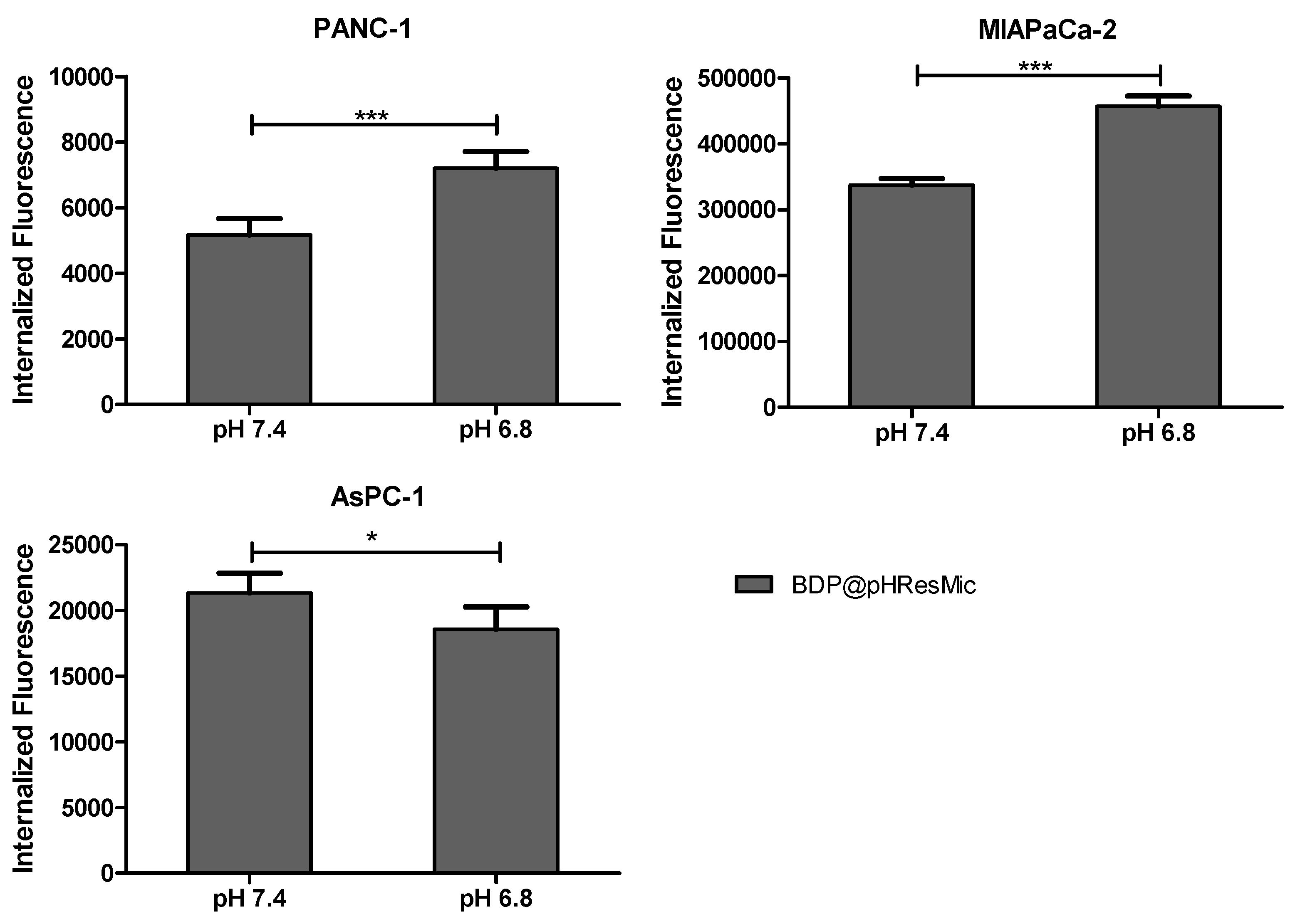
3.4.2. Evaluation of pH-Mediated Internalization of pH-Responsive Micelles
3.4.3. Evaluation of uPAR-Mediated Internalization of Targeted pH-Responsive micelles
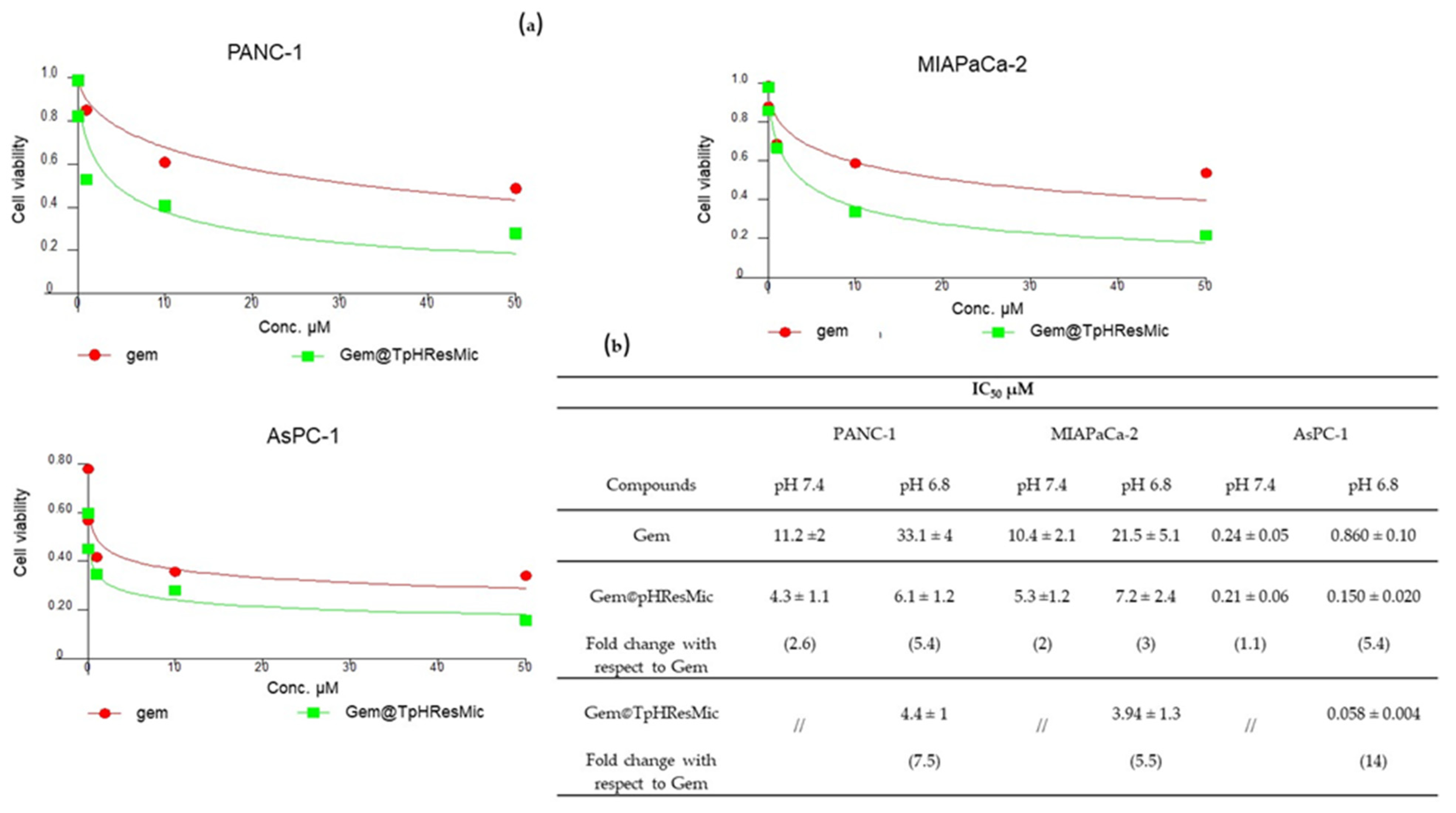
3.4.4. Evaluation of the Antiproliferative Effect of TpHResMic and pHResMic in 2D Models of Pancreatic Cancer
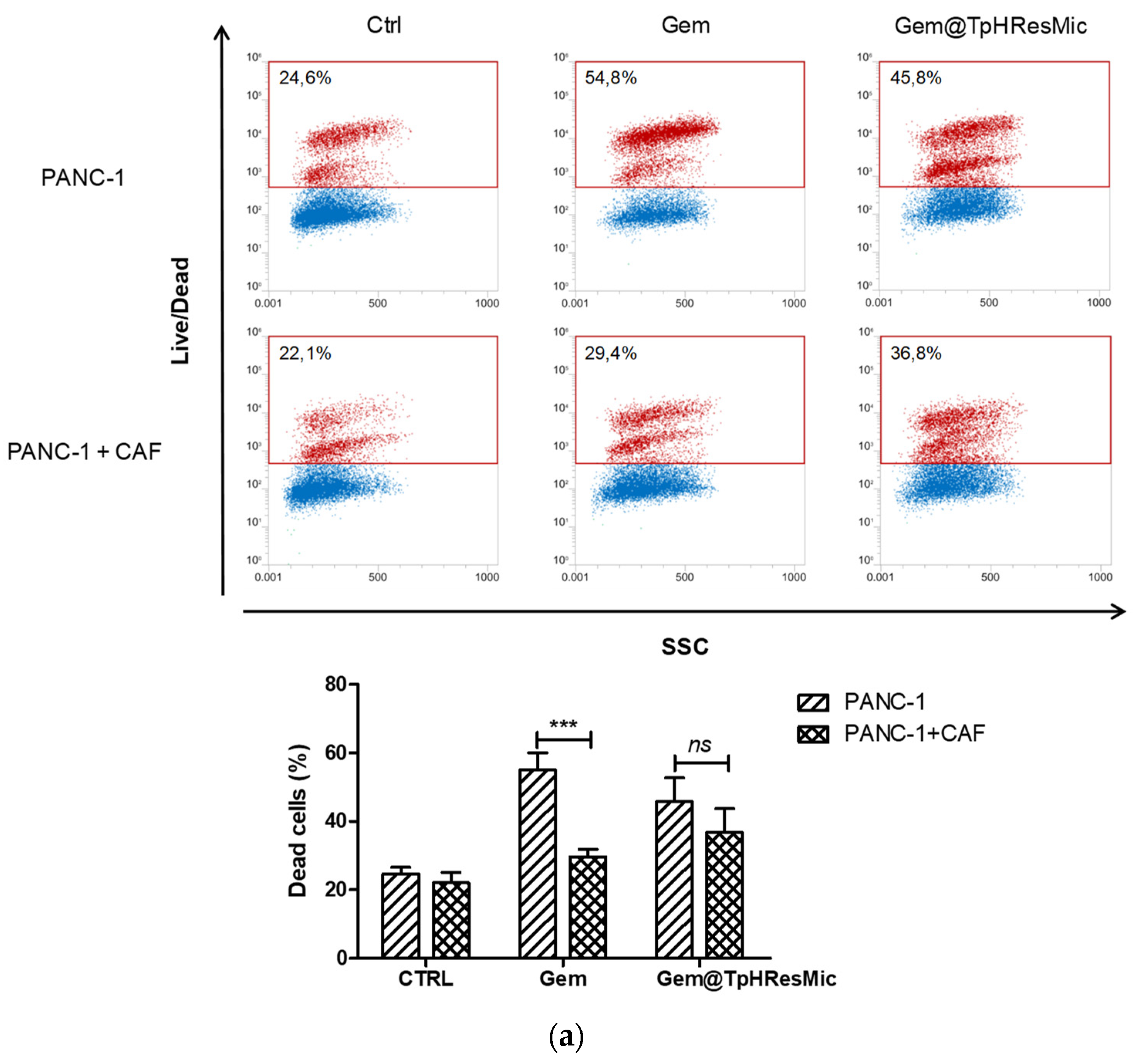
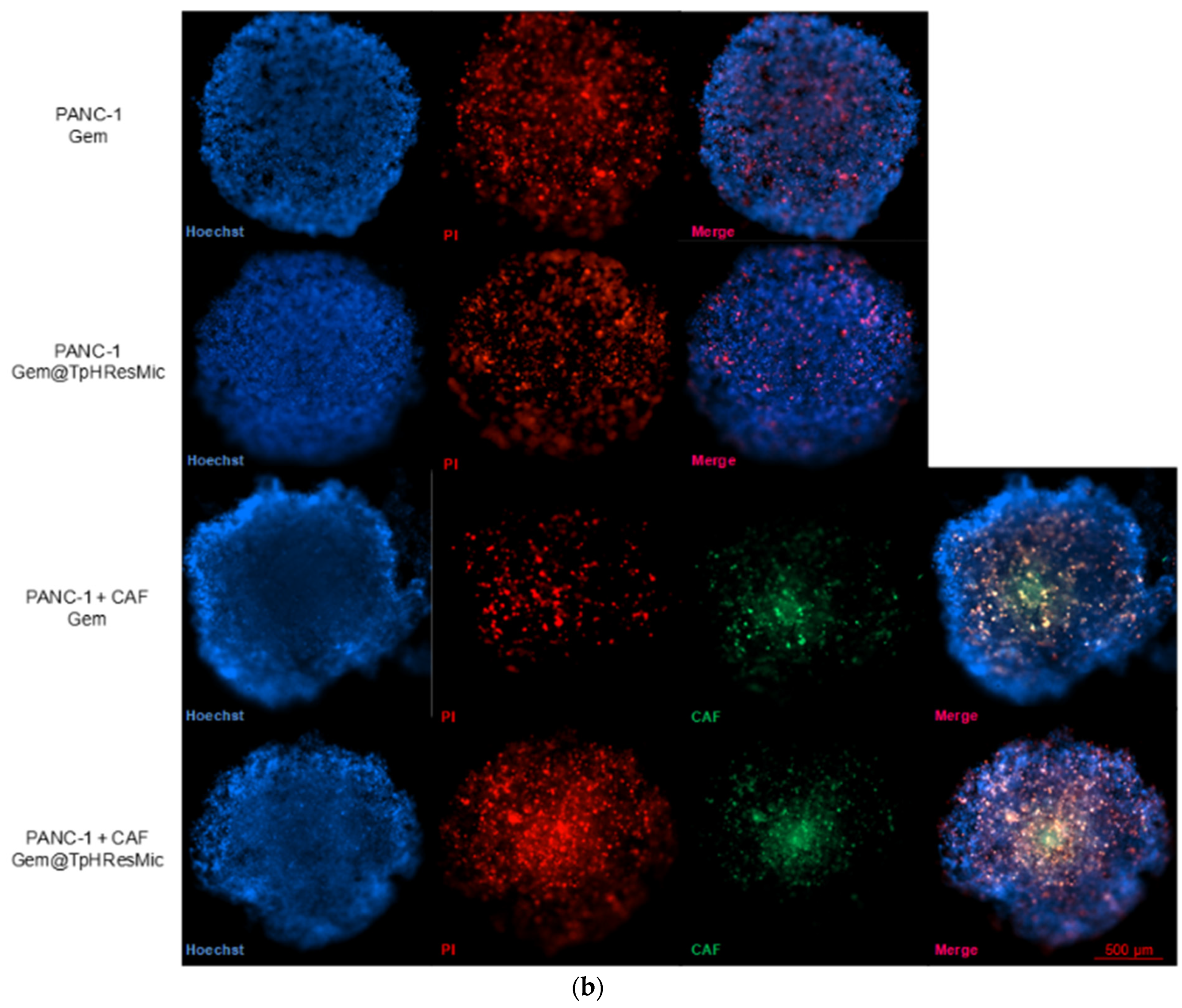
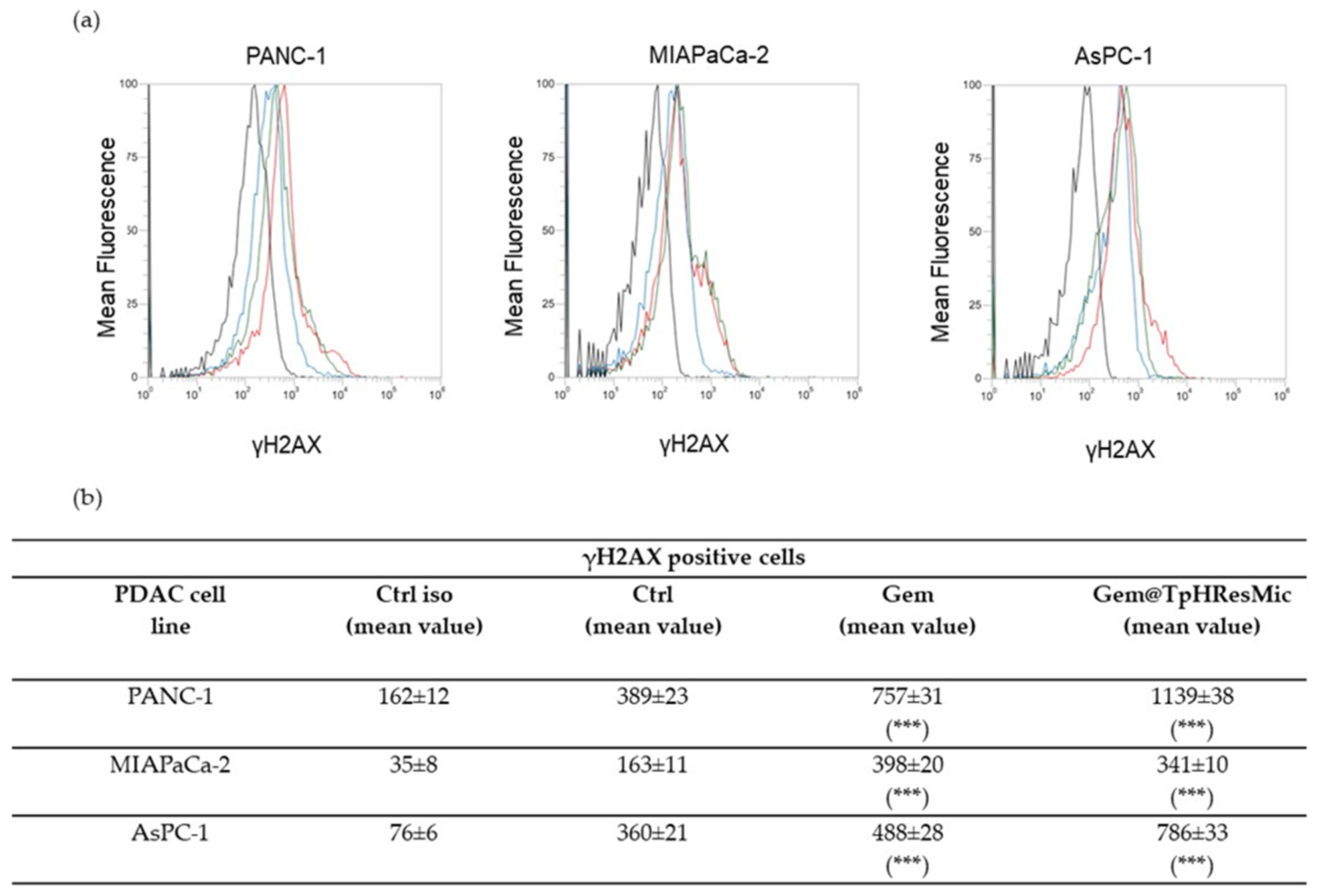
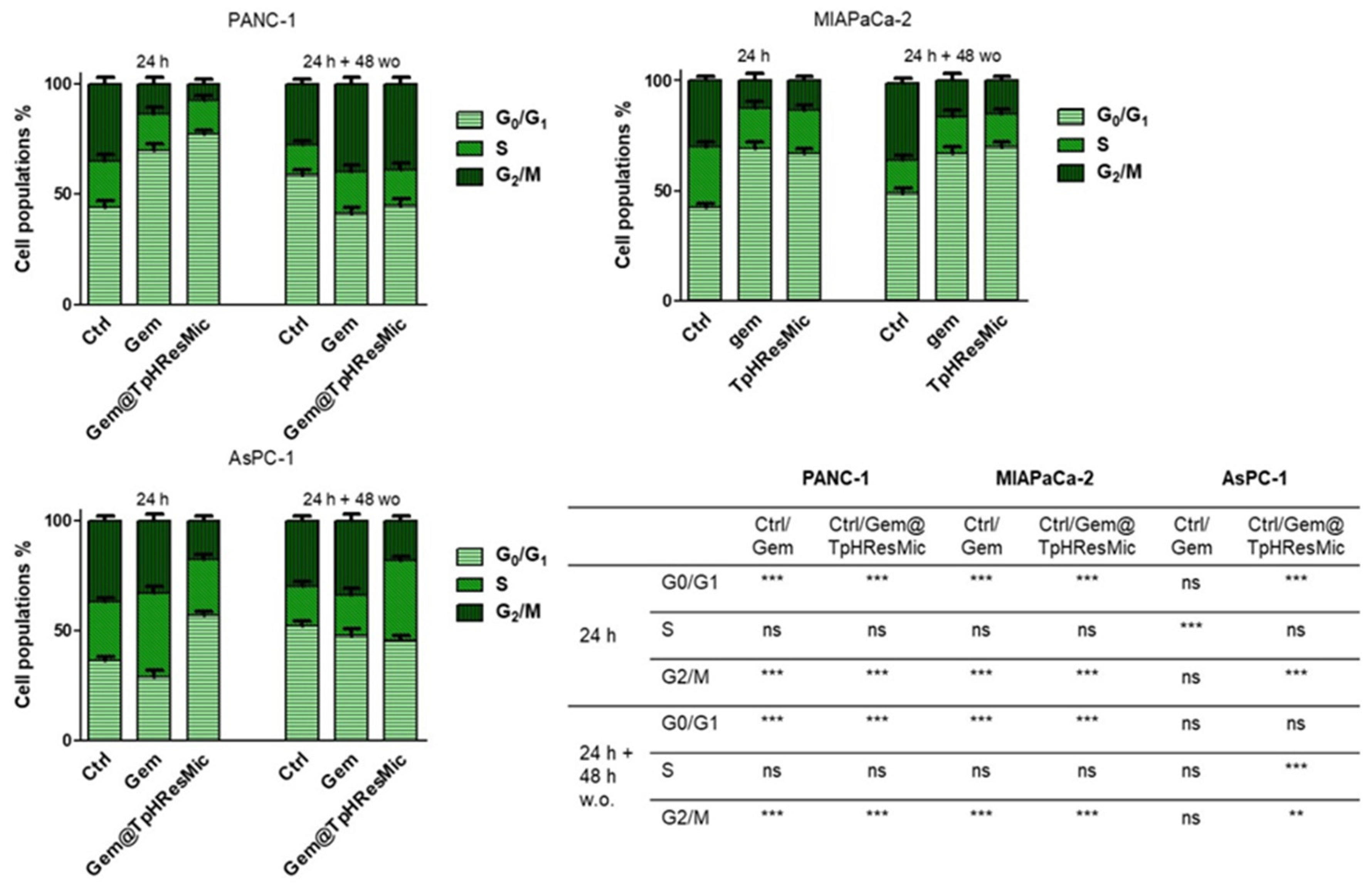
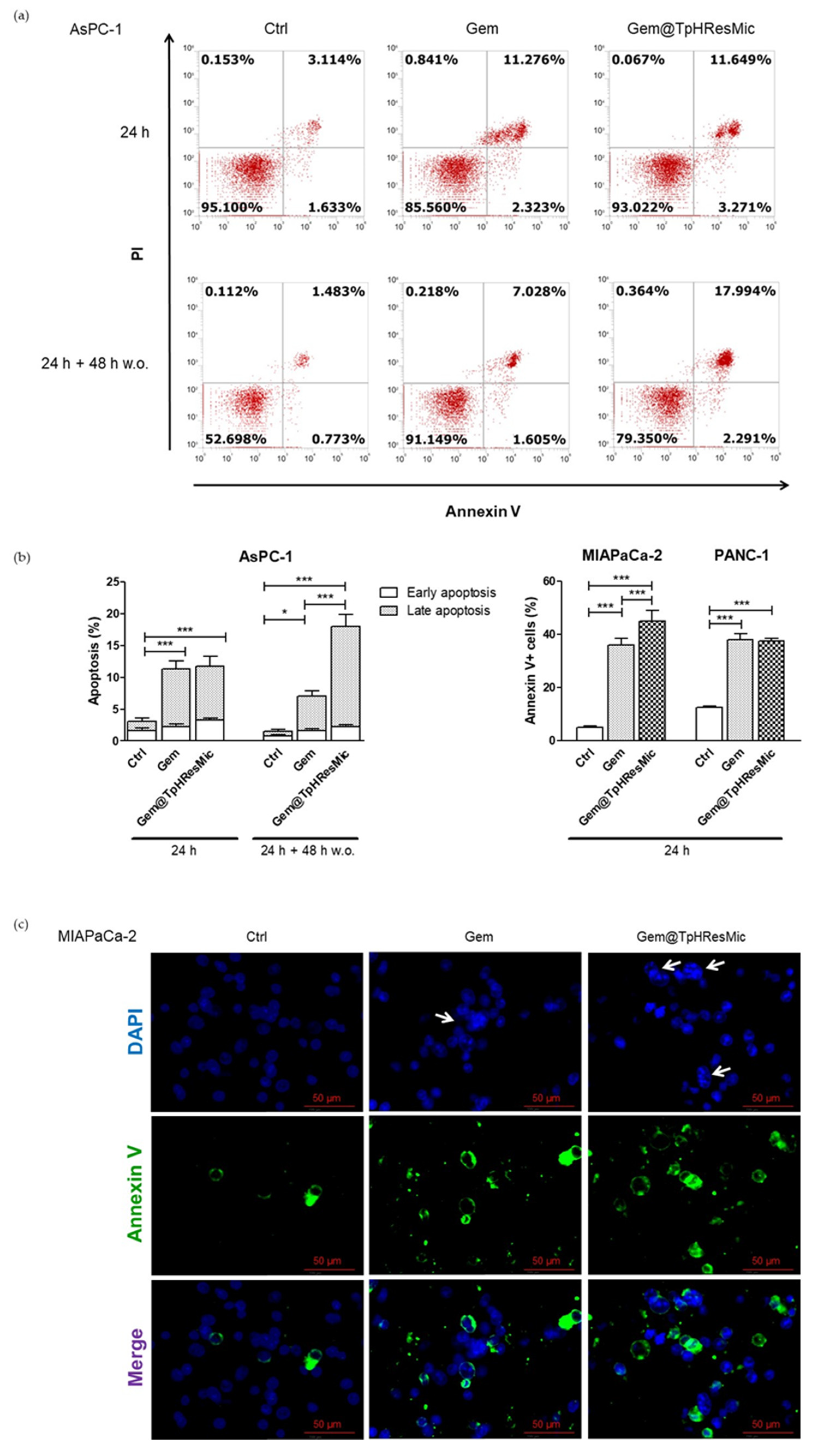
3.4.5. Evaluation of the Antitumor Activity of Free Gem and Gem@TPHResMic through the Analysis of γH2AX, the Cell Cycle Perturbation and the Apoptosis Induction
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- AIOM. Available online: https://www.aiom.it/wp-content/uploads/2020/10/2020_LG_AIOM_Pancreas.pdf (accessed on 15 September 2021).
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2020. CA A Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Porcelli, L.; Iacobazzi, R.M.; Di Fonte, R.; Serratì, S.; Intini, A.; Solimando, A.G.; Brunetti, O.; Calabrese, A.; Leonetti, F.; Azzariti, A.; et al. CAFs and TGF-β Signaling Activation by Mast Cells Contribute to Resistance to Gemcitabine/Nabpaclitaxel in Pancreatic Cancer. Cancers 2019, 11, 330. [Google Scholar] [CrossRef] [Green Version]
- Qin, C.; Yang, G.; Yang, J.; Ren, B.; Wang, H.; Chen, G.; Zhao, F.; You, L.; Wang, W.; Zhao, Y. Metabolism of Pancreatic Cancer: Paving the Way to Better Anticancer Strategies. Mol. Cancer 2020, 19, 50. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Zhou, W.; Liang, C.; Shi, S.; Yu, X.; Chen, Q.; Sun, T.; Lu, Y.; Zhang, Y.; Guo, Q.; et al. Codelivery Nanosystem Targeting the Deep Microenvironment of Pancreatic Cancer. Nano Lett. 2019, 19, 3527–3534. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, Q.; Jurisica, I.; Do, T.; Hedley, D. Hypoxia Predicts Aggressive Growth and Spontaneous Metastasis Formation from Orthotopically Grown Primary Xenografts of Human Pancreatic Cancer. Cancer Res. 2011, 71, 3110–3120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandana, S.; Babiker, H.M.; Mahadevan, D. Therapeutic Trends in Pancreatic Ductal Adenocarcinoma (PDAC). Expert Opin. Investig. Drugs 2018, 28, 161–177. [Google Scholar] [CrossRef]
- Conroy, T.; Hammel, P.; Hebbar, M.; Ben Abdelghani, M.; Wei, A.C.; Raoul, J.-L.; Choné, L.; Francois, E.; Artru, P.; Biagi, J.J.; et al. FOLFIRINOX or Gemcitabine as Adjuvant Therapy for Pancreatic Cancer. N. Engl. J. Med. 2018, 379, 2395–2406. [Google Scholar] [CrossRef]
- Shukla, S.K.; Purohit, V.; Mehla, K.; Gunda, V.; Chaika, N.V.; Vernucci, E.; King, R.J.; Abrego, J.; Goode, G.D.; Dasgupta, A.; et al. MUC1 and HIF-1alpha Signaling Crosstalk Induces Anabolic Glucose Metabolism to Impart Gemcitabine Resistance to Pancreatic Cancer. Cancer Cell 2017, 32, 71–87.e7. [Google Scholar] [CrossRef] [Green Version]
- Dalin, S.; Sullivan, M.R.; Lau, A.N.; Grauman-Boss, B.; Mueller, H.S.; Kreidl, E.; Fenoglio, S.; Luengo, A.; Lees, J.A.; Heiden, M.G.V.; et al. Deoxycytidine Release from Pancreatic Stellate Cells Promotes Gemcitabine Resistance. Cancer Res. 2019, 79, 5723–5733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mashayekhi, V.; Mocellin, O.; Fens, M.H.; Krijger, G.C.; Brosens, L.A.; Oliveira, S. Targeting of Promising Transmembrane Proteins for Diagnosis and Treatment of Pancreatic Ductal Adenocarcinoma. Theranostics 2021, 11, 9022–9037. [Google Scholar] [CrossRef]
- Lai, E.; Puzzoni, M.; Ziranu, P.; Pretta, A.; Impera, V.; Mariani, S.; Liscia, N.; Soro, P.; Musio, F.; Persano, M.; et al. New Therapeutic Targets in Pancreatic Cancer. Cancer Treat. Rev. 2019, 81, 101926. [Google Scholar] [CrossRef]
- Han, W.; Ke, J.; Guo, F.; Meng, F.; Li, H.; Wang, L. Construction and Antitumor Properties of a Targeted Nano-Drug Carrier System Responsive to the Tumor Microenvironment. Int. J. Pharm. 2021, 608, 121066. [Google Scholar] [CrossRef]
- Kashkooli, F.M.; Soltani, M.; Souri, M. Controlled Anti-Cancer drug Release through Advanced Nano-Drug Delivery Systems: Static and Dynamic Targeting Strategies. J. Control. Release 2020, 327, 316–349. [Google Scholar] [CrossRef]
- Chen, Y.; Du, Q.; Zou, Y.; Guo, Q.; Huang, J.; Tao, L.; Shen, X.; Peng, J. Co-Delivery of Doxorubicin and Epacadostat via Heparin coated pH-Sensitive Liposomes to Suppress the Lung Metastasis of Melanoma. Int. J. Pharm. 2020, 584, 119446. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, X.; Zhang, J.; Wang, L.; Ou, C.; Shu, Y.; Wu, Q.; Ma, G.; Gong, C. Gambogic Acid-Encapsulated Polymeric Micelles Improved Therapeutic Effects on Pancreatic Cancer. Chin. Chem. Lett. 2019, 30, 885–888. [Google Scholar] [CrossRef]
- Gurka, M.K.; Pender, D.; Chuong, P.; Fouts, B.L.; Sobelov, A.; McNally, M.W.; Mezera, M.; Woo, S.Y.; McNally, L.R. Identification of Pancreatic Tumors in Vivo with Ligand-Targeted, pH Responsive Mesoporous Silica Nanoparticles by Multispectral Optoacoustic Tomography. J. Control. Release 2016, 231, 60–67. [Google Scholar] [CrossRef] [Green Version]
- Skovgaard, D.; Persson, M.; Kjaer, A. Urokinase Plasminogen Activator Receptor–PET with 68 Ga-NOTA-AE105. PET Clin. 2017, 12, 311–319. [Google Scholar] [CrossRef]
- Gao, G.; Li, Y.; Lee, D.S. Environmental pH-Sensitive Polymeric Micelles for Cancer Diagnosis and Targeted Therapy. J. Control. Release 2012, 169, 180–184. [Google Scholar] [CrossRef]
- Cheng, W.; Gu, L.; Ren, W.; Liu, Y. Stimuli-Responsive Polymers for Anti-Cancer Drug Delivery. Mater. Sci. Eng. C 2014, 45, 600–608. [Google Scholar] [CrossRef]
- Wu, W.; Luo, L.; Wang, Y.; Wu, Q.; Dai, H.-B.; Li, J.-S.; Durkan, C.; Wang, N.; Wang, G.-X. Endogenous pH-Responsive Nanoparticles with Programmable Size Changes for Targeted Tumor Therapy and Imaging Applications. Theranostics 2018, 8, 3038–3058. [Google Scholar] [CrossRef]
- Yu, G.; Ning, Q.; Mo, Z.; Tang, S. Intelligent Polymeric Micelles for Multidrug Co-Delivery and Cancer Therapy. Artif. Cells Nanomed. Biotechnol. 2019, 47, 1476–1487. [Google Scholar] [CrossRef]
- Martins, C.; Araújo, F.; Gomes, M.J.; Fernandes, C.; Nunes, R.; Li, W.; Santos, H.A.; Borges, F.; Sarmento, B. Using Microfluidic Platforms to Develop CNS-Targeted Polymeric Nanoparticles for HIV Therapy. Eur. J. Pharm. Biopharm. 2019, 138, 111–124. [Google Scholar] [CrossRef]
- Arduino, I.; Liu, Z.; Rahikkala, A.; Figueiredo, P.; Correia, A.; Cutrignelli, A.; Denora, N.; Santos, H.A. Preparation of Cetyl Palmitate-Based PEGylated Solid Lipid Nanoparticles by Microfluidic Technique. Acta Biomater. 2021, 121, 566–578. [Google Scholar] [CrossRef]
- Arduino, I.; Liu, Z.; Iacobazzi, R.M.; Lopedota, A.A.; Lopalco, A.; Cutrignelli, A.; Laquintana, V.; Porcelli, L.; Azzariti, A.; Franco, M.; et al. Microfluidic Preparation and in Vitro Evaluation of iRGD-Functionalized Solid Lipid Nanoparticles for Targeted Delivery of Paclitaxel to Tumor Cells. Int. J. Pharm. 2021, 610, 121246. [Google Scholar] [CrossRef]
- Tiboni, M.; Benedetti, S.; Skouras, A.; Curzi, G.; Perinelli, D.R.; Palmieri, G.F.; Casettari, L. 3D-Printed Microfluidic Chip for the Preparation of Glycyrrhetinic Acid-Loaded Ethanolic Liposomes. Int. J. Pharm. 2020, 584, 119436. [Google Scholar] [CrossRef]
- Hussain, M.T.; Tiboni, M.; Perrie, Y.; Casettari, L. Microfluidic Production of Protein Loaded Chimeric Stealth Liposomes. Int. J. Pharm. 2020, 590, 119955. [Google Scholar] [CrossRef]
- Xu, Z.; Lu, C.; Riordon, J.; Sinton, D.; Moffitt, M.G. Microfluidic Manufacturing of Polymeric Nanoparticles: Comparing Flow Control of Multiscale Structure in Single-Phase Staggered Herringbone and Two-Phase Reactors. Langmuir 2016, 32, 12781–12789. [Google Scholar] [CrossRef]
- Chastek, T.Q.; Iida, K.; Amis, E.J.; Fasolka, M.J.; Beers, K.L. A Microfluidic Platform for Integrated Synthesis and Dynamic Light Scattering Measurement of Block Copolymer Micelles. Lab A Chip 2008, 8, 950–957. [Google Scholar] [CrossRef]
- Capretto, L.; Mazzitelli, S.; Colombo, G.; Piva, R.; Penolazzi, L.; Vecchiatini, R.; Zhang, X.; Nastruzzi, C. Production of Polymeric Micelles by Microfluidic Technology for Combined Drug Delivery: Application to Osteogenic Differentiation of Human Periodontal Ligament Mesenchymal Stem Cells (hPDLSCs). Int. J. Pharm. 2013, 440, 195–206. [Google Scholar] [CrossRef]
- Capretto, L.; Chengx, W.; Carugo, D.; Hill, M.; Zhang, X. Microfluidic Reactors for Controlled Synthesis of Polymeric Micelles. J. Control. Release 2010, 148, e25–e26. [Google Scholar] [CrossRef]
- Lu, Y.; Chowdhury, D.; Vladisavljević, G.T.; Koutroumanis, K.; Georgiadou, S. Production of Fluconazole-Loaded Polymeric Micelles Using Membrane and Microfluidic Dispersion Devices. Membranes 2016, 6, 29. [Google Scholar] [CrossRef] [Green Version]
- Nastruzzi, C.; Capretto, L.; Mazzitelli, S.; Brognara, E.; Lampronti, I.; Carugo, D.; Hill, M.; Zhang, X.; Gambari, R. Mithramycin Encapsulated in Polymeric Micelles by Microfluidic Technology as Novel Therapeutic Protocol for Beta-Thalassemia. Int. J. Nanomed. 2012, 7, 307–324. [Google Scholar] [CrossRef] [Green Version]
- Capretto, L.; Carugo, D.; Cheng, W.; Hill, M.; Zhang, X. Continuous-Flow Production of Polymeric Micelles in Microreactors: Experimental and Computational Analysis. J. Colloid Interface Sci. 2011, 357, 243–251. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, S.; Machado, A.; Lecommandoux, S.; Sandre, O.; Gu, F.; Colin, A. Controllable Microfluidic Production of Drug-Loaded PLGA Nanoparticles Using Partially Water-Miscible Mixed Solvent Microdroplets as a Precursor. Sci. Rep. 2017, 7, 4794. [Google Scholar] [CrossRef]
- Feng, Q.; Liu, J.; Li, X.; Chen, Q.; Sun, J.; Shi, X.; Ding, B.; Yu, H.; Li, Y.; Jiang, X. One-Step Microfluidic Synthesis of Nanocomplex with Tunable Rigidity and Acid-Switchable Surface Charge for Overcoming Drug Resistance. Small 2016, 13, 1603109. [Google Scholar] [CrossRef]
- Albuquerque, L.J.C.; Sincari, V.; Jäger, A.; Konefał, R.; Pánek, J.; Černoch, P.; Pavlova, E.; Štěpánek, P.; Giacomelli, F.C.; Jäger, E. Microfluidic-Assisted Engineering of Quasi-Monodisperse pH-Responsive Polymersomes toward Advanced Platforms for the Intracellular Delivery of Hydrophilic Therapeutics. Langmuir 2019, 35, 8363–8372. [Google Scholar] [CrossRef]
- Brown, L.; McArthur, S.L.; Wright, P.C.; Lewis, A.; Battaglia, G. Polymersome Production on a Microfluidic Platform Using pH Sensitive Block Copolymers. Lab A Chip 2010, 10, 1922–1928. [Google Scholar] [CrossRef]
- Amphoterics, N.; Einstein, S.; Dls, C.; Nano, T.Z. Surfactant Micelle Characterization Using Dynamic Light Scattering. J. Phys. Chem. B 2006, MRK809-01, 1–5. [Google Scholar]
- Zdziennicka, A.; Szymczyk, K.; Krawczyk, J.; Jańczuk, B. Critical Micelle Concentration of Some Surfactants and Thermodynamic Parameters of Their Micellization. Fluid Phase Equilibria 2012, 322–323, 126–134. [Google Scholar] [CrossRef]
- Iacobazzi, R.M.; Cutrignelli, A.; Stefanachi, A.; Porcelli, L.; Lopedota, A.A.; Di Fonte, R.; Lopalco, A.; Serratì, S.; Laquintana, V.; Silvestris, N.; et al. Hydroxy-Propil-β-Cyclodextrin Inclusion Complexes of Two Biphenylnicotinamide Derivatives: Formulation and Anti-Proliferative Activity Evaluation in Pancreatic Cancer Cell Models. Int. J. Mol. Sci. 2020, 21, 6545. [Google Scholar] [CrossRef]
- Iacobazzi, R.M.; Porcelli, L.; Lopedota, A.A.; Laquintana, V.; Lopalco, A.; Cutrignelli, A.; Altamura, E.; Di Fonte, R.; Azzariti, A.; Franco, M.; et al. Targeting Human Liver Cancer Cells with Lactobionic Acid-G(4)-PAMAM-FITC Sorafenib Loaded Dendrimers. Int. J. Pharm. 2017, 528, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Azzariti, A.; Colabufo, N.A.; Berardi, F.; Porcelli, L.; Niso, M.; Simone, G.M.; Perrone, R.; Paradiso, A. Cyclohexylpiperazine Derivative PB28, a σ2 Agonist and σ1 Antagonist Receptor, Inhibits Cell Growth, Modulates P-Glycoprotein, and Synergizes with Anthracyclines in Breast Cancer. Mol. Cancer Ther. 2006, 5, 1807–1816. [Google Scholar] [CrossRef] [Green Version]
- Porcelli, L.; Iacobazzi, R.M.; Quatrale, A.E.; Bergamini, C.; Denora, N.; Crupi, P.; Antonacci, D.; Mangia, A.; Simone, G.; Silvestris, N.; et al. Grape Seed Extracts Modify the Outcome of Oxaliplatin in Colon Cancer Cells by Interfering with Cellular Mechanisms of Drug Cytotoxicity. Oncotarget 2017, 8, 50845–50863. [Google Scholar] [CrossRef] [Green Version]
- Yallapu, M.M.; Gupta, B.K.; Jaggi, M.; Chauhan, S.C. Fabrication of Curcumin Encapsulated PLGA Nanoparticles for Improved Therapeutic Effects in Metastatic Cancer Cells. J. Colloid Interface Sci. 2010, 351, 19–29. [Google Scholar] [CrossRef]
- Locatelli, E.; Franchini, M.C. Biodegradable PLGA-b-PEG Polymeric Nanoparticles: Synthesis, Properties, and Nanomedical Applications as Drug Delivery System. J. Nanoparticle Res. 2012, 14, 1316. [Google Scholar] [CrossRef]
- Liu, P.; Yu, H.; Sun, Y.; Zhu, M.; Duan, Y. A mPEG-PLGA-b-PLL Copolymer Carrier for Adriamycin and siRNA Delivery. Biomaterials 2012, 33, 4403–4412. [Google Scholar] [CrossRef]
- Huo, Q.; Zhu, J.; Niu, Y.; Shi, H.; Gong, Y.; Li, Y.; Song, H. pH-Triggered Surface Charge-Switchable Polymer Micelles for the Co-Delivery of Paclitaxel/Disulfiram and Overcoming Multidrug Resistance in Cancer. Int. J. Nanomed. 2017, 12, 8631–8647. [Google Scholar] [CrossRef]
- Chen, S.; Rong, L.; Lei, Q.; Cao, P.-X.; Qin, S.-Y.; Zheng, D.-W.; Jia, H.-Z.; Zhu, J.-Y.; Cheng, S.-X.; Zhuo, R.-X.; et al. A Surface Charge-Switchable and Folate Modified System for Co-Delivery of Proapoptosis Peptide and p53 Plasmid in Cancer Therapy. Biomaterials 2016, 77, 149–163. [Google Scholar] [CrossRef]
- Li, X.-X.; Chen, J.; Shen, J.-M.; Zhuang, R.; Zhang, S.-Q.; Zhu, Z.-Y.; Ma, J.-B. pH-Sensitive Nanoparticles as Smart Carriers for Selective Intracellular Drug Delivery to Tumor. Int. J. Pharm. 2018, 545, 274–285. [Google Scholar] [CrossRef]
- Jeong, J.H.; Park, T.G. Poly(L-Lysine)-g-poly(D,L-Lactic-Co-Glycolic Acid) Micelles for Low Cytotoxic Biodegradable Gene Delivery Carriers. J. Control. Release 2002, 82, 159–166. [Google Scholar] [CrossRef]
- Margiotta, N.; Savino, S.; Denora, N.; Marzano, C.; Laquintana, V.; Cutrignelli, A.; Hoeschele, J.D.; Gandin, V.; Natile, G. Encapsulation of Lipophilic Kiteplatin Pt(iv) Prodrugs in PLGA-PEG Micelles. Dalton Trans. 2016, 45, 13070–13081. [Google Scholar] [CrossRef] [PubMed]
- Annese, C.; Abbrescia, D.I.; Catucci, L.; D’Accolti, L.; Denora, N.; Fanizza, I.; Fusco, C.; La Piana, G. Site-Dependent Biological Activity of Valinomycin Analogs Bearing Derivatizable Hydroxyl Sites. J. Pept. Sci. 2013, 19, 751–757. [Google Scholar] [CrossRef]
- Tahir, N.; Madni, A.; Li, W.; Correia, A.; Khan, M.M.; Rahim, M.A.; Santos, H.A. Microfluidic Fabrication and Characterization of Sorafenib-Loaded Lipid-Polymer Hybrid Nanoparticles for Controlled Drug Delivery. Int. J. Pharm. 2020, 581, 119275. [Google Scholar] [CrossRef]
- Owens, D.E., III; Peppas, N.A. Opsonization, Biodistribution, and Pharmacokinetics of Polymeric Nanoparticles. Int. J. Pharm. 2006, 307, 93–102. [Google Scholar] [CrossRef]
- Moghimi, S.M.; Porter, C.; Muir, I.; Illum, L.; Davis, S. Non-Phagocytic Uptake of Intravenously Injected Microspheres in Rat spleen: Influence of Particle Size and Hydrophilic Coating. Biochem. Biophys. Res. Commun. 1991, 177, 861–866. [Google Scholar] [CrossRef]
- Corbet, C.; Bastien, E.; De Jesus, J.P.S.; Dierge, E.; Martherus, R.; Linden, C.V.; Doix, B.; Degavre, C.; Guilbaud, C.; Petit, L.; et al. TGFβ2-Induced Formation of Lipid Droplets Supports Acidosis-Driven EMT and the Metastatic Spreading of Cancer Cells. Nat. Commun. 2020, 11, 454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denora, N.; Laquintana, V.; Lopalco, A.; Iacobazzi, R.M.; Lopedota, A.; Cutrignelli, A.; Iacobellis, G.; Annese, C.; Cascione, M.; Leporatti, S.; et al. In Vitro Targeting and Imaging the Translocator Protein TSPO 18-kDa through G(4)-PAMAM–FITC Labeled Dendrimer. J. Control. Release 2013, 172, 1111–1125. [Google Scholar] [CrossRef]
- Wang, F.; Liu, H.; Hu, L.; Liu, Y.; Duan, Y.; Cui, R.; Tian, W. The Warburg Effect in Human Pancreatic Cancer Cells Triggers Cachexia in Athymic Mice Carrying the Cancer Cells. BMC Cancer 2018, 18, 360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haschka, M.; Karbon, G.; Fava, L.; Villunger, A. Perturbing mitosis for Anti-Cancer Therapy: Is Cell Death the Only Answer? EMBO Rep. 2018, 19, e45440. [Google Scholar] [CrossRef]


| Micelles | PLGA-PEG | PLGA-PLL | PLGA-PLL-DMA | PLGA-PEG-AE105 |
|---|---|---|---|---|
| NoResMic (Non-responsive micelles) | + | + | − | − |
| pHResMic (pH-responsive micelles) | + | − | + | − |
| TpHResMic (Targeted pH-responsive micelles) | + | − | + | + |
| Micelles Samples | dmean | PdI | ζ-Potential | EE% | |||
|---|---|---|---|---|---|---|---|
| pH 7.4 | pH 6.8 | pH 7.4 | pH 6.8 | pH 7.4 | pH 6.8 | ||
| NoResMic | 102.70 ± 5.64 | 92.24 ± 4.54 | 0.16 ± 0.02 | 0.14 ± 0.05 | 16.60 ± 4.53 | 20.60 ± 3.32 | / |
| pHResMic | 90.20 ± 1.26 | 86.47 ± 1.22 | 0.18 ± 0.03 | 0.15 ± 0.05 | −19.30 ± 3.33 | 25.50 ± 1.59 | / |
| TpHResMic | 165.60 ± 1.24 | 130.60 ± 1.08 | 0.23 ± 0.01 | 0.26 ± 0.02 | −15.64 ± 0.86 | 13.70 ± 1.87 | / |
| Gem@pHResMic | 92.10 ± 1.12 | 104.30 ± 1.23 | 0.15 ± 0.02 | 0.16 ± 0.04 | −15.20 ± 2.09 | 22.80 ± 1.50 | 22.00% ± 2.10 |
| Gem@TpHResMic | 134.30 ± 3.74 | 124.40 ± 3.47 | 0.22 ± 0.05 | 0.19 ± 0.05 | −13.80 ± 1.69 | 16.80 ± 0.70 | 29.00% ± 1.22 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iacobazzi, R.M.; Arduino, I.; Di Fonte, R.; Lopedota, A.A.; Serratì, S.; Racaniello, G.; Bruno, V.; Laquintana, V.; Lee, B.-C.; Silvestris, N.; et al. Microfluidic-Assisted Preparation of Targeted pH-Responsive Polymeric Micelles Improves Gemcitabine Effectiveness in PDAC: In Vitro Insights. Cancers 2022, 14, 5. https://doi.org/10.3390/cancers14010005
Iacobazzi RM, Arduino I, Di Fonte R, Lopedota AA, Serratì S, Racaniello G, Bruno V, Laquintana V, Lee B-C, Silvestris N, et al. Microfluidic-Assisted Preparation of Targeted pH-Responsive Polymeric Micelles Improves Gemcitabine Effectiveness in PDAC: In Vitro Insights. Cancers. 2022; 14(1):5. https://doi.org/10.3390/cancers14010005
Chicago/Turabian StyleIacobazzi, Rosa Maria, Ilaria Arduino, Roberta Di Fonte, Angela Assunta Lopedota, Simona Serratì, Giuseppe Racaniello, Viviana Bruno, Valentino Laquintana, Byung-Chul Lee, Nicola Silvestris, and et al. 2022. "Microfluidic-Assisted Preparation of Targeted pH-Responsive Polymeric Micelles Improves Gemcitabine Effectiveness in PDAC: In Vitro Insights" Cancers 14, no. 1: 5. https://doi.org/10.3390/cancers14010005
APA StyleIacobazzi, R. M., Arduino, I., Di Fonte, R., Lopedota, A. A., Serratì, S., Racaniello, G., Bruno, V., Laquintana, V., Lee, B.-C., Silvestris, N., Leonetti, F., Denora, N., Porcelli, L., & Azzariti, A. (2022). Microfluidic-Assisted Preparation of Targeted pH-Responsive Polymeric Micelles Improves Gemcitabine Effectiveness in PDAC: In Vitro Insights. Cancers, 14(1), 5. https://doi.org/10.3390/cancers14010005














