Review of the Geant4-DNA Simulation Toolkit for Radiobiological Applications at the Cellular and DNA Level
Abstract
:Simple Summary
Abstract
1. Monte Carlo Radiation Track Simulations
| Code | Particles | Energy Range | Target Materials | Chemical Stage | Reference |
|---|---|---|---|---|---|
| CPA100 | e− | Thermalization–256 keV | Water (l), DNA | Yes | Terrisol and Beaudré (1990) [17] |
| DELTA | e− | ≥10 eV–10 keV | Water (v) | Yes | Zaider et al. (1983) [18] |
| EPOTRAN | e−, e+ | ≥7.4 eV–10 keV | Water (l,v) | No | Champion et al. (2012) [19] |
| ETRACK | e−, p, α | ≥10 eV–10 keV | Water (v) | Yes | Ito (1987) [20] |
| ETS | e− | ≥10 eV–10 keV | Water (l,v) | Yes | Hill and Smith (1994) [21] |
| Geant4-DNA | e−, p, H, α, ions | Thermalization–1MeV e−, 100 eV–100 MeV p, H, 1 keV–400 MeVα, 0.5MeV/u−106MeV/u ions | Water (l), DNA, Gold | Yes | Incerti et al. (2010, 2018), Bernal et al. (2015) [14,22,23,24] |
| IONLYS/IONLYS-IRT | e−, p, ions | 0.2 eV–150 keV e−, p, 0.1 MeV-300 MeV ions | Water (l) | Yes | Cobut et al. (1998) [25] |
| KAPLAN | e− | ≥1–10 keV | Water (l,v) | Yes | Kaplan (1990) [26] |
| KITrack | e−, ions | ≥10 eV–100 keV | Water (l) | No | Wiklund et al. (2011) [27] |
| KURBUC (KURBUC/LEAHIST/LEPHIST/CHEM-KURBUC) | e−, p, α, C | 10 eV–10 MeV (10keV, liq.) e−,1 keV–300 MeV p, 1keV/u-2MeV/u α, 1 keV/u–10 MeV/u carbon | Water (l,v) | Yes | Nikjoo et al. (2016) [10] |
| LEEPS | e−, e+ | 0.1–100 keV | All materials | Yes | Fernández-Varea et al. (1996) [28] |
| LEPTS | e−, e+, p | Thermalization–10 keV e−, Thermalization–10 MeV p | Water (v), CH4, C2H4, C4H8O, SF6, C4H4N2 | No | Sanz et al. (2012), Blanco et al. (2013) [29,30] |
| Lion Track | e−, p, ions | >50 eV e−, 0.5 MeV/u–300 MeV/u p, ions | Water (l) | No | Bäckström et al. (2013) [31] |
| MC4 | e−, ions | ≥10 eV e−, ≥0.3 MeV/u ions | Water (l,v) | No | Emfietzoglou et al. (2017) [32] |
| MOCA8B | e− | 10 eV–100 keV | Water (v) | Yes | Paretzke (1970) [33] |
| NASIC | e− | Thermalization–1 MeV e− | Water (l) | Yes | Li et al. (2015) [34] |
| NOTRE DAME | e−, ions | ≥ 10 eV e−, ≥0.3 MeV/u ions | Water (l,v) | Yes | Pimblott et al. (1990) [35] |
| OREC/NOREC | e− | 7.4 eV–1 MeV e− | Water (l) | No | Semenenko et al. (2003) [8] |
| PARTRAC | e−, e+, p, H, α, ions | 1 eV–10 MeV e−, 1 keV–1 GeV p, H, α, 1 MeV/u–1 GeV/u ions | Water (l), DNA | Yes | Friedland et al. (2003) [36] |
| PITS04 | e−, ions | ≥ 10 eV e−, ≥ 0,3 MeV/u ions | Water (l) | No | Wilson et al. (2004) [37] |
| PITS99 | e−, ions | ≥ 10 eV e−, ≥ 0,3 MeV/u ions | Water (v) | Yes | Wilson and Nikjoo (1999) [38] |
| PTra | e−, p, α | 1 eV–10 keV e−, 1–10 MeV α, 300 keV-10 MeV p | Water (l,v), DNA | No | Grosswendt and Pszona (2002) [39] |
| RITRACKS/RETRACKS | e−, ions | 0.1 eV–100 MeV e−, 10−1MeV/u–104MeV/u ions | Water (l,v) | Yes | Plante and Cucinotta (2009) [40] |
| SHERBROOKE | e−, ions | ≥ 10 eV e−, ≥ 0,3 MeV/u ions | Water (l,v) | Yes | Cobut et al. (2004) [41] |
| STBRGEN | e−, ions | ≥ 10 eV e−, ≥ 0,3 MeV/u ions | Water (l,v) | Yes | Chatterjee and Holley (1993) [42] |
| TILDA-V | e−, p, H, ions | ≥ 7,4 eV e−, 10 keV/u–100 MeV/u ions | Water (l,v), DNA | No | Champion et al. (2005) [43] |
| TRAX | e−, p, ions | 1 eV–few MeV e−, 10 eV–few hundred MeV/u ions | Water (v) | Yes | Krämer and Kraft (1994) [44] |
| RADAMOL (TRIOL/STOCHECO) | e−, ions | ≥7.4 eV–2 MeV e−, ≥0.3–200 MeV/u ions | Water (l) | Yes | Bigildeev and Michalik (1996) [45] |
| TRION | e−, ions | ≥10 eV e−, ≥0.3 MeV/u ions | Water (l,v) | No | Lappa et al. (1993) [46] |
| TRACEL/RADYIE/RADIFF | e−, ions | ≥10 eV e−, ≥0.3 MeV/u ions | Water (l,v) | Yes | Tomita et al. (1997) [47] |
2. The Geant4-DNA Extension
3. Physical Interactions in Geant4-DNA: Energy-Loss Models
4. The Physico-Chemical and Chemical Stages of Water Radiolysis
4.1. The Physico-Chemical Stage
4.2. The Chemical Stage
4.2.1. Step by Step Method
4.2.2. Independent Reaction Time
5. Towards the Modelling of Early DNA Damage
5.1. History
5.2. “FullSim” Complete Simulation Chain for DSBs Calculations
5.3. Review of “MolecularDNA”Application
6. Geant4-DNA Extended Examples
6.1. Physics Examples
- The “clustering” example calculates the energy deposition with a dedicated clustering algorithm to assess DNA strand breaks in a simple liquid water geometry [14];
- “dnaphysics” is a general example that enables track-structure simulation of charged particles in a liquid water geometry and allows for the automatic combination between Geant4-DNA physics models and condensed-history models at higher energies (i.e., above 1 MeV) and can be used for benchmarking simulations that are related to track-structure characteristics [23];
- “icsd”, that stands for ionization cluster size distribution, calculates the number of ionizations for each simulated track in a cylinder mimicking a piece of chromatin and uses DNA-like material’s cross sections that were obtained experimentally or by simulations [50];
- “mfp” stands for mean free path and allows the calculation of the aforementioned distance and related distance quantities for a charged particle in a sphere geometry of liquid water [23];
- “microdosimetry” simulates lineal and specific energy distributions and related quantities in liquid water spheres that are randomly placed along the particle track [59];
- “microprox” is another microdosimetric example that calculates proximity functions from energy depositions scored in liquid water spherical shells from randomly selected hits [60];
- “range” example performs a simulation of penetration distances in liquid water [70];
- “slowing” enables simulation of the slowing down spectra of electrons in a cube of liquid water [136];
- “splitting” uses variance reduction techniques to improve the efficiency of the calculation of ionization cluster size distributions. This is done in a nm sized cylinder as in the case of the icsd example and aims to separate secondaries that are generated within the cylinder to avoid the overlapping of tracks [137];
- “spower” allows for stopping power simulations of particles in liquid water with the use of specific physics modules that enable the use of a stationary mode for appropriate computation [23];
- “svalue” calculates the dose to a target volume per unit of cumulated activity in a source volume, called S-value [138,139]. The source and target volumes can be different cell compartments or an entire cell of a simple spherical geometry which can be modified to account for more complex cell geometries, as has been done in many studies i.e. [140,141];
- “wvalue” serves to simulate the mean energy that is expended to form an ion pair known as W-value. It also provides information on the total number of ionizations in a liquid water volume and its penetration details. It is a useful benchmark simulation for the inelastic models given that elastic interactions are indifferent in this simulation scheme [23,58].
6.2. Chemical Examples
- “chem1” aims to show how to activate or deactivate physicochemical and chemical stage after physical stage. Chemical reactions are printed and the step-by-step model is used by default.
- “chem2” provides a user-class “TimeStepAction” which allows users to change Minimum Time Steps. These parameters constrain the minimum time-step that is allowed for each reactant pair using the step-by-step model. The user-class also shows how to print reaction information such as reactants and products as well as their positions.
- “chem3” illustrates how to implement user actions in the chemistry module using the step-by-step model. Users can also visualize the trajectories of the chemical species in time and space using the graphical user interface.
- “chem4” provides scorer classes to compute radiochemical yields (“G”) versus time using the step-by-step model, including a dedicated ROOT graphical interface. The G-value is useful for benchmark simulations in comparing with other MC codes and experimental data [80].
- “chem5” computes radiochemical yields (“G”) versus time using alternative physics and chemical reaction lists using the step-by-step model [142].
6.3. The dnadamage1 Example
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Chetty, I.J.; Curran, B.; Cygler, J.E.; DeMarco, J.J.; Ezzell, G.; Faddegon, B.A.; Kawrakow, I.; Keall, P.J.; Liu, H.; Ma, C.C.-M.; et al. Issues associated with clinical implementation of Monte Carlo based photon and electron external beam treatment planning. Med. Phys. 2007, 34, 4818–4853. [Google Scholar] [CrossRef] [PubMed]
- Briesmeister, J.F. MCNP-A General Monte Carlo mN-Particle Transport Code; Report LA-12625-M; Los Alamos National Laboratory: Los Alamos, NM, USA, 1993. [Google Scholar]
- Liu, J.C.; Bong, P.; Gray, B.; Mao, X.S.; Nelson, G.; Nelson, W.R.; Schultz, D.; Seeman, J. Radiation Safety System of The B-Factory at The Stanford Linear Accelerator Center. Health Phys. 1999, 77, 588–594. [Google Scholar] [CrossRef] [PubMed]
- Allison, J.; Amako, K.; Apostolakis, J.; Arce, P.; Asai, M.; Aso, T.; Bagli, E.; Bagulya, A.; Banerjee, S.; Barrand, G.; et al. Recent developments in Geant4. Nucl. Instrum. Methods Phys. Res. Sect. A Accel. Spectrometers Detect. Assoc. Equip. 2016, 835, 186–225. [Google Scholar] [CrossRef]
- Ferrari, A.; Ferrari, A.; Ranft, J.; Sala, P.R. FLUKA: A Multi-Particle Transport Code; Report CERN-2005-10; European Organization for Nuclear Research: Geneva, Switzerland, 2005. [Google Scholar]
- Baró, J.; Sempau, J.; Fernández-Varea, J.M.; Salvat, F. PENELOPE: An algorithm for Monte Carlo simulation of the penetration and energy loss of electrons and positrons in matter. Nucl. Instrum. Methods Phys. Res. Sect. B Beam Interactions Mater. Atoms 1995, 100, 31–46. [Google Scholar] [CrossRef]
- El Naqa, I.; Pater, P.; Seuntjens, J. Monte Carlo role in radiobiological modelling of radiotherapy outcomes. Phys. Med. Biol. 2012, 57, R75–R97. [Google Scholar] [CrossRef]
- Semenenko, V.A.; Turner, J.E.; Borak, T.B. NOREC, a Monte Carlo code for simulating electron tracks in liquid water. Radiat. Environ. Biophys. 2003, 42, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Friedland, W.; Schmitt, E.; Kundrát, P.; Dingfelder, M.; Baiocco, G.; Barbieri, S.; Ottolenghi, A. Comprehensive track-structure based evaluation of DNA damage by light ions from radiotherapy-relevant energies down to stopping. Sci. Rep. 2017, 7, srep45161. [Google Scholar] [CrossRef]
- Nikjoo, H.; Emfietzoglou, D.; Liamsuwan, T.; Taleei, R.; Liljequist, D.; Uehara, S. Radiation track, DNA damage and response-a review. Rep. Prog. Phys. 2016, 79, 116601. [Google Scholar] [CrossRef]
- Shin, W.-G.; Ramos-Mendez, J.; Faddegon, B.; Tran, H.N.; Villagrasa, C.; Perrot, Y.; Okada, S.; Karamitros, M.; Emfietzoglou, D.; Kyriakou, I.; et al. Evaluation of the influence of physical and chemical parameters on water radiolysis simulations under MeV electron irradiation using Geant4-DNA. J. Appl. Phys. 2019, 126, 114301. [Google Scholar] [CrossRef]
- Lindborg, L.; Nikjoo, H. Microdosimetry and radiation quality determinations in radiation protection and radiation therapy. Radiat. Prot. Dosim. 2011, 143, 402–408. [Google Scholar] [CrossRef]
- Goorley, T.; James, M.; Booth, T.; Brown, F.; Bull, J.; Cox, L.; Durkee, J.; Elson, J.; Fensin, M.; Forster, R.; et al. Features of MCNP6. Ann. Nucl. Energy 2016, 87, 772–783. [Google Scholar] [CrossRef] [Green Version]
- Bernal, M.A.; Bordage, M.; Brown, J.; Davídková, M.; Delage, E.; El Bitar, Z.; Enger, S.; Francis, Z.; Guatelli, S.; Ivanchenko, V.; et al. Track structure modeling in liquid water: A review of the Geant4-DNA very low energy extension of the Geant4 Monte Carlo simulation toolkit. Phys. Med. 2015, 31, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Varea, J.M.; González-Muñoz, G.; Galassi, M.E.; Wiklund, K.; Lind, B.K.; Ahnesjö, A.; Tilly, N. Limitations (and merits) of PENELOPE as a track-structure code. Int. J. Radiat. Biol. 2011, 88, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Niita, K.; Sato, T.; Iwase, H.; Nose, H.; Nakashima, H.; Sihver, L. PHITS—A particle and heavy ion transport code system. Radiat. Meas. 2006, 41, 1080–1090. [Google Scholar] [CrossRef]
- Terissol, M.; Beaudre, A. A simulation of space and time evolution of radiolytic species induced by electrons in water. Radiat. Prot. Dosim. 1990, 31, 175–177. [Google Scholar] [CrossRef]
- Zaider, M.; Brenner, D.J.; Wilson, W.E. The Applications of Track Calculations to Radiobiology I. Monte Carlo Simulation of Proton Tracks. Radiat. Res. 1983, 95, 231. [Google Scholar] [CrossRef]
- Champion, C.; Le Loirec, C.; Stosic, B. EPOTRAN: A full-differential Monte Carlo code for electron and positron transport in liquid and gaseous water. Int. J. Radiat. Biol. 2011, 88, 54–61. [Google Scholar] [CrossRef]
- Ito, A. Calculation of double strand break probability of DNA for low LET radiations based on track structure analysis. In Nuclear and Atomic Data for Radiotherapy and Related Radiobiology; IAEA: Vienna, Austria, 1987. [Google Scholar]
- Hill, M.; Smith, F. Calculation of initial and primary yields in the radiolysis of water. Radiat. Phys. Chem. 1994, 43, 265–280. [Google Scholar] [CrossRef]
- Incerti, S.; Baldacchino, G.; Bernal, M.A.; Capra, R.; Champion, C.; Francis, Z.; Guèye, P.; Mantero, A.; Mascialino, B.; Moretto, P.; et al. The Geant4-DNA project. Int. J. Model. Simulat. Sci. Comput. 2010, 1, 157–178. [Google Scholar] [CrossRef]
- Incerti, S.; Kyriakou, I.; Bernal, M.A.; Bordage, M.C.; Francis, Z.; Guatelli, S.; Ivanchenko, V.; Karamitros, M.; Lampe, N.; Lee, S.B.; et al. Geant4-DNA example applications for track structure simulations in liquid water: A report from the Geant4-DNA Project. Med. Phys. 2018, 45, e722–e739. [Google Scholar] [CrossRef] [Green Version]
- Incerti, S.; Ivanchenko, A.; Karamitros, M.; Mantero, A.; Moretto, P.; Tran, N.H.; Mascialino, B.; Champion, C.; Ivanchenko, V.; Bernal, M.A.; et al. Comparison of Geant4 very low energy cross section models with experimental data in water. Med. Phys. 2010, 37, 4692–4708. [Google Scholar] [CrossRef]
- Cobut, V.; Frongillo, Y.; Patau, J.P.; Goulet, T.; Fraser, M.J.; Jay-Gerin, J.P. Monte Carlo simulation of fast electron and proton tracks in liquid water-I. Physical and physicochemical aspects. Radiat. Phys. Chem. 1998, 51, 229–243. [Google Scholar] [CrossRef]
- Kaplan, I.; Miterev, A.; Sukhonosov, V. Simulation of the primary stage of liquid water radiolysis. Int. J. Radiat. Appl. Instrumentation. Part C. Radiat. Phys. Chem. 1990, 36, 493–498. [Google Scholar] [CrossRef]
- Wiklund, K.; Fernández-Varea, J.M.; Lind, B.K. A Monte Carlo program for the analysis of low-energy electron tracks in liquid water. Phys. Med. Biol. 2011, 56, 1985–2003. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Varea, J.M.; Liljequist, D.; Csillag, S.; Räty, R.; Salvat, F. Monte Carlo simulation of 0.1–100 keV electron and positron transport in solids using optical data and partial wave methods. Nucl. Instrum. Methods Phys. Res. Sect. B Beam Interact. Mater. Atoms 1996, 108, 35–50. [Google Scholar] [CrossRef]
- Sanz, A.G.; Fuss, M.C.; Muñoz, A.; Blanco, F.; Limão-Vieira, P.; Brunger, M.J.; Buckman, S.J.; Garcia, G. Modelling low energy electron and positron tracks for biomedical applications. Int. J. Radiat. Biol. 2011, 88, 71–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanco, F.; Muñoz, A.; Almeida, D.; Da Silva, F.F.; Limão-Vieira, P.; Fuss, M.C.; Sanz, A.G.; Garcia, G. Modelling low energy electron and positron tracks in biologically relevant media. Eur. Phys. J. D 2013, 67, 99. [Google Scholar] [CrossRef]
- Bäckstrom, G.; Galassi, M.E.; Tilly, N.; Ahnesjo, A.; Fernández-Varea, J.M. Track structure of protons and other light ions in liquid water: Applications of the LIonTrack code at the nanometer scale. Med. Phys. 2013, 40, 064101. [Google Scholar] [CrossRef]
- Emfietzoglou, D.; Papamichael, G.; Nikjoo, H. Monte Carlo Electron Track Structure Calculations in Liquid Water Using a New Model Dielectric Response Function. Radiat. Res. 2017, 188, 355–368. [Google Scholar] [CrossRef]
- Paretzke, H.G. Simulation von Elektronenspuren im Energiebereich 0.01–10keV in Wasserdampf; GSF-Berich 24/88 Gesellshaft für Strahlen-und Umwelt-Forschung: München, Germany, 1970. [Google Scholar]
- Li, J.; Li, C.; Qiu, R.; Yan, C.; Xie, W.; Wu, Z.; Zeng, Z.; Tung, C. DNA strand breaks induced by electrons simulated with Nanodosimetry Monte Carlo Simulation Code: NASIC. Radiat. Prot. Dosim. 2015, 166, 38–43. [Google Scholar] [CrossRef]
- Pimblott, S.; LaVerne, J.A.; Mozumder, A.; Green, N.J.B. Structure of electron tracks in water. 1. Distribution of energy deposition events. J. Phys. Chem. 1990, 94, 488–495. [Google Scholar] [CrossRef]
- Friedland, W.; Jacob, P.; Bernhardt, P.; Paretzke, H.G.; Dingfelder, M. Simulation of DNA Damage after Proton Irradiation. Radiat. Res. 2003, 159, 401–410. [Google Scholar] [CrossRef]
- Wilson, W.E.; Miller, J.H.; Lynch, D.J.; Lewis, R.R.; Batdorf, M. Analysis of low-energy electron track structure in liquid water. Radiat. Res. 2004, 161, 591–596. [Google Scholar] [CrossRef] [PubMed]
- Wilson, W.E.; Nikjoo, H. A Monte Carlo code for positive ion track simulation. Radiat. Environ. Biophys. 1999, 38, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Grosswendt, B.; Pszona, S. The track structure of α-particles from the point of view of ionization-cluster formation in “nanometric” volumes of nitrogen. Radiat. Environ. Biophys. 2002, 41, 91–102. [Google Scholar] [CrossRef]
- Plante, I.; Cucinotta, F.A. Cross sections for the interactions of 1 eV–100 MeV electrons in liquid water and application to Monte-Carlo simulation of HZE radiation tracks. New J. Phys. 2009, 11, 063047. [Google Scholar] [CrossRef]
- Cobut, V.; Cirioni, L.; Patau, J.P. Accurate transport simulation of electron tracks in the energy range 1keV–4MeV. Nucl. Intrum. Meth. B 2004, 215, 57–68. [Google Scholar] [CrossRef]
- Chatterjee, A.; Holley, W.R. Computer Simulation of Initial Events in the Biochemical Mechanisms of DNA Damage. Adv. Radiat. Biol. 1993, 17, 181–226. [Google Scholar] [CrossRef]
- Champion, C.; L’Hoir, A.; Politis, M.F.; Fainstein, P.D.; Rivarola, R.D.; Chetioui, A. A Monte Carlo Code for the Simulation of Heavy-Ion Tracks in Water. Radiat. Res. 2005, 163, 222–231. [Google Scholar] [CrossRef]
- Krämer, M.; Kraft, G. Calculations of heavy ion track structure. Radiat. Environ. Biophys. 1994, 33, 91–109. [Google Scholar] [CrossRef]
- Bigildeev, E.; Michalik, V. Charged particle tracks in water of different phases. Monte Carlo simulation of electron tracks. Radiat. Phys. Chem. 1996, 47, 197–207. [Google Scholar] [CrossRef]
- Lappa, A.V.; Bigildeev, E.A.; Burmistrov, D.S.; Vasilyev, O.N. “Trion” code for radiation action calculations and its application in microdosimetry and radiobiology. Radiat. Environ. Biophys. 1993, 32, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Tomita, H.; Kai, M.; Kusama, T.; Ito, A. Monte Carlo simulation of physicochemical processes of liquid water radiolysis. Radiat. Environ. Biophys. 1997, 36, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Dingfelder, M. Track-structure Simulations for Charged Particles. Health Phys. 2012, 103, 590–595. [Google Scholar] [CrossRef] [Green Version]
- LaVerne, J.A.; Pimblott, S.M. Electron Energy-Loss Distributions in Solid, Dry DNA. Radiat. Res. 1995, 141, 208. [Google Scholar] [CrossRef]
- Bug, M.U.; Baek, W.Y.; Rabus, H.; Villagrasa, C.; Meylan, S.; Rosenfeld, A. An electron-impact cross section data set (10 eV–1 keV) of DNA constituents based on consistent experimental data: A requisite for Monte Carlo simulations. Radiat. Phys. Chem. 2017, 130, 459–479. [Google Scholar] [CrossRef]
- Zein, S.A.; Bordage, M.-C.; Francis, Z.; Macetti, G.; Genoni, A.; Cappello, C.D.; Shin, W.-G.; Incerti, S. Electron transport in DNA bases: An extension of the Geant4-DNA Monte Carlo toolkit. Nucl. Instrum. Methods Phys. Res. Sect. B Beam Interact. Mater. Atoms 2021, 488, 70–82. [Google Scholar] [CrossRef]
- Bernhardt, P.; Paretzke, H.G. Calculation of electron impact ionization cross sections of DNA using the Deutch-Mark and Binary-Encounter Bethe formalisms. Int. J. Mass Spectrom. 2003, 223, 599–611. [Google Scholar] [CrossRef]
- Tan, Z.; Xia, X.; Liu, M.; Zhao, M.; Ji, Y.; Li, F.; Huang, B. Cross sections of electron elastic interactions in DNA. Radiat. Environ. Biophys. 2004, 43, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Moejko, P.; Sanche, L. Cross section calculations for electron scattering from DNA and RNA bases. Radiat. Environ. Biophys. 2003, 42, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Abril, I.; Garcia-Molina, R.; Denton, C.D.; Kyriakou, I.; Emfietzoglou, D. Energy loss of hydrogen- and helium-ion beams in DNA: Calculations based on a realistic energy-loss function of the target. Radiat. Res. 2011, 175, 247–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abril, I.; Garcia-Molina, R.; De Vera, P.; Kyriakou, I.; Emfietzoglou, D. Inelastic Collisions of Energetic Protons in Biological Media. Quantum Boundaries Life 2013, 65, 129–164. [Google Scholar] [CrossRef]
- Garcia-Molina, R.; Abril, I.; Kyriakou, I.; Emfietzoglou, D. Inelastic scattering and energy loss of swift electron beams in biologically relevant materials. Surf. Interface Anal. 2017, 49, 11–17. [Google Scholar] [CrossRef]
- Kyriakou, I.; Incerti, S.; Francis, Z. Technical Note: Improvements in Geant4 energy-loss model and the effect on low-energy electron transport in liquid water. Med. Phys. 2015, 42, 3870–3876. [Google Scholar] [CrossRef]
- Kyriakou, I.; Emfietzoglou, D.; Ivanchenko, V.; Bordage, M.C.; Guatelli, S.; Lazarakis, P.; Tran, N.H.; Incerti, S. Microdosimetry of electrons in liquid water using the low-energy models of Geant4. J. Appl. Phys. 2017, 122, 024303. [Google Scholar] [CrossRef]
- Incerti, S.; Kyriakou, I.; Bordage, M.C.; Guatelli, S.; Ivanchenko, V.; Emfietzoglou, D. Track structure simulations of proximity functions in iquid water using the Geant4-DNA toolkit. J. Appl. Phys. 2019, 125, 104301. [Google Scholar] [CrossRef] [Green Version]
- Lazarakis, P.; Incerti, S.; Ivanchenko, V.; Kyriakou, I.; Emfietzoglou, D.; Corde, S.; Rosenfeld, A.; Lerch, M.; Tehei, M.; Guatelli, S. Investigation of track structure and condensed history physics models for applications in radiation dosimetry on a micro and nano scale in Geant4. Biomed. Phys. Eng. Express 2018, 4, 024001. [Google Scholar] [CrossRef] [Green Version]
- Kyriakou, I.; Ivanchenko, V.; Sakata, D.; Bordage, M.; Guatelli, S.; Incerti, S.; Emfietzoglou, D. Influence of track structure and condensed history physics models of Geant4 to nanoscale electron transport in liquid water. Phys. Med. 2019, 58, 149–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engels, E.; Bakr, S.; Bolst, D.; Sakata, D.; Li, N.; Lazarakis, P.; McMahon, S.J.; Ivanchenko, V.N.; Rosenfeld, A.B.; Incerti, S.; et al. Advances in modelling gold nanoparticle radiosensitization using new Geant4-DNA physics models. Phys. Med. Biol. 2020, 65, 225017. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-K.; Rudd, M.E. Binary-encounter-dipole model for electron-impact ionization. Phys. Rev. A 1994, 50, 3954–3967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emfietzoglou, D.; Karava, K.; Papamichael, G.; Moscovitch, M. Monte Carlo simulation of the energy loss of low-energy electrons in liquid water. Phys. Med. Biol. 2003, 48, 2355–2371. [Google Scholar] [CrossRef]
- Emfietzoglou, D.; Kyriakou, I.; Garcia-Molina, R.; Abril, I. Inelastic mean free path of low-energy electrons in condensed media: Beyond the standard models. Surf. Interface Anal. 2017, 49, 4–10. [Google Scholar] [CrossRef]
- Dingfelder, M.; Ritchie, R.H.; Turner, J.E.; Friedland, W.; Paretzke, H.G.; Hamm, R.N. Comparisons of Calculations with PARTRAC and NOREC: Transport of Electrons in Liquid Water. Radiat. Res. 2008, 169, 584–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emfietzoglou, D.; Kyriakou, I.; Abril, I.; Garcia-Molina, R.; Nikjoo, H. Inelastic scattering of low-energy electrons in liquid water computed from optical-data models of the Bethe surface. Int. J. Radiat. Biol. 2011, 88, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Heller, J.M.; Hamm, R.N.; Birkhoff, R.D.; Painter, L.R. Collective oscillation in liquid water. J. Chem. Phys. 1974, 60, 3483–3486. [Google Scholar] [CrossRef]
- Kyriakou, I.; Šefl, M.; Nourry, V.; Incerti, S. The impact of new Geant4-DNA cross section models on electron track structure simulations in liquid water. J. Appl. Phys. 2016, 119, 194902. [Google Scholar] [CrossRef]
- Lampe, N.; Karamitros, M.; Breton, V.; Brown, J.M.; Sakata, D.; Sarramia, D.; Incerti, S. Mechanistic DNA damage simulations in Geant4-DNA Part 2: Electron and proton damage in a bacterial cell. Phys. Med. 2018, 48, 146–155. [Google Scholar] [CrossRef]
- Margis, S.; Magouni, M.; Kyriakou, I.; Georgakilas, A.G.; Incerti, S.; Emfietzoglou, D. Microdosimetric calculations of the direct DNA damage induced by low energy electrons using the Geant4-DNA Monte Carlo code. Phys. Med. Biol. 2020, 65, 045007. [Google Scholar] [CrossRef] [PubMed]
- Emfietzoglou, D.; Kyriakou, I.; Garcia-Molina, R.; Abril, I.; Nikjoo, H. Inelastic Cross Sections for Low-Energy Electrons in Liquid Water: Exchange and Correlation Effects. Radiat. Res. 2013, 180, 499–513. [Google Scholar] [CrossRef] [PubMed]
- Emfietzoglou, D.; Kyriakou, I.; Garcia-Molina, R.; Abril, I. The effect of static many-body local-field corrections to inelastic electron scattering in condensed media. J. Appl. Phys. 2013, 114, 144907. [Google Scholar] [CrossRef]
- Bordage, M.; Bordes, J.; Edel, S.; Terrissol, M.; Franceries, X.; Bardiès, M.; Lampe, N.; Incerti, S. Implementation of new physics models for low energy electrons in liquid water in Geant4-DNA. Phys. Med. 2016, 32, 1833–1840. [Google Scholar] [CrossRef]
- Peudon, A.; Edel, S.; Terrissol, M. Molecular basic data calculation for radiation transport in chromatin. Radiat. Prot. Dosim. 2006, 122, 128–135. [Google Scholar] [CrossRef]
- Edel. S. Modélisation du Transport des Protons et des Électrons Dans l’AND Plasmide. Ph.D. Thesis, Toulouse III-Paul Sabatier University, Toulouse, France, 2006.
- Dingfelder, M.; Hantke, D.; Inokuti, M.; Paretzke, H.G. Electron inelastic-scattering cross sections in liquid water. Radiat. Phys. Chem. 1998, 53, 1–18. [Google Scholar] [CrossRef]
- Karamitros, M.; Luan, S.; Bernal, M.; Allison, J.; Baldacchino, G.; Davidkova, M.; Francis, Z.; Friedland, W.; Ivantchenko, V.; Mantero, A.; et al. Diffusion-controlled reactions modeling in Geant4-DNA. J. Comput. Phys. 2014, 274, 841–882. [Google Scholar] [CrossRef]
- Karamitros, M.; Mantero, A.; Incerti, S.; Friedland, W.; Baldacchino, G.; Barberet, P.; Bernal, M.; Capra, R.; Champion, C.; El Bitar, Z.; et al. Modeling Radiation Chemistry in the Geant4 Toolkit. Prog. Nucl. Sci. Technol. 2011, 2, 503–508. [Google Scholar] [CrossRef]
- Clifford, P.; Green, N.J.B.; Oldfield, M.J.; Pilling, M.J.; Pimblott, S.M. Stochastic Models of Multi-species Kinetics in Radiation-induced Spurs. J. Chem. Soc. Faraday Trans. Phys. Chem. Condens. Phases 1986, 82, 2673–2689. [Google Scholar] [CrossRef]
- Kreipl, M.S.; Friedland, W.; Paretzke, H.G. Time- and space-resolved Monte Carlo study of water radiolysis for photon, electron and ion irradiation. Radiat. Environ. Biophys. 2009, 48, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Plante, I.; Devroye, L. Considerations for the independent reaction times and step-by-step methods for radiation chemistry simulations. Radiat. Phys. Chem. 2017, 139, 157–172. [Google Scholar] [CrossRef]
- Uehara, S.; Nikjoo, H. Monte Carlo Simulation of Water Radiolysis for Low-energy Charged Particles. J. Radiat. Res. 2006, 47, 69–81. [Google Scholar] [CrossRef] [Green Version]
- LaVerne, J.A. OH Radicals and Oxidizing Products in the Gamma Radiolysis of Water. Radiat. Res. 2000, 153, 196–200. [Google Scholar] [CrossRef]
- Jay-Gerin, J.-P.; Ferradini, C. A new estimate of the radical yield at early times in the radiolysis of liquid water. Chem. Phys. Lett. 2000, 317, 388–391. [Google Scholar] [CrossRef]
- El Omar, A.K.; Schmidhammer, U.; Jeunesse, P.; Larbre, J.-P.; Lin, M.; Muroya, Y.; Katsumura, Y.; Pernot, P.; Mostafavi, M. Time-Dependent Radiolytic Yield of OH• Radical Studied by Picosecond Pulse Radiolysis. J. Phys. Chem. A 2011, 115, 12212–12216. [Google Scholar] [CrossRef] [PubMed]
- Shiraishi, H.; Katsumura, Y.; Hiroishi, D.; Ishigure, K.; Washio, M. Pulse-Radiolysis Study on the Yield of Hydrated Electron at Elevated Temperaturess. J. Phys. Chem. 1988, 92, 3011–3017. [Google Scholar] [CrossRef]
- Sumiyoshi, T.; Katayama, M. The yield of hydrated electrons at 30 picoseconds. Chem. Lett. 1982, 11, 1887–1890. [Google Scholar] [CrossRef]
- Hunt, J.W.; Wolff, R.K.; Bronskill, M.J.; Jonah, C.D.; Hart, E.J.; Matheson, M.S. Radiolytic yields of hydrated electrons at 30 to 1000 picoseconds after energy absorption. J. Phys. Chem. 1973, 77, 425–426. [Google Scholar] [CrossRef]
- Wolff, R.K.; Bronskill, M.J.; Aldrich, J.E.; Hunt, J.W. Picosecond pulse radiolysis. IV. Yield of the solvated electron at 30 picoseconds. J. Phys. Chem. 1973, 77, 1350–1355. [Google Scholar] [CrossRef]
- Buxton, G.V. Nanosecond Pulse Radiolysis of Aqueous Solutions Containing Proton and Hydroxyl Radical Scavengers. Proc. Math. Phys. Eng. Sci. 1972, 328, 9–21. [Google Scholar] [CrossRef]
- Muroya, Y.; Lin, M.; Wu, G.; Iijima, H.; Yoshii, K.; Ueda, T.; Kudo, H.; Katsumura, Y. A re-evaluation of the initial yield of the hydrated electron in the picosecond time range. Radiat. Phys. Chem. 2005, 72, 169–172. [Google Scholar] [CrossRef]
- Pikaev, A.K.; Kabakchi, S.A.; Zansokhova, A.A. Yields and reactions of hydrogen ions on radiolysis of water and aqueous solutions. Faraday Discuss. Chem. Soc. 1977, 63, 112–123. [Google Scholar] [CrossRef]
- Cercek, B.; Kongshaug, M. Hydrogen ion yields in the radiolysis of neutral aqueous solutions. J. Phys. Chem. 1969, 73, 2056–2058. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.F.; Vojnovic, B.; Michael, B.D. The radiation-chemical yields of H3O+ and OH− as determined by nanosecond conductimetric measurements. Radiat. Phys. Chem. 1977, 26, 301–303. [Google Scholar] [CrossRef]
- Schmidt, K.H.; Ander, S.M. Formation and recombination of the hydronium ion (H3O+) and hydroxide in irradiated water. J. Phys. Chem. 1969, 73, 2846–2852. [Google Scholar] [CrossRef]
- Draganić, Z.; Draganić, I. Formation of primary reducing yields (Geaq- and GH2) in the radiolysis of aqueous solutions of some positive ions. Int. J. Radiat. Phys. Chem. 1975, 7, 381–386. [Google Scholar] [CrossRef]
- LaVerne, J.A.; Pimblott, S. Scavenger and time dependences of radicals and molecular products in the electron radiolysis of water: Examination of experiments and models. J. Phys. Chem. 1991, 95, 3196–3206. [Google Scholar] [CrossRef]
- Draganic, Z.D.; Draganic, I.G. Formation of primary hydrogen atom yield (GH) in the.gamma. radiolysis of water. J. Phys. Chem. 1972, 76, 2733–2737. [Google Scholar] [CrossRef]
- Chatzipapas, K.P.; Papadimitroulas, P.; Emfietzoglou, D.; Kalospyros, S.A.; Hada, M.; Georgakilas, A.G.; Kagadis, G.C. Ionizing Radiation and Complex DNA Damage: Quantifying the Radiobiological Damage Using Monte Carlo Simulations. Cancers 2020, 12, 799. [Google Scholar] [CrossRef] [Green Version]
- Chatzipappas, K.P.; Papadimitroulas, P.; Obeitat, M.A.; McConnell, K.A.; Kirby, N.; Loudos, G.; Kagadis, G.C.; Papanikolaou, N. Quantification of DNA Double Strand Breaks using Geant4-DNA. Med. Phys. 2019, 46, 405–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karamitros, M.; Brown, J.M.C.; Lampe, N.; Sakata, D.; Tran, N.H.; Shin, W.G.; LaVerne, J.A.; Mendez, J.R.; Guatelli, S.; Incerti, S. Implementing the Independent Reaction Time method in Geant4 for radiation chemistry simulations. arXiv 2020, arXiv:2006.14225. [Google Scholar]
- Ramos-Méndez, J.; Shin, W.; Karamitros, M.; Domínguez-Kondo, J.; Tran, N.H.; Incerti, S.; Villagrasa, C.; Perrot, Y.; Štěpán, V.; Okada, S.; et al. Independent reaction times method in Geant4-DNA: Implementation and performance. Med. Phys. 2020, 47, 5919–5930. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.N.; Mendez, J.R.; Shin, W.-G.; Perrot, Y.; Faddegon, B.; Okada, S.; Karamitros, M.; Davídková, M.; Štěpán, V.; Incerti, S.; et al. Assessment of DNA damage with a new Independent Reaction Time approach implemented in Geant4-DNA for the simulation of diffusion-controlled reactions between radio-induced reactive species and a chromatin fiber. Med. Phys. 2021, 48, 890–901. [Google Scholar] [CrossRef] [PubMed]
- Shin, W.G. Development and Application of the Geant4-DNA Toolkit for the Simulation of Radiobiological Effects at the Sub-Cellular Scale. Bordeaux University, France. 2020. Available online: https://tel.archives-ouvertes.fr/tel-03161030/document (accessed on 10 November 2021).
- Jorgensen, T.J. Enhancing radiosensitivity: Targeting the DNA repair pathways. Cancer Biol. Ther. 2009, 8, 665–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banáth, J.P.; Olive, P.L. Expression of phosphorylated histone H2AX as a surrogate of cell killing by drugs that create DNA double-strand breaks. Cancer Res. 2003, 63, 4347–4350. [Google Scholar] [PubMed]
- Chatzipapas, K.P.; Papadimitroulas, P.; Loudos, G.; Papanikolaou, N.; Kagadis, G.C. IDDRRA: A novel platform, based on Geant4-DNA to quantify DNA damage by ionizing radiation. Med. Phys. 2021, 48, 2624–2636. [Google Scholar] [CrossRef] [PubMed]
- Charlton, D.; Nikjoo, H.; Humm, J. Calculation of Initial Yields of Single- and Double-strand Breaks in Cell Nuclei from Electrons, Protons and Alpha Particles. Int. J. Radiat. Biol. 1989, 56, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Holley, W.R.; Chatterjee, A.; Magee, J.L. Production of DNA Strand Breaks by Direct Effects of Heavy Charged Particles. Radiat. Res. 1990, 121, 161. [Google Scholar] [CrossRef]
- Chatterjee, A.; Magee, J.L. Theoretical investigation of the production of strand breaks in DNA by water radicals. Radiat. Prot. Dosim. 1985, 13, 137–140. [Google Scholar] [CrossRef]
- Tomita, H.; Kai, M.; Kusama, T.; Aoki, Y.; Ito, A. Monte Carlo simulation of DNA strand breaks induced by monoenergetic electrons using higher-order structure models of DNA. Int. J. Radiat. Biol. 1994, 66, 669–682. [Google Scholar] [PubMed]
- Nikjoo, H.; Terrissol, M. Computational modelling of low-energy electron-induced DNA damage by early physical and chemical events. Int. J. Radiat. Biol. 1997, 71, 467–483. [Google Scholar] [CrossRef]
- Garty, G.; Schulte, R.; Shchemelinin, S.; Leloup, C.; Assaf, G.; Breskin, A.; Chechik, R.; Bashkirov, V.; Milligan, J.; Grosswendt, B. A nanodosimetric model of radiation-induced clustered DNA damage yields. Phys. Med. Biol. 2010, 55, 761–781. [Google Scholar] [CrossRef]
- Meylan, S.; Incerti, S.; Karamitros, M.; Tang, N.; Bueno, M.; Clairand, I.; Villagrasa, C. Simulation of early DNA damage after the irradiation of a fibroblast cell nucleus using Geant4-DNA. Sci. Rep. 2017, 7, 11923. [Google Scholar] [CrossRef]
- Lampe, N.; Karamitros, M.; Breton, V.; Brown, J.M.; Kyriakou, I.; Sakata, D.; Sarramia, D.; Incerti, S. Mechanistic DNA damage simulations in Geant4-DNA part 1: A parameter study in a simplified geometry. Phys. Med. 2018, 48, 135–145. [Google Scholar] [CrossRef]
- Friedland, W.; Dingfelder, M.; Kundrát, P.; Jacob, P. Track structures, DNA targets and radiation effects in the biophysical Monte Carlo simulation code PARTRAC. Mutat. Res. Mol. Mech. Mutagen. 2011, 711, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Bernal, M.A.; Sikansi, D.; Cavalcante, F.; Incerti, S.; Champion, C.; Ivanchenko, V.; Francis, Z.; Karamitros, M. Performance of a new atomistic geometrical model of the B-DNA configuration for DNA-radiation interaction simulations. J. Phys. Conf. Ser. 2014, 490, 012150. [Google Scholar] [CrossRef] [Green Version]
- Meylan, S.; Vimont, U.; Incerti, S.; Clairand, I.; Villagrasa, C. Geant4-DNA simulations using complex DNA geometries generated by the DnaFabric tool. Comput. Phys. Commun. 2016, 204, 159–169. [Google Scholar] [CrossRef]
- Tang, N.; Bueno, M.; Meylan, S.; Incerti, S.; Clairand, I.; Villagrasa, C. Simulation of early radiation-induced DNA damage on different types of cell nuclei. Radiat. Prot. Dosim. 2018, 183, 26–31. [Google Scholar] [CrossRef]
- Elia, M.C.; Bradley, M.O. Influence of chromatin structure on the induction of DNA double strand breaks by ionizing radiation. Cancer Res. 1992, 52, 1580–1586. [Google Scholar]
- Magnander, K.; Hultborn, R.; Claesson, K.; Elmroth, K. Clustered DNA Damage in Irradiated Human Diploid Fibroblasts: Influence of Chromatin Organization. Radiat. Res. 2010, 173, 272–282. [Google Scholar] [CrossRef]
- Tang, N.; Bueno, M.; Meylan, S.; Incerti, S.; Tran, N.H.; Vaurijoux, A.; Gruel, G.; Villagrasa, C. Influence of chromatin compaction on simulated early radiation-induced DNA damage using Geant4-DNA. Med. Phys. 2019, 46, 1501–1511. [Google Scholar] [CrossRef] [Green Version]
- Tang, N.; Bueno, M.; Meylan, S.; Perrot, Y.; Tran, H.N.; Freneau, A.; Villagrasa, C.; Dos Santos, M.; Gruel, G.; Incerti, S.; et al. Assessment of the biological response of endothelial cells irradiated with 40 kVp, 220 kVp and 4 MV X-rays by means of micro and nanodosimetric calculations. Int. J. Mol. Sci. 2019, 20, 6204. [Google Scholar] [CrossRef] [Green Version]
- Lampe, N. The Long Term Impact of Ionising Radiation on Living Systems; Université Clermont Auvergne: Clermont, France, 2017; Available online: https://tel.archives-ouvertes.fr/tel-01626614/document (accessed on 10 November 2021).
- Sakata, D.; Lampe, N.; Karamitros, M.; Kyriakou, I.; Belov, O.; Bernal, M.A.; Bolst, D.; Bordage, M.-C.; Breton, V.; Brown, J.M.; et al. Evaluation of early radiation DNA damage in a fractal cell nucleus model using Geant4-DNA. Phys. Med. 2019, 62, 152–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakata, D.; Belov, O.; Bordage, M.-C.; Emfietzoglou, D.; Guatelli, S.; Inaniwa, T.; Ivanchenko, V.; Karamitros, M.; Kyriakou, I.; Lampe, N.; et al. Fully integrated Monte Carlo simulation for evaluating radiation induced DNA damage and subsequent repair using Geant4-DNA. Sci. Rep. 2020, 10, 20788. [Google Scholar] [CrossRef]
- Nikjoo, H.; O’Neill, P.; Terrissol, M.; Goodhead, D. Modelling of Radiation-induced DNA Damage: The Early Physical and Chemical Event. Int. J. Radiat. Biol. 1994, 66, 453–457. [Google Scholar] [CrossRef]
- Milligan, J.R.; Aguilera, J.A.; Ward, J.F. Variation of single-strand break yield with scavenger concentration for plasmid DNA irradiated in aqueous solution. Radiat. Res. 1997, 133, 151–157. [Google Scholar] [CrossRef]
- Udovicić, L.; Mark, F.; Bothe, E. Yields of single-strand breaks in double-stranded calf thymus DNA irradiated in aqueous solution in the presence of oxygen and scavengers. Radiat. Res. 1994, 140, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Belov, O.V.; Krasavin, E.A.; Lyashko, M.S.; Batmunkh, M.; Sweilam, N.H. A quantitative model of the major pathways for radiation-induced DNA double-strand break repair. J. Theor. Biol. 2015, 366, 115–130. [Google Scholar] [CrossRef]
- Shin, W.-G.; Ramos-Mendez, J.; Tran, N.H.; Okada, S.; Perrot, Y.; Villagrasa, C.; Incerti, S. Geant4-DNA simulation of the pre-chemical stage of water radiolysis and its impact on initial radiochemical yields. Phys. Med. 2021, 88, 86–90. [Google Scholar] [CrossRef]
- Arce, P.; Bolst, D.; Bordage, M.; Brown, J.M.C.; Cirrone, P.; Cortés-Giraldo, M.A.; Cutajar, D.; Cuttone, G.; Desorgher, L.; Dondero, P.; et al. Report on G4-Med, a Geant4 benchmarking system for medical physics applications developed by the Geant4 Medical Simulation Benchmarking Group. Med. Phys. 2021, 48, 19–56. [Google Scholar] [CrossRef]
- Geant-Val Web Interface. Available online: https://geant-val.cern.ch/layouts (accessed on 10 November 2021).
- Incerti, S.; Kyriakou, I.; Tran, N.H. Geant4-DNA simulation of electron slowing-down spectra in liquid water. Nucl. Instrum. Methods Phys. Res. Sect. B Beam Interact. Mater. Atoms 2017, 397, 45–50. [Google Scholar] [CrossRef]
- Ramos-Méndez, J.; Schuemann, J.; Incerti, S.; Paganetti, H.; Schulte, R.; Faddegon, B. Flagged uniform particle splitting for variance reduction in proton and carbon ion track-structure simulations. Phys. Med. Biol. 2017, 62, 5908–5925. [Google Scholar] [CrossRef]
- Goddu, S.M.; Howell, R.; Bouchet, L.; Bolch, W.E.; Rao, D.V. MIRD Cellular S Values; Society of Nuclear Medicine: Reston, VA, USA, 1997. [Google Scholar]
- Sgouros, G. Dosimetry of internal emitters. J. Nucl. Med. 2005, 46, 18S–27S. [Google Scholar]
- Šefl, M.; Incerti, S.; Papamichael, G.; Emfietzoglou, D. Calculation of cellular S-values using Geant4-DNA: The effect of cell geometry. Appl. Radiat. Isot. 2015, 104, 113–123. [Google Scholar] [CrossRef]
- André, T.; Morini, F.; Karamitros, M.; Delorme, R.; Le Loirec, C.; Campos, L.; Champion, C.; Groetz, J.-E.; Fromm, M.; Bordage, M.-C.; et al. Comparison of Geant4-DNA simulation of S-values with other Monte Carlo codes. Nucl. Instrum. Methods Phys. Res. Sect. B Beam Interact. Mater. Atoms 2014, 319, 87–94. [Google Scholar] [CrossRef] [Green Version]
- Ramos-Mendez, J.; Perl, J.; Schuemann, J.; McNamara, A.; Paganetti, H.; Faddegon, B. Monte Carlo simulation of chemistry following radiolysis with TOPAS-nBio. Phys. Med. Biol. 2018, 63, 105014. [Google Scholar] [CrossRef]
- Sakata, D.; Incerti, S.; Bordage, M.C.; Lampe, N.; Okada, S.; Emfietzoglou, D.; Kyriakou, I.; Murakami, K.; Sasaki, T.; Tran, N.H.; et al. An implementation of discrete electron transport models for gold in the Geant4 simulation toolkit. J. Appl. Phys. 2016, 120, 244901. [Google Scholar] [CrossRef] [Green Version]
- Sakata, D.; Kyriakou, I.; Okada, S.; Tran, N.H.; Lampe, N.; Guatelli, S.; Bordage, M.; Ivanchenko, V.; Murakami, K.; Sasaki, T.; et al. Geant4-DNA track-structure simulations for gold nanoparticles: The importance of electron discrete models in nanometer volumes. Med. Phys. 2018, 45, 2230–2242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuemann, J.; Bagley, A.F.; Berbeco, R.; Bromma, K.; Butterworth, K.T.; Byrne, H.L.; Chithrani, B.D.; Cho, S.H.; Cook, J.R.; Favaudon, V.; et al. Roadmap for metal nanoparticles in radiation therapy: Current status, translational challenges, and future directions. Phys. Med. Biol. 2020, 65, 21RM02. [Google Scholar] [CrossRef]
- Li, W.B.; Stangl, S.; Klapproth, A.; Shevtsov, M.; Hernandez, A.; Kimm, M.A.; Schuemann, J.; Qiu, R.; Michalke, B.; Bernal, M.A.; et al. Application of High-Z Gold Nanoparticles in Targeted Cancer Radiotherapy—Pharmacokinetic Modeling, Monte Carlo Simulation and Radiobiological Effect Modeling. Cancers 2021, 13, 5370. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.; Chappuis, F.; Incerti, S.; Bochud, F.; Desorgher, L. Geant4-DNA Modeling of Water Radiolysis beyond the Microsecond: An On-Lattice Stochastic Approach. Int. J. Mol. Sci. 2021, 22, 6023. [Google Scholar] [CrossRef] [PubMed]
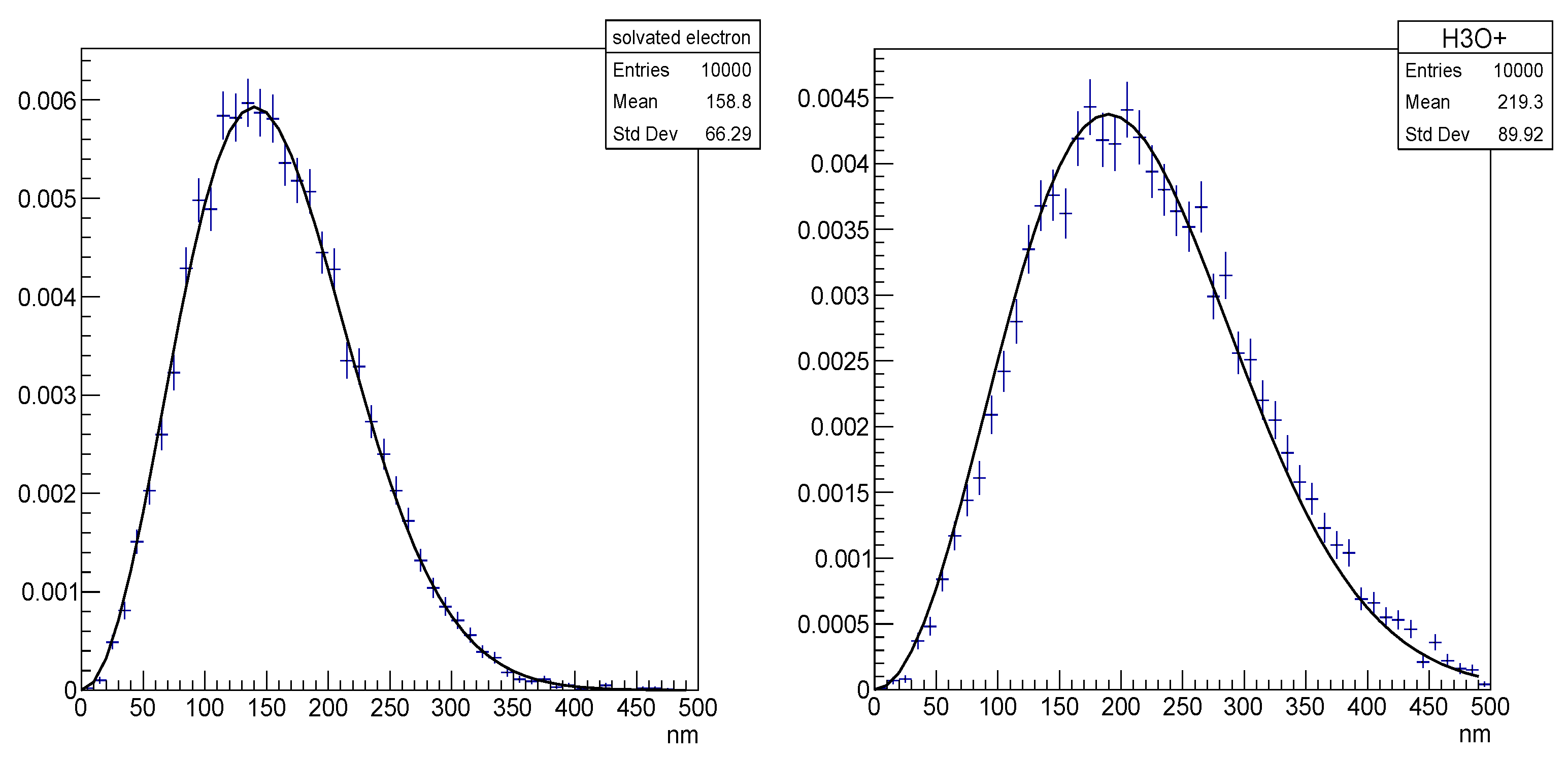
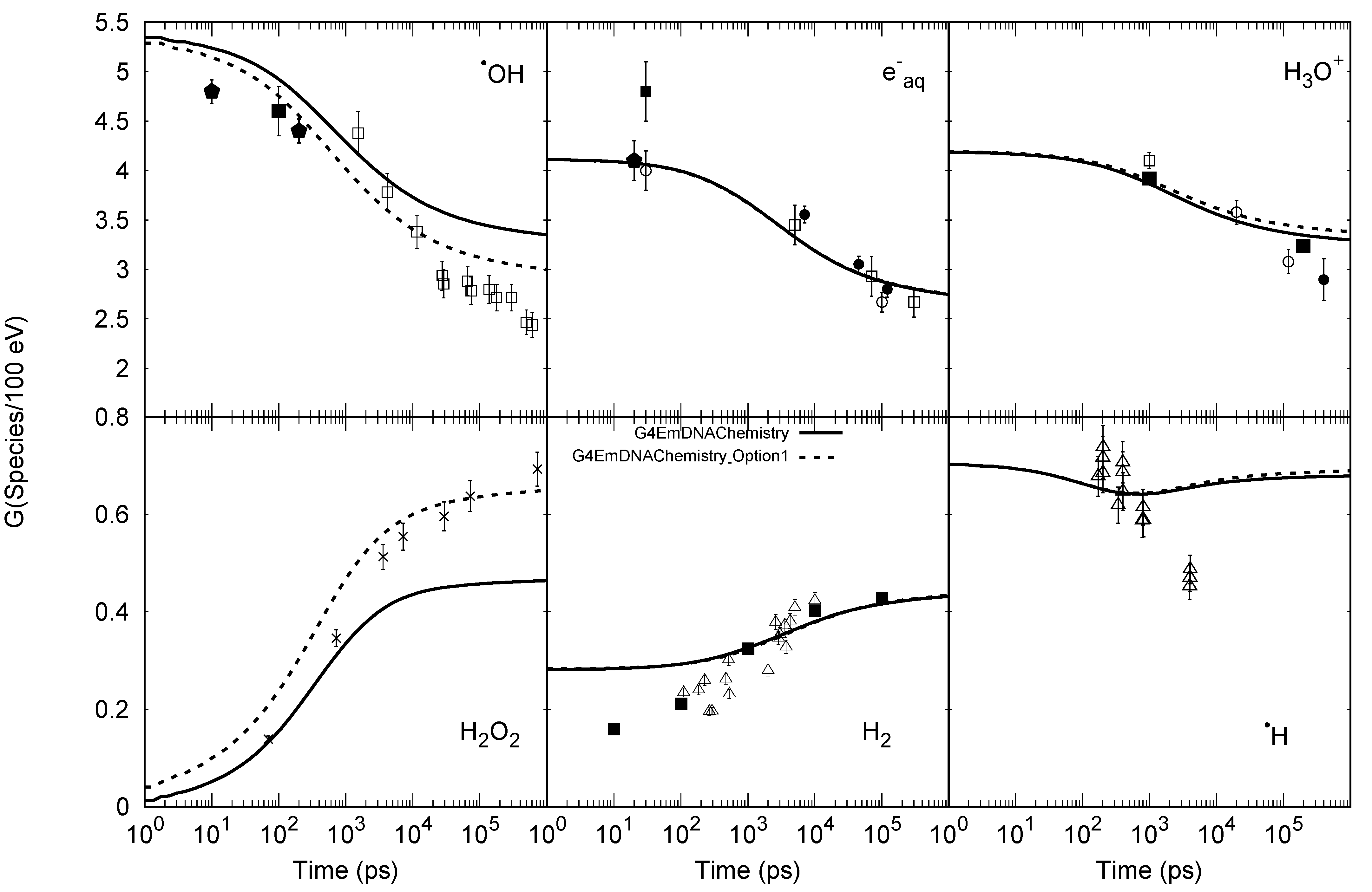
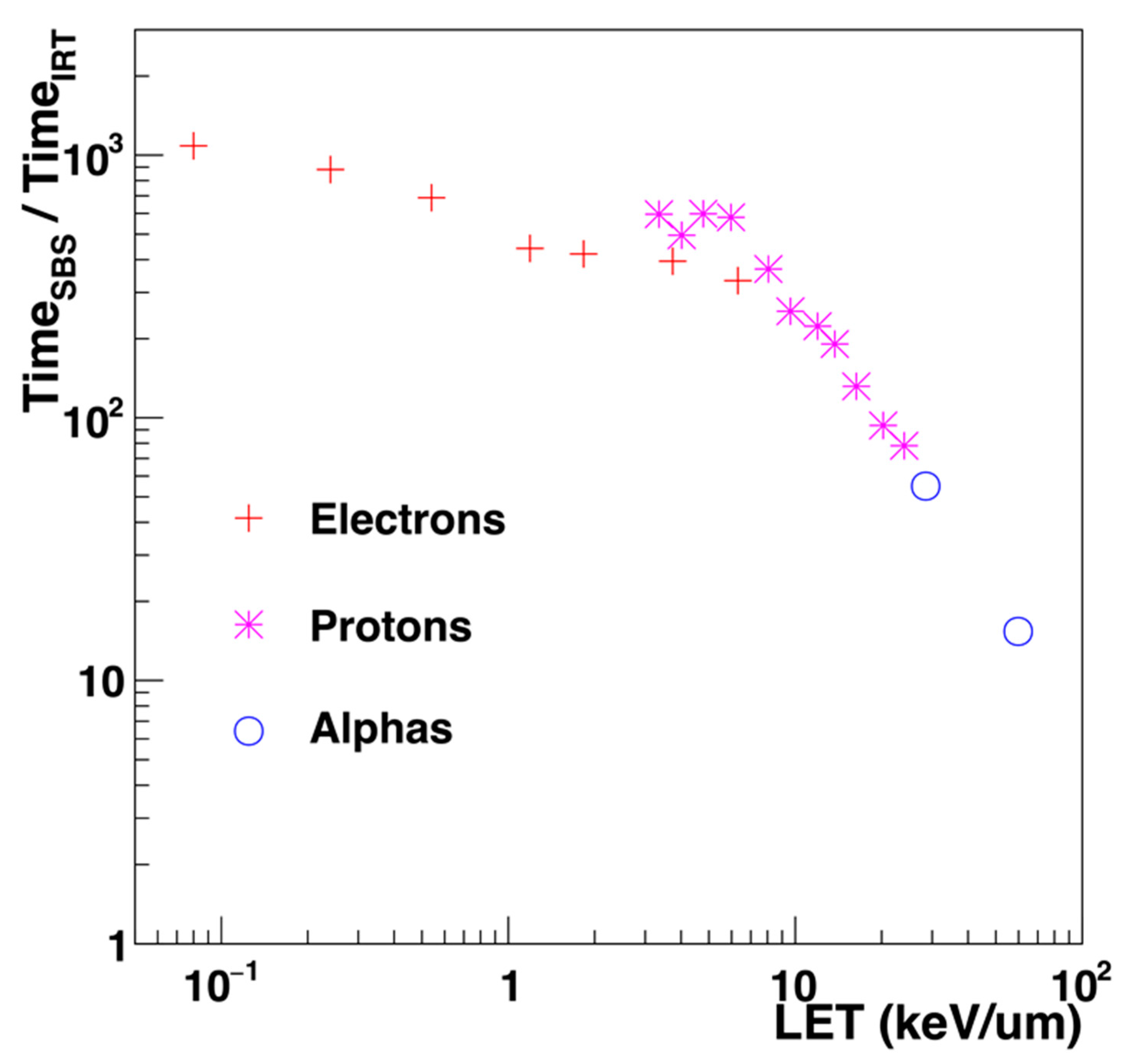
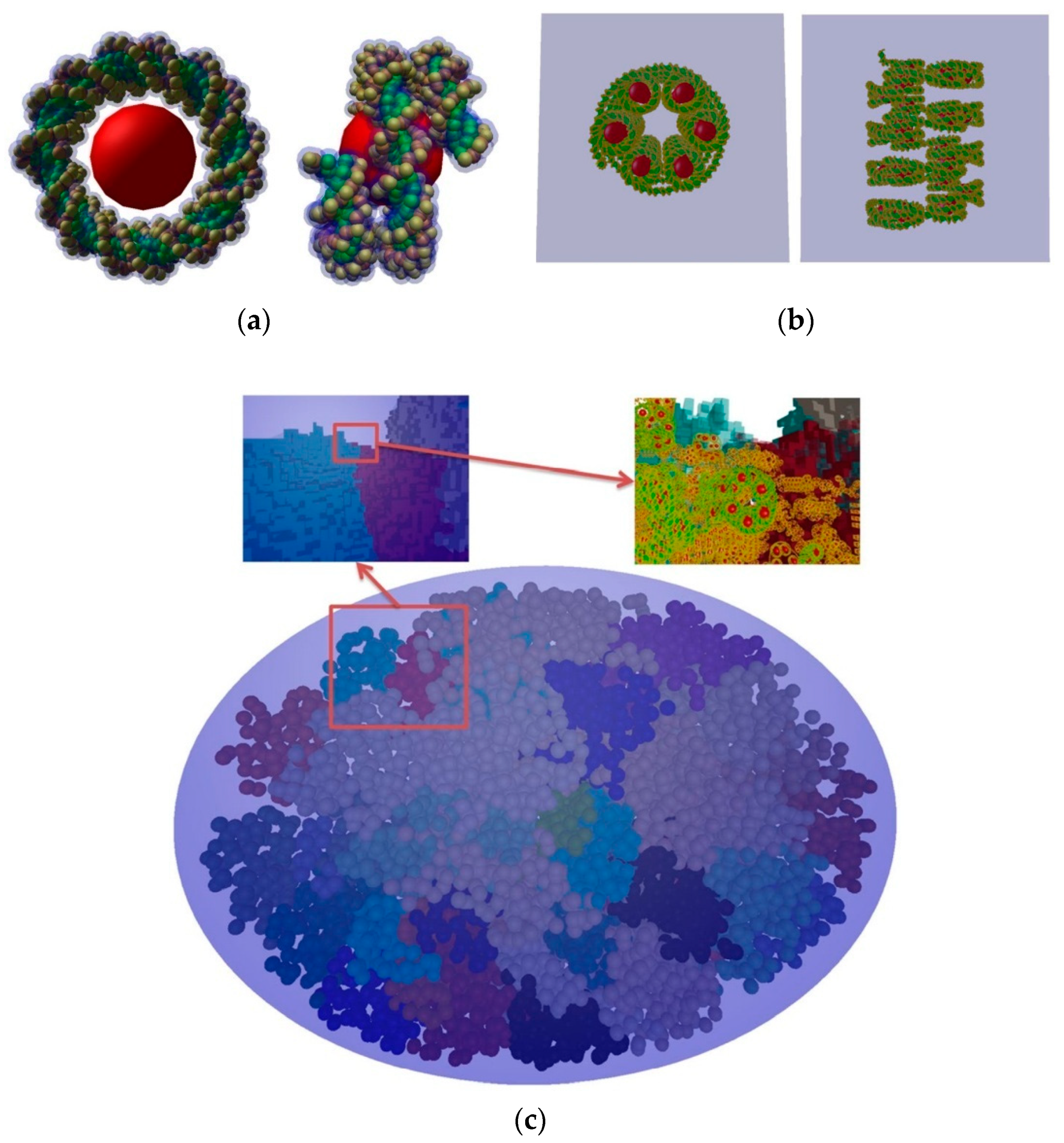
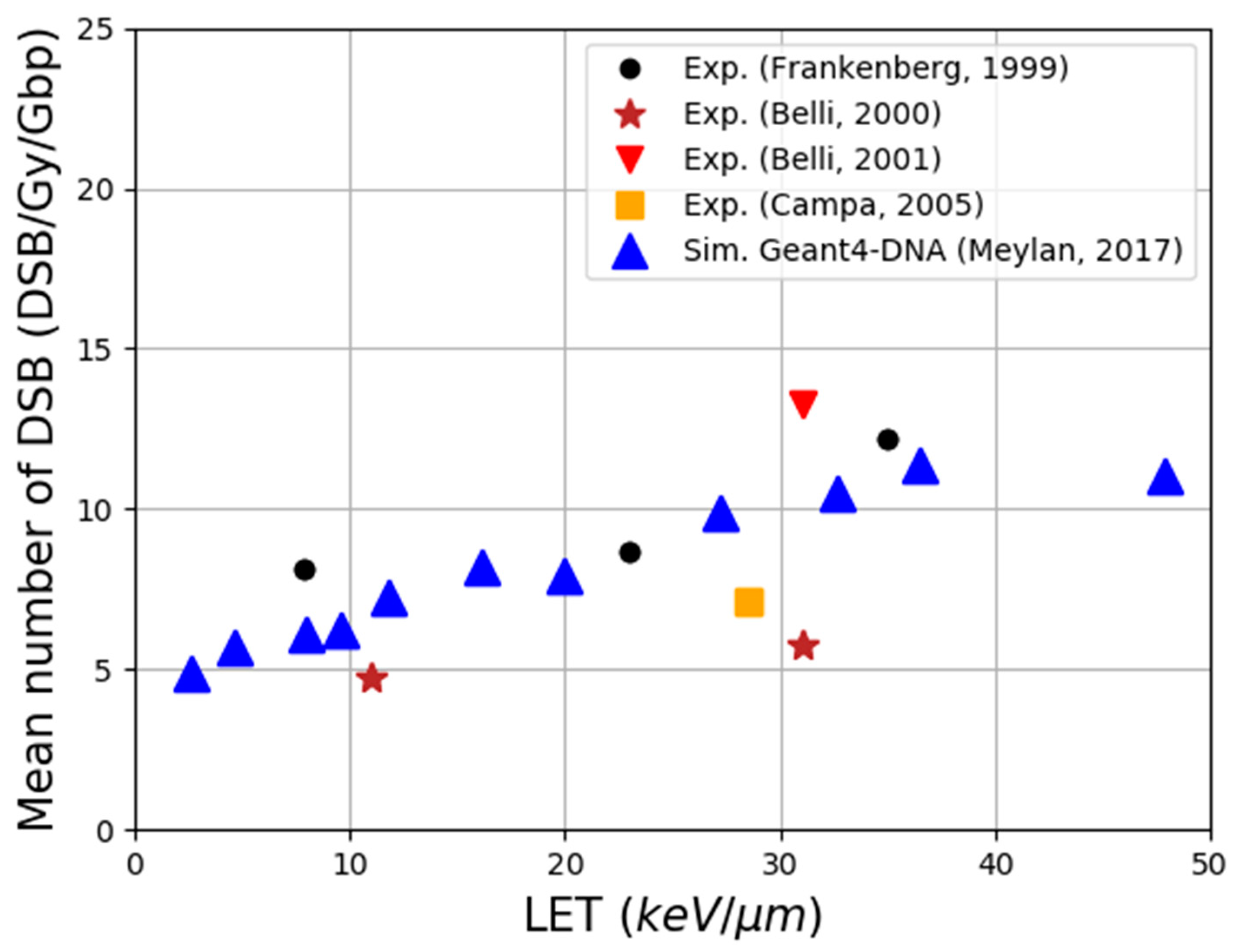
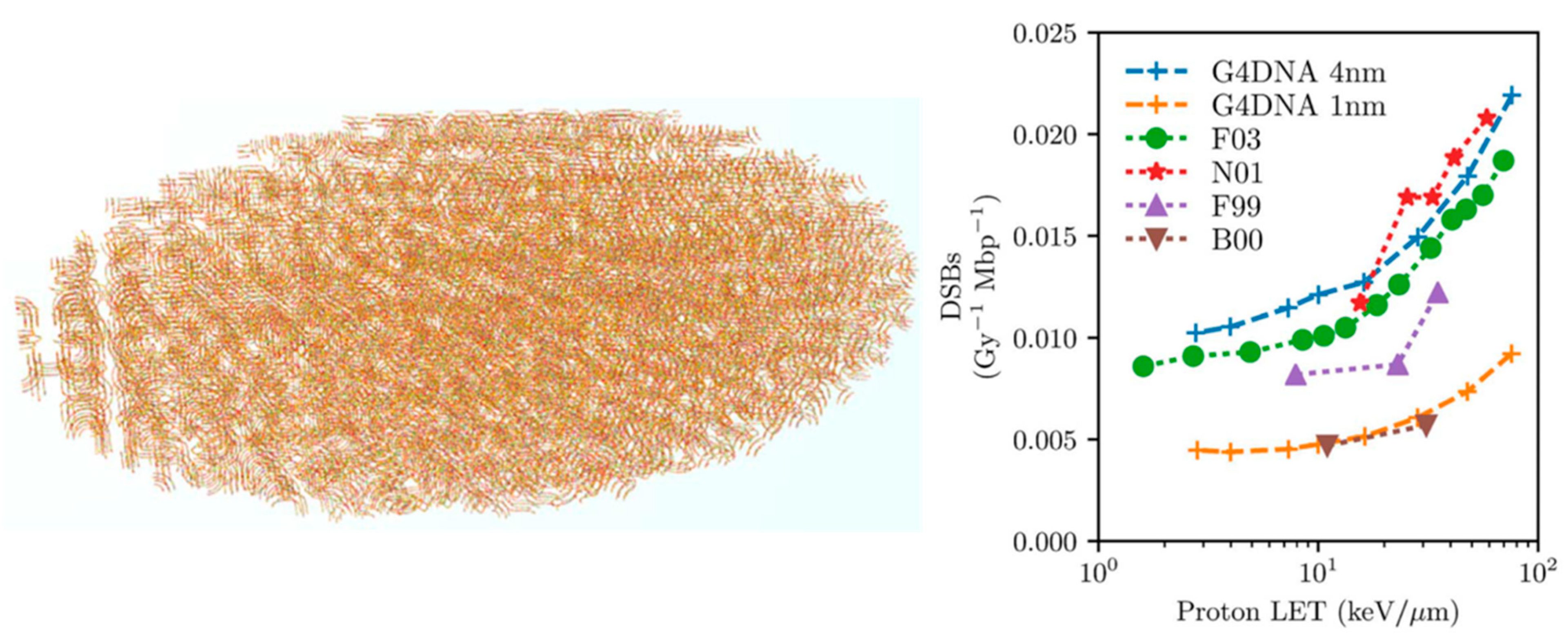
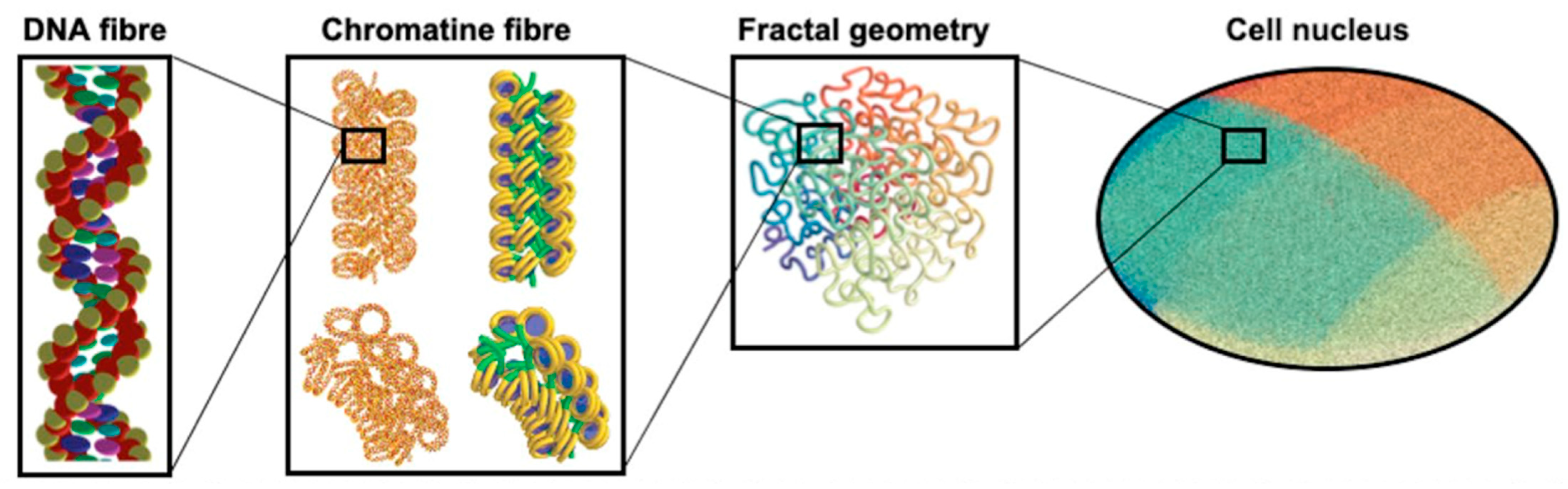
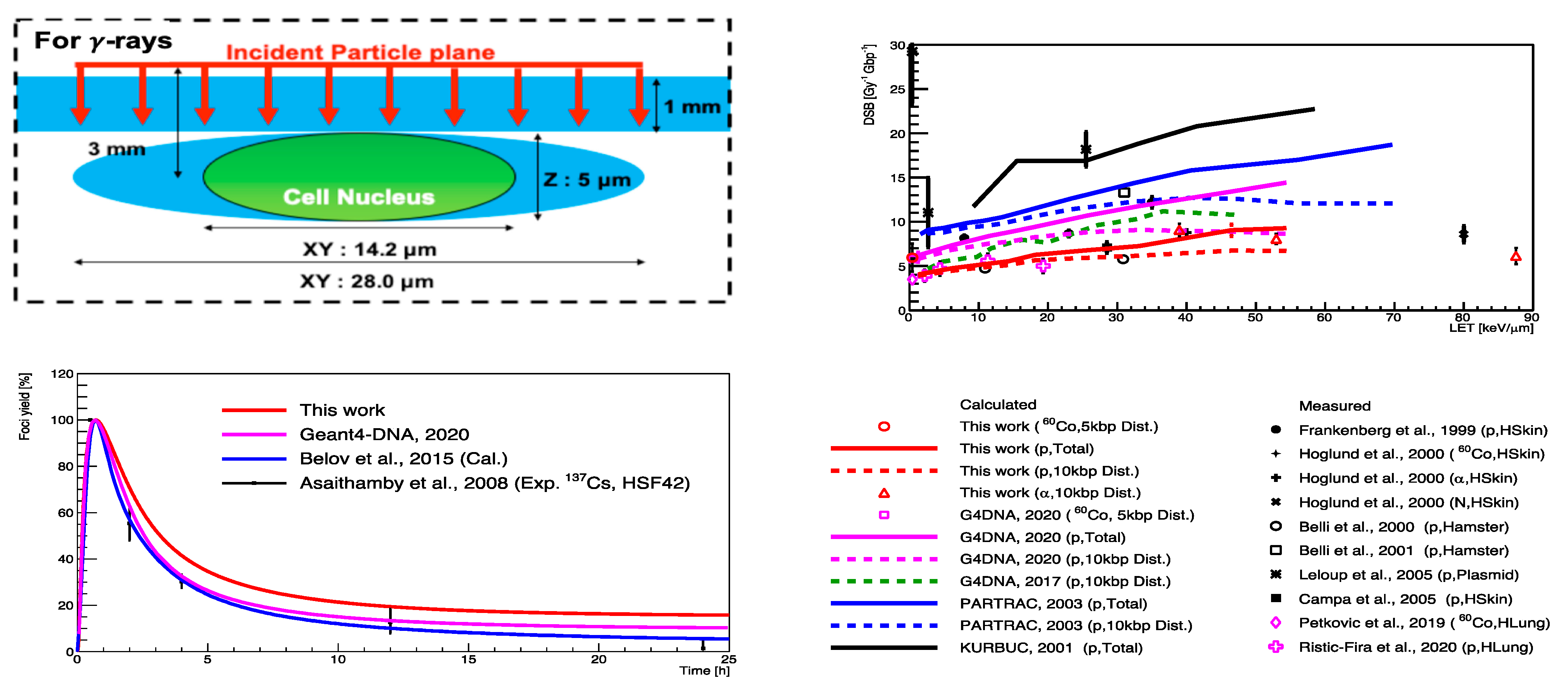
| Electronic State | Decay Channel | Fraction |
|---|---|---|
| All ionization states | H3O+ + •OH (through proton transfer) | 100% |
| Excitation state A1B1: (4a1/3s) | •OH + H• H2O + ∆E | 65% 35% |
| Excitation state B1A1: (4a1/3s) | HO++ •OH + e−aq •OH + •OH + H2 H2O + ∆E | 55% 15% 30% |
| Excitation state: Rydberg, diffusion bands | HO++ •OH + e−aq H2O + ∆E | 50% 50% |
| Electron attachment | OH− + •OH + H2 | 100 |
| Electron-hole recombination | •OH + H• | 55% |
| H2 + 2•OH | 15% | |
| H2O + ∆E | 30% |
| Reaction | ||
|---|---|---|
| G4EmDNAChemistry | G4EmDNAChemistry_ Option1 | |
| 0.5 | 0.636 | |
| 2.65 | 2.5 | |
| 2.95 | 2.95 | |
| 2.11 | 2.11 | |
| 1.41 | 1.10 | |
| 0.44 | 0.550 | |
| 1.44 | 1.55 | |
| 1.2 | 0.503 | |
| 14.3 | 11.3 | |
| Parameters | FullSim | MolecularDNA | |
|---|---|---|---|
| Physical parameters | Rdir (Å) | VDWR + hydration shells * | 3.5 |
| Elower(eV) | 17.5 | 5.0 | |
| Ehigher(eV) | 17.5 | 37.5 | |
| Chemical parameters | POH | 0.4 | 0.405 |
| Tchem (ns) | 2.5 | 5.0 | |
| dkill (nm) | N/A | 9.0 | |
| Simulated DSBs and Experimental Foci at 1 Gy | 40 kVp X-rays | 220 kVp X-rays | 4 MV X-rays |
|---|---|---|---|
| Sim. mean number of DSBs per nucleus | 21.0 ± 0.3 | 21.0 ± 0.3 | 16.8 ± 0.3 |
| Exp. mean number of γ-H2AX foci per nucleus | 18.59 ± 0.43 | 18.64 ± 2.33 | 16.46 ± 1.63 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kyriakou, I.; Sakata, D.; Tran, H.N.; Perrot, Y.; Shin, W.-G.; Lampe, N.; Zein, S.; Bordage, M.C.; Guatelli, S.; Villagrasa, C.; et al. Review of the Geant4-DNA Simulation Toolkit for Radiobiological Applications at the Cellular and DNA Level. Cancers 2022, 14, 35. https://doi.org/10.3390/cancers14010035
Kyriakou I, Sakata D, Tran HN, Perrot Y, Shin W-G, Lampe N, Zein S, Bordage MC, Guatelli S, Villagrasa C, et al. Review of the Geant4-DNA Simulation Toolkit for Radiobiological Applications at the Cellular and DNA Level. Cancers. 2022; 14(1):35. https://doi.org/10.3390/cancers14010035
Chicago/Turabian StyleKyriakou, Ioanna, Dousatsu Sakata, Hoang Ngoc Tran, Yann Perrot, Wook-Geun Shin, Nathanael Lampe, Sara Zein, Marie Claude Bordage, Susanna Guatelli, Carmen Villagrasa, and et al. 2022. "Review of the Geant4-DNA Simulation Toolkit for Radiobiological Applications at the Cellular and DNA Level" Cancers 14, no. 1: 35. https://doi.org/10.3390/cancers14010035
APA StyleKyriakou, I., Sakata, D., Tran, H. N., Perrot, Y., Shin, W.-G., Lampe, N., Zein, S., Bordage, M. C., Guatelli, S., Villagrasa, C., Emfietzoglou, D., & Incerti, S. (2022). Review of the Geant4-DNA Simulation Toolkit for Radiobiological Applications at the Cellular and DNA Level. Cancers, 14(1), 35. https://doi.org/10.3390/cancers14010035







