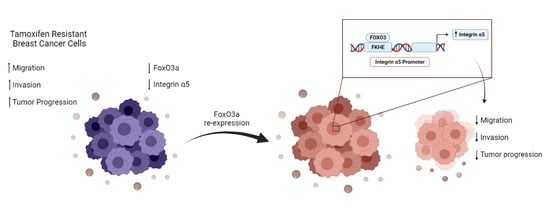
FoxO3a Inhibits Tamoxifen-Resistant Breast Cancer Progression by Inducing Integrin α5 Expression
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture Conditions and Treatments
2.2. Generation of Tamoxifen-Resistant Cell Lines
2.3. Generation of FoxO3a Inducible Stable Cell Lines
2.4. Plasmids and Transient Transfections
2.5. siRNA-Mediated RNA Interference
2.6. Growth Assay
2.7. Spheroid Assay
2.8. Colony-Forming Assay
2.9. Wound-Healing Scratch Assay
2.10. Boyden Chambers Transmigration and Invasion Assays
2.11. RNA Extraction, Reverse Transcription, and Real-Time (RT)-PCR
2.12. Western Blotting (WB) Assay
2.13. Cloning and Luciferase Assay
2.14. Chromatin Immunoprecipitation (ChIP) Assay
2.15. cBioPortal Database
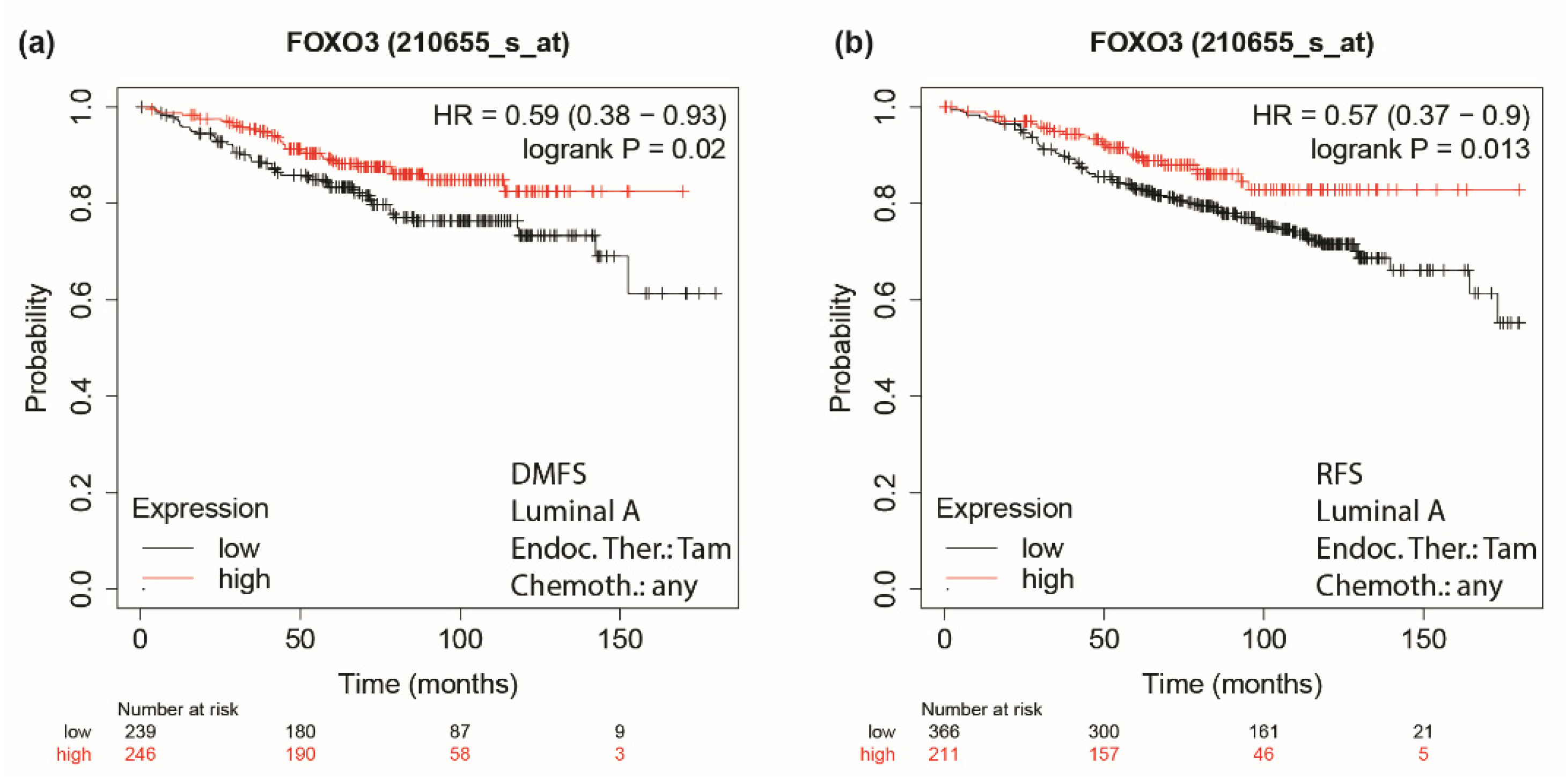
2.16. Kaplan–Meier Analysis
2.17. Statistical Analysis
3. Results
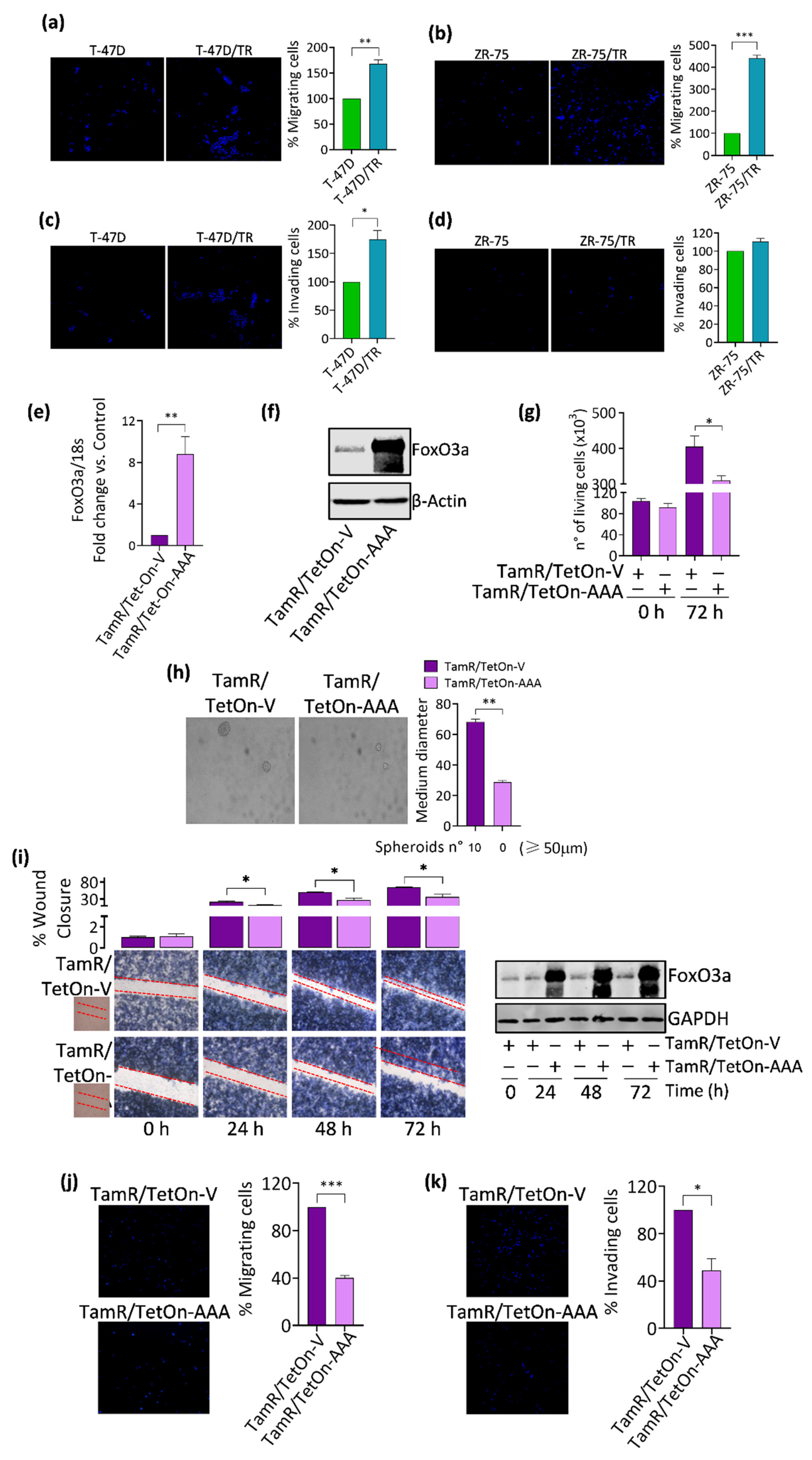
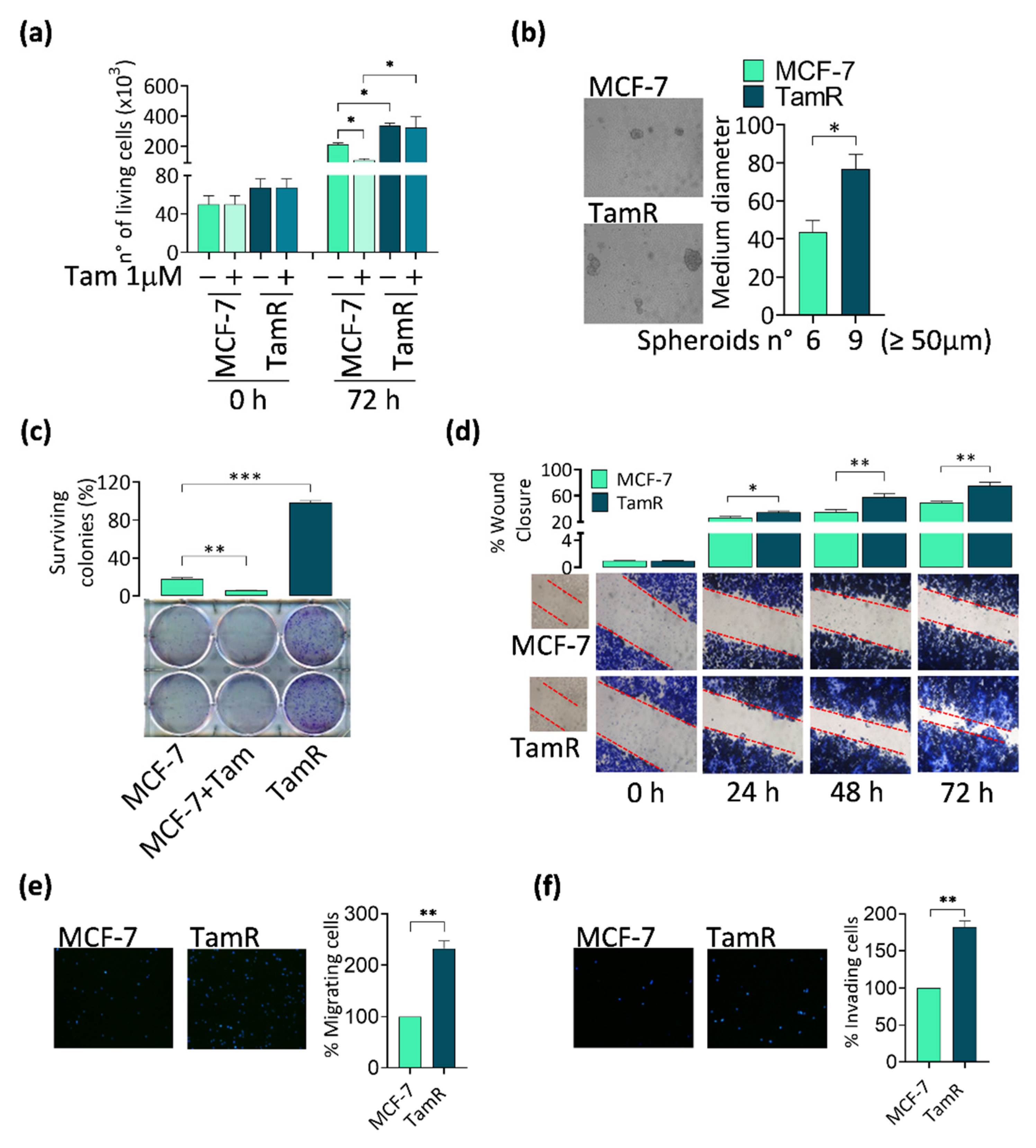
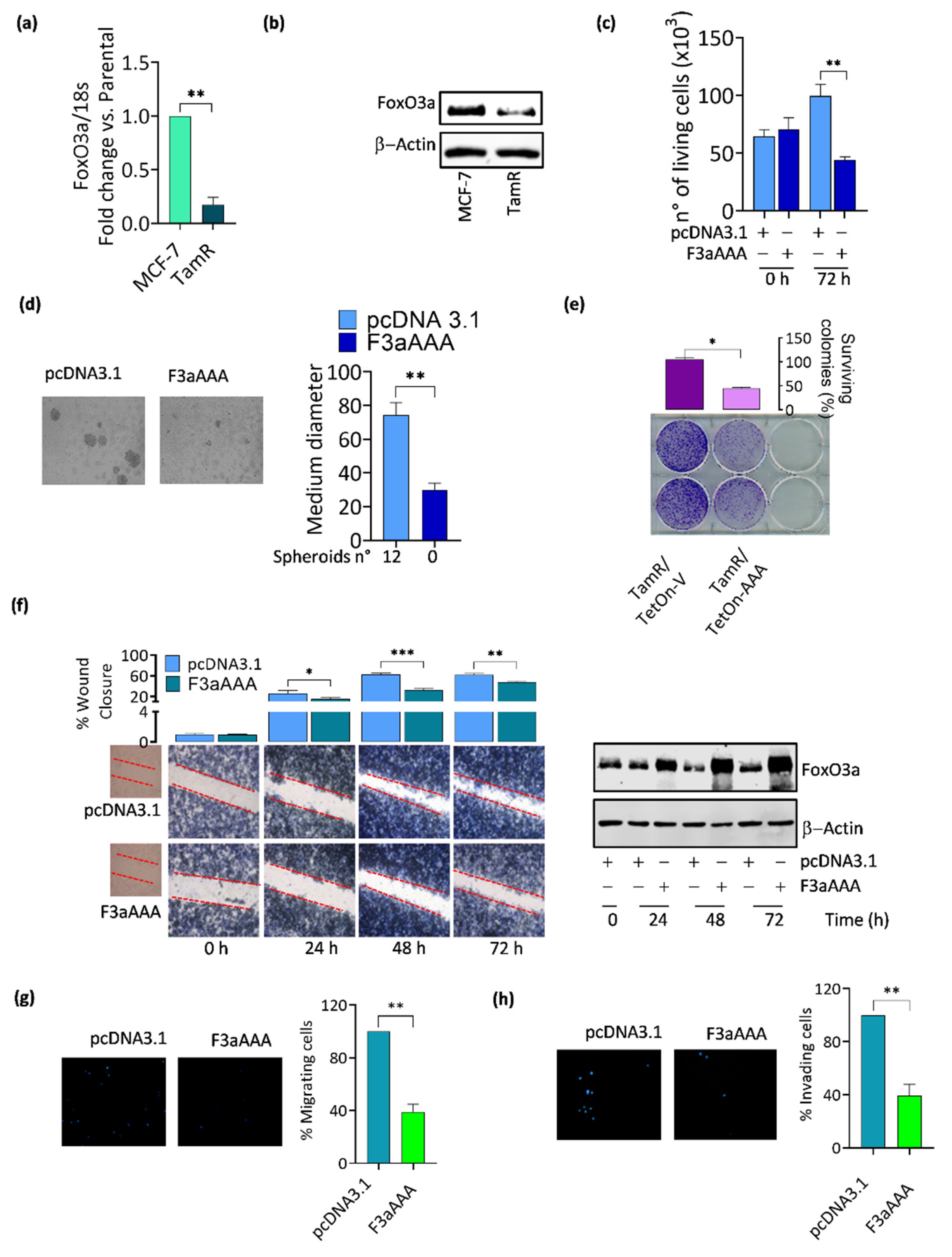
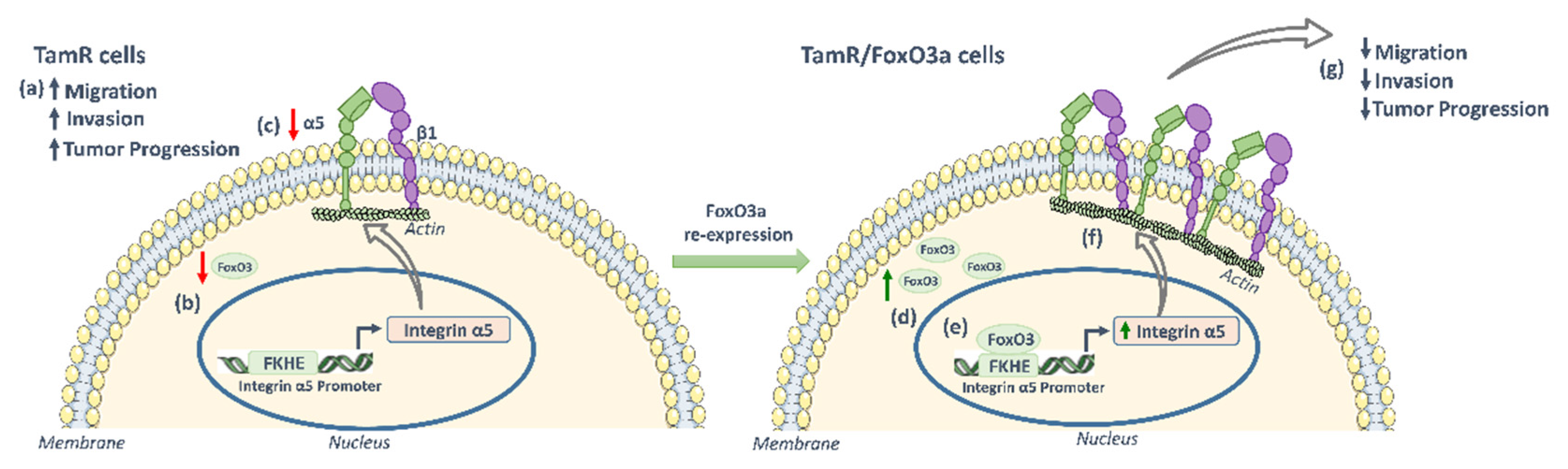
3.1. FoxO3a Re-Expression Reduces the Aggressiveness of Tamoxifen-Resistant BCCs
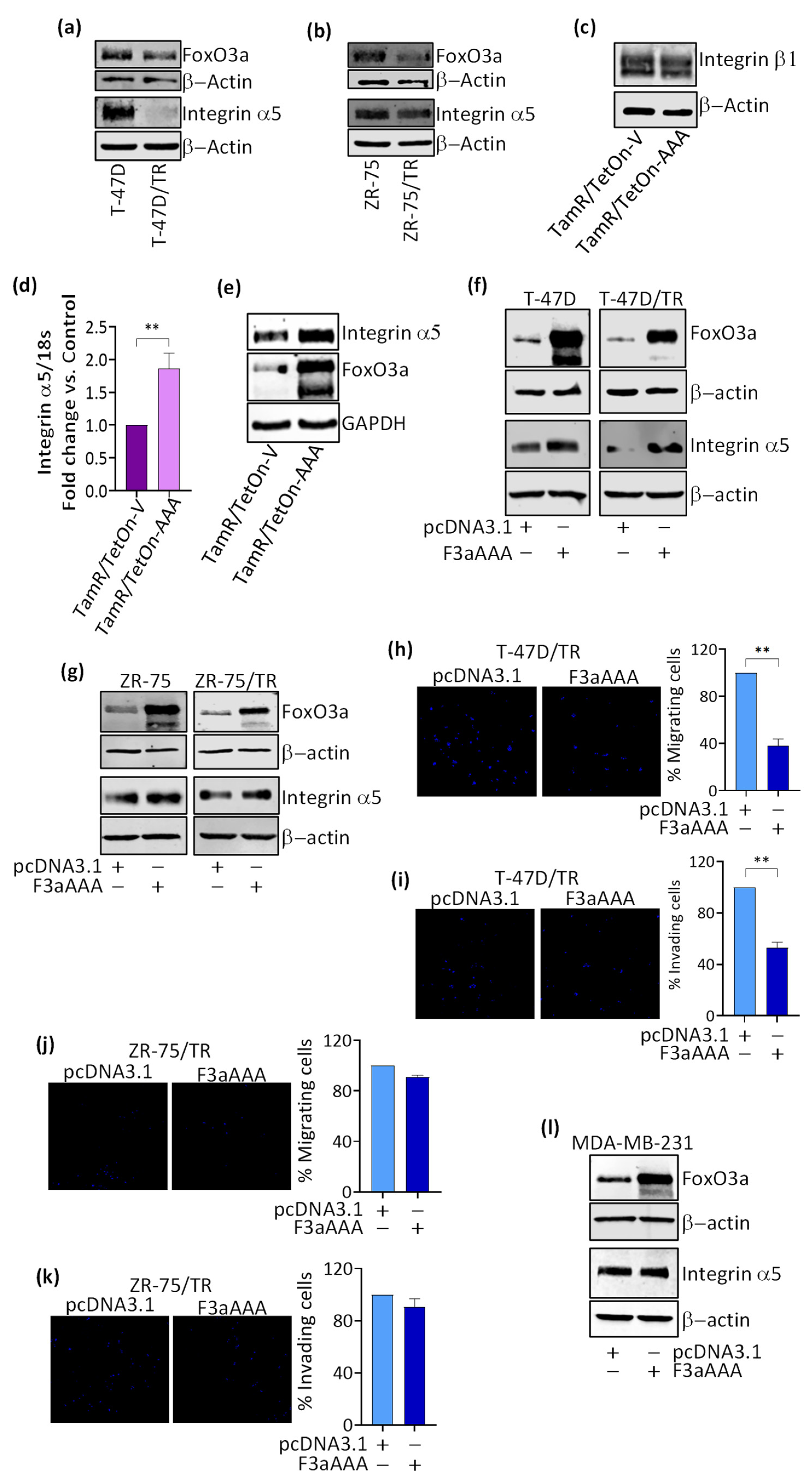
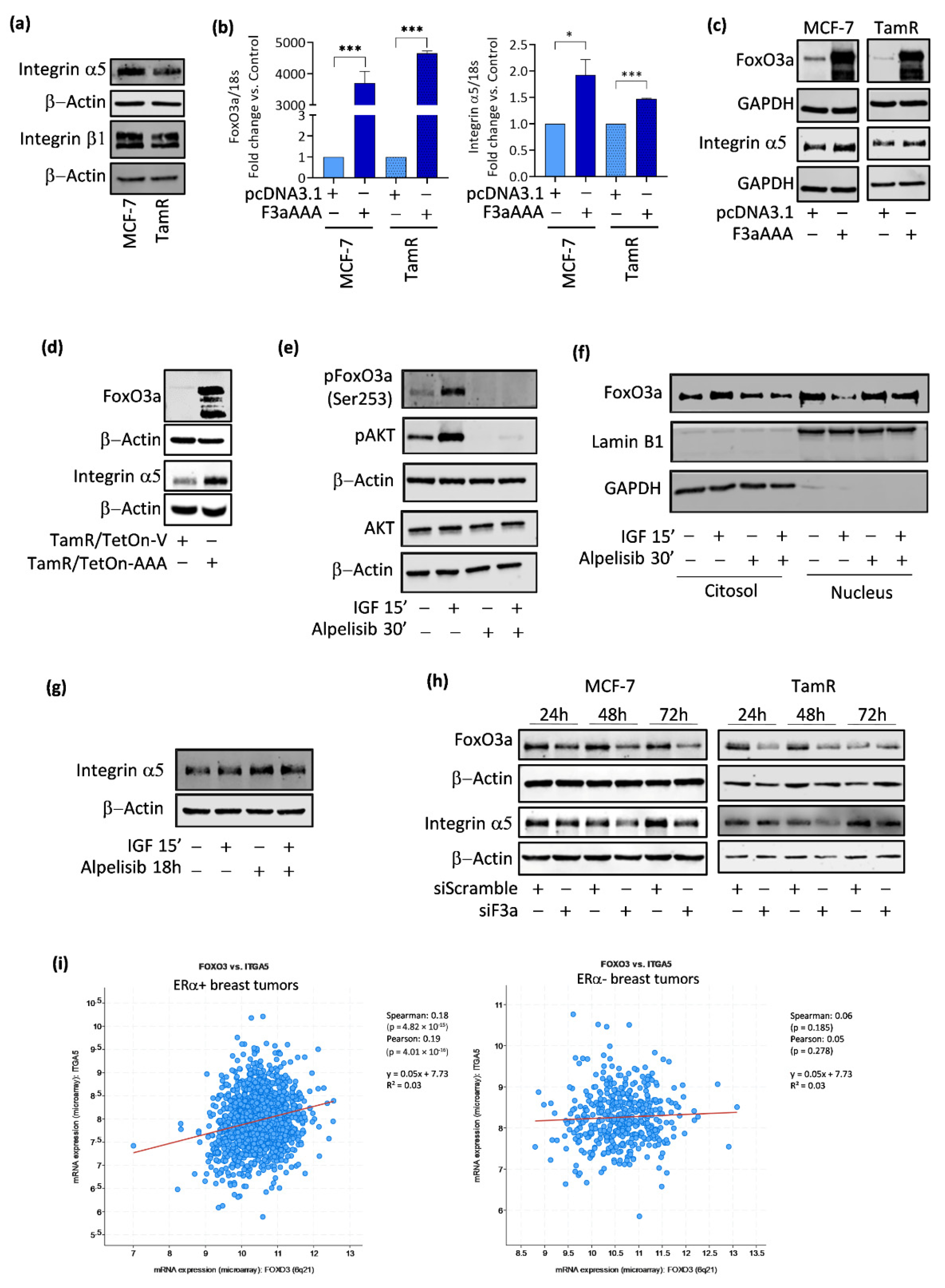
3.2. FoxO3a Modulates the Expression of the Integrin α5 Subunit of the Fibronectin Receptor
3.3. FoxO3a and Integrin α5 Are Positively Correlated in ERα+ BC
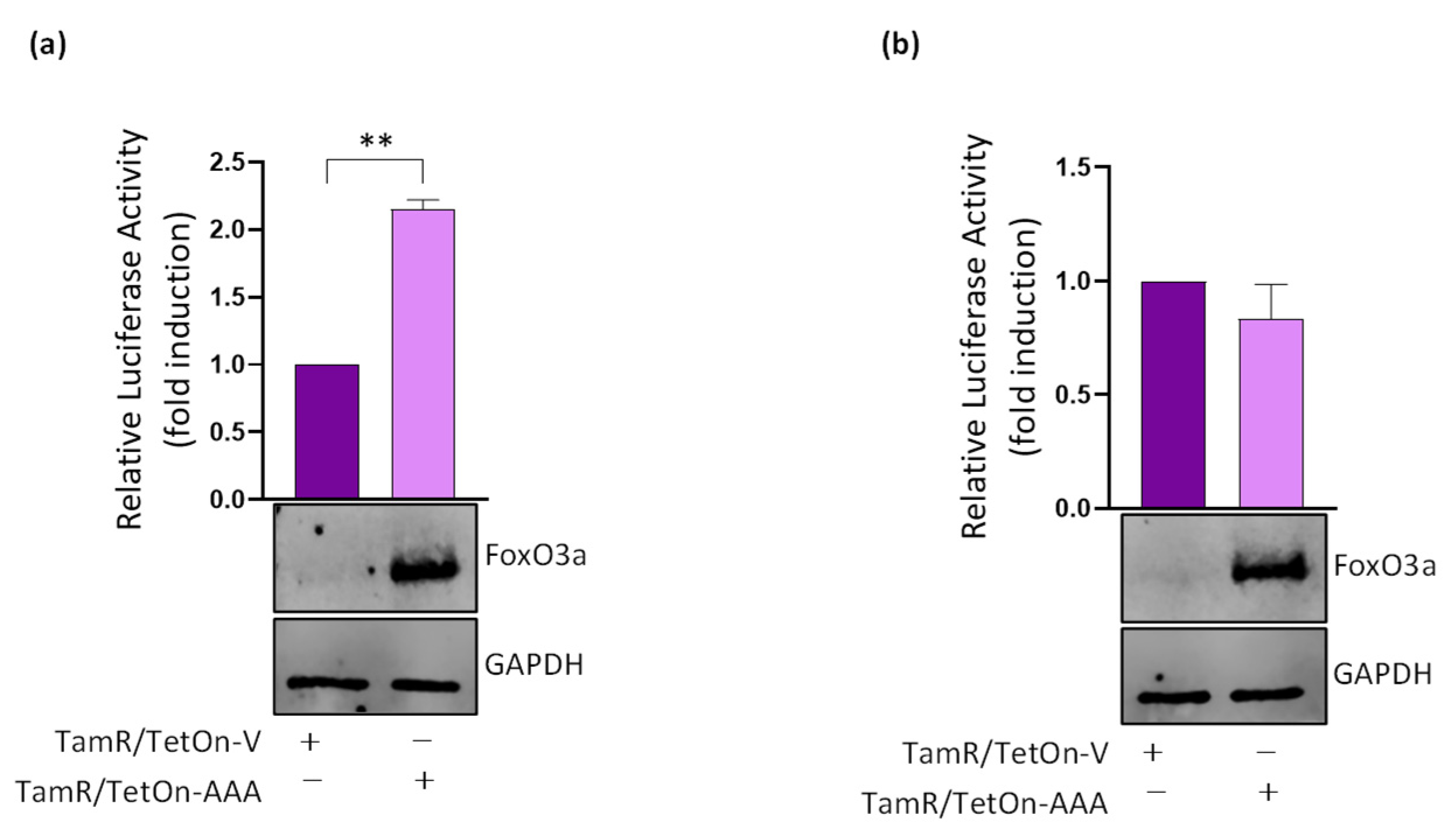
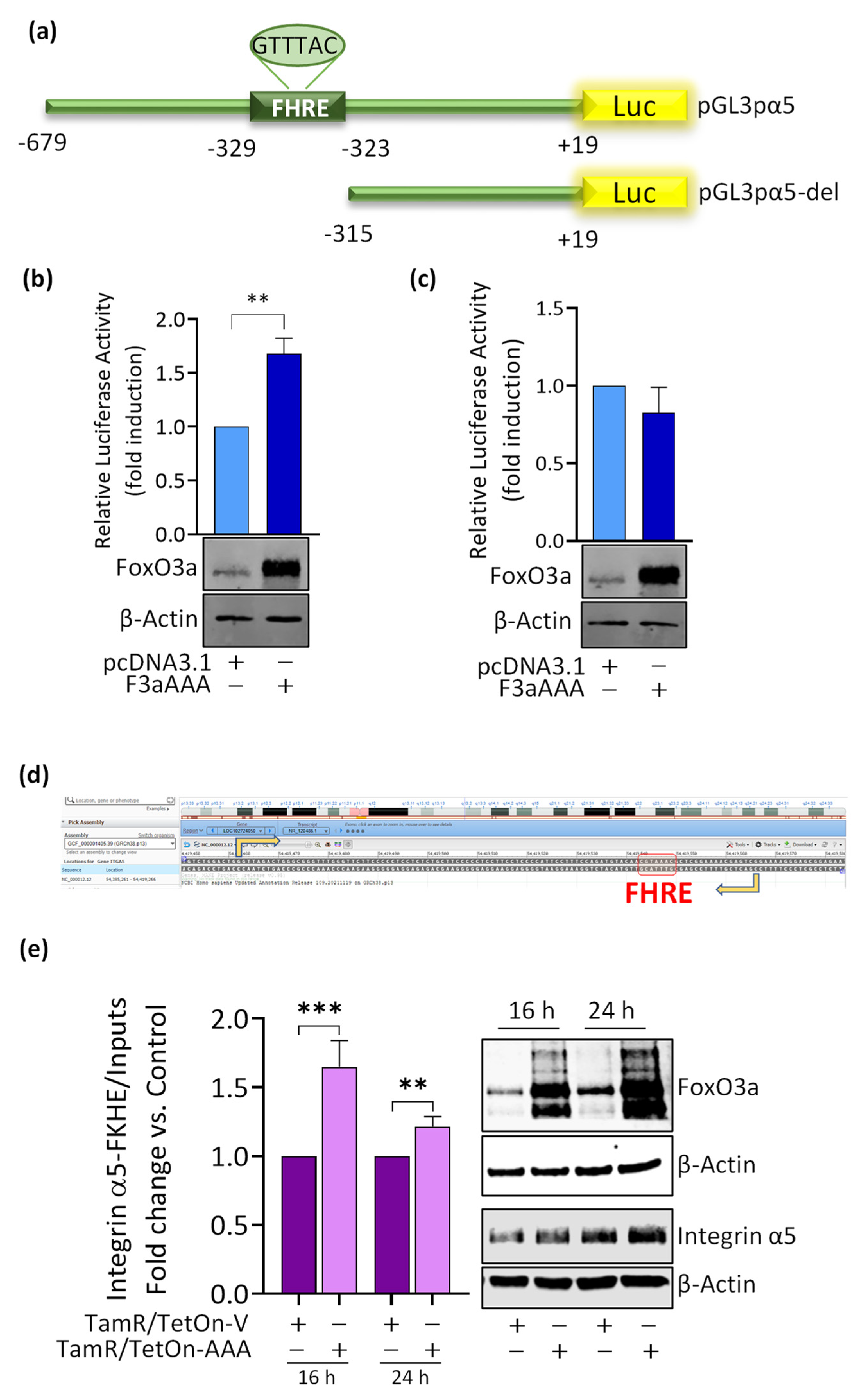
3.4. FoxO3a Binds to and Transactivates the Human Integrin alpha5 Promoter
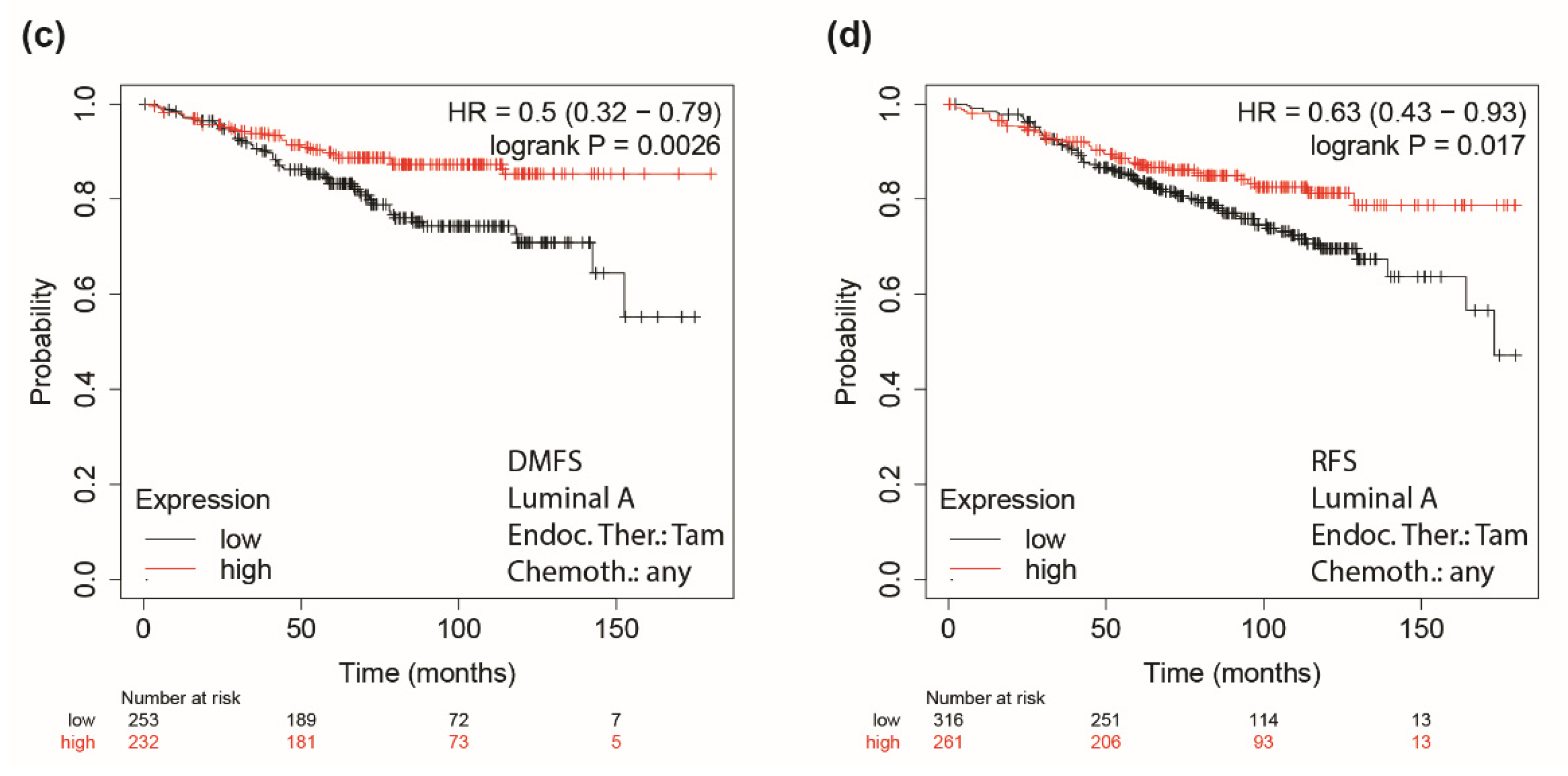
3.5. Role of Integrin a5 in FoxO3a-Dependent Inhibition of TamR Cells Migration and Invasion and Clinical Implications
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
Appendix B
Appendix C
Appendix D
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Shah, R.; O’Regan, R.M. Adjuvant Endocrine Therapy. Cancer Treat. Res. 2018, 173, 15–29. [Google Scholar] [CrossRef]
- Andrahennadi, S.; Sami, A.; Manna, M.; Pauls, M.; Ahmed, S. Current Landscape of Targeted Therapy in Hormone Receptor-Positive and HER2-Negative Breast Cancer. Curr. Oncol. 2021, 28, 1803–1822. [Google Scholar] [CrossRef]
- Brufsky, A.M.; Dickler, M.N. Estrogen Receptor-Positive Breast Cancer: Exploiting Signaling Pathways Implicated in Endocrine Resistance. Oncologyst 2018, 23, 528–539. [Google Scholar] [CrossRef] [Green Version]
- Araki, K.; Miyoshi, Y. Mechanism of resistance to endocrine therapy in breast cancer: The important role of PI3K/Akt/mTOR in estrogen receptor-positive, HER2-negative breast cancer. Breast Cancer 2018, 25, 392–401. [Google Scholar] [CrossRef]
- Dong, C.; Wu, J.; Chen, Y.; Nie, J.; Chen, C. Activation of PI3K/AKT/mTOR Pathway Causes Drug Resistance in Breast Cancer. Front. Pharmacol. 2021, 12, 628690. [Google Scholar] [CrossRef] [PubMed]
- Burgering, B.M.; Medema, R.H. Decisions on life and death: FOXO Forkhead transcription factors are in command when PKB/Akt is off duty. J. Leukoc. Biol. 2003, 73, 689–701. [Google Scholar] [CrossRef]
- Calissi, G.; Lam, E.W.; Link, W. Therapeutic strategies targeting FOXO transcription factors. Nat. Rev. Drug Discov. 2021, 20, 21–38. [Google Scholar] [CrossRef] [PubMed]
- Pellegrino, M.; Rizza, P.; Dona, A.; Nigro, A.; Ricci, E.; Fiorillo, M.; Perrotta, I.; Lanzino, M.; Giordano, C.; Bonofiglio, D.; et al. FoxO3a as a Positive Prognostic Marker and a Therapeutic Target in Tamoxifen-Resistant Breast Cancer. Cancers 2019, 11, 1858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.Y.; Zong, C.S.; Xia, W.Y.; Yamaguchi, H.; Ding, Q.Q.; Xie, X.M.; Lang, J.Y.; Lai, C.C.; Chang, C.J.; Huang, W.C.; et al. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat. Cell Biol. 2008, 10, 138-U122. [Google Scholar] [CrossRef] [Green Version]
- Dansen, T.B.; Burgering, B.M.T. Unravelling the tumor-suppressive functions of FOXO proteins. Trends Cell Biol. 2008, 18, 421–429. [Google Scholar] [CrossRef]
- Liu, Y.; Ao, X.; Ding, W.; Ponnusamy, M.; Wu, W.; Hao, X.; Yu, W.; Wang, Y.; Li, P.; Wang, J. Critical role of FOXO3a in carcinogenesis. Mol. Cancer 2018, 17, 104. [Google Scholar] [CrossRef] [Green Version]
- Lanzino, M.; Morelli, C.; Garofalo, C.; Panno, M.L.; Mauro, L.; Ando, S.; Sisci, D. Interaction between Estrogen Receptor Alpha and Insulin/IGF Signaling in Breast Cancer. Curr. Cancer Drug Targets 2008, 8, 597–610. [Google Scholar] [CrossRef]
- Zou, Y.Y.; Tsai, W.B.; Cheng, C.J.; Hsu, C.; Chung, Y.M.; Li, P.C.; Lin, S.H.; Hu, M.C.T. Forkhead box transcription factor FOXO3a suppresses estrogen-dependent breast cancer cell proliferation and tumorigenesis. Breast Cancer Res. 2008, 10, R21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, H.H.; Herrera, R.E.; Coronado-Heinsohn, E.; Yang, M.C.; Ludes-Meyers, J.H.; Seybold-Tilson, K.J.; Nawaz, Z.; Yee, D.; Barr, F.G.; Diab, S.G.; et al. Forkhead homologue in rhabdomyosarcoma functions as a bifunctional nuclear receptor-interacting protein with both coactivator and corepressor functions. J. Biol. Chem. 2001, 276, 27907–27912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morelli, C.; Lanzino, M.; Garofalo, C.; Maris, P.; Brunelli, E.; Casaburi, I.; Catalano, S.; Bruno, R.; Sisci, D.; Ando, S. Akt2 Inhibition Enables the Forkhead Transcription Factor FoxO3a to Have a Repressive Role in Estrogen Receptor alpha Transcriptional Activity in Breast Cancer Cells. Mol. Cell Biol. 2010, 30, 857–870. [Google Scholar] [CrossRef] [Green Version]
- van der Vos, K.E.; Coffer, P.J. FOXO-binding partners: It takes two to tango. Oncogene 2008, 27, 2289–2299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Habashy, H.O.; Rakha, E.A.; Aleskandarany, M.; Ahmed, M.A.; Green, A.R.; Ellis, I.O.; Powe, D.G. FOXO3a nuclear localisation is associated with good prognosis in luminal-like breast cancer. Breast Cancer Res. Treat. 2011, 129, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Gomes, A.R.; Monteiro, L.J.; Wong, S.Y.; Wu, L.H.; Ng, T.T.; Karadedou, C.T.; Millour, J.; Ip, Y.C.; Cheung, Y.N.; et al. Constitutively nuclear FOXO3a localization predicts poor survival and promotes Akt phosphorylation in breast cancer. PLoS ONE 2010, 5, e12293. [Google Scholar] [CrossRef] [Green Version]
- Rehman, A.; Kim, Y.; Kim, H.; Sim, J.; Ahn, H.; Chung, M.S.; Shin, S.J.; Jang, K. FOXO3a expression is associated with lymph node metastasis and poor disease-free survival in triple-negative breast cancer. J. Clin. Pathol. 2018, 71, 806–813. [Google Scholar] [CrossRef]
- Storz, P.; Doppler, H.; Copland, J.A.; Simpson, K.J.; Toker, A. FOXO3a Promotes Tumor Cell Invasion through the Induction of Matrix Metalloproteinases. Mol. Cell Biol. 2009, 29, 4906–4917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sisci, D.; Maris, P.; Cesario, M.G.; Anselmo, W.; Coroniti, R.; Trombino, G.E.; Romeo, F.; Ferraro, A.; Lanzino, M.; Aquila, S.; et al. The estrogen receptor alpha is the key regulator of the bifunctional role of FoxO3a transcription factor in breast cancer motility and invasiveness. Cell Cycle 2013, 12, 3405–3420. [Google Scholar] [CrossRef]
- Gomes, A.R.; Zhao, F.; Lam, E.W.F. Role and regulation of the forkhead transcription factors FOXO3a and FOXM1 in carcinogenesis and drug resistance. Chin. J. Cancer 2013, 32, 365–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bullock, M. FOXO factors and breast cancer: Outfoxing endocrine resistance. Endocr.-Relat. Cancer 2016, 23, R113–R130. [Google Scholar] [CrossRef] [Green Version]
- Sang, Y.T.; Chen, B.; Song, X.J.; Li, Y.M.; Liang, Y.R.; Han, D.W.; Zhang, N.; Zhang, H.W.; Liu, Y.; Chen, T.; et al. circRNA_0025202 Regulates Tamoxifen Sensitivity and Tumor Progression via Regulating the miR-182-5p/FOXO3a Axis in Breast Cancer. Mol. Ther. 2019, 27, 1638–1652. [Google Scholar] [CrossRef] [PubMed]
- Pellegrino, M.; Rizza, P.; Nigro, A.; Ceraldi, R.; Ricci, E.; Perrotta, I.; Aquila, S.; Lanzino, M.; Ando, S.; Morelli, C.; et al. FoxO3a Mediates the Inhibitory Effects of the Antiepileptic Drug Lamotrigine on Breast Cancer Growth. Mol. Cancer Res. 2018, 16, 923–934. [Google Scholar] [CrossRef] [Green Version]
- Sisci, D.; Middea, E.; Morelli, C.; Lanzino, M.; Aquila, S.; Rizza, P.; Catalano, S.; Casaburi, I.; Maggiolini, M.; Ando, S. 17beta-estradiol enhances alpha(5) integrin subunit gene expression through ERalpha-Sp1 interaction and reduces cell motility and invasion of ERalpha-positive breast cancer cells. Breast Cancer Res. Treat. 2010, 124, 63–77. [Google Scholar] [CrossRef] [Green Version]
- Cerami, E.; Gao, J.J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [Green Version]
- Hiscox, S.; Morgan, L.; Barrow, D.; Dutkowskil, C.; Wakeling, A.; Nicholson, R.I. Tamoxifen resistance in breast cancer cells is accompanied by an enhanced motile and invasive phenotype: Inhibition by gefitinib (‘Iressa’, ZD1839). Clin. Experim. Metastas. 2004, 21, 201–212. [Google Scholar] [CrossRef]
- Al Saleh, S.; Sharaf, L.H.; Luqmani, Y.A. Signalling pathways involved in endocrine resistance in breast cancer and associations with epithelial to mesenchymal transition. Int. J. Oncol. 2011, 38, 1197–1217. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Shenouda, S.; Baranwal, S.; Rathinam, R.; Jain, P.; Bao, L.; Hazari, S.; Dash, S.; Alahari, S.K. Integrin subunits alpha5 and alpha6 regulate cell cycle by modulating the chk1 and Rb/E2F pathways to affect breast cancer metastasis. Mol. Cancer 2011, 10, 84. [Google Scholar] [CrossRef] [Green Version]
- Lobert, V.H.; Brech, A.; Pedersen, N.M.; Wesche, J.; Oppelt, A.; Malerod, L.; Stenmark, H. Ubiquitination of alpha 5 beta 1 integrin controls fibroblast migration through lysosomal degradation of fibronectin-integrin complexes. Dev. Cell 2010, 19, 148–159. [Google Scholar] [CrossRef] [Green Version]
- You, D.; Jung, S.P.; Jeong, Y.; Bae, S.Y.; Lee, J.E.; Kim, S. Fibronectin expression is upregulated by PI-3K/Akt activation in tamoxifen-resistant breast cancer cells. BMB Rep. 2017, 50, 615–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.H.; Man, H.T.; Zhao, X.D.; Dong, N.; Ma, S.L. Estrogen receptor-positive breast cancer molecular signatures and therapeutic potentials. Biomed. Rep. 2014, 2, 41–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piscuoglio, S.; Ng, C.K.Y.; Weigelt, B.; Chandarlapaty, S.; Reis-Filho, J.S. ESR1 and endocrine therapy resistance: More than just mutations. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2018, 29, 787–789. [Google Scholar] [CrossRef] [PubMed]
- Early Breast Cancer Trialists’ Collaborative Group. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: An overview of the randomised trials. Lancet 2005, 365, 1687–1717. [Google Scholar] [CrossRef]
- Ali, S.; Rasool, M.; Chaoudhry, H.; Pushparaj, P.N.; Jha, P.; Hafiz, A.; Mahfooz, M.; Abdus Sami, G.; Azhar Kamal, M.; Bashir, S.; et al. Molecular mechanisms and mode of tamoxifen resistance in breast cancer. Bioinformation 2016, 12, 135–139. [Google Scholar] [CrossRef]
- Yao, J.; Deng, K.; Huang, J.; Zeng, R.; Zuo, J. Progress in the Understanding of the Mechanism of Tamoxifen Resistance in Breast Cancer. Front. Pharmacol. 2020, 11, 592912. [Google Scholar] [CrossRef]
- Silva, J.; Cavazos, D.A.; Donzis, E.; Friedrichs, W.E.; Marcinialk, R.; Degraffenried, L.A. Akt-induced tamoxifen resistance is associated with altered FKHR regulation. Cancer Investig. 2007, 25, 569–573. [Google Scholar] [CrossRef]
- Vaziri-Gohar, A.; Zheng, Y.; Houston, K.D. IGF-1 Receptor Modulates FoxO1-Mediated Tamoxifen Response in Breast Cancer Cells. Mol. Cancer Res. 2017, 15, 489–497. [Google Scholar] [CrossRef] [Green Version]
- Essaghir, A.; Dif, N.; Marbehant, C.Y.; Coffer, P.J.; Demoulin, J.B. The Transcription of FOXO Genes Is Stimulated by FOXO3 and Repressed by Growth Factors. J. Biol. Chem. 2009, 284, 10334–10342. [Google Scholar] [CrossRef] [Green Version]
- McGovern, U.B.; Francis, R.E.; Peck, B.; Guest, S.K.; Wang, J.; Myatt, S.S.; Krol, J.; Kwok, J.M.M.; Polychronis, A.; Coombes, R.C.; et al. Gefitinib (Iressa) represses FOXM1 expression via FOXO3a in breast cancer. Mol. Cancer 2009, 8, 582–591. [Google Scholar] [CrossRef] [Green Version]
- Bergamaschi, A.; Madak-Erdogan, Z.; Kim, Y.J.; Choi, Y.L.; Lu, H.L.; Katzenellenbogen, B.S. The forkhead transcription factor FOXM1 promotes endocrine resistance and invasiveness in estrogen receptor-positive breast cancer by expansion of stem-like cancer cells. Breast Cancer Res. 2014, 16, 436. [Google Scholar] [CrossRef] [Green Version]
- Millour, J.; Constantinidou, D.; Stavropoulou, A.V.; Wilson, M.S.C.; Myatt, S.S.; Kwok, J.M.M.; Sivanandan, K.; Coombes, R.C.; Medema, R.H.; Hartman, J.; et al. FOXM1 is a transcriptional target of ER alpha and has a critical role in breast cancer endocrine sensitivity and resistance. Oncogene 2010, 29, 2983–2995. [Google Scholar] [CrossRef] [Green Version]
- Seguin, L.; Desgrosellier, J.S.; Weis, S.M.; Cheresh, D.A. Integrins and cancer: Regulators of cancer stemness, metastasis, and drug resistance. Trends Cell Biol. 2015, 25, 234–240. [Google Scholar] [CrossRef] [Green Version]
- Campbell, P.S.; Mavingire, N.; Khan, S.; Rowland, L.K.; Wooten, J.V.; Opoku-Agyeman, A.; Guevara, A.; Soto, U.; Cavalli, F.; Loaiza-Perez, A.I.; et al. AhR ligand aminoflavone suppresses alpha6-integrin-Src-Akt signaling to attenuate tamoxifen resistance in breast cancer cells. J. Cell. Physiol. 2018, 234, 108–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pontiggia, O.; Sampayo, R.; Raffo, D.; Motter, A.; Xu, R.; Bissell, M.J.; Joffe, E.B.; Simian, M. The tumor microenvironment modulates tamoxifen resistance in breast cancer: A role for soluble stromal factors and fibronectin through beta1 integrin. Breast Cancer Res. Treat. 2012, 133, 459–471. [Google Scholar] [CrossRef] [Green Version]
- Yuan, J.; Liu, M.; Yang, L.; Tu, G.; Zhu, Q.; Chen, M.; Cheng, H.; Luo, H.; Fu, W.; Li, Z.; et al. Acquisition of epithelial-mesenchymal transition phenotype in the tamoxifen-resistant breast cancer cell: A new role for G protein-coupled estrogen receptor in mediating tamoxifen resistance through cancer-associated fibroblast-derived fibronectin and beta1-integrin signaling pathway in tumor cells. Breast Cancer Res. 2015, 17, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnawi, R.; Al-Khaldi, S.; Colak, D.; Tulbah, A.; Al-Tweigeri, T.; Fallatah, M.; Monies, D.; Ghebeh, H.; Al-Alwan, M. beta 1 Integrin is essential for fascin-mediated breast cancer stem cell function and disease progression. Int. J. Cancer 2019, 145, 830–841. [Google Scholar] [CrossRef]
- Gui, G.P.; Wells, C.A.; Browne, P.D.; Yeomans, P.; Jordan, S.; Puddefoot, J.R.; Vinson, G.P.; Carpenter, R. Integrin expression in primary breast cancer and its relation to axillary nodal status. Surgery 1995, 117, 102–108. [Google Scholar] [CrossRef]
- Alday-Parejo, B.; Stupp, R.; Ruegg, C. Are Integrins Still Practicable Targets for Anti-Cancer Therapy? Cancers 2019, 11, 978. [Google Scholar] [CrossRef] [Green Version]
- Zhao, F.; Lam, E.W. Role of the forkhead transcription factor FOXO-FOXM1 axis in cancer and drug resistance. Front. Med. 2012, 6, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Kushwaha, P.P.; Gupta, S. Emerging targets in cancer drug resistance. Cancer Drug Resist. 2019, 2, 161–177. [Google Scholar] [CrossRef] [Green Version]
- Robertson, J.F.R.; Coleman, R.E.; Cheung, K.L.; Evans, A.; Holcombe, C.; Skene, A.; Rea, D.; Ahmed, S.; Jahan, A.; Horgan, K.; et al. Proliferation and AKT Activity Biomarker Analyses after Capivasertib (AZD5363) Treatment of Patients with ER(+) Invasive Breast Cancer (STAKT). Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2020, 26, 1574–1585. [Google Scholar] [CrossRef] [Green Version]
- Taylor, S.; Lam, M.; Pararasa, C.; Brown, J.E.; Carmichael, A.R.; Griffiths, H.R. Evaluating the evidence for targeting FOXO3a in breast cancer: A systematic review. Cancer Cell Int. 2015, 15, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ricci, E.; Fava, M.; Rizza, P.; Pellegrino, M.; Bonofiglio, D.; Casaburi, I.; Lanzino, M.; Giordano, C.; Bruno, R.; Sirianni, R.; et al. FoxO3a Inhibits Tamoxifen-Resistant Breast Cancer Progression by Inducing Integrin α5 Expression. Cancers 2022, 14, 214. https://doi.org/10.3390/cancers14010214
Ricci E, Fava M, Rizza P, Pellegrino M, Bonofiglio D, Casaburi I, Lanzino M, Giordano C, Bruno R, Sirianni R, et al. FoxO3a Inhibits Tamoxifen-Resistant Breast Cancer Progression by Inducing Integrin α5 Expression. Cancers. 2022; 14(1):214. https://doi.org/10.3390/cancers14010214
Chicago/Turabian StyleRicci, Elena, Mariarosa Fava, Pietro Rizza, Michele Pellegrino, Daniela Bonofiglio, Ivan Casaburi, Marilena Lanzino, Cinzia Giordano, Rosalinda Bruno, Rosa Sirianni, and et al. 2022. "FoxO3a Inhibits Tamoxifen-Resistant Breast Cancer Progression by Inducing Integrin α5 Expression" Cancers 14, no. 1: 214. https://doi.org/10.3390/cancers14010214
APA StyleRicci, E., Fava, M., Rizza, P., Pellegrino, M., Bonofiglio, D., Casaburi, I., Lanzino, M., Giordano, C., Bruno, R., Sirianni, R., Barone, I., Sisci, D., & Morelli, C. (2022). FoxO3a Inhibits Tamoxifen-Resistant Breast Cancer Progression by Inducing Integrin α5 Expression. Cancers, 14(1), 214. https://doi.org/10.3390/cancers14010214