Identification of Let-7 miRNA Activity as a Prognostic Biomarker of SHH Medulloblastoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Preprocessing of Gene Expression, miRNA Expression, Methylation Data, and CNV Data
2.2. Identifying Cis-Regulatory Genes by Integrating Gene Expression, Methylation Data, and CNV Data
2.3. Identifying Subgroups in GSE42658
2.4. Inferring Tumor Purity of SHH-MB Tumors
2.5. Inferring miRNA Activity with ActMiR
2.6. Survival Association of miRNA Activity or Gene Expression
2.7. Interaction between Activity and Gene Expression
2.8. Identification of Functional Target Genes Enriched for Canonical Pathways
2.9. Detecting Small Molecules That Might Be Effective to SHH Subtypes with Poor Prognosis
2.10. Validation
2.11. Computational Methods
3. Results
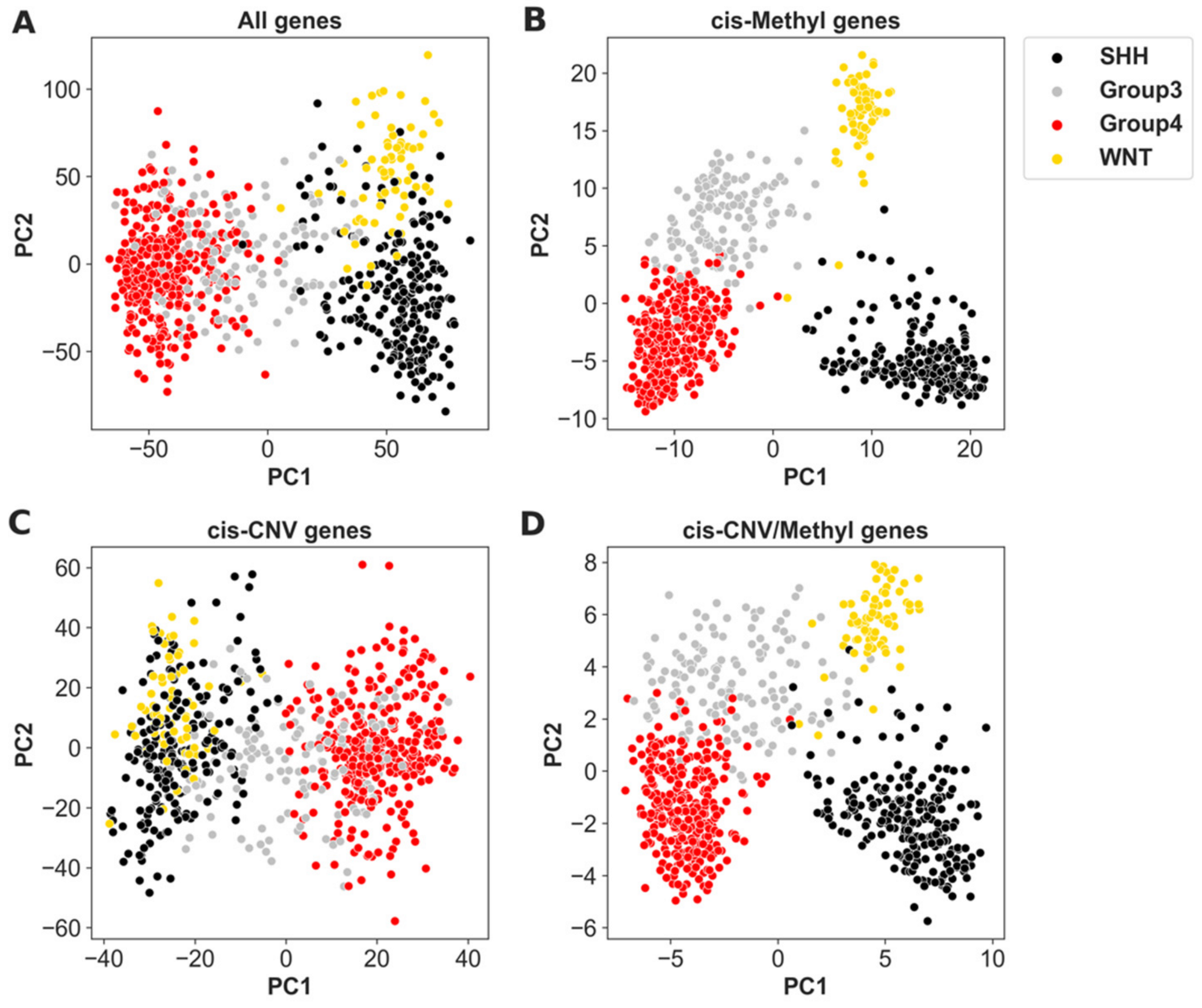
3.1. Inter- and Intra-Tumor Heterogenity of MB Accessed by Cis-Methylation Regulated Genes
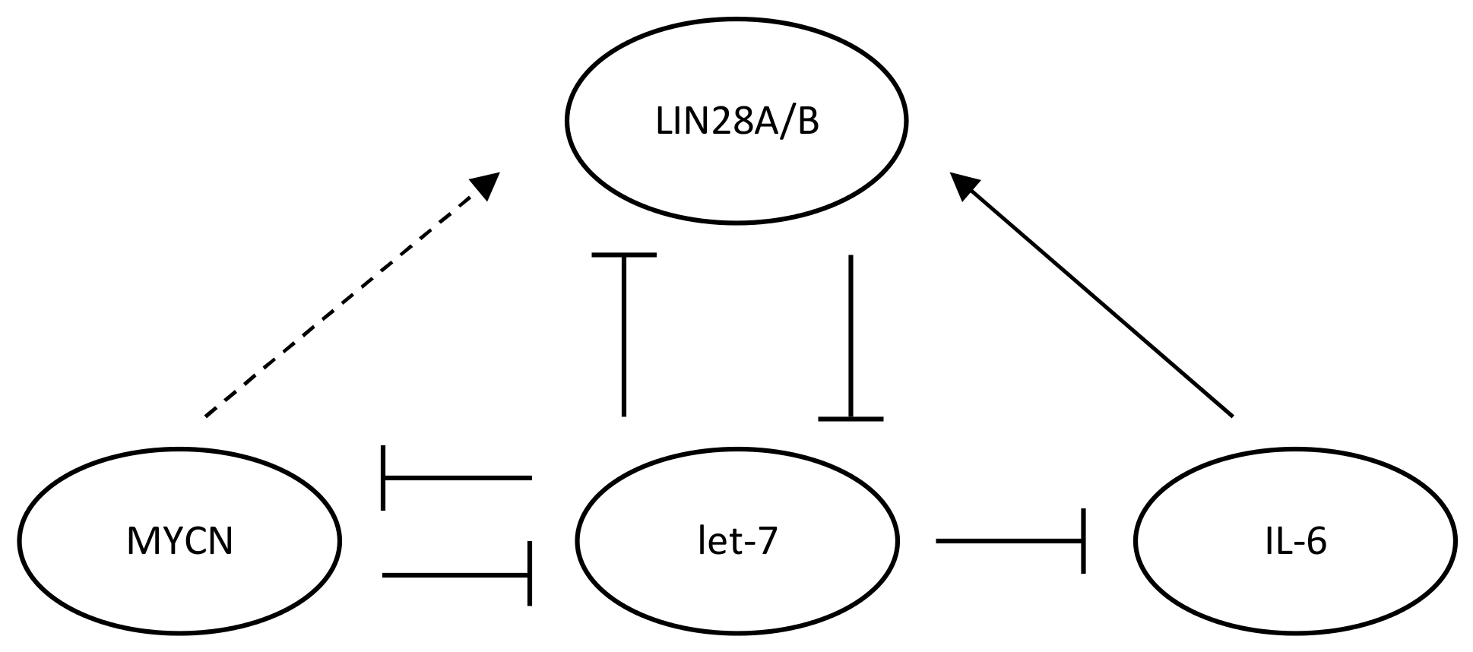
3.2. MYCN/LIN28/Let-7 Axis
3.3. MB Has Higher Let-7 Activity Than Other Brain Tissues and Subgroup-Specific Activity
3.4. Stratifying SHH-MB by MYCN Expression and Let-7 Activity
3.5. Validation in an Independent MB Cohort
3.6. Drug Repurposing to Identify Potential Therapeutic Treatment for SHH Subgroups
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Khanna, V.; Achey, R.L.; Ostrom, Q.T.; Block-Beach, H.; Kruchko, C.; Barnholtz-Sloan, J.S.; de Blank, P.M. Incidence and survival trends for medulloblastomas in the United States from 2001 to 2013. J. Neurooncol. 2017, 135, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Thompson, E.M.; Hielscher, T.; Bouffet, E.; Remke, M.; Luu, B.; Gururangan, S.; McLendon, R.E.; Bigner, D.D.; Lipp, E.S.; Perreault, S.; et al. Prognostic value of medulloblastoma extent of resection after accounting for molecular subgroup: A retrospective integrated clinical and molecular analysis. Lancet Oncol. 2016, 17, 484–495. [Google Scholar] [CrossRef]
- Hughes, E.N.; Shillito, J.; Sallan, S.E.; Loeffler, J.S.; Cassady, J.R.; Tarbell, N.J. Medulloblastoma at the joint center for radiation therapy between 1968 and 1984. The influence of radiation dose on the patterns of failure and survival. Cancer 1988, 61, 1992–1998. [Google Scholar] [CrossRef]
- Gajjar, A.; Chintagumpala, M.; Ashley, D.; Kellie, S.; Kun, L.E.; Merchant, T.E.; Woo, S.; Wheeler, G.; Ahern, V.; Krasin, M.J.; et al. Risk-adapted craniospinal radiotherapy followed by high-dose chemotherapy and stem-cell rescue in children with newly diagnosed medulloblastoma (St Jude Medulloblastoma-96): Long-term results from a prospective, multicentre trial. Lancet Oncol. 2006, 7, 813–820. [Google Scholar] [CrossRef]
- Tarbell, N.J.; Friedman, H.; Polkinghorn, W.R.; Yock, T.; Zhou, T.; Chen, Z.; Burger, P.; Barnes, P.; Kun, L. High-risk medulloblastoma: A pediatric oncology group randomized trial of chemotherapy before or after radiation therapy (POG 9031). J. Clin. Oncol. 2013, 31, 2936–2941. [Google Scholar] [CrossRef]
- Polkinghorn, W.R.; Tarbell, N.J. Medulloblastoma: Tumorigenesis, current clinical paradigm, and efforts to improve risk stratification. Nat. Clin. Pract. Oncol. 2007, 4, 295–304. [Google Scholar] [CrossRef]
- Northcott, P.A.; Dubuc, A.M.; Pfister, S.; Taylor, M.D. Molecular subgroups of medulloblastoma. Expert Rev. Neurother. 2012, 12, 871–884. [Google Scholar] [CrossRef]
- Juraschka, K.; Taylor, M.D. Medulloblastoma in the age of molecular subgroups. J. Neurosurg. Pediatr. 2019, 24, 353–363. [Google Scholar] [CrossRef]
- Taylor, M.D.; Northcott, P.A.; Korshunov, A.; Remke, M.; Cho, Y.J.; Clifford, S.C.; Eberhart, C.G.; Parsons, D.W.; Rutkowski, S.; Gajjar, A.; et al. Molecular subgroups of medulloblastoma: The current consensus. Acta Neuropathol. 2012, 123, 465–472. [Google Scholar] [CrossRef]
- El Doussouki, M.; Gajjar, A.; Chamdine, O. Molecular genetics of medulloblastoma in children: Diagnostic, therapeutic and prognostic implications. Future Neurol. 2019, 14, 20–33. [Google Scholar] [CrossRef]
- Northcott, P.A.; Buchhalter, I.; Morrissy, A.S.; Hovestadt, V.; Weischenfeldt, J.; Ehrenberger, T.; Gröbner, S.; Segura-Wang, M.; Zichner, T.; Rudneva, V.A.; et al. The whole-genome landscape of medulloblastoma subtypes. Nature 2017, 547, 311–317. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Bavle, A.; Parsons, D.W. From One to Many: Further Refinement of Medulloblastoma Subtypes Offers Promise for Personalized Therapy. Cancer Cell 2017, 31, 727–729. [Google Scholar] [CrossRef]
- Gibson, P.; Tong, Y.; Robinson, G.; Thompson, M.C.; Currle, D.S.; Eden, C.; Kranenburg, T.A.; Hogg, T.; Poppleton, H.; Martin, J.; et al. Subtypes of medulloblastoma have distinct developmental origins. Nature 2010, 468, 1095–1099. [Google Scholar] [CrossRef]
- Cavalli, F.M.G.; Remke, M.; Rampasek, L.; Peacock, J.; Shih, D.J.H.; Luu, B.; Garzia, L.; Torchia, J.; Nor, C.; Morrissy, A.S.; et al. Intertumoral Heterogeneity within Medulloblastoma Subgroups. Cancer Cell 2017, 31, 737–754.e6. [Google Scholar] [CrossRef]
- Northcott, P.A.; Shih, D.J.H.; Peacock, J.; Garzia, L.; Morrissy, S.; Zichner, T.; Stütz, A.M.; Korshunov, A.; Reimand, J.; Steven, E.; et al. Subgroup specific structural variation across 1000 medulloblastoma genomes. Nature 2012, 488, 49–56. [Google Scholar] [CrossRef]
- Kool, M.; Jones, D.T.W.; Jäger, N.; Northcott, P.A.; Pugh, T.J.; Hovestadt, V.; Piro, R.M.; Esparza, L.A.; Markant, S.L.; Remke, M.; et al. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell 2014, 25, 393–405. [Google Scholar] [CrossRef]
- Lee, Y.; Kawagoe, R.; Sasai, K.; Li, Y.; Russell, H.R.; Curran, T.; McKinnon, P.J. Loss of suppressor-of-fused function promotes tumorigenesis. Oncogene 2007, 26, 6442–6447. [Google Scholar] [CrossRef]
- Kool, M.; Koster, J.; Bunt, J.; Hasselt, N.E.; Lakeman, A.; van Sluis, P.; Troost, D.; Meeteren, N.S.; Caron, H.N.; Cloos, J.; et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS ONE 2008, 3, e3088. [Google Scholar] [CrossRef]
- Northcott, P.A.; Hielscher, T.; Dubuc, A.; Mack, S.; Shih, D.; Remke, M.; Al-Halabi, H.; Albrecht, S.; Jabado, N.; Eberhart, C.G.; et al. Pediatric and adult sonic hedgehog medulloblastomas are clinically and molecularly distinct. Acta Neuropathol. 2011, 122, 231–240. [Google Scholar] [CrossRef]
- Ramaswamy, V.; Nör, C.; Taylor, M.D. p53 and Meduloblastoma. Cold Spring Harb. Perspect. Med. 2015, 6, a026278. [Google Scholar] [CrossRef]
- Zhukova, N.; Ramaswamy, V.; Remke, M.; Pfaff, E.; Shih, D.J.H.; Martin, D.C.; Castelo-Branco, P.; Baskin, B.; Ray, P.N.; Bouffet, E.; et al. Subgroup-specific prognostic implications of TP53 mutation in medulloblastoma. J. Clin. Oncol. 2013, 31, 2927–2935. [Google Scholar] [CrossRef]
- Ramaswamy, V.; Remke, M.; Bouffet, E.; Faria, C.C.; Perreault, S.; Cho, Y.-J.; Shih, D.J.; Luu, B.; Dubuc, A.M.; Northcott, P.A.; et al. Recurrence patterns across medulloblastoma subgroups: An integrated clinical and molecular analysis. Lancet Oncol. 2013, 14, 1200–1207. [Google Scholar] [CrossRef]
- Ramaswamy, V.; Remke, M.; Adamski, J.; Bartels, U.; Tabori, U.; Wang, X.; Huang, A.; Hawkins, C.; Mabbott, D.; Laperriere, N.; et al. Medulloblastoma subgroup-specific outcomes in irradiated children: Who are the true high-risk patients? Neuro. Oncol. 2016, 18, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Mollashahi, B.; Aghamaleki, F.S.; Movafagh, A. The Roles of miRNAs in Medulloblastoma: A Systematic Review. J. Cancer Prev. 2019, 24, 79–90. [Google Scholar] [CrossRef]
- Northcott, P.A.; Jones, D.T.W.; Kool, M.; Robinson, G.W.; Gilbertson, R.J.; Cho, Y.J.; Pomeroy, S.L.; Korshunov, A.; Lichter, P.; Taylor, M.D.; et al. Medulloblastomics: The end of the beginning. Nat. Rev. Cancer 2012, 12, 818–834. [Google Scholar] [CrossRef]
- Thompson, M.C.; Fuller, C.; Hogg, T.L.; Dalton, J.; Finkelstein, D.; Lau, C.C.; Chintagumpala, M.; Adesina, A.; Ashley, D.M.; Kellie, S.J.; et al. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2006, 24, 1924–1931. [Google Scholar] [CrossRef]
- Hovestadt, V.; Jones, D.T.W.; Picelli, S.; Wang, W.; Kool, M.; Northcott, P.A.; Sultan, M.; Stachurski, K.; Ryzhova, M.; Warnatz, H.-J.; et al. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature 2014, 510, 537–541. [Google Scholar] [CrossRef]
- Molenaar, J.J.; Domingo-Fernández, R.; Ebus, M.E.; Lindner, S.; Koster, J.; Drabek, K.; Mestdagh, P.; van Sluis, P.; Valentijn, L.J.; van Nes, J.; et al. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat. Genet. 2012, 44, 1199–1206. [Google Scholar] [CrossRef]
- Ruiz-Pérez, M.V.; Henley, A.B.; Arsenian-Henriksson, M. The MYCN Protein in Health and Disease. Genes 2017, 8, 113. [Google Scholar] [CrossRef] [PubMed]
- Meyer, N.; Penn, L.Z. Reflecting on 25 years with MYC. Nat. Rev. Cancer 2008, 8, 976–990. [Google Scholar] [CrossRef] [PubMed]
- Scotting, P.J.; Walker, D.A.; Perilongo, G. Childhood solid tumours: A developmental disorder. Nat. Rev. Cancer 2005, 5, 481–488. [Google Scholar] [CrossRef]
- Zimmerman, K.A.; Yancopoulos, G.D.; Collum, R.G.; Smith, R.K.; Kohl, N.E.; Denis, K.A.; Nau, M.M.; Witte, O.N.; Toran-Allerand, D.; Gee, C.E. Differential expression of myc family genes during murine development. Nature 1986, 319, 780–783. [Google Scholar] [CrossRef]
- Balzeau, J.; Menezes, M.R.; Cao, S.; Hagan, J.P. The LIN28/let-7 pathway in cancer. Front. Genet. 2017, 8, 31. [Google Scholar] [CrossRef]
- Wu, L.; Wang, Q.; Yao, J.; Jiang, H.; Xiao, C.; Wu, F. MicroRNA let-7g and let-7i inhibit hepatoma cell growth concurrently via downregulation of the anti-apoptoticprotein B-cell lymphoma-extra large. Oncol. Lett. 2015, 9, 213–218. [Google Scholar] [CrossRef][Green Version]
- Sampson, V.B.; Rong, N.H.; Han, J.; Yang, Q.; Aris, V.; Soteropoulos, P.; Petrelli, N.J.; Dunn, S.P.; Krueger, L.J. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007, 67, 9762–9770. [Google Scholar] [CrossRef]
- Viswanathan, S.R.; Daley, G.Q.; Gregory, R.I. Selective blockade of microRNA processing by Lin28. Science 2008, 320, 97–100. [Google Scholar] [CrossRef]
- Hagan, J.P.; Croce, C.M. MicroRNAs in carcinogenesis. Cytogenet. Genome Res. 2007, 118, 252–259. [Google Scholar] [CrossRef]
- Farazi, T.A.; Hoell, J.I.; Morozov, P.; Tuschl, T. MicroRNAs in human cancer BT. In MicroRNA Cancer Regulation: Advanced Concepts, Bioinformatics and Systems Biology Tools; Schmitz, U., Wolkenhauer, O., Vera, J., Eds.; Springer: Dordrecht, The Netherlands, 2013; pp. 1–20. ISBN 978-94-007-5590-1. [Google Scholar]
- Jansson, M.D.; Lund, A.H. MicroRNA and cancer. Mol. Oncol. 2012, 6, 590–610. [Google Scholar] [CrossRef]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Ebert, M.S.; Neilson, J.R.; Sharp, P.A. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 2007, 4, 721–726. [Google Scholar] [CrossRef] [PubMed]
- Arvey, A.; Larsson, E.; Sander, C.; Leslie, C.S.; Marks, D.S. Target mRNA abundance dilutes microRNA and siRNA activity. Mol. Syst. Biol. 2010, 6, 363. [Google Scholar] [CrossRef] [PubMed]
- Mullokandov, G.; Baccarini, A.; Ruzo, A.; Jayaprakash, A.D.; Tung, N.; Israelow, B.; Evans, M.J.; Sachidanandam, R.; Brown, B.D. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat. Methods 2012, 9, 840–846. [Google Scholar] [CrossRef]
- Lee, E.; Ito, K.; Zhao, Y.; Schadt, E.E.; Irie, H.Y.; Zhu, J. Inferred miRNA activity identifies miRNA-mediated regulatory networks underlying multiple cancers. Bioinformatics 2016, 32, 96–105. [Google Scholar] [CrossRef]
- Ocasio, J.; Babcock, B.; Malawsky, D.; Weir, S.J.; Loo, L.; Simon, J.M.; Zylka, M.J.; Hwang, D.; Dismuke, T.; Sokolsky, M.; et al. scRNA-seq in medulloblastoma shows cellular heterogeneity and lineage expansion support resistance to SHH inhibitor therapy. Nat. Commun. 2019, 10, 5829. [Google Scholar] [CrossRef]
- Ramaswamy, V.; Taylor, M.D. Bioinformatic Strategies for the Genomic and Epigenomic Characterization of Brain Tumors. Methods Mol. Biol. 2019, 1869, 37–56. [Google Scholar] [CrossRef]
- Henriquez, N.V.; Forshew, T.; Tatevossian, R.; Ellis, M.; Richard-Loendt, A.; Rogers, H.; Jacques, T.S.; Reitboeck, P.G.; Pearce, K.; Sheer, D.; et al. Comparative expression analysis reveals lineage relationships between human and murine gliomas and a dominance of glial signatures during tumor propagation in vitro. Cancer Res. 2013, 73, 5834–5844. [Google Scholar] [CrossRef]
- Olshen, A.B.; Venkatraman, E.S.; Lucito, R.; Wigler, M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 2004, 5, 557–572. [Google Scholar] [CrossRef]
- Suzuki, M.M.; Bird, A. DNA methylation landscapes: Provocative insights from epigenomics. Nat. Rev. Genet. 2008, 9, 465–476. [Google Scholar] [CrossRef]
- DiNardo, D.N.; Butcher, D.T.; Robinson, D.P.; Archer, T.K.; Rodenhiser, D.I. Functional analysis of CpG methylation in the BRCA1 promoter region. Oncogene 2001, 20, 5331–5340. [Google Scholar] [CrossRef]
- Yoo, S.; Huang, T.; Campbell, J.D.; Lee, E.; Tu, Z.; Geraci, M.W.; Powell, C.A.; Schadt, E.E.; Spira, A.; Zhu, J. MODMatcher: Multi-Omics Data Matcher for Integrative Genomic Analysis. PLoS Comput. Biol. 2014, 10, e1003790. [Google Scholar] [CrossRef]
- Seabold, S.; Perktold, J. Statsmodels: Econometric and statistical modeling with python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010. [Google Scholar]
- Hovestadt, V.; Smith, K.S.; Bihannic, L.; Filbin, M.G.; Shaw, M.L.; Baumgartner, A.; DeWitt, J.C.; Groves, A.; Mayr, L.; Weisman, H.R.; et al. Resolving medulloblastoma cellular architecture by single-cell genomics. Nature 2019, 572, 74–79. [Google Scholar] [CrossRef]
- Lee, E.; Collazo-Lorduy, A.; Castillo-Martin, M.; Gong, Y.; Wang, L.; Oh, W.K.; Galsky, M.D.; Cordon-Cardo, C.; Zhu, J. Identification of microR-106b as a prognostic biomarker of p53-like bladder cancers by ActMir. Oncogene 2018, 37, 5858–5872. [Google Scholar] [CrossRef]
- Kaplan, E.L.; Meier, P. Nonparametric Estimation from Incomplete Observations. J. Am. Stat. Assoc. 1958, 53, 457–481. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Lamb, J. The Connectivity Map: A new tool for biomedical research. Nat. Rev. Cancer 2007, 7, 54–60. [Google Scholar] [CrossRef]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171, 1437–1452.e17. [Google Scholar] [CrossRef]
- Cho, Y.-J.; Tsherniak, A.; Tamayo, P.; Santagata, S.; Ligon, A.; Greulich, H.; Berhoukim, R.; Amani, V.; Goumnerova, L.; Eberhart, C.G.; et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2011, 29, 1424–1430. [Google Scholar] [CrossRef]
- Fattet, S.; Haberler, C.; Legoix, P.; Varlet, P.; Lellouch-Tubiana, A.; Lair, S.; Manie, E.; Raquin, M.-A.; Bours, D.; Carpentier, S.; et al. Beta-catenin status in paediatric medulloblastomas: Correlation of immunohistochemical expression with mutational status, genetic profiles, and clinical characteristics. J. Pathol. 2009, 218, 86–94. [Google Scholar] [CrossRef]
- Park, A.K.; Lee, J.Y.; Cheong, H.; Ramaswamy, V.; Park, S.H.; Kool, M.; Phi, J.H.; Choi, S.A.; Cavalli, F.; Taylor, M.D.; et al. Subgroup-specific prognostic signaling and metabolic pathways in pediatric medulloblastoma. BMC Cancer 2019, 19, 571. [Google Scholar] [CrossRef] [PubMed]
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Fertig, E.J.; Jaffe, A.E.; Storey, J.D.; Zhang, Y.; Torres, L.C. Sva: Surrogate Variable Analysis 2019. Available online: https://bioconductor.org/packages/release/bioc/html/sva.html (accessed on 3 June 2021).
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Davidson-Pilon, C.; Kalderstam, J.; Jacobson, N.; Reed, S.; Kuhn, B.; Zivich, P.; Williamson, M.; Abdeali, J.K.; Datta, D.; Fiore-Gartland, A.; et al. CamDavidsonPilon/lifelines: v0.25.11. 2021. Available online: https://zenodo.org/record/4136578#.YcrJ-2hBxPY (accessed on 3 June 2021).
- Therneau, T.M.; Grambsch, P.M. Modeling Survival Data: Extending the Cox Model; Springer: New York, NY, USA, 2000; ISBN 0-387-98784-3. [Google Scholar]
- Waskom, M. Seaborn: Statistical Data Visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]
- Hunter, J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Lei, H.; Oh, S.P.; Okano, M.; Jüttermann, R.; Goss, K.A.; Jaenisch, R.; Li, E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 1996, 122, 3195–3205. [Google Scholar] [CrossRef]
- Gifford, C.A.; Ziller, M.J.; Gu, H.; Trapnell, C.; Donaghey, J.; Tsankov, A.; Shalek, A.K.; Kelley, D.R.; Shishkin, A.A.; Issner, R.; et al. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell 2013, 153, 1149–1163. [Google Scholar] [CrossRef]
- Xie, W.; Schultz, M.D.; Lister, R.; Hou, Z.; Rajagopal, N.; Ray, P.; Whitaker, J.W.; Tian, S.; Hawkins, R.D.; Leung, D.; et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell 2013, 153, 1134–1148. [Google Scholar] [CrossRef]
- Yang, Z.-J.; Ellis, T.; Markant, S.L.; Read, T.-A.; Kessler, J.D.; Bourboulas, M.; Schüller, U.; Machold, R.; Fishell, G.; Rowitch, D.H.; et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell 2008, 14, 135–145. [Google Scholar] [CrossRef]
- Wagner, A.J.; Meyers, C.; Laimins, L.A.; Hay, N. c-Myc induces the expression and activity of ornithine decarboxylase. Cell Growth Differ. Mol. Biol. J. Am. Assoc. Cancer Res. 1993, 4, 879–883. [Google Scholar]
- Bello-Fernandez, C.; Packham, G.; Cleveland, J.L. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc. Natl. Acad. Sci. USA 1993, 90, 7804–7808. [Google Scholar] [CrossRef]
- Hogarty, M.D.; Norris, M.D.; Davis, K.; Liu, X.; Evageliou, N.F.; Hayes, C.S.; Pawel, B.; Guo, R.; Zhao, H.; Sekyere, E.; et al. ODC1 is a critical determinant of MYCN oncogenesis and a therapeutic target in neuroblastoma. Cancer Res. 2008, 68, 9735–9745. [Google Scholar] [CrossRef]
- Boyerinas, B.; Park, S.-M.; Hau, A.; Murmann, A.E.; Peter, M.E. The role of let-7 in cell differentiation and cancer. Endocr. Relat. Cancer 2010, 17, F19–F36. [Google Scholar] [CrossRef]
- Borgenvik, A.; Čančer, M.; Hutter, S.; Swartling, F.J. Targeting MYCN in Molecularly Defined Malignant Brain Tumors. Front. Oncol. 2020, 10, 626751. [Google Scholar] [CrossRef]
- Sholler, G.L.S.; Ferguson, W.; Bergendahl, G.; Bond, J.P.; Neville, K.; Eslin, D.; Brown, V.; Roberts, W.; Wada, R.K.; Oesterheld, J.; et al. Maintenance DFMO Increases Survival in High Risk Neuroblastoma. Sci. Rep. 2018, 8, 14445. [Google Scholar] [CrossRef]
- Koomoa, D.-L.T.; Yco, L.P.; Borsics, T.; Wallick, C.J.; Bachmann, A.S. Ornithine decarboxylase inhibition by alpha-difluoromethylornithine activates opposing signaling pathways via phosphorylation of both Akt/protein kinase B and p27Kip1 in neuroblastoma. Cancer Res. 2008, 68, 9825–9831. [Google Scholar] [CrossRef]
- Santhana Kumar, K.; Neve, A.; Guerreiro Stucklin, A.S.; Kuzan-Fischer, C.M.; Rushing, E.J.; Taylor, M.D.; Tripolitsioti, D.; Behrmann, L.; Kirschenbaum, D.; Grotzer, M.A.; et al. TGF-β Determines the Pro-migratory Potential of bFGF Signaling in Medulloblastoma. Cell Rep. 2018, 23, 3798–3812.e8. [Google Scholar] [CrossRef]
- Holzhauser, S.; Lukoseviciute, M.; Andonova, T.; Ursu, R.G.; Dalianis, T.; Wickström, M.; Kostopoulou, O.N. Targeting Fibroblast Growth Factor Receptor (FGFR) and Phosphoinositide 3-kinase (PI3K) Signaling Pathways in Medulloblastoma Cell Lines. Anticancer Res. 2020, 40, 53–66. [Google Scholar] [CrossRef]
- Croce, C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009, 10, 704–714. [Google Scholar] [CrossRef]
- Powers, J.T.; Tsanov, K.M.; Pearson, D.S.; Roels, F.; Spina, C.S.; Ebright, R.; Seligson, M.; de Soysa, Y.; Cahan, P.; Theißen, J.; et al. Multiple mechanisms disrupt the let-7 microRNA family in neuroblastoma. Nature 2016, 535, 246–251. [Google Scholar] [CrossRef]
- Rodini, C.O.; Suzuki, D.E.; Saba-Silva, N.; Cappellano, A.; de Souza, J.E.S.; Cavalheiro, S.; Toledo, S.R.C.; Okamoto, O.K. Expression analysis of stem cell-related genes reveal OCT4 as a predictor of poor clinical outcome in medulloblastoma. J. Neurooncol. 2012, 106, 71–79. [Google Scholar] [CrossRef]
- Monti, P.; Menichini, P.; Speciale, A.; Cutrona, G.; Fais, F.; Taiana, E.; Neri, A.; Bomben, R.; Gentile, M.; Gattei, V.; et al. Heterogeneity of TP53 Mutations and P53 Protein Residual Function in Cancer: Does It Matter? Front. Oncol. 2020, 10, 593383. [Google Scholar] [CrossRef]
- Skowron, P.; Farooq, H.; Cavalli, F.M.G.; Morrissy, A.S.; Ly, M.; Hendrikse, L.D.; Wang, E.Y.; Djambazian, H.; Zhu, H.; Mungall, K.L.; et al. The transcriptional landscape of Shh medulloblastoma. Nat. Commun. 2021, 12, 1749. [Google Scholar] [CrossRef]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-Seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]





| Dataset | Subgroup | Subgroup Number | Age | Number |
|---|---|---|---|---|
| Training (GSE85218) | SHH | 223 | 0–3 | 62 |
| 4–10 | 55 | |||
| 10–17 | 29 | |||
| 18+ | 69 | |||
| WNT | 70 | 0–3 | 1 | |
| 4–10 | 23 | |||
| 10–17 | 27 | |||
| 18+ | 13 | |||
| Group 3 | 144 | 0–3 | 24 | |
| 4–10 | 90 | |||
| 10–17 | 17 | |||
| 18+ | 5 | |||
| Group 4 | 326 | 0–3 | 11 | |
| 4–10 | 181 | |||
| 10–17 | 108 | |||
| 18+ | 14 | |||
| Validation | SHH | 46 | <3 | 23 |
| ≥3 | 22 | |||
| WNT | 21 | <3 | 0 | |
| ≥3 | 21 | |||
| Group 3 | 37 | <3 | 9 | |
| ≥3 | 27 | |||
| Group 4 | 74 | <3 | 3 | |
| ≥3 | 71 |
| Subgroup | Gene | Hazard Ratio | Wald p | Log-Rank p |
|---|---|---|---|---|
| Training All Samples | MYCN | 0.86 | 1.9 × 10−2 | 0.019 |
| MYC | 1.1 | 4.9 × 10−3 | 4.6 × 10−3 | |
| LIN28A | 1.0 | 0.98 | 0.98 | |
| LIN28B | 1.4 | 1.1 × 10−6 | 8.0 × 10−7 | |
| Training SHH | MYCN | 1.3 | 0.25 | 0.26 |
| MYC | 0.97 | 0.91 | 0.91 | |
| LIN28A | 1.1 | 0.79 | 0.79 | |
| LIN28B | 1.5 | 0.01 | 9.2 × 10−3 | |
| Training WNT | MYCN | 2.8 | 0.45 | 0.43 |
| MYC | 0.40 | 0.26 | 0.27 | |
| LIN28A | 1.8 | 0.70 | 0.70 | |
| LIN28B | 0.82 | 0.92 | 0.92 | |
| Training Group 3 | MYCN | 0.74 | 0.028 | 0.029 |
| MYC | 1.2 | 0.020 | 0.019 | |
| LIN28A | 0.71 | 0.29 | 0.29 | |
| LIN28B | 1.3 | 0.11 | 0.11 | |
| Training Group 4 | MYCN | 1.1 | 0.49 | 0.49 |
| MYC | 1.3 | 1.01 × 10−3 | 8.4 × 10−4 | |
| LIN28A | 0.96 | 0.83 | 0.83 | |
| LIN28B | 0.97 | 0.87 | 0.87 |
| Test | Variable | Hazard Ratio | Wald p | Log-Rank p |
|---|---|---|---|---|
| Surv~MYCN | MYCN | 1.3 | 0.25 | 0.26 |
| Surv~LIN28A | LIN28A | 1.1 | 0.79 | 0.79 |
| Surv~LIN28B | LIN28B | 1.5 | 0.011 | 0.0092 |
| Surv~let-7 | let-7 | 169.5 | 0.32 | 0.32 |
| Surv~purity | purity | 3.3 | 0.11 | 0.11 |
| Surv~age | age | 0.99 | 0.55 | 0.55 |
| Surv~MYCN + let-7 | MYCN | 1.7 | 0.066 | 0.069 |
| let-7 | 2.9 × 105 | |||
| Surv~purity + MYCN + let-7 | purity | 1.4 × 10−3 | 0.025 | 0.024 |
| MYCN | 1.7 | |||
| let-7 | 7.7 × 103 |
| Subgroup | Gene | Hazard Ratio | Wald p | Log-Rank p |
|---|---|---|---|---|
| Validation all | MYCN | 1.3 | 0.019 | 0.019 |
| MYC | 1.0 | 0.58 | 0.58 | |
| LIN28A | 1.7 | 0.041 | 0.044 | |
| Validation SHH | MYCN | 1.4 | 0.14 | 0.14 |
| MYC | 0.77 | 0.49 | 0.49 | |
| LIN28A | 5.3 | 0.074 | 3.5 × 10−5 | |
| Validation WNT | MYCN | 5.2 | 0.15 | 0.076 |
| MYC | 0.20 | 0.23 | 0.17 | |
| LIN28A | 65.8 | 0.27 | 0.25 | |
| Validation Group3 | MYCN | 0.90 | 0.82 | 0.82 |
| MYC | 1.2 | 0.30 | 0.29 | |
| LIN28A | 2.0 | 0.28 | 0.28 | |
| Validation Group4 | MYCN | 1.7 | 1.1 × 10−3 | 4.3 × 10−4 |
| MYC | 1.1 | 0.64 | 0.64 | |
| LIN28A | 0.66 | 0.39 | 0.39 |
| Test | Variable | Hazard Ratio | Wald p | Log-Rank p |
|---|---|---|---|---|
| Surv~MYCN | MYCN | 1.4 | 0.14 | 0.14 |
| Surv~LIN28A | LIN28A | 5.3 | 0.074 | 3.5 × 10−5 |
| Surv~LIN28B | LIN28B | N/A | N/A | N/A |
| Surv~let-7 | let-7 | 11.4 | 0.55 | 0.55 |
| Surv~purity | purity | 0.20 | 0.81 | 0.81 |
| Surv~age | age <3 | 1.2 | 0.79 | 0.79 |
| age ≥3 | N/A | |||
| Surv~MYCN + let-7 | MYCN | 1.8 | 0.039 | 0.025 |
| let-7 | 7.7 × 103 | |||
| Surv~purity + MYCN + let-7 | purity | 0.98 | 0.090 | 0.057 |
| MYCN | 1.8 | |||
| let-7 | 7.7 × 103 |
| Score 1 | Name | Description | Number of Drugs 2 | Enrichment Score 3 |
|---|---|---|---|---|
| −99.26 | cobalt(II)-chloride | HSP inducer | 1 (3) | - |
| −98.8 | amonafide | Topoisomerase inhibitor | 1 (16) | - |
| −98.64 | embelin | HCV inhibitor | 1 (3) | - |
| −97.22 | hyperforin | Cyclooxygenase inhibitor | 1 (57) | - |
| −97.02 | parthenolide | NF-kB pathway inhibitor | 1 (12) | - |
| −95.75 | dapsone | Bacterial antifolate | 1 (3) | - |
| −95.6 | brivanib | FGFR inhibitor | 3 (3) | −97.12 |
| −94.79 | ketoconazole | Sterol demethylase inhibitor | 1 (6) | - |
| −94.68 | piperacillin | Bacterial cell wall synthesis inhibitor | 1 (29) | - |
| −94.61 | ziprasidone | Dopamine receptor antagonist | 1 (65) | - |
| −92.54 | tienilic-acid | Sodium/potassium/chloride transporter inhibitor | 1 (4) | - |
| −92.07 | sitagliptin | Dipeptidyl peptidase inhibitor | 1 (3) | - |
| −91.88 | orantinib | FGFR inhibitor | 3 (3) | −97.12 |
| −91.59 | FCCP | Mitochondrial oxidative phosphorylation uncoupler | 1 (2) | - |
| −91.52 | XMD-885 | Leucine rich repeat kinase inhibitor | 1 (3) | - |
| −91.3 | geldanamycin | HSP inhibitor | 1 (13) | - |
| −90.67 | harpagoside | Acetylcholinesterase inhibitor | 1 (17) | - |
| −90.59 | tyrphostin-AG-1478 | EGFR inhibitor | 1 (42) | - |
| −90.43 | AG-370 | PDGFR receptor inhibitor | 1 (9) | - |
| −90.06 | PD-173074 | FGFR inhibitor | 3 (3) | −97.12 |
| −90.05 | kinetin-riboside | Apoptosis stimulant | 1 (10) | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Westphal, M.S.; Lee, E.; Schadt, E.E.; Sholler, G.S.; Zhu, J. Identification of Let-7 miRNA Activity as a Prognostic Biomarker of SHH Medulloblastoma. Cancers 2022, 14, 139. https://doi.org/10.3390/cancers14010139
Westphal MS, Lee E, Schadt EE, Sholler GS, Zhu J. Identification of Let-7 miRNA Activity as a Prognostic Biomarker of SHH Medulloblastoma. Cancers. 2022; 14(1):139. https://doi.org/10.3390/cancers14010139
Chicago/Turabian StyleWestphal, Maximillian S., Eunjee Lee, Eric E. Schadt, Giselle S. Sholler, and Jun Zhu. 2022. "Identification of Let-7 miRNA Activity as a Prognostic Biomarker of SHH Medulloblastoma" Cancers 14, no. 1: 139. https://doi.org/10.3390/cancers14010139
APA StyleWestphal, M. S., Lee, E., Schadt, E. E., Sholler, G. S., & Zhu, J. (2022). Identification of Let-7 miRNA Activity as a Prognostic Biomarker of SHH Medulloblastoma. Cancers, 14(1), 139. https://doi.org/10.3390/cancers14010139






