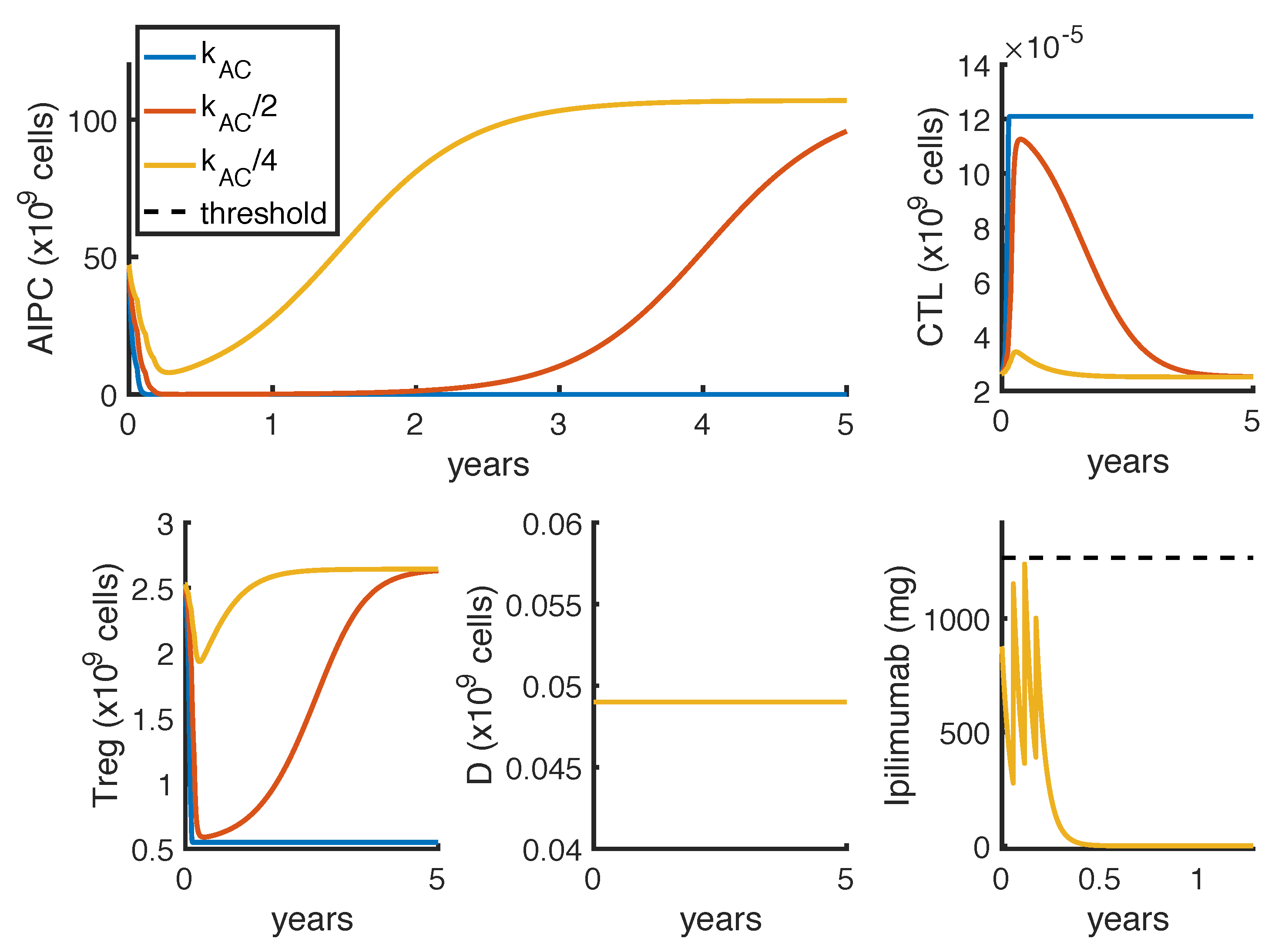1. Introduction
Prostate cancer (PCa) is one of the most common tumor in male population [
1]. Although organ-confined tumors are successfully controlled by surgery or radiotherapy [
2], a quote of patients experiences biochemical progression and most of them ultimately develop distant metastases. Androgen deprivation therapy represents the treatment cornerstone of advanced disease and provides a variable period of disease control [
3]. Nevertheless, all the patients show progressive disease and achieve a condition of castration-resistance. In this disease state several agents demonstrated to be able to significantly prolong the patients’ survival: chemotherapeutics (docetaxel, cabazitaxel), androgen-receptos signaling inhibitors (ARSI), radiopharmaceuticals (radium 223) [
4]. In addition, new generation ARSI are tested for advanced prostate cancer, such as apalutamide, darolutamide, and enzalutamide in non metastatic castration resistant prostate cancer (CRPC) [
5], while abiraterone, apalutamide, and enzalutamide have been employed in metastatic castration sensitive prostate cancer (CSPC) treatments [
6]. However, there are no efficacious therapeutic options for the treatment of metastatic CRPC. Immunotherapy represents an innovative anticancer strategy which recently led to unprecedented improvements in the prognosis of several tumors, such as lung cancer, renal cancer, melanoma, head and neck cancer [
7]. The most widely used immunotherapies are based on the administration of checkpoint inhibitors which exert their anticancer activity by targeting immune checkpoints as PD1 or PDL1. In addition, anti-CTLA4 drugs, such as
ipilimumab, demonstrated their efficacy in some tumors, such as melanoma [
8].
PCa is usually considered as a “cold” tumor, with an immuno-suppressive microenvironment. Nevertheless, different immunotherapy-based strategies have been tested in prostate cancer patients, including vaccine-based therapies and CTLA-4 and PD1-PDL1 inhibition [
9]. To date,
sipuleucel-T is the only immunotherapy agent approved for the tratment of PCa by Food and Drug Administration (FDA), but not by European Medicines Agency (EMA). On the other hand, conflicting results were obtained with
ipilimumab. However, this drug remains a potentially active agent in this disease, and therefore clinical trials are investigating its efficacy in the treatment of PCa [
10].
Mathematical models describing tumor-immune interactions can support clinical decisions. In particular, they can be employed to predict the efficacy of an immunotherapy and, therefore, can help in identifying mechanisms that need to be further investigated. There are many mathematical models of PCa. A comprehensive review [
11], published in 2020, collects the main models describing PCa evolution and its interaction with immune system. Most of the models in literature include androgen-deprivation therapy, with the aim of investigating the effect of this treatment on PCa, or to find the optimal-drug delivery [
12]. There are also works focusing on PCa immunotherapy, but they include only the dendritic cell vaccine
sipuleucel-T. The first model considering more than one immunotherapy has been proposed by Peng et al. [
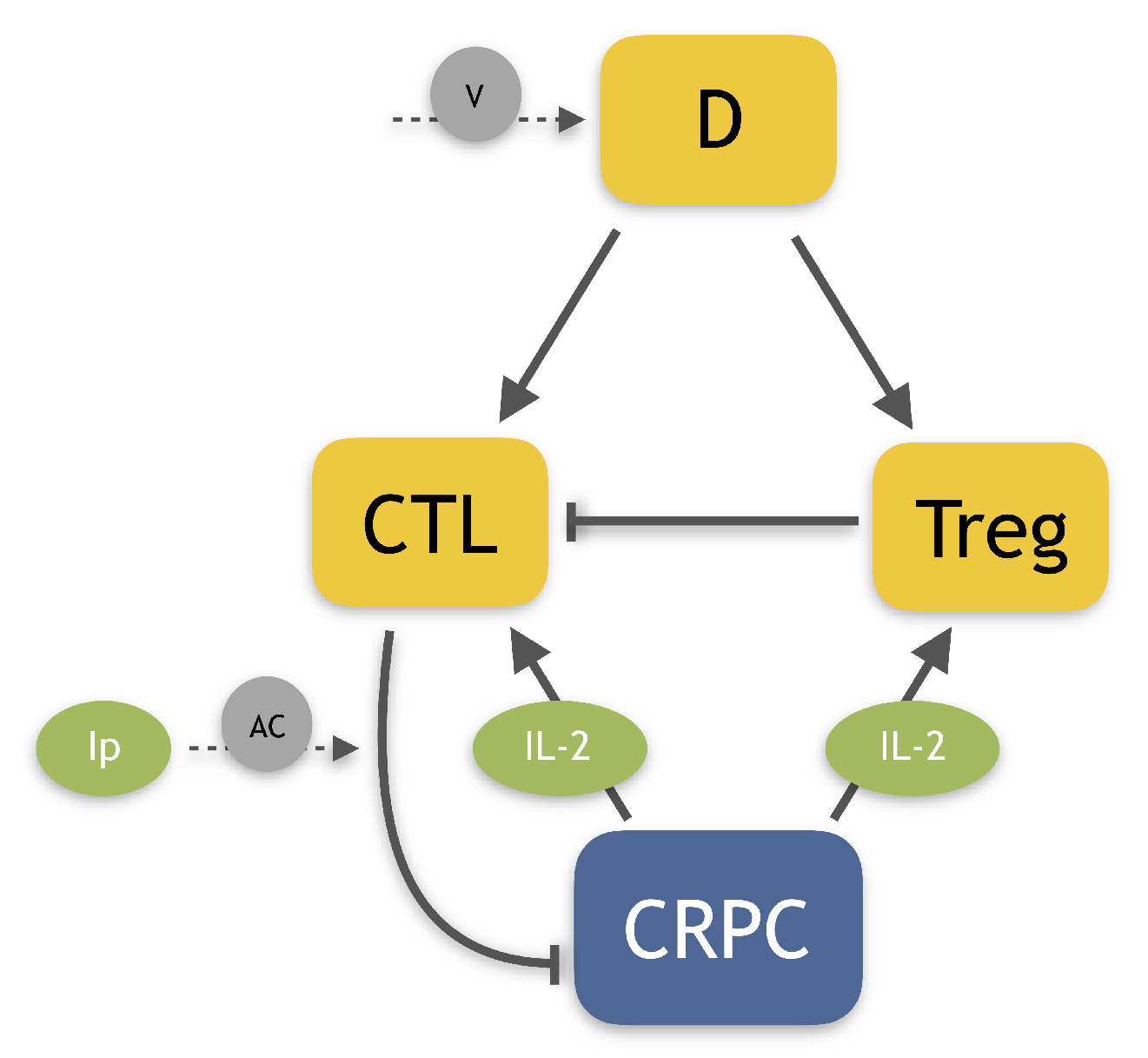
13]. The authors developed a pre-clinical model of PCa including three different immunotherapies: the dendritic cell vaccine, an anti-Treg drug, and anti-IL-2. In a previous work, we extended this model [
14], by including a more detailed description of the tumor micro-environment and two other immunotherapies, the infusion of Natural Killer cells (NK) and an Immune-Checkpoint Inhibitor (ICB). These extensions allowed us to test a large variety of possible combination therapies, by considering both their efficacy in reducing the tumor and their synergy. We also developed a model of human castration-resistant PCa including dendritic cell vaccine
sipuleucel-T and the anti-CTLA4
ipilimumab [
15]. This work aimed at investigating the effect of the immunotherapies on the model steady-states, in order to evaluate if they could lead to tumor eradication. Our results showed that therapies involving the drug
ipilimumab are potentially able to make the no-tumor steady-state attractive, for some reasonable parameter values. Given these promising results, we decide to employ this mathematical model in the present work. The goal of this project is to identify the optimal administration protocol of the drug
ipilimumab, which, administered as mono-therapy or in combination with the vaccine
sipuleucel-T, is able to control or eradicate the tumor. To this end, we need to integrate our previous model by including information about drug toxicity, which is crucial to define a reliable administration protocol. Indeed, when patients are subject to drug administration, adverse events (AEs) can occur.
Side effects due to drug administration are often classified as moderate, i.e., grade 1 and 2, and severe, with grade ≥3. The latter are generally more intense, and they can have a longer resolution time. The AEs observed in a clinical trial are always collected and analyzed in order to estimate the drug toxicity. Sometimes, promising clinical trials have been interrupted due to unexpected side effects, and therefore, this information cannot be ignored. In the literature, there are several examples of PK/PD mathematical models defining the drug toxicity also from a molecular point of view. For example, the work by El-Masri et al. [
16] describes the toxicity by considering the effect of a mixture of drugs on the human body. Similar methods have been employed also in the context of prostate cancer [
17]. Even if this approach have been previously used to define optimal control problems [
18], for our scope, we do not need such a detailed description, which would increase the model complexity both from mathematical and computational point of view. Another possible approach for taking into account drug toxicity in optimal control problems is based on connecting the toxicity with the drug concentration, by defining a maximum acceptable drug exposure and a critical concentration threshold, over which the drug becomes not tolerated [
19,
20]. However, the
ipilimumab has not yet been approved for PCa and there are not enough information about maximal tolerated doses and drug exposure. Therefore, we introduce a toxicity function depending on
ipilimumab concentration.
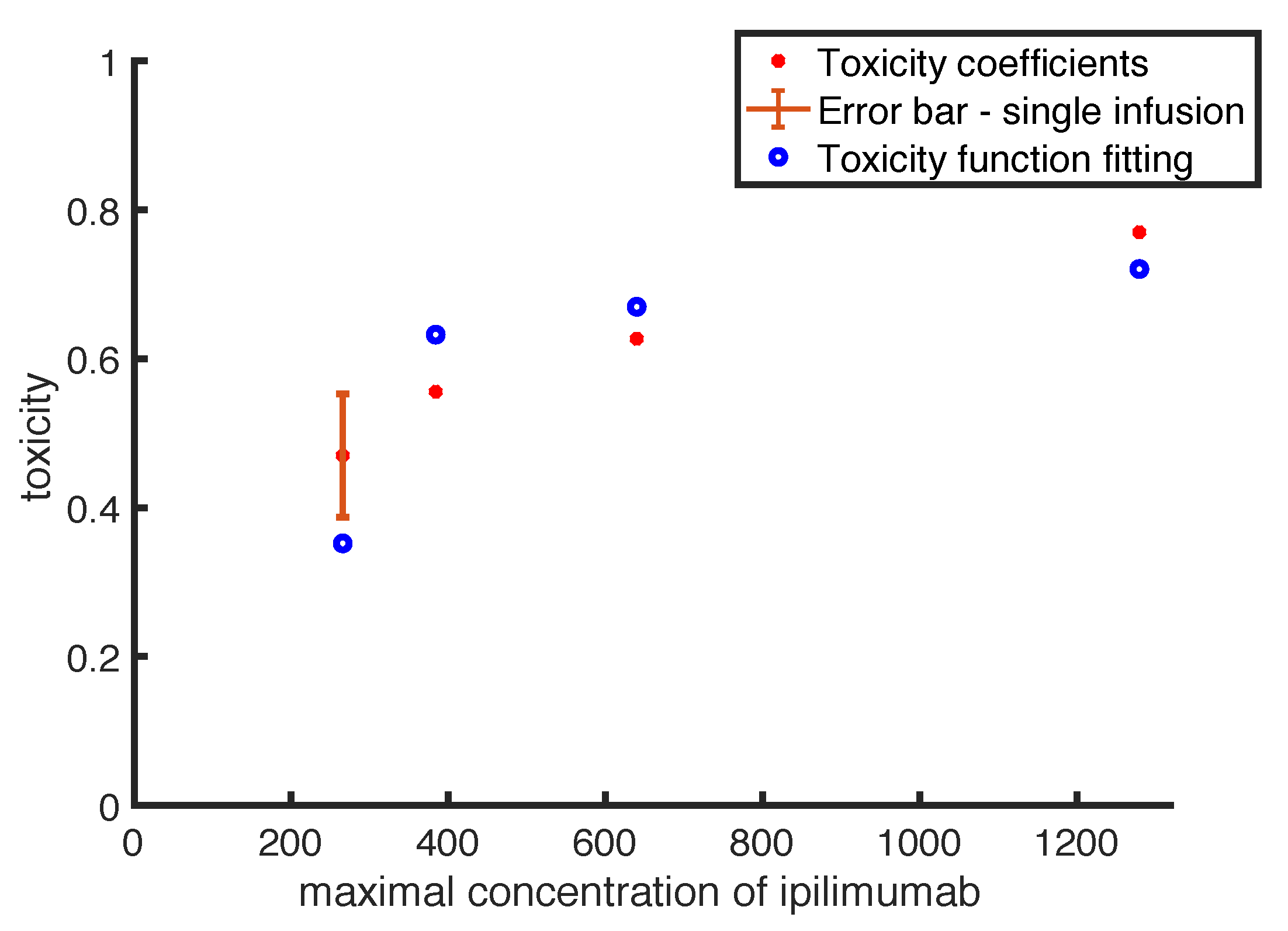
Up to our knowledge, in literature there are few mathematical models describing drug toxicity function, and the most detailed has been introduced by Hadjiandreou et al. [
21]. The authors described the toxicity as a linear function of the drug concentration multiplied by a coefficient, representing the side effect magnitude. This coefficient has been defined by considering the side effects registered into the patient population, weighted by their severity. However, often adverse events can be considered negligible for low drug doses, while they become more frequent and strong in case of high doses, showing a non-linear relation with drug concentration. Therefore, we use a non-linear toxicity to fit toxicity coefficients, computed from literature experimental observations. For a given clinical protocol, the corresponding toxicity coefficient is defined by the percentage of patients showing moderate and severe adverse events, weighted by a function of the average resolution times of the side effects. The occurrence of side effects can produce a stop of the treatment affecting its efficacy. The need of side effects recovery leads to either a delay or a definitive stop of the treatment delivery. Thus the evaluation of the side effects recovery time is essential in evaluating the drug toxicity. Indeed, in patient populations with equivalent adverse events and frequencies, the life quality of those patients having lower average resolution times considerably improves, and therefore we can take the resolution time as an indication of a a lower drug toxicity.
The paper is organized as follows. In
Section 2, we describe the experimental data used to estimate the toxicity function, we present the mathematical model and some technical aspects related to the optimization. We show the model results in
Section 3, including the estimates of the toxicity function and the proposed optimal administration protocols, obtained by fixing different constraints. In
Appendix B, we also analyze the synergy between the
ipilimumab and
sipuleucel-T, by means of a modified Bliss combination index, in order to further investigate how the two immunotherapies work in combination. In the Discussion we summarize the main contributions, linking the mathematical results with biological observations.
4. Discussion
In this work, we employ a mathematical model of human prostate cancer in order to determine optimal
ipilimumab administration protocols able to reduce/eradicate tumor by balancing the treatment ability in reducing tumor and the drug toxicity.
Ipilimumab is an anti-CTLA4 approved for the treatment of several tumors, and tested in metastatic castration-resistant prostate cancer [
10]. We consider patients previously treated with androgen deprivation therapy, who develop the castration-resistant prostate cancer form, and we investigate the efficacy of immunotherapies on those tumor cells. We examine the effect of
ipilimumab administered as mono-therapy and in combination with the dendritic cell vaccine
sipuleucel-T, administered following the FDA recommendations. Our results highlight that the administration of
ipilimumab is potentially able to control or eradicate the tumor. In particular, the optimal administration protocols seem to be feasible, and the corresponding toxicity profile is comparable with those observed in the clinical trials (
Table 2,
Table 4 and
Table 5), suggesting that the proposed therapy could be well-tolerated. Moreover, the result obtained by fixing
weeks for the optimal administration of
ipilimumab as mono-therapy can be compared to the high-dose tested protocol [
23,
28].
The combination with the vaccine is predicted to improve the efficacy of the mono-therapy, since the corresponding optimal protocol provides a faster tumor reduction, while administering a lower amount of anti-CTLA4, with a consequent reduction in
ipilimumab-related toxicity. This effect is probably due to the increase in CTLs proliferation induced by the dendritic cell vaccine [
31], which, coupled with a drug aiming at increasing the CTL tumor-killing activity, causes a stronger tumor suppression. However, clinical studies highlight that only few patients show an evident positive effect as a consequence of
ipilimumab therapies. In the study by Small et al., only 2 out of 14 patients treated with a single dose of
ipilimumab had a significant PSA reduction (>50%), while Slovin et al. [
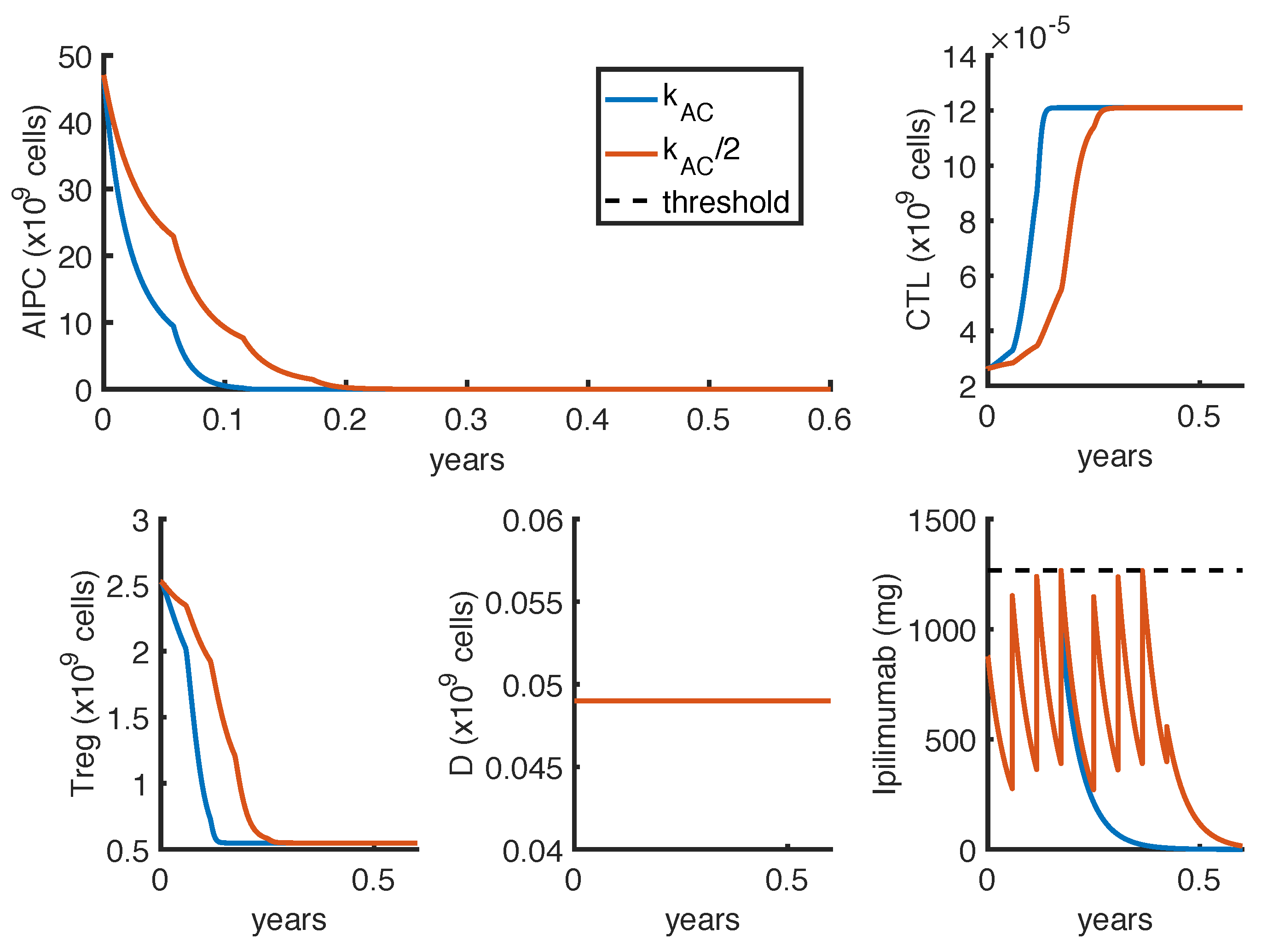
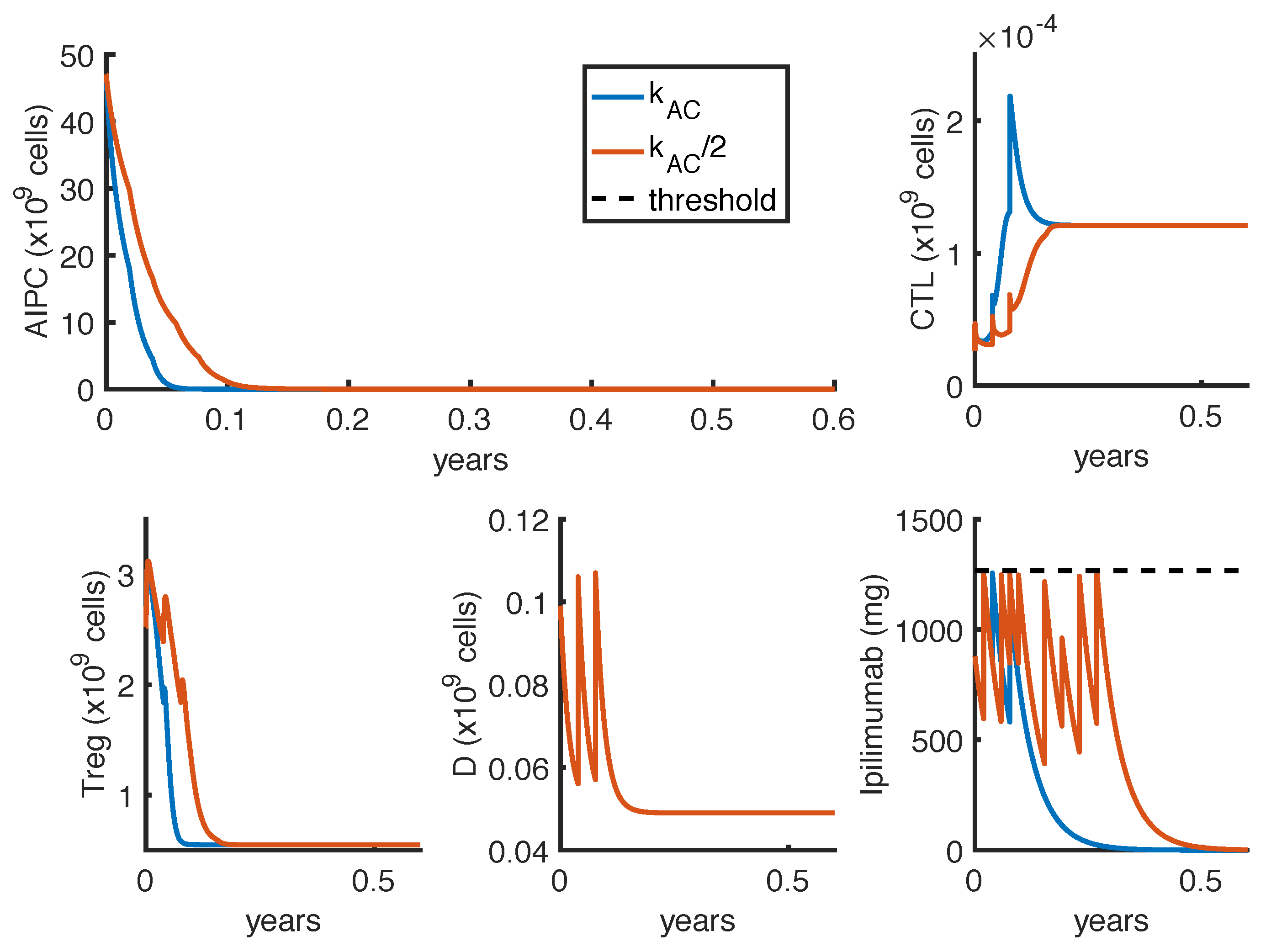
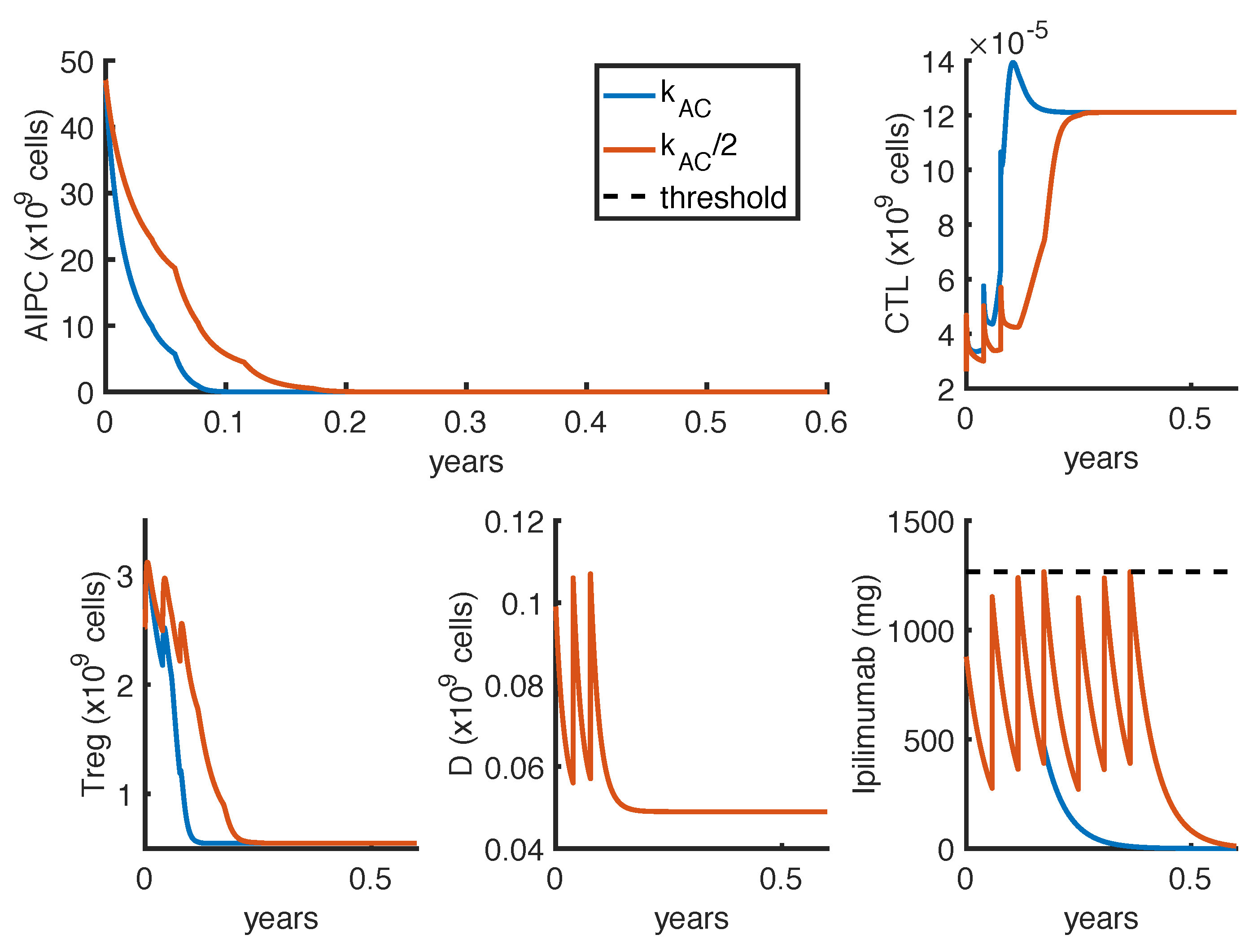
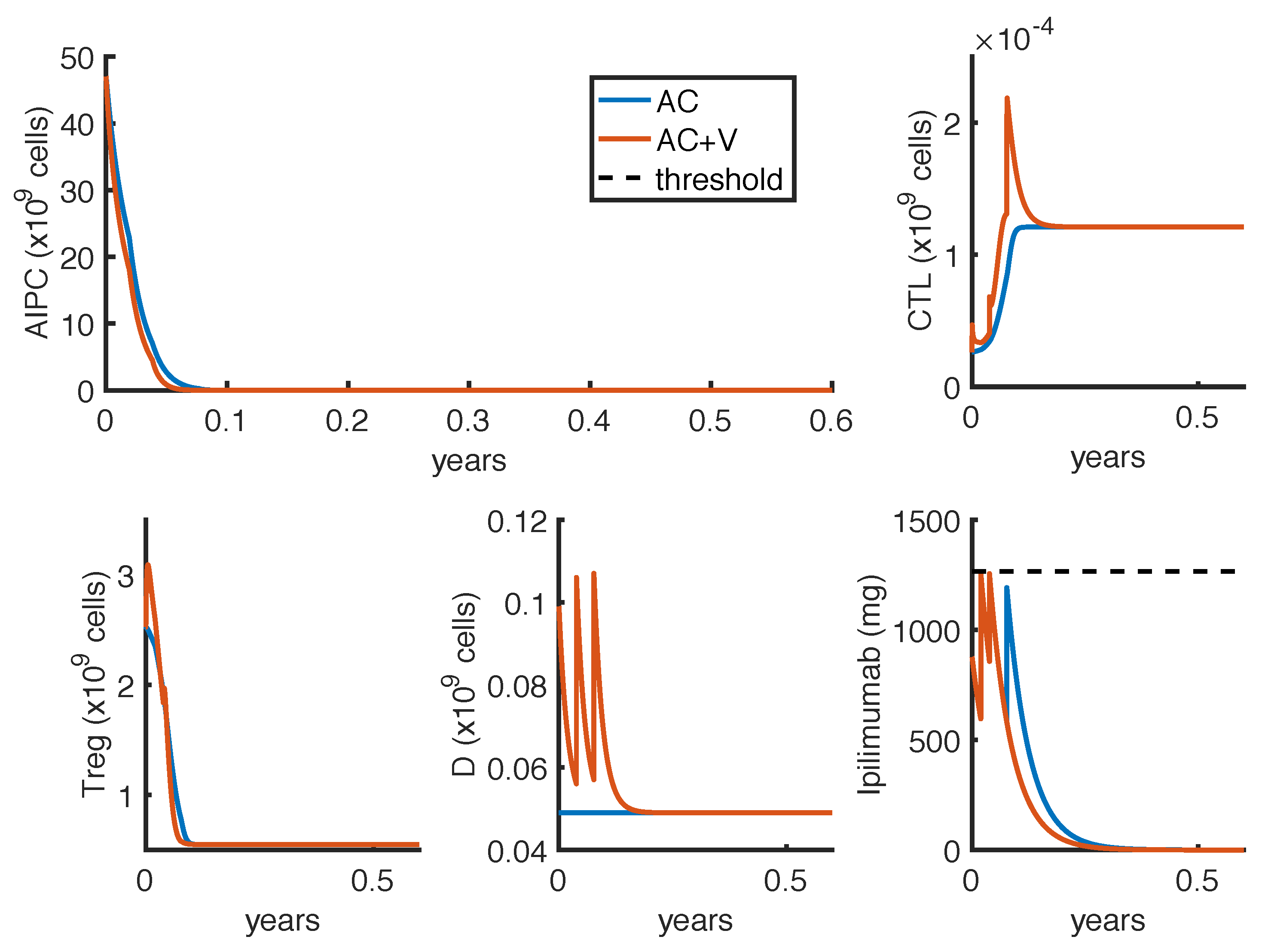
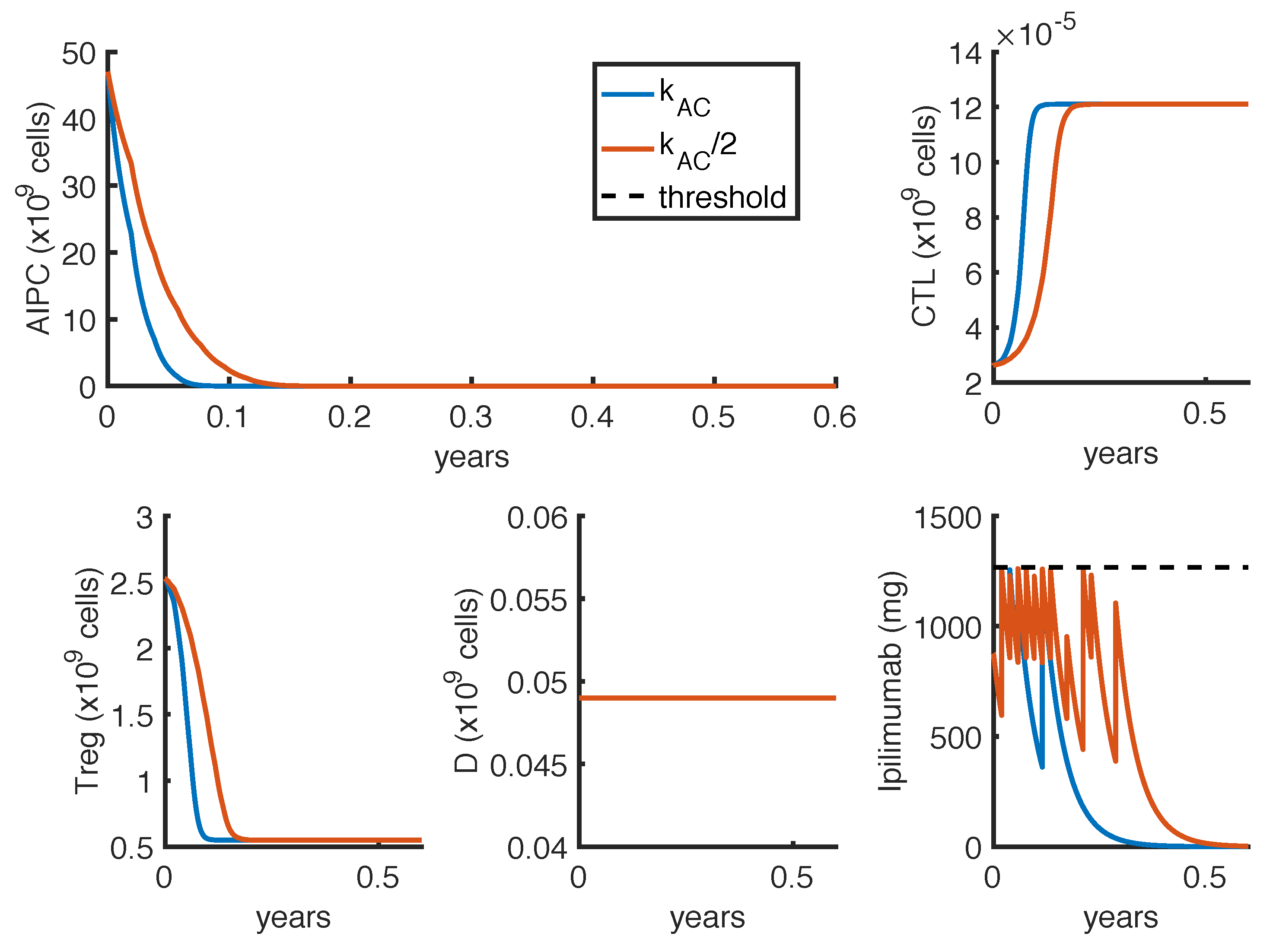
23] observed a complete and a partial response in 2 out of 16 patients, treated with intermittent protocols of high-doses. These different outcomes can be explained by patient heterogeneity, since the success of a therapy depends on patient-specific parameters. One of these is the parameter
, which measures how much the
ipilimumab can improve the tumor-killing activity of CTLs. As shown in
Figure 7, a perturbation of this parameter can qualitatively change the model outcomes, and the optimal protocol becomes unable to even control the tumor growth.
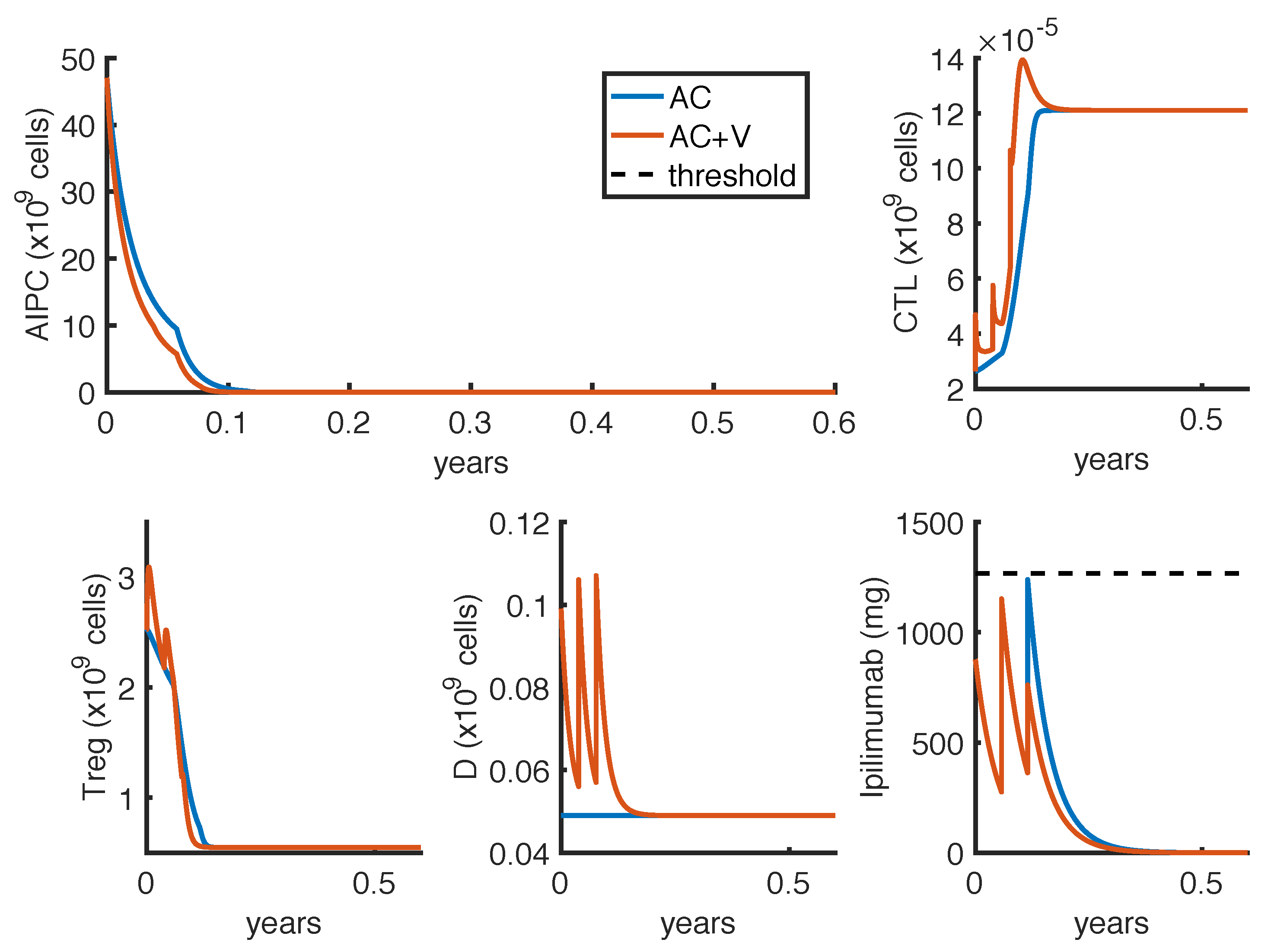
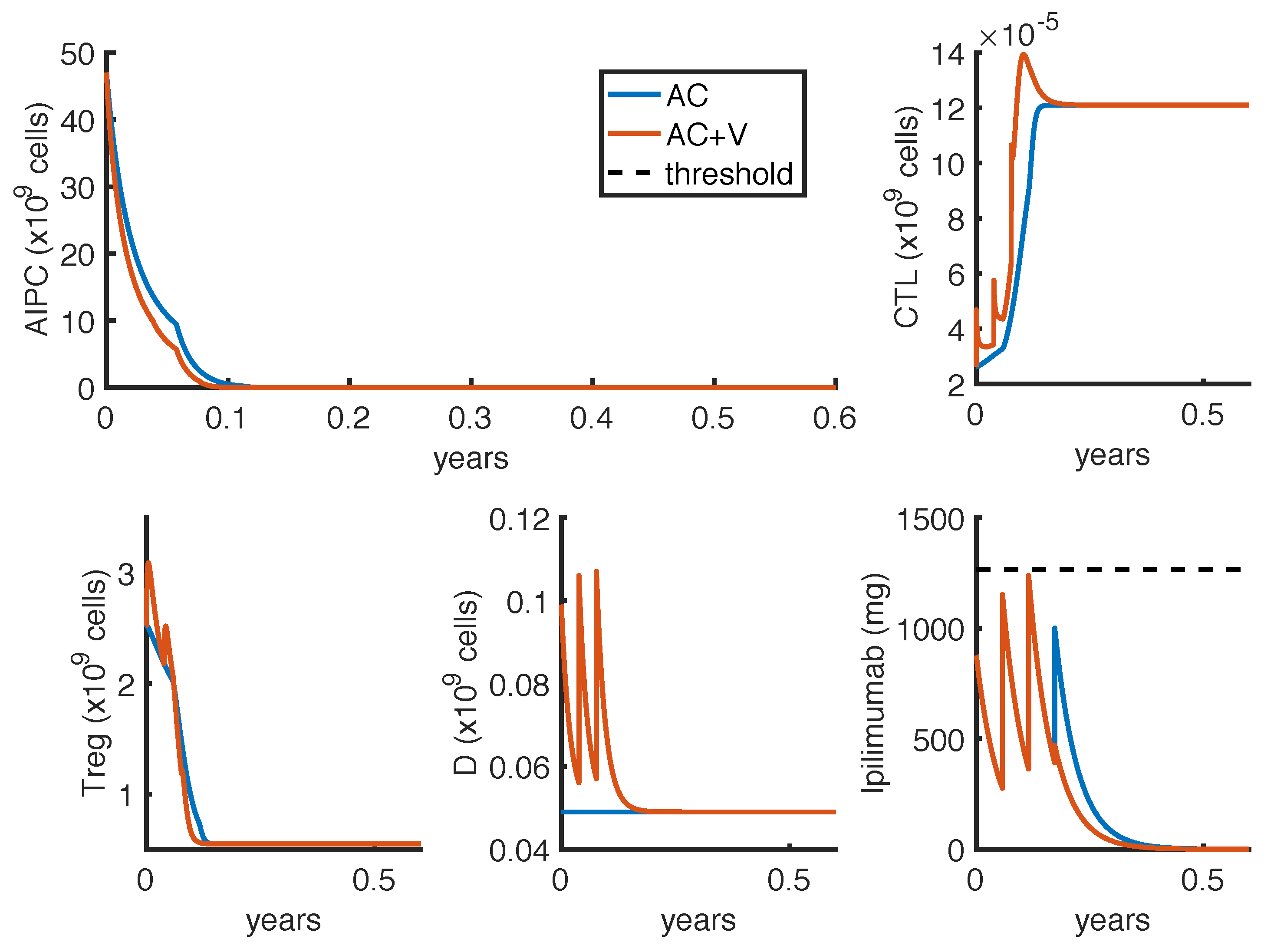
To further investigate if the tumor eradication could be possible even for those patients who respond less well to the standard therapies, we find the optimal protocol for lower values of
. By analyzing the optimal administration protocols we obtained under different model constraints on
, i.e., the minimum time interval between two infusions, we see that the model exhibits two different administration approaches: either one with frequent infusions at lower dosage, or another one consisting in two repeated standard protocols (one infusion every 3 weeks for 4 cycles) with maximal doses. Both the values of
(tumor density) and
(toxicity) corresponding to the choice
weeks are higher than the ones obtained by imposing
week (
Table 6). Similar results hold for the reference value of
(
Table 4 and
Table 5), although the difference is lower. However, we cannot conclude that the optimal protocol with
weeks is less suitable than the one with
week, since our results need to be validated from the clinical point of view and our analysis does not consider side effects and practical constraints. On the one hand, we do not have any information about close-range
ipilimumab infusions, and therefore the numerical results obtained with
week could not be reliable, since a patient could be affected by an increased toxicity depending on frequent infusions, which are not taken into account. Furthermore, we neglect the cost of the drug administration, which is, however, a crucial information to determine the feasibility of a therapy. For instance, comparing the mono-therapy protocols (rows 1 and 3 in
Table 6), the one associated to
week consists in administering 12 infusions of
ipilimumab, while the other one with
weeks consists in 8 infusions. As every infusion of
ipilimumab costs around
$30,000 [
32], the predicted optimal administration protocol with
week, in reality, could not be feasible.
Our results point out the synergy between
ipilimumab and
sipuleucel-T (see
Appendix B). By looking at the optimal protocols, the vaccine allows one to reduce the anti-CTLA4 dosage while reaching the goal of tumor eradication. The vaccine administered alone shows poor performances, while, coupled with the
ipilimumab, its effectiveness considerably improves. The limited effect of the vaccine has been observed in other clinical studies [
31], which highlight a few months increase in life expectancy, but not a PSA reduction. This could suggest to administer the dendritic cell vaccine in combination with other immunotherapies. Moreover, the results showed in
Table A1 and
Table A3 predict a stronger synergy for lower values of
. This seems indicate that a combination therapy could provide best performances for those patients who do not have a good response to
ipilimumab mono-therapy. The potential synergy between
ipilimumab and
sipuleucel-T has also been observed in clinical studies, such as [
33], which report an increase of the median survival in patients with metastatic-progressive prostate cancer. A phase 1 clinical trial is currently investigating the efficacy of this combination immunotherapy in patients with advanced prostate cancer [
34]. Thought a sensitivity analysis on a toxicity function parameter, we also investigate the reliability of the model prediction, by evaluating if the model outcomes can be highly influenced by the uncertainty in computing drug toxicity. Our analysis indicates that the computed optimal administration protocols seems not be dramatically affected by parameter perturbation, by highlighting a general robustness of the analysis herein reported.
In conclusion, the presented work is a theoretical analysis of the effect of
ipilimumab under optimal protocols of administration. Additional efforts are needed to make this approach suitable to clinical applications. The definition we used for the toxicity function could be more accurate if more detailed experimental data were available. For example, we assumed that the resolution times of AEs of
ipilimumab in PCa can be compared with the ones registered in melanoma. However, the median age of melanoma patients is usually lower than the one of PCa patients, and this could affect the resolution times. Moreover, the resolution time corresponding to the single infusion of
ipilimumab was supposed to be the same as for the intermittent administration protocol, due to the lack of this information in the study by Small et al. [
22]. When patient’s treatment time series and associated clinical data will be publicly available, it would be also interesting to generate an in silico population of patients to compare the outcomes of the optimal control problem with clinical trial results, in terms of individual patient responses. It is important to note that our results depend on the initial state used for the model simulations. In the perspective of proposing a personalized protocol, the initial state should be set in accordance with the patient condition. The model can also be calibrated to patient-specific parameters, such as the tumor proliferation rate. In this perspective, the model outcomes can be used not only to find the optimal protocol for a personalized therapy, but also to evaluate if the patient responds well to the treatment. Indeed, by comparing the model predicted tumor progression with the patient prostate cancer evolution, the model could indicate, in almost real time, whether the treatment is working as expected, and therefore could be helpful in supporting clinical decisions.