Simple Summary
Small cell lung cancer (SCLC) is an aggressive tumor, which can occur either de novo or from the histologic transformation of non-small cell lung cancer. Liquid biopsy has demonstrated its capability to detect, characterize and monitor different cancers. The aim of this systematic review was to assess the potential added value of liquid biopsy, in terms of circulating tumor DNA (ctDNA) and circulating tumor cells (CTCs), in the management of SCLC, either de novo or transformed. We found ctDNA analysis as the most valuable and feasible technology to be integrated into clinical for disease monitoring (response, relapse, transformation) or for genomic profiling of SCLC, with a potential use also for prognostic stratification. CTCs hold a strong prognostic significance, as confirmed by our meta-analysis (even if potentially biased), but the heterogeneity of available data, the lack of agreed cut-offs, and the less affordable technology make CTCs more difficult to be integrated into present clinical practice.
Abstract
Background: The potential added value of liquid biopsy (LB) is not well determined in the case of small cell lung cancer (SCLC), an aggressive tumor that can occur either de novo or from the histologic transformation of non-small cell lung cancer (NSCLC). Methods: A systematic review of studies adopting LB in patients with SCLC have been performed to assess the clinical utility of circulating tumor DNA (ctDNA) or circulating tumor cells (CTCs). Results: After a screening of 728 records, 62 studies (32 evaluating CTCs, 27 ctDNA, and 3 both) met predetermined eligibility criteria. Only four studies evaluated LB in the diagnostic setting for SCLC, while its prognostic significance was evaluated in 38 studies and prominently supported by both ctDNA and CTCs. A meta-analysis of 11 studies as for CTCs enumeration showed an HR for overall survival of 2.63 (1.71–4.05), with a potential publication bias. The feasibility of tumor genomic profiling and the predictive role of LB in terms of response/resistance to chemotherapy was assessed in 11 and 24 studies, respectively, with greater consistency for those regarding ctDNA. Intriguingly, several case reports suggest that LB can indirectly capture the transition to SCLC in NSCLC treated with EGFR tyrosine kinase inhibitors. Conclusions: While dedicated trials are needed, LB holds potential clinical roles in both de novo and transformed SCLC. CtDNA analysis appears the most valuable and practicable tool for both disease monitoring and genomic profiling.
1. Background
Small cell lung cancer (SCLC) is an aggressive lung cancer with neuroendocrine features, characterized by a strong association with tobacco smoke exposure, high cell growth fraction, and early and extensive metastatic propensity. Despite the initial high rate of responsivity to cytotoxic chemotherapy and radiation therapy, the rapid development of resistance and the high rate of relapse justify the poor outcome overall [1]. Only 6% of 290,000 patients worldwide diagnosed annually with SCLC will survive 5 years from diagnosis, mostly those with the limited stage of disease (LS-SCLC) [2]. In addition, a subset of patients affected by oncogene-driven non-small-cell lung cancer (NSCLC) experiences a histologic transformation in SCLC, commonly under the therapeutic pressure of tyrosine kinase inhibitors (TKIs). Small-cell transformation represents an acquired mechanism of resistance to TKIs reported in 5–15% of EGFR mutated NSCLC. In these cases, transformed cells usually maintain the original activating EGFR mutation and present other genetic alterations resembling classical SCLC (e.g., mutations or loss of RB1 and TP53, mutations of PIK3CA) [3,4]. While liquid biopsy (LB) is extensively used in patients with NSCLC to detect on-target resistance mechanisms, tissue re-biopsy appears crucial for identifying histologic transformations.
Given that nearly all SCLC patients are not treated with surgical resections, the shortage of banked tumor tissue has been an obstacle for comprehensive genomic characterization, hindering the understanding of the biology of SCLC and possibly slowing down preclinical drug development [5]. LB represents nowadays a minimally invasive tool for obtaining tumor-derived components such as circulating tumor DNA (ctDNA) or circulating tumor cells (CTCs) with different applications across many cancer types [6,7,8,9]. SCLC ctDNA can be identified and profiled by detecting either a single gene of interest (e.g., TP53) or a panel of SCLC-associated genes with technologies widely available in clinical laboratories [10,11]. The amount of ctDNA in a sample of circulating free DNA (cfDNA) can vary enormously between patients with lung cancer, reflecting the histologic type, the tumor burden, and the sites of disease [12,13]. CTCs are very rare in the bloodstream, but they can be isolated using marker-dependent approaches (e.g., by antibodies against epithelial proteins) or exploiting biophysical differences of CTCs from other blood cells [5]. The most adopted and the only FDA-approved system is the CellSearch platform, in which the epithelial cell adhesion molecule (EpCAM/CD326) is used for CTC detection [14]. CTCs derived from patients with SCLC maintain their tumorigenic properties in immunocompromised mice, also forming patient CTC-derived explants (CDX) [15], and several studies have demonstrated their enumeration as an independent prognostic biomarker for survival [16,17,18], albeit, a consensus on the optimal cut-off threshold is lacking. Other potential roles of ctDNA and CTCs are less clear, even though a number of studies have reported fragmented data on their diagnostic, genomic profiling, predictive, or prognostic value.
We performed a systematic review of the studies available in the literature evaluating LB as a clinical implement in the management of SCLC, including transformed SCLC from NSCLC.
2. Methods
2.1. Definition of the Outcome
The purpose is to evaluate the current potential added value of LB, in terms of circulating tumor DNA (ctDNA) and circulating tumor cells (CTCs), in patients with de novo SCLC or transformed SCLC.
2.2. Data Source and Search Strategy
A systematic review of the literature was performed according to PRISMA statement criteria [19,20] on 6 February 2021. The Medline and EMBASE databases were searched for relevant records between 2000 and 2021 that met the study inclusion/exclusion criteria. Unpublished data presented as abstracts in relevant international congresses (American Society of Clinical Oncology (ASCO), European Society of Medical Oncology (ESMO), and International Association for the Study of Lung Cancer (IASLC)) were included. Hand searches were performed to identify further potentially eligible studies, as well. The decision to include a study for review was made by consensus between two authors (EGP and MP). The research criteria were limited to human studies published only in the English language. The search terms were (“small cell” OR “small-cell” OR “SCLC” NOT “non-small”) OR (“transformed small cell” OR “transformed small-cell” OR “transformed SCLC” OR “histologic transformation”) AND (“ctDNA” OR “cfDNA” OR “circulating free DNA” OR “circulating tumor DNA” OR “CTC” OR “liquid biopsy”).
Main study inclusion criteria:
- Analysis of cfDNA/ctDNA or CTCs in plasma/serum of patients with SCLC included histologically transformed SCLC from NSCLC;
- Genomic profiling, diagnosis, treatment response, and/or survival data collected and correlated with cf/ctDNA or CTCs in humans.
Study exclusion criteria:
- Not specific for SCLC, except in the cases of small cell transformation;
- Analysis of tumoral circulating components different from ctDNA or CTCs.
For the quantitative analysis [21], we included in a meta-analysis of prognostic value those studies with comparable techniques used for ctDNA detection or CTC enrichment. The quality of studies was evaluated through the Nottingham–Ottawa Scale (NOS). The total scores ranged from 0 (worst) to 9 (best), with a score of at least seven indicative of high quality. Included studies reported:
- -
- Hazard ratios (HR) with 95% confidence intervals (CI);
- -
- Sample size;
- -
- Cut-off of CTC number.
2.3. Statistical Analysis
Methods described by Tierney et al. [22] were adopted for the prognostic meta-analysis and for the collection of time-to-event data. All statistical analyses were performed using R software (R version 4.0.5) (R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/ accessed on 5 March 2021) with metafor package (Viechtbauer, W+010 https://www.jstatsoft.org/v36/i03/ accessed on 5 March 2021), dmetar package (Harrer, M., Cuijpers, P. et al. 2019 http://dmetar.protectlab.org accessed on 5 March 2021), and meta package (Schwarzer, G. 2020 https://CRAN.R-project.org/package=meta accessed on 5 March 2021). Tau-squared and I2 index were used to evaluate heterogeneity. Funnel plots with Egger’s regression tests were used to examine publication bias.
3. Results
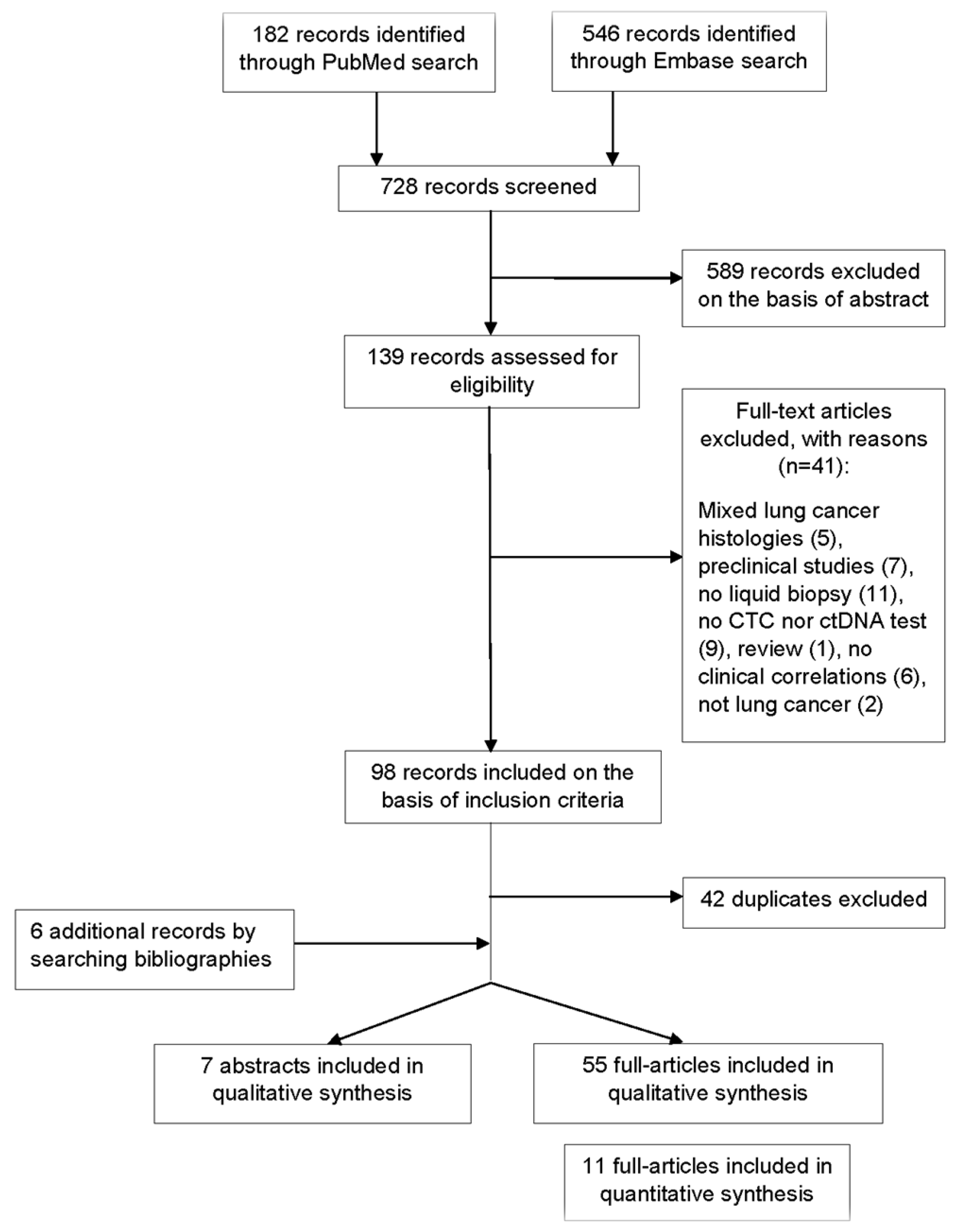
A total of 728 records were screened to be included in the systematic review (Figure 1). According to selection criteria, we identified 56 records found through database searching (PUBMED and EMBASE) and 6 records by searching bibliographies. As a result, 62 records were eligible and included in the systematic review: 55 full-text articles studies and 7 abstracts presented at international congresses. Fifteen studies concerned transformed SCLC. Overall, identified records included mainly clinical trials with exploratory endpoints assessing the value of LB, observational studies, and case series or case reports for transformed SCLC. A total of 32 studies evaluated CTCs, 27 studies evaluated the role of circulating DNA, and 3 studies explored both (Tables S1 and S4).
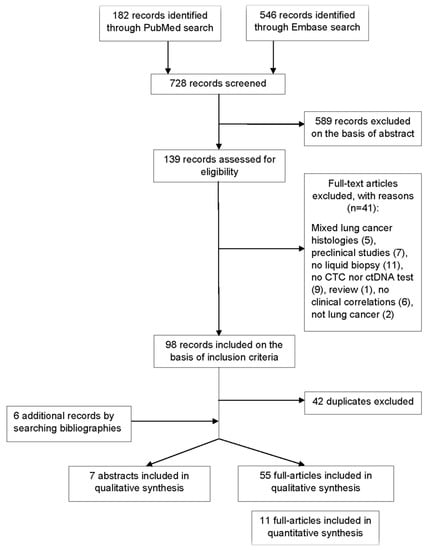
Figure 1.
Flow diagram representing the systematic review process performed according to PRISMA statement.
The detection rate of ctDNA in patients harboring de novo SCLC varied between 49% and 100%, with a median of 91%. Among six studies in which next-generation sequencing (NGS) technology was adopted with panels of 5–430 genes, the detection rate was 71–100%, with a median of 91% (Table S2).
The detection rate of CTCs at baseline in 20 studies varied between 49% and 96%, with a median of 85%, with different assays. The median number of CTCs and the median detection rate with different assays, among studies with available data, in patients with extensive-stage SCLC (ES-SCLC) versus limited-stage SCLC (LS-SCLC), were 38 vs. 2, and 95% vs. 65%, respectively (Table S3).
We subdivided our results into five categories according to the investigated role of ctDNA or CTCs: diagnostic (studies including negative controls), genomic profiling, predictive of treatment response, and prognostic (in terms of disease recurrence or survival) for SCLC; the last category was related to the application of LB in cases of small-cell transformation of NSCLC.
3.1. Diagnostic
Four studies (Table S1) compared the detectability of ctDNA or CTCs from patients with SCLC and from healthy volunteers. With overall 172 SCLC patients and 176 non-cancer controls, these works described in the plasma of patients the presence of a higher DNA concentration (with longer fragments), more frequent TP53 mutations (36% of patients with early-stage SCLCand 11% of non-cancer controls), detectable tumor-related mutations and CNAs by NGS in 84–94% of cases versus 0 in non-cancer controls, and detectable CTCs in 86% of cases versus 0% in healthy donors [10,11,23,24].
3.2. Genomic Profiling
Overall, 11 studies reported data of genomic profiling of SCLC by LB (Table S1). Different authors presented results of ctDNA analysis by NGS [10,12,25,26,27,28,29]. A higher concordance (median of 94% of paired mutations) between ctDNA and tissue was reported to be reachable by means of a deep sequencing [28,30], also showing a correlation in allelic frequency (AF) of gene mutations between plasma and tissue [28]. Some mutations were exclusively detected in ctDNA, and, comparing samples collected at baseline and after treatment, new mutations appeared or became dominant in the post-treatment samples [28,30]. Mohan and colleagues identified potential therapeutic targets in >50% of patients [10]. Devarakonda et al. provided the widest study of genomic profiling of SCLC using Guardant360 platform on ctDNA from 564 patients and highlighted the identification of potentially targetable alterations involving the androgen receptor gene (AR), the RTK/RAS pathway, or genes involved in DNA repair [26]. The most frequent genetic alterations detected by means of ctDNA analysis in seven studies through different assays [10,12,25,26,27,28,29] are reported in Table 1 and Table 2.

Table 1.
Most frequent (median throughout all studies ≥5%) mutated genes detected by ctDNA in 6 studies through different assays. Frequencies of genetic alterations researched in at least 2 studies are reported. N.: number of patients; Ref.: reference; -: not evaluated in gene panel.

Table 2.
Most frequent (median throughout all studies ≥5%) genes with copy number variation (CNV, namely amplification, and deletion) detected by ctDNA in 5 studies through different assays. Frequencies of genetic alterations researched in at least 2 studies are reported. The assay by Owonikoko could detect amplification events of MYC and AURKA only, but no results were reported by authors [25], then it is not included in the table. N.: number of patients; Ref.: reference; -: not evaluated in gene panel.
An association between CTC count and quantity of circulating free DNA (cfDNA) has been described in a cohort of 12 patients; moreover, identical genetic alterations were identifiable from both CTC-derived DNA and cfDNA [31]. Su et al. demonstrated the feasibility of genomic profiling by means of single-cell sequencing of CTCs in SCLC patients, with 68–99% of mutations observed in tissues detectable in CTCs. The authors observed that CTCs mainly disseminated from the primary tumor with which they shared the majority of the mutations, while metastatic sites formed minor clones unobserved in CTCs [32].
3.3. Predictive
We found 24 studies reporting data about the predictive potential of LB (Table S1). Several investigators reported a correlation between qualitative and quantitative changes in ctDNA and variation in tumor burden under treatments [10,12,28,29,33,34]. Hence, a rise in detection rate and also in AFs or copy number of DNA alterations may precede clinical evidence of disease progression; on the other hand, a reduction can be observed in cases of response to chemotherapies [10,12,28]. Herbreteau et al. reported a significantly lower activity of atezolizumab in those patients with detectable ctDNA in II line setting, while no differences existed with chemotherapy [29]. Thomas et al. described a case of BRCA1-mutated SCLC treated within a phase II trial with olaparib and durvalumab. The patient achieved a complete response with a decline in the frequency of BRCA1 mutation in ctDNA [34]. One group found distinct qualitative features by ctDNA analysis in patients with chemosensitive or chemorefractory SCLC. Alterations of TP53, ATM, and FLCN were higher in the chemorefractory group and related with shorter PFS, while APC abnormalities occurred more frequently among chemosensitive patients [35]. Owonikoko et al., in a negative randomized phase II trial with paclitaxel in addiction either to alisertib (inhibitor of Aurora A kinase, a regulator of mitosis) or placebo in relapsed SCLC, retrospectively reported better survival in patients with alterations of the genes involved in cell cycle regulation among patients treated with alisertib [25].
A lack of significance in the predictive role of the CTC number assessed by CellSearch at baseline was described in four out of five studies [17,36,37,38,39]. Most of the studies exploring the significance of CTC count modification using CellSearch, reported a reduction in enumeration after chemotherapy [17,18,40,41], but no statistically significant differences among groups showing different responses to treatment [17,18,37,38,39,42]. A correlation between tumor burden in response to therapy and CTC number by CellSearch was observed only in II-III line setting among patients treated, respectively, with temozolomide +/− veliparib and with pazopanib [43,44,45]. Conversely, adopting assays different from CellSearch, a significant correlation with disease response and changes in CTC count can be found. Incremental CTC count has been reported using a method based on the specific telomerase activation of cancer cells (OBP-401) [46], while folate receptor-positive CTCs [47], CK-19 mRNA-positive CTC [48], or DLL3+/CD45− CTCs [49], decrease during response. Finally, different profiles of copy number aberrations (CNAs) in CTCs and in ctDNA have been associated with different responsiveness to first-line chemotherapy by three groups [32,35,50].
3.4. Prognostic
This was the most explored purpose of LB with overall 38 clinical reports available (9 by ctDNA, 27 by CTC, 2 by both) (Table S1). Two studies, with overall 49 patients, reported a significant association between a higher abundance of cfDNA/ctDNA at baseline and shorter OS and PFS [12,28]. Palma presented data of survival benefit among patients with a continuous drop in ctDNA levels during chemotherapy [51]. Contrasting prognostic connotations have been reported for total mutation burden, mutations of specific genes, or AFs of certain mutations, while the CNA signature measured by ctDNA sequencing could predict survival [10,25,28,33,52,53]. Alterations in SETBP1, PBRM1, ATRX, EP300, ATM, PIK3CA/G, or NOTCH1 have been associated with OS or PFS in patients treated with platinum-based chemotherapy or with chemoradiation by two small studies [27,52]. A French group observed a worse OS among patients with relapsed SCLC and detectable mutations through an NGS panel limited to NOTCH1-3, TP53, and RB1 genes [29]. After the first description of prognostic significance hold by CTCs detected by flow cytometry [54], several groups reported a significant correlation between survival and the number of CTCs found by CellSearch, regardless of the statistical method applied. Baseline pretreatment CTC number consistently correlated with OS in 15 studies (including ES and/or LS-SCLC) with different cut-offs (from 2 CTCs up to 282 CTCs) [10,16,17,36,37,38,40,42,55,56,57,58,59,60,61]. A higher number of CTCs after the first cycle of chemotherapy was also prognostic for worse outcomes in six studies [16,18,42,57,58,60]. Overall, six studies reported a prognostic significance of CTCs enumeration post-second cycle, post-treatment, and at the time of relapse [17,18,36,37,59,61]. Some authors observed a prognostic significance not only in the absolute number of CTCs but also in its reduction during chemotherapy [16,17,60]. Gadgeel et al., evaluating pembrolizumab as maintenance treatment after platinum and etoposide, collected CTCs before the first three cycles of pembrolizumab. No correlation with PFS and OS was found with any CTC count before or during immunotherapy [62]. Two groups described a strong prognostic value of CTCs in LS-SCLC treated with CRT [55,56]. In the second-line setting, Aggarwal presented data showing a lack of association between CTC count and survival in a small cohort of patients with relapsed/refractory SCLC [31]. Two groups, evaluating the activity of second-line pazopanib and second-line temozolomide in combination with veliparib or with placebo, respectively, reported a correlation between a number of CTCs ≥5 both before treatment and after the first cycle with a significantly shorter OS [43,45]. Different groups explored the prognostic value of distinct phenotypes of CTCs. Detectable BCL2+ or DLL3+ or chromosome 8 centromere probe (CEP8)+ and CD45- CTCs correlated with worse outcome at baseline and/or after first cycle of first-line chemotherapy [49,57,63]. No correlation was found with TTF-1+, or CD56+, or pancytokeratin+, and CD45- CTCs [23,36]. Igawa and colleagues observed a prolonged OS in patients with <2 CTCs at baseline, using the aforementioned OBP-401 assay [46]. Shen et al., by means of an LT-PCR to detect folate receptor-positive CTCs, found a trend for longer OS (p = 0.056) in patients with low CTC level at baseline, while no differences were evident between patients with positive or negative CTC count [47]. Shi et al. described a strong prognostic value of CK19 mRNA-positive CTCs (HR for OS 3.31 when detectable after treatment) [48]. One group also reported a correlation between the presence of CK+/Vim+ CTCs after one cycle of second-line pazopanib and shorter survival [44]. In addition, copy number aberrations (CNA) signatures measured in CTCs from pretreatment blood samples could predict PFS and OS [32,50].
Meta-Analysis
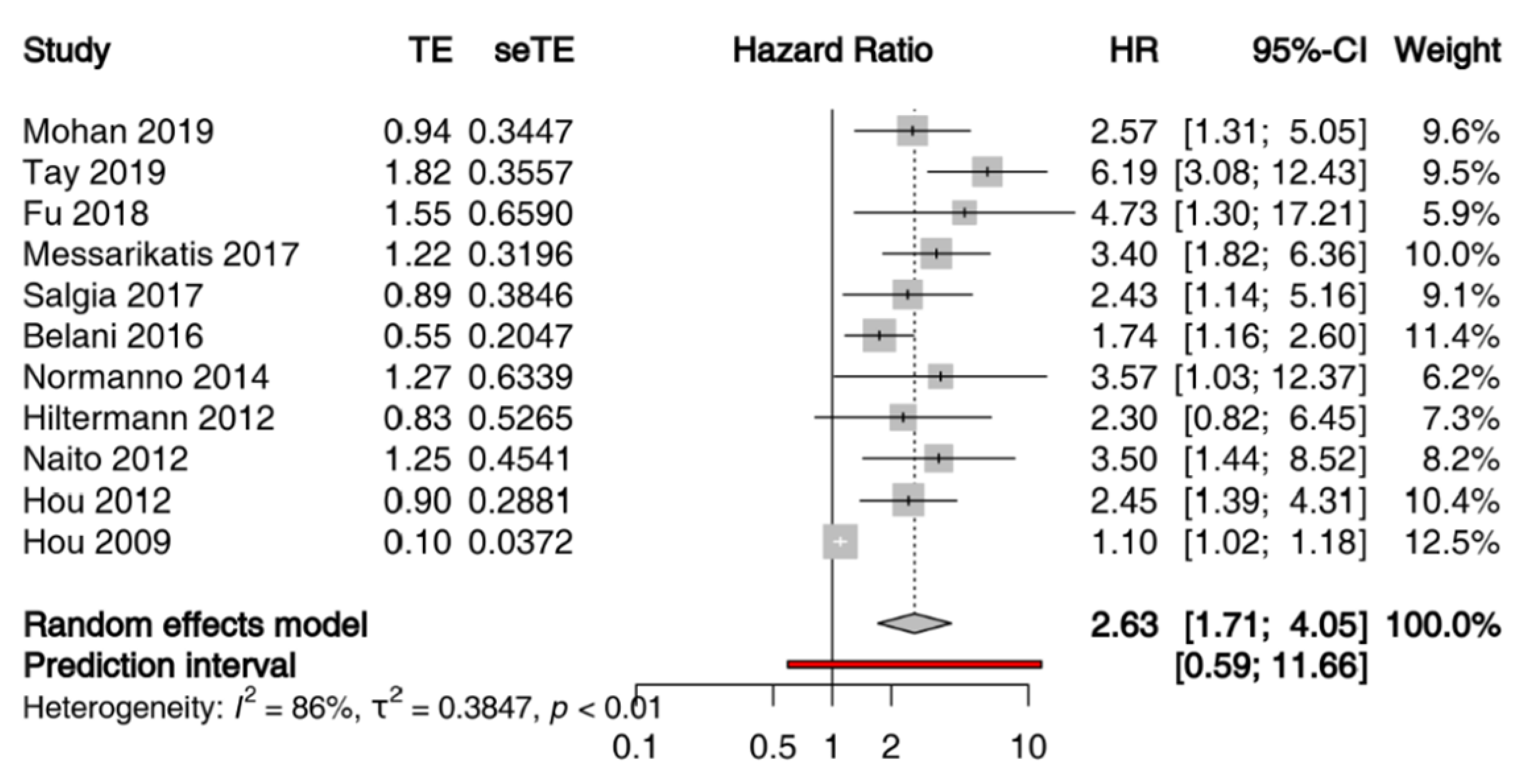
Given the heterogeneity of techniques adopted for quantitatively and qualitatively evaluation of ctDNA and of time points of sample collection, a meta-analysis of the prognostic value of ctDNA in patients with SCLC was not feasible. Conversely, 11 studies comprising 861 patients met the eligibility criteria for a meta-analysis of the prognostic value of CTCs assessed by CellSearch (Table S4). We performed a meta-analysis of HR for OS of the number of CTCs at baseline, with different cut-offs across studies. When multiple cut-offs were explored in the same study, we chose those with the better operating characteristics throughout ROC analysis for the meta-analysis. All studies included patients with LS-SCLC, or ES-SCLC treated with chemoradiation or first-line platinum etoposide (+/− experimental drug if it did not significantly affected survival) [38,42]. The sample size per study ranged from 51 to 120 patients, and studies were published between 2009 and 2019. Because the heterogeneity across the studies was high (I2 = 86%, p < 0.01), the estimated pooled HR was calculated using a random-effect model. The pooled HR showed that the presence of CTCs correlated with reduced OS (HR = 2.63; 95% CI: 1.71–4.05) (Figure 2). Moreover, the visual inspection of the funnel plot and the Egger test demonstrated the presence of a potential publication bias (Figure S1).
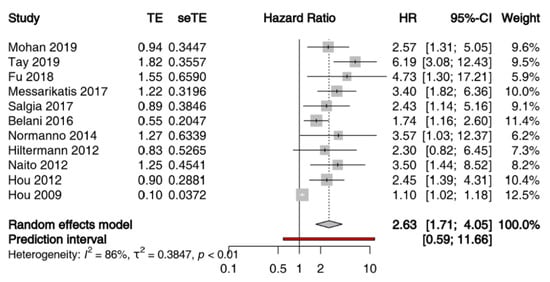
Figure 2.
Prognostic significance of CTCs assessed by CellSearch in patients with SCLC at baseline: forest plot of hazard ratios for overall survival.
3.5. Small-Cell Transformation of NSCLC
Our group and others described cases of patients with progressive NSCLC under TKI treatment with evidence of sharp elevation of AF of EGFR activating mutation in ctDNA measured with ddPCR, concurrent with evidence of histologic transformation to SCLC [64,65,66]. Performing NGS on ctDNA, other researchers also reported a significant elevation in EGFR mutation AF concomitant with the increase in AF of TP53, RB1, or PIK3CA alterations before or at the moment of the histologic transformation, after TKI failure [67,68,69,70,71]. A subsequent reduction in the allelic abundance of these mutations after an SCLC-directed treatment has been reported, as well [69,70]. Two groups presented data supporting the utility of ctDNA in detecting the EGFR resistant mutation T790M even after SCLC transformation, uncovering the spatial heterogeneity of the tumor [72,73], while one group described the disappearance of acquired G1202R ALK mutation, concurrent with SCLC transformation [74]. We also found two studies demonstrating that CTC phenotyping and single-cell CTC sequencing could suggest a histologic transformation. Ni et al., through a single-cell exome sequencing, identified EGFR, PIK3CA, RB1, and TP53 mutations in CTCs of a patient with transformed SCLC, with higher abundance than in the original NSCLC [75]. More recently, a Chinese group applied a new assay allowing rapid CTC isolation with different aptamers and characterization by immunocytochemistry. They firstly reported a significant reduction in mean nuclear size and a phenotype shift of CTCs concurrent with SCLC transformation in 14 patients [76] (Table 3).

Table 3.
ctDNA and CTCs changes in cases of SCLC transformation of EGFR mutated or ALK-rearranged NSCLC. ABS: abstract; NA: data not available. PD: progressive disease. tSCLC: transformed SCLC.
4. Discussion
To our knowledge, this is the first systematic review concerning the applications of LB (considering both ctDNA and CTCs) focused on SCLC, also innovatively including patients affected by transformed SCLC. The limitations of data retrieved are mostly related to the variety of techniques adopted for LB, diversity of patient populations (ES or LS-SCLC, different lines of treatment) or limited sample size, the timing of plasma samples (e.g., at baseline, during, or after treatments) and heterogeneous approaches to statistical analyses. With such considerations, we found interesting potential applications for each purpose of the five explored.
4.1. Diagnosis
Among the potential applications of LB, cancer diagnosis and screening represent the greatest challenge in oncology. Even though SCLC presents relatively higher concentrations of tumor-derived components in plasma, the presence of mutations in cfDNA potentially associated with non-malignant processes and the elevated growth rate with an early metastatic propensity of this cancer type add obstacles to the feasibility of a screening program. Limited data support this application for LB in SCLC to date. Mohan and colleagues recently demonstrated high sensitivity and specificity with multi-gene NGS of ctDNA, in contrast with previous data on single gene sequencing [11], describing detectable tumor-related CNAs in 84% and non-synonymous mutations in 94% of patients (77% and 91% in LS-SCLC), versus 0% in non-cancer controls [10]. No other studies have focused on the diagnostic role of ctDNA in SCLC. Moreover, two studies have recently demonstrated that multicancer blood tests (with multi-gene or methylation analysis, respectively) could detect a broad range of cancer types, SCLC included [78,79]. CTC enumeration could be a more accurate, even if more expensive, strategy for early SCLC diagnosis. About a half of patients with LS-SCLC present detectable CTCs with an assay based on immunomagnetic enrichment and immunocytochemistry, with no CTC detectable with the same method in healthy donors [23]. By means of CellSearch assay, 3% of healthy cases [80] and 60–85% of patients with LS-SCLC may present detectable CTCs (Table S3). Preliminary data brace a potential application of LB also in the differential diagnosis between SCLC and NSCLC or other carcinomas. More abundant CNV changes and mutations, also with higher AFs, have been detected in ctDNA of patients with SCLC than with other tumor types, even with NSCLC [12,27]. This could reflect the higher proliferation rate with a tendency to early hematogenous spread of SCLC. In addition, the enumeration and phenotyping of CTCs [76,81] could suggest a diagnosis of SCLC rather than other cancer types.
4.2. Genomic Profiling
Different authors demonstrated that genomic characterization of SCLC is feasible by LB, with high detection rates for ctDNA (>90%) and CTCs (median of 85%). Analysis of ctDNA could provide both qualitative and quantitative data on tumor-related genomic alterations. A good concordance with tissue-based profiling has been reported with the sequencing of ctDNA by different groups [28,30,82]. As expected, TP53 and RB1 have been the most frequently altered genes detected on ctDNA [12,25,26]. Recurrent amplification of SOX2, MYC family genes, or FGFR1 has also been reported by LB (Table 2), consistently with previous tissue-based analysis [83,84]. The differences in gene alteration rates reported by different authors (Table 2 and Table 3) may be related to limited sample sizes, to the variety of assays, gene panels, and depth of sequencing, to differences in tumor burden, ongoing therapies, or timing of sample collection (for example, alterations in APC and AR genes have been reported more frequently in relapse samples [52,54]), and to the possibility of hematopoietic variants (clonal hematopoiesis of indeterminate potential—CHIP) being misclassified as tumor derived [28,31,85]. Moreover, loss of heterozygosity events is difficult to detect at low AFs in ctDNA with common assays, and this could justify the lower frequencies of TP53 or RB1 deletions compared to tissue sequencing in previous studies [83]. In some cases, ctDNA analysis could reveal genomic alterations not detected in tissue, mirroring the spatial heterogeneity of the tumor [28]. One group performed single-cell sequencing of CTCs from SCLC patients, demonstrating that also CTCs constitute a reliable source for genomic profiling [75]. Moreover, this sort of information is not yet directly useful in the clinic today. SCLC has historically been treated as a homogenous cancer with no targetable driver genetic alterations. Recent data are diametrically changing this paradigm toward a critically heterogeneous disease, with the identification of different transcriptional subtypes [86] and also a variety of subpopulations of cells emerging with treatment resistance [87,88]. Such heterogeneity limits the reliability of tissue biopsies, enhancing the potential role of both ctDNA and CTCs, which could help a rational development of new therapeutic options [89,90].
4.3. Predictive Value
We found several studies reporting a correlation between tumor burden modifications in response to chemotherapy and quantitative changes in ctDNA, both in terms of ctDNA abundance and in terms of AF of specific gene mutations [10,12,28,33,34,82]. Moreover, specific qualitative molecular features have been associated with chemosensitivity (APC alterations) and chemoresistance (TP53, ATM, and FLCN alterations), respectively, but larger-scale studies are required to validate these findings [35]. A negative predictive value of detectable ctDNA at baseline has been reported for patients treated with atezolizumab. The reason is not clear, but higher levels of ctDNA may reflect a higher tumor burden, which has been already linked with the lower efficacy of immune-checkpoint inhibitors in lung cancer [91]. Even if these studies are based on small cohorts of patients, all together, these data support ctDNA monitoring in patients with SCLC as a potential tool for better management of the disease. Moreover, the best frequency of blood collection is not yet determined, and prospective studies are necessary to fully assess the reliability of ctDNA for clinical decisions. Moreover, predictive biomarkers for targeted therapies are still lacking in SCLC, albeit initial data are emerging from clinical trials exploring targeted agents, and ctDNA analysis could be helpful in such cases [25].
The association between CTC enumeration and treatment response is less clear. Different groups reported data showing a reduction in CTCs after treatments in SCLC patients [36,40,41,54]. Such reduction was also documented in other cancer types [92]. Moreover, no statistically significant correlation between the type of response and CTC count (both at baseline and after chemotherapy, assessed by CellSearch) was observed in studies by Naito, Hiltermann, Wang, Salgia, Aggarwal, and Belani [17,18,37,38,39,42]. Three different groups, on the other hand, observed significantly fewer CTCs among patients achieving a disease control compared to patients with PD as the best response. Moreover, these studies were conducted, respectively, in a small phase I cohort of patients, in a second-line setting with an antiangiogenetic agent, and in a mixed group of LS and ES-SCLC patients [36,45,58]. Different assays may detect different subpopulations of CTCs, which can show diverse behavior in the outflow from the tumor, not always mirroring the changes in the tumor size. For example, tumor cells down-modulate the epithelial markers undergoing epithelial mesenchymal transition (EMT); thus, CellSearch fails to capture these EpCAM-negative CTCs, while the OBP-401 assay for CTC identification, which is based on the telomerase expression, -could be more suitable for the detection of tumor cells involved in EMT process [46]. A Greek group widely studied CTCs with diverse phenotypes, and they observed a close relationship between changes in tumor burden and count of DLL3+, CD45− CTCs [49]. DLL3 has been known to be highly expressed in SCLC. More recently, Gay et al. described different transcriptional subtypes of SCLC, and DLL3 appeared to be more expressed in platinum-sensitive subtypes [86]. While the technology for CTC detection is not accessible in the majority of the laboratories, the novel identification of transcriptional subtypes and the development of chimeric antigen receptor (CAR) T cells or of antibody-drug conjugates may add new value to CTCs (i.e., for transcriptome analysis and for characterization of the proteins expressed on the surface of cancerous cells, the surfaceome).
4.4. Prognostic Value
The available data show an association between a higher quantity of cfDNA/ctDNA and shorter survival [12,28], with better outcomes in cases of reduction in ctDNA levels over therapy [51]. The prognostic significance of tumor mutational load, which appears higher in cases of mutations of cell cycle regulation genes [25], is not clear since conflicting data have been reported in small studies [25,28]. From a qualitative point of view, mutations of different genes have been associated with shorter OS by a single small study [27]. No other study reported such association, neither previous data derived from tissue analysis [93]. Mohan et al. reported a prognostic significance of mutations of any gene, included TP53, only when they are present in ctDNA with high AFs [10], suggesting a relationship with enhanced shedding of ctDNA rather than with specific genetic mutations. Anyway, the great heterogeneity in gene panels, depth of sequencing, and the quantity of blood collected and evaluated are major variables that prevent a reliable comparison among studies.
Several studies independently demonstrated a prognostic significance of CTC count in patients with SCLC. Moreover, the huge heterogeneity of these analyses did not allow us to reach a consensus on the optimal cut-off threshold of CTC number and on the time of plasma collection. One reason that could also hinder the achievement of a strict cut-off is the huge variability in the number of CTCs existing at baseline among ES-SCLC (median number: 9.5–91 per 7.5 mL), while the range appears less wide among LS-SCLC (median number: 1–21 per 7.5 mL) (Table S3). Baseline CTC count seems to be associated with the number of organs involved by disease [37,40,46,60,63], and in particular with liver metastases, but not with brain or bone metastasis [16,17,40,58]. Another hurdle could derive from the subpopulation of CTCs detected by a specific assay, as mentioned above. So the question may be if CTC numbers should be integrated by CTC phenotype as a criterion for prognostic definition and therapy decisions. Other prognostic information could derive from the enumeration of CTCs at different time points or from their longitudinal quantification, as demonstrated by different authors [16,17,18,36,37,59,60]. In our meta-analysis, a pooled HR for death of 2.63 was found considering the CTC population detected by CellSearch at baseline among SCLC patients at any stage. Anyway, a relevant risk of publication bias is suggested by the funnel plot (Figure S1). Moreover, the choice of ROC analysis for the identification of the optimal cut-off adopted in these studies creates a methodologic bias toward a higher HR. The highest HRs were reported by two studies assessing CTCs exclusively in patients with LS-SCLC [55,56], where the abundance of CTCs could reveal a micrometastatic disease. In 2014, Zhang and colleagues meta-analyzed results from seven studies and calculated a pooled HR of 1.9 for OS [94]. They found no publication bias; moreover, they included papers where CTCs were detected by different assays (five out of seven with CellSearch) and at different time points; furthermore, Normanno et al. [60] reported an HR with switched groups, but the value included in the meta-analysis was not adequately inverted [94].
4.5. Value of CTCs and ctDNA Changes in Small-Cell Transformation of NSCLC
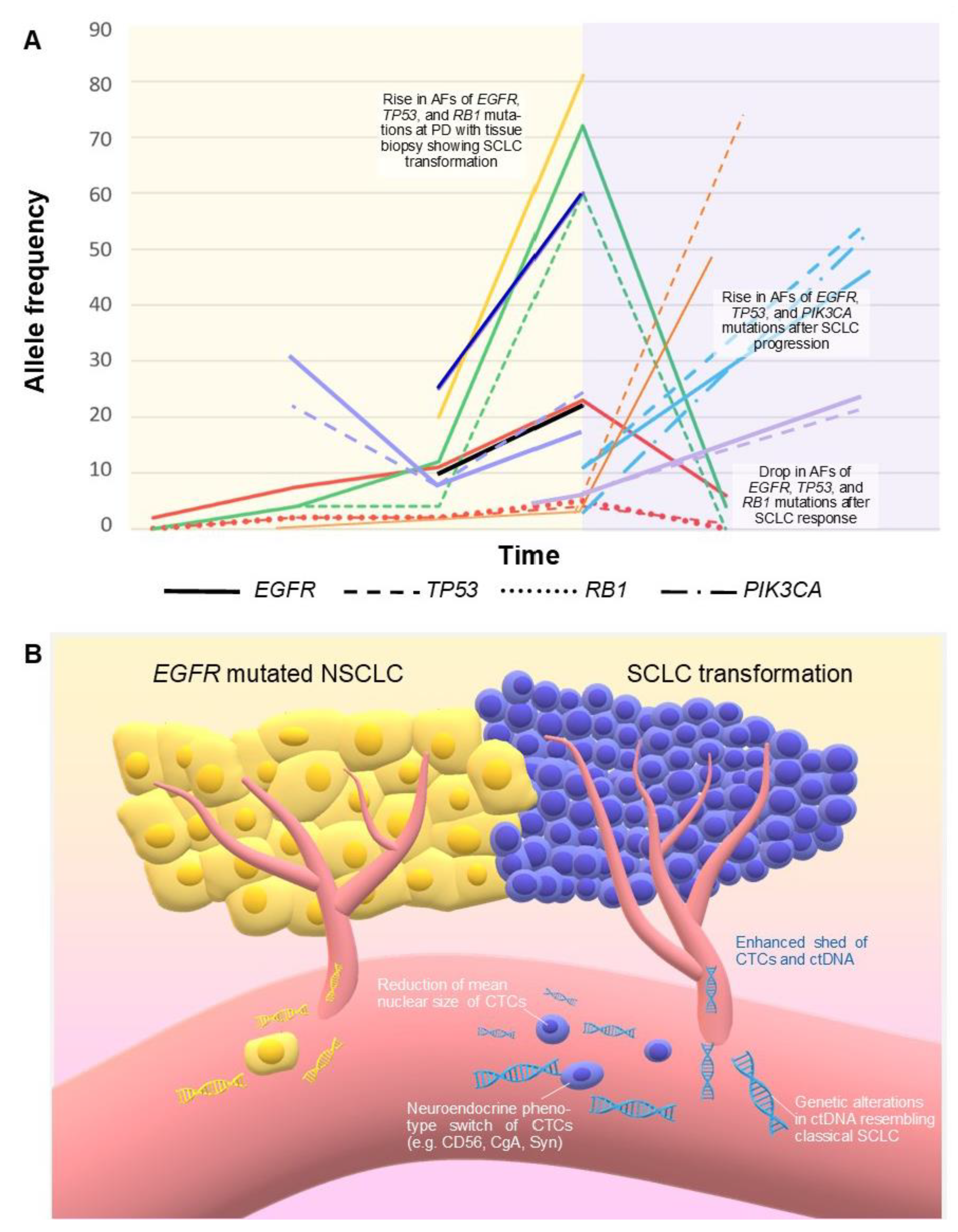
In some patients affected by EGFR mutated NSCLC, EGFR mutations become undetectable through LB after the failure of a TKI. In other cases, mutations in ctDNA could persist with similar or higher AF, even with secondary resistance mechanisms [95]. Among 67 patients with EGFR mutated NSCLC after PD to EGFR TKIs, and increased EGFR mutation abundance on ctDNA was evident in 37% of cases, while in the majority of cases, the allelic fraction of EGFR activating mutation was either stable (19%) or reduced (43%). Patients with stable or increasing EGFR mutant AF at PD showed worse PFS and OS [96]. Interestingly, increasing levels of EGFR activating mutation in ctDNA were described in all the case reports (8 patients overall) available in literature in which LB was performed after TKI failure with evidence of small cell transformation [64,65,66,67,69,71]. In two other cases, the EGFR activating mutation was not retained in neuroendocrine cells, or ctDNA analysis was performed after chemotherapies with platinum and docetaxel, and the EGFR mutant AF was not elevated [70]. Among the eight evaluable patients, the extent of the absolute increase in AFs of activating EGFR mutation at the moment of small cell transformation after TKIs, ranged from +2% up to +61%, with a median of +35% (Table 3, Figure 3). Transformed SCLCs usually retain original activating EGFR mutation, and the sharp increase in EGFR mutant AF at the moment of small cell transformation could be a consequence of the higher growth rate, the more aggressive behavior with increased tumor burden, and the enhanced DNA tumor shed observed in SCLC compared with NSCLC [12]. Typical CNAs observed in classical SCLC (Table 2) have also been reported in ctDNA in cases of histologic transformation, such as amplifications of MYCL1, SOX2, or CNTN3 [70]. Furthermore, LB could allow a successful treatment with a third-generation EGFR TKI in those cases where the resistance mutation T790M appears on ctDNA after a previous small-cell transformation [72,73].
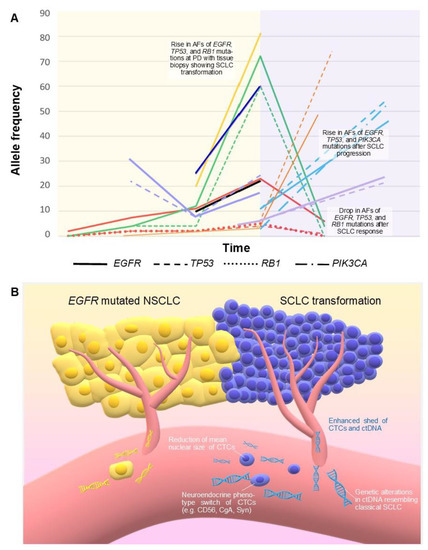
Figure 3.
(A) Correlation between SCLC transformation of EGFR mutated NSCLC and changes in mutation allele frequencies over time detected by ctDNA in 9 patients, as reported in 7 evaluable studies (Table 3). Each patient corresponds to a different color. Time not in scale. Red: Schmid 2020 [69]; dark blue: Pizzutilo 2019 [64]; yellow and black: 2 patients by Minari 2018 [66]; green: Iijima 2018 [67]; orange: 1 patient by Tsui 2018, the other 2 patients were not reported because the diagnosis of histologic transformation was performed under chemotherapy potentially active against SCLC [70]; light blue: Mooradian 2017 [68]; purple: 2 patients by Vendrell 2020; a third patient was not represented because AF at the moment of transformation was not reported [71]. The yellow background represents the EGFR mutated NSCLC; the pink background represents the histologic transformation. (B) Enhanced DNA tumor shed and a higher number of CTCs have been reported in SCLC compared with NSCLC. These features, together with qualitative changes in ctDNA and CTCs, could also characterize transformed SCLC. AF: allele frequency. CTCs: circulating tumor cells. ctDNA: circulating tumor DNA. PD: progressive disease.
Marked higher counts of CTCs have been observed in SCLC in comparison with other malignancies, including NSCLC [15,81], and a correlation exists between CTC count and quantity of cfDNA [10,31]. In two studies, single-cell exome sequencing was performed in CTCs collected from patients presenting SCLC transformation. Mutations in EGFR, PIK3CA, RB1, and TP53 were identified [75,76]. Zhu et al. demonstrated that a histological transformation can be reflected by a significant reduction in the mean nuclear size of CTCs, with phenotypic changes rapidly assessable with immunocytochemistry [76]. Given the continuous flow of CTCs, their “real-time” characterization may become a promising method for timely monitoring of the clonal evolution of solid tumors. Tissue re-biopsy appears now irreplaceable for detecting histologic transformation; furthermore, a ctDNA analysis could suggest a neuroendocrine transversion when it shows an increased EGFR muation AF, especially if together with typical SCLC-associated genetic alterations (Table 1 and Table 2), in patients with aggressive progression. CTC detection by morphology-based enrichment methods [81] could potentially become an alternative to tissue re-biopsy for detection of histologic transformation since these CTCs could undergo cytopathological analysis (i.e., positivity for CD56, CgA, or Syn in case of small cell carcinoma).
While these preliminary data deserve confirmations in wider studies, SCLC transformation may increase its frequency with more targeted anticancer treatments available, and its detection could be easier with the suggestions of an LB. The next step could be the design of studies dedicated to this subset of patients in order to expand their treatment options. Recently, the first clinical trial of durvalumab and olaparib for patients with EGFR mutated transformed SCLC has been launched (NCT04538378).
5. Conclusions
In conclusion, LB provides a huge amount of data in patients with SCLC, exploitable in diverse settings. Qualitative and quantitative changes in CTCs and ctDNA hold a diagnostic potential for both de novo and transformed SCLC, where a phenotypic switch of CTCs and changes in ctDNA could suggest a neuroendocrine transformation of NSCLC. In particular, ctDNA alterations (e.g., EGFR mutant AF) are easily evaluable also in non-academic laboratories. Despite this, a diagnosis of SCLC cannot rely only on LB findings. In addition, genomic profiling of SCLC is feasible by ctDNA analysis, even if results are affected by the adopted assay, tumor burden, or ongoing therapies. Such information may be precious in clinical trials exploring the targetability of molecular flaws. Monitoring response to treatment appears the most mature potential added value of ctDNA for the management of SCLC. CTCs hold a prognostic significance and a strong translational potential for SCLC, but the heterogeneity of available data and the less affordable technology for CTC detection in clinical laboratories hinder their implementation in present daily practice. Comprehensive integration of data derived from ctDNA and different CTCs subpopulations could provide accurate prognostic definitions, decisive biomarkers for therapy decisions, and a fertile field for translational research.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cancers13092265/s1. Figure S1: Prognostic significance of CTCs assessed by CellSearch in patients with SCLC at baseline: funnel plot showing hazard ratios and standard errors for overall survival; Table S1: Studies of liquid biopsy in de novo SCLC, included in the systematic review, with diagnostic, genomic profiling, predictive and/or prognostic findings. Studies are classified in three groups on the basis of the tumor-derived component analyzed (CTCs, ctDNA, or both); Table S2: Detection of ctDNA in SCLC. Table S3: Detection of CTCs in SCLC at baseline; Table S4: Studies included in meta-analysis of prognostic value of baseline CTCs assessed by CellSearch.
Author Contributions
E.G.P.: conception and design of the study, acquisition, analysis and interpretation of data, drafting of the article. M.P.: acquisition, analysis and interpretation of data, drafting of the article. A.A.: analysis and interpretation of data, critical revision of the draft. L.R.: acquisition and interpretation of data, critical revision of the draft. C.L.: analysis and interpretation of data, critical revision of the draft. S.M.V.: interpretation of data, critical revision of the draft. D.S.: interpretation of data, critical revision of the draft. G.C.: interpretation of data, critical revision of the draft. L.G.: interpretation of data, critical revision of the draft. S.S.: conception and design of the study, analysis and interpretation of data, critical revision of the draft, final approval of the version to be submitted. A.S.-B.: conception and design of the study, analysis and interpretation of data, critical revision of the draft, final approval of the version to be submitted. All authors have read and agreed to the published version of the manuscript.
Funding
No one for this paper.
Acknowledgments
Authors are supported by grants from Associazione Italiana Ricerca Cancro grant AIRC 5 × mille [Project ID 51000] Special Program Molecular Clinical Oncology, AIRC Investigator Grant [Project ID 20685 and 16788], and AIRC Special Program 5 per mille metastases [Project ID 21091]; IANG-CRC grant from Fondazione Regionale Ricerca Biomedica (FRRB) of Regione Lombardia; Fondazione Oncologia Niguarda Onlus, grant Terapia Molecolare dei Tumori and grant Studies to Develop Therapies Against Colorectal Cancer in Young Adults.
Conflicts of Interest
S.M.V. received honoraria from Bayer, Janssen Cilag, and Astrazeneca. A.S.-B. has acted as a consultant/advisory member for Amgen, Bayer, Lilly, and Merck-Serono. S.S. is an advisory board member for Amgen, Bayer, BMS, Celgene, Incyte, Merck, Novartis, Roche, Seattle Genetics.
References
- Pietanza, M.C.; Byers, L.A.; Minna, J.D.; Rudin, C.M. Small Cell Lung Cancer: Will Recent Progress Lead to Improved Outcomes? Clin. Cancer Res. 2015, 21, 2244–2255. [Google Scholar] [CrossRef]
- American Cancer Society. Cancer Facts & Figures 2020; American Cancer Society: Atlanta, GA, USA, 2020. [Google Scholar]
- Oser, M.G.; Niederst, M.J.; Sequist, L.V.; Engelman, J.A. Transformation from Non-Small-Cell Lung Cancer to Small-Cell Lung Cancer: Molecular Drivers and Cells of Origin. Lancet Oncol. 2015, 16, e165–e172. [Google Scholar] [CrossRef]
- Niederst, M.J.; Sequist, L.V.; Poirier, J.T.; Mermel, C.H.; Lockerman, E.L.; Garcia, A.R.; Katayama, R.; Costa, C.; Ross, K.N.; Moran, T.; et al. RB Loss in Resistant EGFR Mutant Lung Adenocarcinomas That Transform to Small-Cell Lung Cancer. Nat. Commun. 2015, 6, 6377. [Google Scholar] [CrossRef]
- Blackhall, F.; Frese, K.K.; Simpson, K.; Kilgour, E.; Brady, G.; Dive, C. Will Liquid Biopsies Improve Outcomes for Patients with Small-Cell Lung Cancer? Lancet Oncol. 2018, 19, e470–e481. [Google Scholar] [CrossRef]
- Siravegna, G.; Mussolin, B.; Buscarino, M.; Corti, G.; Cassingena, A.; Crisafulli, G.; Ponzetti, A.; Cremolini, C.; Amatu, A.; Lauricella, C.; et al. Clonal Evolution and Resistance to EGFR Blockade in the Blood of Colorectal Cancer Patients. Nat. Med. 2015, 21, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Rolfo, C.; Mack, P.C.; Scagliotti, G.V.; Baas, P.; Barlesi, F.; Bivona, T.G.; Herbst, R.S.; Mok, T.S.; Peled, N.; Pirker, R.; et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer (NSCLC): A Statement Paper from the IASLC. J. Thorac. Oncol. 2018, 13, 1248–1268. [Google Scholar] [CrossRef]
- Massihnia, D.; Pizzutilo, E.G.; Amatu, A.; Tosi, F.; Ghezzi, S.; Bencardino, K.; Di Masi, P.; Righetti, E.; Patelli, G.; Scaglione, F.; et al. Liquid Biopsy for Rectal Cancer: A Systematic Review. Cancer Treat. Rev. 2019, 79, 101893. [Google Scholar] [CrossRef] [PubMed]
- Buono, G.; Gerratana, L.; Bulfoni, M.; Provinciali, N.; Basile, D.; Giuliano, M.; Corvaja, C.; Arpino, G.; Mastro, L.D.; Placido, S.D.; et al. Circulating Tumor DNA Analysis in Breast Cancer: Is It Ready for Prime-Time? Cancer Treat. Rev. 2019, 73, 73–83. [Google Scholar] [CrossRef]
- Mohan, S.; Foy, V.; Ayub, M.; Leong, H.S.; Schofield, P.; Sahoo, S.; Descamps, T.; Kilerci, B.; Smith, N.K.; Carter, M.; et al. Profiling of Circulating Free DNA Using Targeted and Genome-Wide Sequencing in Patients with SCLC. J. Thorac. Oncol. 2020, 15, 216–230. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Cuesta, L.; Perdomo, S.; Avogbe, P.H.; Leblay, N.; Delhomme, T.M.; Gaborieau, V.; Abedi-Ardekani, B.; Chanudet, E.; Olivier, M.; Zaridze, D.; et al. Identification of Circulating Tumor DNA for the Early Detection of Small-Cell Lung Cancer. EBioMedicine 2016, 10, 117–123. [Google Scholar] [CrossRef]
- Almodovar, K.; Iams, W.T.; Meador, C.B.; Zhao, Z.; York, S.; Horn, L.; Yan, Y.; Hernandez, J.; Chen, H.; Shyr, Y.; et al. Longitudinal Cell-Free DNA Analysis in Patients with Small Cell Lung Cancer Reveals Dynamic Insights into Treatment Efficacy and Disease Relapse. J. Thorac. Oncol. 2018, 13, 112–123. [Google Scholar] [CrossRef]
- Wakelee, H.A.; Gadgeel, S.M.; Goldman, J.W.; Reckamp, K.L.; Karlovich, C.A.; Melnikova, V.; Jean-Charles, S.; Yu, H.A.; Solomon, B.J.; Perol, M.; et al. Epidermal Growth Factor Receptor (EGFR) Genotyping of Matched Urine, Plasma and Tumor Tissue from Non-Small Cell Lung Cancer (NSCLC) Patients (Pts) Treated with Rociletinib. J. Clin. Oncol. 2016, 34 (Suppl. 15), 9001. [Google Scholar] [CrossRef]
- CELLSEARCH®. Available online: https://www.cellsearchctc.com/ (accessed on 5 July 2020).
- Hodgkinson, C.L.; Morrow, C.J.; Li, Y.; Metcalf, R.L.; Rothwell, D.G.; Trapani, F.; Polanski, R.; Burt, D.J.; Simpson, K.L.; Morris, K.; et al. Tumorigenicity and Genetic Profiling of Circulating Tumor Cells in Small-Cell Lung Cancer. Nat. Med. 2014, 20, 897–903. [Google Scholar] [CrossRef]
- Hou, J.-M.; Krebs, M.G.; Lancashire, L.; Sloane, R.; Backen, A.; Swain, R.K.; Priest, L.J.C.; Greystoke, A.; Zhou, C.; Morris, K.; et al. Clinical Significance and Molecular Characteristics of Circulating Tumor Cells and Circulating Tumor Microemboli in Patients with Small-Cell Lung Cancer. J. Clin. Oncol. 2012, 30, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Naito, T.; Tanaka, F.; Ono, A.; Yoneda, K.; Takahashi, T.; Murakami, H.; Nakamura, Y.; Tsuya, A.; Kenmotsu, H.; Shukuya, T.; et al. Prognostic Impact of Circulating Tumor Cells in Patients with Small Cell Lung Cancer. J. Thorac. Oncol. 2012, 7, 512–519. [Google Scholar] [CrossRef] [PubMed]
- Hiltermann, T.J.N.; Pore, M.M.; van den Berg, A.; Timens, W.; Boezen, H.M.; Liesker, J.J.W.; Schouwink, J.H.; Wijnands, W.J.A.; Kerner, G.S.M.A.; Kruyt, F.A.E.; et al. Circulating Tumor Cells in Small-Cell Lung Cancer: A Predictive and Prognostic Factor. Ann. Oncol. 2012, 23, 2937–2942. [Google Scholar] [CrossRef] [PubMed]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gøtzsche, P.C.; Ioannidis, J.P.A.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA Statement for Reporting Systematic Reviews and Meta-Analyses of Studies That Evaluate Health Care Interventions: Explanation and Elaboration. PLoS Med. 2009, 6, e1000100. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. J. Clin. Epidemio. 2009, 62, 1006–1012. [Google Scholar] [CrossRef] [PubMed]
- Stroup, D.F.; Berlin, J.A.; Morton, S.C.; Olkin, I.; Williamson, G.D.; Rennie, D.; Moher, D.; Becker, B.J.; Sipe, T.A.; Thacker, S.B.; et al. Meta-Analysis of Observational Studies in Epidemiology: A Proposal for Reporting. JAMA 2000, 283, 2008–2012. [Google Scholar] [CrossRef]
- Tierney, J.F.; Stewart, L.A.; Ghersi, D.; Burdett, S.; Sydes, M.R. Practical Methods for Incorporating Summary Time-to-Event Data into Meta-Analysis. Trials 2007, 8, 16. [Google Scholar] [CrossRef]
- Wang, Y.-L.; Liu, C.-H.; Li, J.; Ma, X.-P.; Gong, P. Clinical Significance of Circulating Tumor Cells in Patients with Small-Cell Lung Cancer. Tumori 2017, 103, 242–248. [Google Scholar] [CrossRef]
- Board, R.E.; Williams, V.S.; Knight, L.; Shaw, J.; Greystoke, A.; Ranson, M.; Dive, C.; Blackhall, F.H.; Hughes, A. Isolation and Extraction of Circulating Tumor DNA from Patients with Small Cell Lung Cancer. Ann. N. Y. Acad. Sci. 2008, 1137, 98–107. [Google Scholar] [CrossRef]
- Owonikoko, T.K.; Niu, H.; Nackaerts, K.; Csoszi, T.; Ostoros, G.; Mark, Z.; Baik, C.; Joy, A.A.; Chouaid, C.; Jaime, J.C.; et al. Randomized Phase II Study of Paclitaxel plus Alisertib versus Paclitaxel plus Placebo as Second-Line Therapy for SCLC: Primary and Correlative Biomarker Analyses. J. Thorac. Oncol. 2020, 15, 274–287. [Google Scholar] [CrossRef]
- Devarakonda, S.; Sankararaman, S.; Herzog, B.H.; Gold, K.A.; Waqar, S.N.; Ward, J.P.; Raymond, V.M.; Lanman, R.B.; Chaudhuri, A.A.; Owonikoko, T.K.; et al. Circulating Tumor DNA Profiling in Small-Cell Lung Cancer Identifies Potentially Targetable Alterations. Clin. Cancer Res. 2019, 25, 6119–6126. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Thompson, J.; Fisher, H.; Zhang, P.; Huang, C.-C.; Wang, L. Genomic Alterations of Plasma Cell-Free DNAs in Small Cell Lung Cancer and Their Clinical Relevance. Lung Cancer 2018, 120, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Nong, J.; Gong, Y.; Guan, Y.; Yi, X.; Yi, Y.; Chang, L.; Yang, L.; Lv, J.; Guo, Z.; Jia, H.; et al. Circulating Tumor DNA Analysis Depicts Subclonal Architecture and Genomic Evolution of Small Cell Lung Cancer. Nat. Commun. 2018, 9, 3114. [Google Scholar] [CrossRef] [PubMed]
- Herbreteau, G.; Langlais, A.; Greillier, L.; Audigier-Valette, C.; Uwer, L.; Hureaux, J.; Moro-Sibilot, D.; Guisier, F.; Carmier, D.; Madelaine, J.; et al. Circulating Tumor DNA as a Prognostic Determinant in Small Cell Lung Cancer Patients Receiving Atezolizumab. J. Clin. Med. 2020, 9, 3861. [Google Scholar] [CrossRef]
- Yaung, S.; Woestmann, C.; Xi, L.; Ju, C.; Hinzmann, B.; Thomas, M.; Lasitschka, F.; Meister, M.; Schneider, M.; Herth, F.J.F.; et al. 1744P-Mutational Profiling of Tumour Tissue and Sequential Plasma Illustrates Emergent Clones during Treatment in Late Stage Small Cell Lung Cancer (SCLC). Ann. Oncol. 2019, 30, v713–v714. [Google Scholar] [CrossRef]
- Aggarwal, C.; Badola, S.; Shin, H.; Bedford, L.; Collins, S.; Derk, B.; Fostel, J.; Ecsedy, J.; Evans, T.; Bauml, J.; et al. PUB140 A Pilot Study to Assess Circulating Tumor Cells, Circulating Tumor Cell DNA and Cell Free DNA in Patients with Small Cell Lung Cancer. J. Thorac. Oncol. 2017, 12, S1527. [Google Scholar] [CrossRef][Green Version]
- Su, Z.; Wang, Z.; Ni, X.; Duan, J.; Gao, Y.; Zhuo, M.; Li, R.; Zhao, J.; Ma, Q.; Bai, H.; et al. Inferring the Evolution and Progression of Small-Cell Lung Cancer by Single-Cell Sequencing of Circulating Tumor Cells. Clin. Cancer Res. 2019, 25, 5049–5060. [Google Scholar] [CrossRef]
- Gonzalez, R.; Silva, J.M.; Sanchez, A.; Dominguez, G.; Garcia, J.M.; Chen, X.Q.; Stroun, M.; Provencio, M.; España, P.; Anker, P.; et al. Microsatellite Alterations and TP53 Mutations in Plasma DNA of Small-Cell Lung Cancer Patients: Follow-up Study and Prognostic Significance. Ann. Oncol. 2000, 11, 1097–1104. [Google Scholar] [CrossRef]
- Thomas, A.; Vilimas, R.; Trindade, C.; Erwin-Cohen, R.; Roper, N.; Xi, L.; Krishnasamy, V.; Levy, E.; Mammen, A.; Nichols, S.; et al. Durvalumab in Combination with Olaparib in Patients with Relapsed SCLC: Results from a Phase II Study. J. Thorac. Oncol. 2019, 14, 1447–1457. [Google Scholar] [CrossRef]
- Zhang, J.; Tian, C.; Lv, F.; Wang, J.; Han, W.; Nie, J.; Dai, L.; Hu, W.; Chen, X.; Ma, X.; et al. Molecular Analysis of Cell-Free DNA Identifies Distinct Molecular Features in Patients with Chemosensitive and Chemorefractory Small Cell Lung Cancer. Cancer Commun. 2019, 39, 20. [Google Scholar] [CrossRef]
- Messaritakis, I.; Stoltidis, D.; Kotsakis, A.; Dermitzaki, E.-K.; Koinis, F.; Lagoudaki, E.; Koutsopoulos, A.; Politaki, E.; Apostolaki, S.; Souglakos, J.; et al. TTF-1- and/or CD56-Positive Circulating Tumor Cells in Patients with Small Cell Lung Cancer (SCLC). Sci. Rep. 2017, 7, 45351. [Google Scholar] [CrossRef]
- Aggarwal, C.; Wang, X.; Ranganathan, A.; Torigian, D.; Troxel, A.; Evans, T.; Cohen, R.B.; Vaidya, B.; Rao, C.; Connelly, M.; et al. Circulating Tumor Cells as a Predictive Biomarker in Patients with Small Cell Lung Cancer Undergoing Chemotherapy. Lung Cancer 2017, 112, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Belani, C.P.; Dahlberg, S.E.; Rudin, C.M.; Fleisher, M.; Chen, H.X.; Takebe, N.; Velasco, M.R.; Tester, W.J.; Sturtz, K.; Hann, C.L.; et al. Vismodegib or Cixutumumab in Combination with Standard Chemotherapy for Patients with Extensive-Stage Small Cell Lung Cancer: A Trial of the ECOG-ACRIN Cancer Research Group (E1508). Cancer 2016, 122, 2371–2378. [Google Scholar] [CrossRef]
- Wang, X.; Ma, K.; Wang, Y.; He, H.; Hu, J.-F.; Li, W. Evaluation of Circulating Tumor Cells in Predicting Therapeutic Response in Small Cell Lung Cancer Patients. Arch. Med. Res. 2016, 47, 454–459. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.-M.; Greystoke, A.; Lancashire, L.; Cummings, J.; Ward, T.; Board, R.; Amir, E.; Hughes, S.; Krebs, M.; Hughes, A.; et al. Evaluation of Circulating Tumor Cells and Serological Cell Death Biomarkers in Small Cell Lung Cancer Patients Undergoing Chemotherapy. Am. J. Pathol. 2009, 175, 808–816. [Google Scholar] [CrossRef] [PubMed]
- Bevilacqua, S.; Gallo, M.; Franco, R.; Rossi, A.; De Luca, A.; Rocco, G.; Botti, G.; Gridelli, C.; Normanno, N. A “Live” Biopsy in a Small-Cell Lung Cancer Patient by Detection of Circulating Tumor Cells. Lung Cancer 2009, 65, 123–125. [Google Scholar] [CrossRef] [PubMed]
- Salgia, R.; Weaver, R.W.; McCleod, M.; Stille, J.R.; Yan, S.B.; Roberson, S.; Polzer, J.; Flynt, A.; Raddad, E.; Peek, V.L.; et al. Prognostic and Predictive Value of Circulating Tumor Cells and CXCR4 Expression as Biomarkers for a CXCR4 Peptide Antagonist in Combination with Carboplatin-Etoposide in Small Cell Lung Cancer: Exploratory Analysis of a Phase II Study. Investig. New Drugs 2017, 35, 334–344. [Google Scholar] [CrossRef]
- Pietanza, M.C.; Waqar, S.N.; Krug, L.M.; Dowlati, A.; Hann, C.L.; Chiappori, A.; Owonikoko, T.K.; Woo, K.M.; Cardnell, R.J.; Fujimoto, J.; et al. Randomized, Double-Blind, Phase II Study of Temozolomide in Combination With Either Veliparib or Placebo in Patients With Relapsed-Sensitive or Refractory Small-Cell Lung Cancer. J. Clin. Oncol. 2018, 36, 2386–2394. [Google Scholar] [CrossRef] [PubMed]
- Messaritakis, I.; Politaki, E.; Koinis, F.; Stoltidis, D.; Apostolaki, S.; Plataki, M.; Dermitzaki, E.-K.; Georgoulias, V.; Kotsakis, A. Dynamic Changes of Phenotypically Different Circulating Tumor Cells Sub-Populations in Patients with Recurrent/Refractory Small Cell Lung Cancer Treated with Pazopanib. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Koinis, F.; Agelaki, S.; Karavassilis, V.; Kentepozidis, N.; Samantas, E.; Peroukidis, S.; Katsaounis, P.; Hartabilas, E.; Varthalitis, I.I.; Messaritakis, I.; et al. Second-Line Pazopanib in Patients with Relapsed and Refractory Small-Cell Lung Cancer: A Multicentre Phase II Study of the Hellenic Oncology Research Group. Br. J. Cancer 2017, 117, 8–14. [Google Scholar] [CrossRef]
- Igawa, S.; Gohda, K.; Fukui, T.; Ryuge, S.; Otani, S.; Masago, A.; Sato, J.; Murakami, K.; Maki, S.; Katono, K.; et al. Circulating Tumor Cells as a Prognostic Factor in Patients with Small Cell Lung Cancer. Oncol. Lett. 2014, 7, 1469–1473. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zhao, J.; Jiang, T.; Li, X.; Zhao, C.; Su, C.; Zhou, C. Predictive and Prognostic Value of Folate Receptor-Positive Circulating Tumor Cells in Small Cell Lung Cancer Patients Treated with First-Line Chemotherapy. Oncotarget 2017, 8, 49044–49052. [Google Scholar] [CrossRef]
- Shi, W.-L.; Li, J.; Du, Y.-J.; Zhu, W.-F.; Wu, Y.; Hu, Y.-M.; Chen, Y.-C. CK-19 MRNA-Positive Cells in Peripheral Blood Predict Treatment Efficacy and Survival in Small-Cell Lung Cancer Patients. Med. Oncol. 2013, 30, 755. [Google Scholar] [CrossRef] [PubMed]
- Messaritakis, I.; Nikolaou, M.; Koinis, F.; Politaki, E.; Koutsopoulos, A.; Lagoudaki, E.; Vetsika, E.-K.; Georgoulias, V.; Kotsakis, A. Characterization of DLL3-Positive Circulating Tumor Cells (CTCs) in Patients with Small Cell Lung Cancer (SCLC) and Evaluation of Their Clinical Relevance during Front-Line Treatment. Lung Cancer 2019, 135, 33–39. [Google Scholar] [CrossRef]
- Carter, L.; Rothwell, D.G.; Mesquita, B.; Smowton, C.; Leong, H.S.; Fernandez-Gutierrez, F.; Li, Y.; Burt, D.J.; Antonello, J.; Morrow, C.J.; et al. Molecular Analysis of Circulating Tumor Cells Identifies Distinct Copy-Number Profiles in Patients with Chemosensitive and Chemorefractory Small-Cell Lung Cancer. Nat. Med. 2017, 23, 114–119. [Google Scholar] [CrossRef]
- Palma, J.F.; Woestmann, C.; McNamara, S.; Hinzmann, B.; Fröhler, S.; Adams, H.-P.; Feldkamp, M.; Siemann, S.; Lange, M.; Blüher, A.; et al. Early Assessment of Therapy Response in Small Cell Lung Cancer via Longitudinal CtDNA Analysis. JCO 2018, 36, 8577. [Google Scholar] [CrossRef]
- Jin, Y.; Chen, Y.M.; Tang, H.R.; Li, Q.; Li, P.S.; Hu, X.; Guan, Y.F.; Xia, X.F.; Yi, X.; Zhang, J.J.; et al. Clinical Potential of CtDNA-Based TMB in Small Cell Lung Cancer Recieving Chemoradiotherapy. J. Clin. Oncol. 2020, 38 (Suppl. 15), 3536. [Google Scholar] [CrossRef]
- Yaung, S.; Xi, L.; Woestmann, C.; McNamara, S.; Hinzmann, B.; Froehler, S.; Tikoo, N.; Ju, C.; Balasubramanyam, A.; Adams, H.-P.; et al. Ecological Diversity Indices as Measurements of Tumor Heterogeneity Correlates with Clinical Outcomes in Late Stage Small Cell Lung Cancer (SCLC). Annal Oncol. 2018, 29, viii54. [Google Scholar] [CrossRef]
- Kularatne, B.Y.; Lorigan, P.; Browne, S.; Suvarna, S.K.; Smith, M.O.; Lawry, J. Monitoring Tumour Cells in the Peripheral Blood of Small Cell Lung Cancer Patients. Cytometry 2002, 50, 160–167. [Google Scholar] [CrossRef]
- Tay, R.Y.; Fernández-Gutiérrez, F.; Foy, V.; Burns, K.; Pierce, J.; Morris, K.; Priest, L.; Tugwood, J.; Ashcroft, L.; Lindsay, C.R.; et al. Prognostic Value of Circulating Tumour Cells in Limited-Stage Small-Cell Lung Cancer: Analysis of the Concurrent Once-Daily versus Twice-Daily Radiotherapy (CONVERT) Randomised Controlled Trial. Ann. Oncol. 2019, 30, 1114–1120. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Zhu, Y.; Jing, W.; Guo, D.; Kong, L.; Yu, J. Incorporation of Circulating Tumor Cells and Whole-Body Metabolic Tumor Volume of 18F-FDG PET/CT Improves Prediction of Outcome in IIIB Stage Small-Cell Lung Cancer. Chin. J. Cancer Res. 2018, 30, 596–604. [Google Scholar] [CrossRef] [PubMed]
- Messaritakis, I.; Nikolaou, M.; Politaki, E.; Koinis, F.; Lagoudaki, E.; Koutsopoulos, A.; Georgoulia, N.; Georgoulias, V.; Kotsakis, A. Bcl-2 Expression in Circulating Tumor Cells (CTCs) of Patients with Small Cell Lung Cancer (SCLC) Receiving Front-Line Treatment. Lung Cancer 2018, 124, 270–278. [Google Scholar] [CrossRef]
- Pietanza, M.C.; Litvak, A.M.; Varghese, A.M.; Krug, L.M.; Fleisher, M.; Teitcher, J.B.; Holodny, A.I.; Sima, C.S.; Woo, K.M.; Ng, K.K.; et al. A Phase I Trial of the Hedgehog Inhibitor, Sonidegib (LDE225), in Combination with Etoposide and Cisplatin for the Initial Treatment of Extensive Stage Small Cell Lung Cancer. Lung Cancer 2016, 99, 23–30. [Google Scholar] [CrossRef]
- Cheng, Y.; Liu, X.-Q.; Fan, Y.; Liu, Y.-P.; Liu, Y.; Liu, Y.; Ma, L.-X.; Liu, X.-H.; Li, H.; Bao, H.-Z.; et al. Circulating Tumor Cell Counts/Change for Outcome Prediction in Patients with Extensive-Stage Small-Cell Lung Cancer. Future Oncol. 2016, 12, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Normanno, N.; Rossi, A.; Morabito, A.; Signoriello, S.; Bevilacqua, S.; Di Maio, M.; Costanzo, R.; De Luca, A.; Montanino, A.; Gridelli, C.; et al. Prognostic Value of Circulating Tumor Cells’ Reduction in Patients with Extensive Small-Cell Lung Cancer. Lung Cancer 2014, 85, 314–319. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.H.; Wick, J.A.; Sittampalam, G.S.; Nirmalanandhan, V.S.; Ganti, A.K.; Neupane, P.C.; Williamson, S.K.; Godwin, A.K.; Schmitt, S.; Smart, N.J.; et al. A Multicenter Pilot Study Examining the Role of Circulating Tumor Cells as a Blood-Based Tumor Marker in Patients with Extensive Small-Cell Lung Cancer. Front Oncol. 2014, 4, 271. [Google Scholar] [CrossRef]
- Gadgeel, S.M.; Pennell, N.A.; Fidler, M.J.; Halmos, B.; Bonomi, P.; Stevenson, J.; Schneider, B.; Sukari, A.; Ventimiglia, J.; Chen, W.; et al. Phase II Study of Maintenance Pembrolizumab in Patients with Extensive-Stage Small Cell Lung Cancer (SCLC). J. Thorac. Oncol. 2018, 13, 1393–1399. [Google Scholar] [CrossRef]
- Wang, P.-P.; Liu, S.-H.; Chen, C.-T.; Lv, L.; Li, D.; Liu, Q.-Y.; Liu, G.-L.; Wu, Y. Circulating Tumor Cells as a New Predictive and Prognostic Factor in Patients with Small Cell Lung Cancer. J. Cancer 2020, 11, 2113–2122. [Google Scholar] [CrossRef] [PubMed]
- Pizzutilo, E.G.; Lauricella, C.; Cerea, G.; Giannetta, L.G.; Tomasello, G.; Stabile, S.; Motta, V.; Alexiadis, S.; Scaglione, F.; Vanzulli, A.; et al. Concurrent Small-Cell Transformation and Emergence of Trans-C797S and T790M Mutations Under Sequential Treatment With EGFR Inhibitors in Lung Adenocarcinoma. JCO Precis. Oncol. 2019, 1–5. [Google Scholar] [CrossRef]
- Piotrowska, Z.; Niederst, M.J.; Karlovich, C.A.; Wakelee, H.A.; Neal, J.W.; Mino-Kenudson, M.; Fulton, L.; Hata, A.N.; Lockerman, E.L.; Kalsy, A.; et al. Heterogeneity Underlies the Emergence of EGFRT790 Wild-Type Clones Following Treatment of T790M-Positive Cancers with a Third-Generation EGFR Inhibitor. Cancer Discov. 2015, 5, 713–722. [Google Scholar] [CrossRef]
- Minari, R.; Bordi, P.; Del Re, M.; Facchinetti, F.; Mazzoni, F.; Barbieri, F.; Camerini, A.; Comin, C.E.; Gnetti, L.; Azzoni, C.; et al. Primary Resistance to Osimertinib Due to SCLC Transformation: Issue of T790M Determination on Liquid Re-Biopsy. Lung Cancer 2018, 115, 21–27. [Google Scholar] [CrossRef]
- Iijima, Y.; Hirotsu, Y.; Mochizuki, H.; Amemiya, K.; Oyama, T.; Uchida, Y.; Kobayashi, Y.; Tsutsui, T.; Kakizaki, Y.; Miyashita, Y.; et al. Dynamic Changes and Drug-Induced Selection of Resistant Clones in a Patient With EGFR-Mutated Adenocarcinoma That Acquired T790M Mutation and Transformed to Small-Cell Lung Cancer. Clin. Lung Cancer 2018, 19, e843–e847. [Google Scholar] [CrossRef]
- Mooradian, M.J.; Piotrowska, Z.; Drapkin, B.J.; Dias-Santagata, D.; Marcoux, N.; Arnaoutakis, K.; Nagy, R.J.; Lanman, R.; Iafrate, A.J.; Farago, A.F.; et al. Clonal Evolution and the Role of Serial Liquid Biopsies in a Case of Small-Cell Lung Cancer–Transformed EGFR Mutant Non–Small-Cell Lung Cancer. JCO Precis. Oncol. 2017, 1, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Schmid, S.; Stewart, E.L.; Martins-Filho, S.N.; Cabanero, M.; Wang, A.; Bao, H.; Wu, X.; Patel, D.; Chen, Z.; Law, J.H.; et al. Early Detection of Multiple Resistance Mechanisms by CtDNA Profiling in a Patient With EGFR-Mutant Lung Adenocarcinoma Treated With Osimertinib. Clin. Lung Cancer 2020. [Google Scholar] [CrossRef]
- Tsui, D.W.Y.; Murtaza, M.; Wong, A.S.C.; Rueda, O.M.; Smith, C.G.; Chandrananda, D.; Soo, R.A.; Lim, H.L.; Goh, B.C.; Caldas, C.; et al. Dynamics of Multiple Resistance Mechanisms in Plasma DNA during EGFR-targeted Therapies in Non-small Cell Lung Cancer. EMBO Mol. Med. 2018, 10. [Google Scholar] [CrossRef]
- Vendrell, J.A.; Quantin, X.; Serre, I.; Solassol, J. Combination of Tissue and Liquid Biopsy Molecular Profiling to Detect Transformation to Small Cell Lung Carcinoma during Osimertinib Treatment. Ther. Adv. Med. Oncol. 2020, 12, 1758835920974192. [Google Scholar] [CrossRef]
- Han, J.; Zhang, Q.; Wang, B.; Zhou, Q.; Yan, L.; Zhang, Z.; Chen, H.; Su, J.; Xie, Z.; Niu, F.; et al. JCES01.24 Molecular Mechanism of Transformation from Adenocarcinoma to Small-Cell Lung Cancer after EGFR-TKI. J. Thorac. Oncol. 2017, 12, S240–S241. [Google Scholar] [CrossRef][Green Version]
- Nishioka, N.; Yamada, T.; Harita, S.; Hirai, S.; Katayama, Y.; Nakano, T.; Okura, N.; Tamiya, N.; Kaneko, Y.; Uchino, J.; et al. Successful Sequential Treatment of Refractory Tumors Caused by Small Cell Carcinoma Transformation and EGFR-T790M Mutation Diagnosed by Repeated Genetic Testing in a Patient with Lung Adenocarcinoma Harboring Epidermal Growth Factor Receptor Mutations: A Case Report. Respir. Med. Case Rep. 2018, 25, 261–263. [Google Scholar] [CrossRef]
- Ou, S.-H.I.; Lee, T.K.; Young, L.; Fernandez-Rocha, M.Y.; Pavlick, D.; Schrock, A.B.; Zhu, V.W.; Milliken, J.; Ali, S.M.; Gitlitz, B.J. Dual Occurrence of ALK G1202R Solvent Front Mutation and Small Cell Lung Cancer Transformation as Resistance Mechanisms to Second Generation ALK Inhibitors without Prior Exposure to Crizotinib. Pitfall of Solely Relying on Liquid Re-Biopsy? Lung Cancer 2017, 106, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Ni, X.; Zhuo, M.; Su, Z.; Duan, J.; Gao, Y.; Wang, Z.; Zong, C.; Bai, H.; Chapman, A.R.; Zhao, J.; et al. Reproducible Copy Number Variation Patterns among Single Circulating Tumor Cells of Lung Cancer Patients. Proc. Natl. Acad. Sci. USA 2013, 110, 21083–21088. [Google Scholar] [CrossRef]
- Zhu, Y.; Zou, C.; Zhang, J.; Jiang, W.; Guan, F.; Tang, K.; Li, S.; Li, G.; Wang, J.; Ke, Z. Dynamically Monitoring the Clonal Evolution of Lung Cancer Based on the Molecular Characterization of Circulating Tumor Cells Using Aptamer Cocktail-Modified Nanosubstrates. ACS Appl. Mater. Interfaces 2020, 12, 5671–5679. [Google Scholar] [CrossRef]
- Alì, G.; Bruno, R.; Giordano, M.; Prediletto, I.; Marconi, L.; Zupo, S.; Fedeli, F.; Ribechini, A.; Chella, A.; Fontanini, G. Small Cell Lung Cancer Transformation and the T790M Mutation: A Case Report of Two Acquired Mechanisms of TKI Resistance Detected in a Tumor Rebiopsy and Plasma Sample of EGFR-Mutant Lung Adenocarcinoma. Oncol. Lett. 2016, 12, 4009–4012. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lennon, A.M.; Buchanan, A.H.; Kinde, I.; Warren, A.; Honushefsky, A.; Cohain, A.T.; Ledbetter, D.H.; Sanfilippo, F.; Sheridan, K.; Rosica, D.; et al. Feasibility of Blood Testing Combined with PET-CT to Screen for Cancer and Guide Intervention. Science 2020, 369. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.C.; Oxnard, G.R.; Klein, E.A.; Swanton, C.; Seiden, M.V.; Liu, M.C.; Oxnard, G.R.; Klein, E.A.; Smith, D.; Richards, D.; et al. Sensitive and Specific Multi-Cancer Detection and Localization Using Methylation Signatures in Cell-Free DNA. Ann. Oncol. 2020, 31, 745–759. [Google Scholar] [CrossRef]
- Menarini Silicon Biosystems. CELLSEARCH® Circulating Tumor Cell Kit (Epithelial) Instructions for Use. Available online: https://documents.cellsearchctc.com/ (accessed on 25 July 2020).
- Yu, N.; Zhou, J.; Cui, F.; Tang, X. Circulating Tumor Cells in Lung Cancer: Detection Methods and Clinical Applications. Lung 2015, 193, 157–171. [Google Scholar] [CrossRef]
- Wang, J.; Gong, Y.; Nong, J.; Yi, Y.; Guan, Y.; Yang, L.; Jia, H.; Zhang, S.; Yi, X.; Liao, Z.; et al. MA 01.03 The Potential of CtDNA Sequencing in Disease Monitoring and Depicting Genomic Evolution of Small-Cell Lung Cancer Under Therapy. J. Thorac. Oncol. 2017, 12, S1800. [Google Scholar] [CrossRef]
- George, J.; Lim, J.S.; Jang, S.J.; Cun, Y.; Ozretić, L.; Kong, G.; Leenders, F.; Lu, X.; Fernández-Cuesta, L.; Bosco, G.; et al. Comprehensive Genomic Profiles of Small Cell Lung Cancer. Nature 2015, 524, 47–53. [Google Scholar] [CrossRef]
- Rudin, C.M.; Durinck, S.; Stawiski, E.W.; Poirier, J.T.; Modrusan, Z.; Shames, D.S.; Bergbower, E.A.; Guan, Y.; Shin, J.; Guillory, J.; et al. Comprehensive Genomic Analysis Identifies SOX2 as a Frequently Amplified Gene in Small-Cell Lung Cancer. Nat. Genet. 2012, 44, 1111–1116. [Google Scholar] [CrossRef]
- Swanton, C.; Venn, O.; Aravanis, A.; Hubbell, E.; Maddala, T.; Beausang, J.F.; Filippova, D.; Gross, S.; Jamshidi, A.; Shen, L.; et al. Prevalence of Clonal Hematopoiesis of Indeterminate Potential (CHIP) Measured by an Ultra-Sensitive Sequencing Assay: Exploratory Analysis of the Circulating Cancer Genome Atlas (CCGA) Study. JCO 2018, 36, 12003. [Google Scholar] [CrossRef]
- Gay, C.M.; Stewart, C.A.; Park, E.M.; Diao, L.; Groves, S.M.; Heeke, S.; Nabet, B.Y.; Fujimoto, J.; Solis, L.M.; Lu, W.; et al. Patterns of Transcription Factor Programs and Immune Pathway Activation Define Four Major Subtypes of SCLC with Distinct Therapeutic Vulnerabilities. Cancer Cell 2021. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.A.; Gay, C.M.; Xi, Y.; Sivajothi, S.; Sivakamasundari, V.; Fujimoto, J.; Bolisetty, M.; Hartsfield, P.M.; Balasubramaniyan, V.; Chalishazar, M.D.; et al. Single-Cell Analyses Reveal Increased Intratumoral Heterogeneity after the Onset of Therapy Resistance in Small-Cell Lung Cancer. Nat. Cancer 2020, 1, 423–436. [Google Scholar] [CrossRef] [PubMed]
- Farago, A.F.; Yeap, B.Y.; Stanzione, M.; Hung, Y.P.; Heist, R.S.; Marcoux, J.P.; Zhong, J.; Rangachari, D.; Barbie, D.A.; Phat, S.; et al. Combination Olaparib and Temozolomide in Relapsed Small Cell Lung Cancer. Cancer Discov. 2019. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, H.; Sen, T.; Rudin, C.M. Targeted Therapies and Biomarkers in Small Cell Lung Cancer. Front. Oncol. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Sabari, J.K.; Lok, B.H.; Laird, J.H.; Poirier, J.T.; Rudin, C.M. Unravelling the Biology of SCLC: Implications for Therapy. Nat. Rev. Clin. Oncol. 2017, 14, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Katsurada, M.; Nagano, T.; Tachihara, M.; Kiriu, T.; Furukawa, K.; Koyama, K.; Otoshi, T.; Sekiya, R.; Hazama, D.; Tamura, D.; et al. Baseline Tumor Size as a Predictive and Prognostic Factor of Immune Checkpoint Inhibitor Therapy for Non-Small Cell Lung Cancer. Anticancer Res. 2019, 39, 815–825. [Google Scholar] [CrossRef]
- Yan, W.-T.; Cui, X.; Chen, Q.; Li, Y.-F.; Cui, Y.-H.; Wang, Y.; Jiang, J. Circulating Tumor Cell Status Monitors the Treatment Responses in Breast Cancer Patients: A Meta-Analysis. Sci. Rep. 2017, 7, 43464. [Google Scholar] [CrossRef]
- Dowlati, A.; Lipka, M.B.; McColl, K.; Dabir, S.; Behtaj, M.; Kresak, A.; Miron, A.; Yang, M.; Sharma, N.; Fu, P.; et al. Clinical Correlation of Extensive-Stage Small-Cell Lung Cancer Genomics. Ann. Oncol. 2016, 27, 642–647. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, H.-T.; Li, B.-G. Prognostic Significance of Circulating Tumor Cells in Small--Cell Lung Cancer Patients: A Meta-Analysis. Asian Pac. J. Cancer Prev. 2014, 15, 8429–8433. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Cui, S.; Ding, J.; Sun, Y.; Zhang, L.; Zhao, Y.; Gu, A.; Chu, T.; Wang, H.; Zhong, H.; et al. Dynamics of EGFR Mutations in Plasma Recapitulates the Clinical Response to EGFR-TKIs in NSCLC Patients. Oncotarget 2017, 8, 63846–63856. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhuo, M.; Ye, X.; Bai, H.; Wang, Z.; Sun, Y.; Zhao, J.; An, T.; Duan, J.; Wu, M.; et al. Quantification of Mutant Alleles in Circulating Tumor DNA Can Predict Survival in Lung Cancer. Oncotarget 2016, 7, 20810–20824. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).