The Multiple Facets of ATRX Protein
Abstract
Simple Summary
Abstract
1. Introduction
2. ATRX Gene and Protein
3. ATRX Interacts with DAXX in PML Nuclear Bodies Promoting H3.3 Deposition
4. ATRX Contributes to Pericentromeric and Telomeric Heterochromatin Formation and Maintenance
4.1. ATRX Role at Pericentromeric Heterochromatin
4.2. ATRX Role in Telomeric Heterochromatin
5. ATRX Participates in DNA Repair Process
5.1. ATRX Prevents Replication Stress in Specific Regions of the Genome
5.2. ATRX Suppress Homologous Recombination at Telomeres
6. ATRX Role in X Chromosome Inactivation
7. ATRX Is Involved in Repression of Transposon, Retrotransposon and Viral DNA
8. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stayton, C.L.; Dabovic, B.; Gulisano, M.; Gecz, J.; Broccoll, V.; Glovanazzl, S.; Bossolasco, M.; Monaco, L.; Rastan, S.; Boncinelli, E.; et al. Cloning and characterization of a new human Xq13 gene, encoding a putative helicase. Hum. Mol. Genet. 1994, 3, 1957–1964. [Google Scholar] [CrossRef] [PubMed]
- Picketts, D.J.; Higgs, D.R.; Bachoo, S.; Blake, D.J.; Quarrell, O.W.J.; Gibbons, R.J. ATRX encodes a novel member of the SNF2 family of proteins: Mutations point to a common mechanism underlying the ATR-X syndrome. Hum. Mol. Genet. 1996, 5, 1899–1907. [Google Scholar] [CrossRef] [PubMed]
- Argentaro, A.; Yang, J.C.; Chapman, L.; Kowalczyk, M.S.; Gibbons, R.J.; Higgs, D.R.; Neuhaus, D.; Rhodes, D. Structural consequences of disease-causing mutations in the ATRX-DNMT3-DNMT3L (ADD) domain of the chromatin-associated protein ATRX. Proc. Natl. Acad. Sci. USA 2007, 104, 11939–11944. [Google Scholar] [CrossRef]
- Watson, L.A.; Goldberg, H.; Berube, N.G. Emerging roles of ATRX in cancer. Epigenomics 2015, 7, 1365–1378. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, R.J.; Picketts, D.J.; Villard, L.; Higgs, D.R. Mutations in a putative global transcriptional regulator cause X-linked mental retardation with α-thalassemia (ATR-X syndrome). Cell 1995, 80, 837–845. [Google Scholar] [CrossRef]
- Liu, X.Y.; Gerges, N.; Korshunov, A.; Sabha, N.; Khuong-Quang, D.A.; Fontebasso, A.M.; Fleming, A.; Hadjadj, D.; Schwartzentruber, J.; Majewski, J.; et al. Frequent ATRX mutations and loss of expression in adult diffuse astrocytic tumors carrying IDH1/IDH2 and TP53 mutations. Acta Neuropathol. 2012, 124, 615–625. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Shi, C.; Edil, B.H.; De Wilde, R.F.; Klimstra, D.S.; Maitra, A.; Schulick, R.D.; Tang, L.H.; Wolfgang, C.L.; Choti, M.A.; et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 2011, 331, 1199–1203. [Google Scholar] [CrossRef]
- Jiao, Y.; Killela, P.J.; Reitman, Z.J.; Rasheed, B.A.; Heaphy, C.M.; de Wilde, R.F.; Rodriguez, F.J.; Rosemberg, S.; Oba-Shinjo, S.M.; Marie, S.K.N.; et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget 2012, 3, 709–722. [Google Scholar] [CrossRef]
- Schwartzentruber, J.; Korshunov, A.; Liu, X.Y.; Jones, D.T.W.; Pfaff, E.; Jacob, K.; Sturm, D.; Fontebasso, A.M.; Quang, D.A.K.; Tönjes, M.; et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 2012, 482, 226–231. [Google Scholar] [CrossRef]
- Kannan, K.; Inagaki, A.; Silber, J.; Gorovets, D.; Zhang, J.; Kastenhuber, E.R.; Heguy, A.; Petrini, J.H.; Chan, T.A.; Huse, J.T. Whole exome sequencing identifies ATRX mutation as a key molecular determinant in lower-grade glioma. Oncotarget 2012, 3, 1194–1204. [Google Scholar] [CrossRef]
- Molenaar, J.J.; Koster, J.; Zwijnenburg, D.A.; Van Sluis, P.; Valentijn, L.J.; Van Der Ploeg, I.; Hamdi, M.; Van Nes, J.; Westerman, B.A.; Van Arkel, J.; et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 2012, 483, 589–593. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Bahrami, A.; Pappo, A.; Easton, J.; Dalton, J.; Hedlund, E.; Ellison, D.; Shurtleff, S.; Wu, G.; Wei, L.; et al. Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep. 2014, 7, 104–112. [Google Scholar] [CrossRef] [PubMed]
- De Vitis, M.; Berardinelli, F.; Sgura, A. Telomere length maintenance in cancer: At the crossroad between telomerase and alternative lengthening of telomeres (ALT). Int. J. Mol. Sci. 2018, 19, 606. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- COSMIC|Catalogue of Somatic Mutations in Cancer. Available online: https://cancer.sanger.ac.uk/cosmic (accessed on 16 April 2021).
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef]
- Villard, L.; Bonino, M.C.; Abidi, F.; Ragusa, A.; Belougne, J.; Lossi, A.M.; Seaver, L.; Bonnefont, J.P.; Romano, C.; Fichera, M.; et al. Evaluation of a mutation screening strategy for sporadic cases of ATR-X syndrome. J. Med. Genet. 1999, 36, 183–186. [Google Scholar] [CrossRef]
- Lee, N.G.; Hong, Y.K.; Yu, S.Y.; Han, S.Y.; Geum, D.; Cho, K.S. dXNP, a Drosophila homolog of XNP/ATRX, induces apoptosis via Jun-N-terminal kinase activation. FEBS Lett. 2007, 581, 2625–2632. [Google Scholar] [CrossRef]
- Gecz, J.; Pollaord, H.; Consalez, G.; Villard, L.; Stayton, C.; Millasseau, P.; Khrestchatisky, M.; Fontes, M. Cloning and expression of the murine homologue of a putative human x-linked nuclear protein gene closely linked to PGK1 in Xq13.3. Hum. Mol. Genet. 1994, 3, 39–44. [Google Scholar] [CrossRef]
- Villard, L.; Lossi, A.M.; Cardoso, C.; Proud, V.; Chiaroni, P.; Colleaux, L.; Schwartz, C.; Fontés, M. Determination of the genomic structure of the XNP/ATRX gene encoding a potential zinc finger helicase. Genomics 1997, 43, 149–155. [Google Scholar] [CrossRef]
- Picketts, D.J.; Tastan, A.O.; Higgs, D.R.; Gibbons, R.J. Comparison of the human and murine ATRX gene identifies highly conserved, functionally important domains. Mamm. Genome 1998, 9, 400–403. [Google Scholar] [CrossRef]
- Garrick, D.; Samara, V.; McDowell, T.L.; Smith, A.J.H.; Dobbie, L.; Higgs, D.R.; Gibbons, R.J. A conserved truncated isoform of the ATR-X syndrome protein lacking the SWI/SNF-homology domain. Gene 2004, 326, 23–34. [Google Scholar] [CrossRef]
- Bérubé, N.G. ATRX in chromatin assembly and genome architecture during development and disease. Biochem. Cell Biol. 2011, 89, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Wu, S.; Liu, H.; Stratt, R.; Barak, O.G.; Shiekhattar, R.; Picketts, D.J.; Yang, X. A Novel Transcription Regulatory Complex Containing Death Domain-associated Protein and the ATR-X Syndrome Protein. J. Biol. Chem. 2004, 279, 20369–20377. [Google Scholar] [CrossRef] [PubMed]
- Hoelper, D.; Huang, H.; Jain, A.Y.; Patel, D.J.; Lewis, P.W. Structural and mechanistic insights into ATRX-dependent and -independent functions of the histone chaperone DAXX. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef]
- Haase, S.; Garcia-fabiani, M.B.; Carney, S.; Altshuler, D.; Núñez, F.J.; Méndez, F.M.; Núñez, F.; Lowenstein, P.R.; Castro, M.G. Mutant ATRX: Uncovering a new therapeutic target for glioma. Expert Opin. Ther. Targets 2018, 22, 599–613. [Google Scholar] [CrossRef]
- Ren, W.; Medeiros, N.; Warneford-Thomson, R.; Wulfridge, P.; Yan, Q.; Bian, J.; Sidoli, S.; Garcia, B.A.; Skordalakes, E.; Joyce, E.; et al. Disruption of ATRX-RNA interactions uncovers roles in ATRX localization and PRC2 function. Nat. Commun. 2020, 11, 1–15. [Google Scholar] [CrossRef]
- UniProt. Available online: https://www.uniprot.org/ (accessed on 26 April 2021).
- Cardoso, C.; Lutz, Y.; Mignon, C.; Compe, E.; Depetris, D.; Mattei, M.G.; Fontes, M.; Colleaux, L. ATR-X mutations cause impaired nuclear location and altered DNA binding properties of the XNP/ATR-X protein. J. Med. Genet. 2000, 37, 746–751. [Google Scholar] [CrossRef] [PubMed]
- Schindler, U.; Beckmann, H.; Cashmore, A.R. HAT3.1, a novel Arabidopsis homeodomain protein containing a conserved cysteine-rich region. Plant. J. 1993, 4, 137–150. [Google Scholar] [CrossRef]
- Gibbons, R.J.; Bachoo, S.; Picketts, D.J.; Aftimos, S.; Asenbauer, B.; Bergoffen, J.; Berry, S.A.; Dahl, N.; Fryer, A.; Keppler, K.; et al. Mutations in transcriptional regulator ATRX establish the functional significance of a PHD-like domain. Nat. Genet. 1997, 17, 146–148. [Google Scholar] [CrossRef]
- Xie, S.; Wang, Z.; Okano, M.; Nogami, M.; Li, Y.; He, W.W.; Okumura, K.; Li, E. Cloning, expression and chromosome locations of the human DNMT3 gene family. Gene 1999, 236, 87–95. [Google Scholar] [CrossRef]
- Aapola, U.; Shibuya, K.; Scott, H.S.; Ollila, J.; Vihinen, M.; Heino, M.; Shintani, A.; Kawasaki, K.; Minoshima, S.; Krohn, K.; et al. Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics 2000, 65, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Ratnakumar, K.; Bernstein, E. ATRX: The case of a peculiar chromatin remodeler. Epigenetics 2013, 8, 3–9. [Google Scholar] [CrossRef]
- Heaphy, C.M.; De Wilde, R.F.; Jiao, Y.; Klein, A.P.; Edil, B.H.; Shi, C.; Bettegowda, C.; Rodriguez, F.J.; Eberhart, C.G.; Hebbar, S.; et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science 2011, 333, 425. [Google Scholar] [CrossRef] [PubMed]
- Dyer, M.A.; Qadeer, Z.A.; Valle-Garcia, D.; Bernstein, E. ATRX and DAXX: Mechanisms and mutations. Cold Spring Harb. Perspect. Med. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Gibbons, R.; Yan, Z.; Yang, D.; McDowell, T.L.; Sechi, S.; Qin, J.; Zhou, S.; Higgs, D.; Wang, W. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc. Natl. Acad. Sci. USA 2003, 100, 10635–10640. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, C.; Timsit, S.; Villard, L.; Khrestchatisky, M.; Fontès, M.; Colleaux, L. Specific interaction between the XNP/ATR-X gene product and the SET domain of the human EZH2 protein. Hum. Mol. Genet. 1998, 7, 679–684. [Google Scholar] [CrossRef]
- Le Douarin, B.; Nielsen, A.L.; Garnier, J.M.; Ichinose, H.; Jeanmougin, F.; Losson, R.; Chambon, P. A possible involvement of TIF1α, and TIF1β in the epigenetic control of transcription by nuclear receptors. EMBO J. 1996, 15, 6701–6715. [Google Scholar] [CrossRef]
- Nan, X.; Hou, J.; Maclean, A.; Nasir, J.; Lafuente, M.J.; Shu, X.; Kriaucionis, S.; Bird, A. Interaction between chromatin proteins MECP2 and ATRX is disrupted by mutations that cause inherited mental retardation. Proc. Natl. Acad. Sci. USA 2007, 104, 2709–2714. [Google Scholar] [CrossRef]
- Raghunandan, M.; Yeo, J.E.; Walter, R.; Saito, K.; Harvey, A.J.; Ittershagen, S.; Lee, E.A.; Yang, J.; Hoatlin, M.E.; Bielinsky, A.K.; et al. Functional cross talk between the Fanconi anemia and ATRX/DAXX histone chaperone pathways promotes replication fork recovery. Hum. Mol. Genet. 2020, 29, 1083–1095. [Google Scholar] [CrossRef]
- Huh, M.S.; Ivanochko, D.; Hashem, L.E.; Curtin, M.; Delorme, M.; Goodall, E.; Yan, K.; Picketts, D.J. Stalled replication forks within heterochromatin require ATRX for protection. Cell Death Dis. 2016, 7. [Google Scholar] [CrossRef]
- Leung, J.W.C.; Ghosal, G.; Wang, W.; Shen, X.; Wang, J.; Li, L.; Chen, J. Alpha thalassemia/mental retardation syndrome X-linked gene product ATRX is required for proper replication restart and cellular resistance to replication stress. J. Biol. Chem. 2013, 288, 6342–6350. [Google Scholar] [CrossRef] [PubMed]
- Juhász, S.; Elbakry, A.; Mathes, A.; Löbrich, M. ATRX Promotes DNA Repair Synthesis and Sister Chromatid Exchange during Homologous Recombination. Mol. Cell 2018, 71, 11–24.e7. [Google Scholar] [CrossRef] [PubMed]
- Ratnakumar, K.; Duarte, L.F.; LeRoy, G.; Hasson, D.; Smeets, D.; Vardabasso, C.; Bönisch, C.; Zeng, T.; Xiang, B.; Zhang, D.Y.; et al. ATRX-mediated chromatin association of histone variant macroH2A1 regulates α-globin expression. Genes Dev. 2012, 26, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorthy, M.; Smith, S. Loss of ATRX Suppresses Resolution of Telomere Cohesion to Control Recombination in ALT Cancer Cells. Cancer Cell 2015, 28, 357–369. [Google Scholar] [CrossRef]
- Baumann, C.; De La Fuente, R. ATRX marks the inactive X chromosome (Xi) in somatic cells and during imprinted X chromosome inactivation in trophoblast stem cells. Chromosoma 2009, 118, 209–222. [Google Scholar] [CrossRef]
- Sarma, K.; Cifuentes-Rojas, C.; Ergun, A.; Del Rosario, A.; Jeon, Y.; White, F.; Sadreyev, R.; Lee, J.T. ATRX directs binding of PRC2 to Xist RNA and Polycomb targets. Cell 2014, 159, 869–883. [Google Scholar] [CrossRef]
- Chu, H.P.; Cifuentes-Rojas, C.; Kesner, B.; Aeby, E.; Lee, H.G.; Wei, C.; Oh, H.J.; Boukhali, M.; Haas, W.; Lee, J.T. TERRA RNA Antagonizes ATRX and Protects Telomeres. Cell 2017, 170, 86–101.e16. [Google Scholar] [CrossRef]
- Park, J.; Lee, H.; Han, N.; Kwak, S.; Lee, H.T.; Kim, J.H.; Kang, K.; Youn, B.H.; Yang, J.H.; Jeong, H.J.; et al. Long non-coding RNA ChRO1 facilitates ATRX/DAXX-dependent H3.3 deposition for transcription-associated heterochromatin reorganization. Nucleic Acids Res. 2018, 46, 11759–11775. [Google Scholar] [CrossRef]
- Iwase, S.; Xiang, B.; Ghosh, S.; Ren, T.; Lewis, P.W.; Cochrane, J.C.; Allis, C.D.; Picketts, D.J.; Patel, D.J.; Li, H.; et al. ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome. Nat. Struct. Mol. Biol. 2011, 18, 769–776. [Google Scholar] [CrossRef]
- Dhayalan, A.; Tamas, R.; Bock, I.; Tattermusch, A.; Dimitrova, E.; Kudithipudi, S.; Ragozin, S.; Jeltsch, A. The ATRX-ADD domain binds to H3 tail peptides and reads the combined methylation state of K4 and K9. Hum. Mol. Genet. 2011, 20, 2195–2203. [Google Scholar] [CrossRef]
- Kernohan, K.D.; Jiang, Y.; Tremblay, D.C.; Bonvissuto, A.C.; Eubanks, J.H.; Mann, M.R.W.; Bérubé, N.G. ATRX Partners with Cohesin and MeCP2 and Contributes to Developmental Silencing of Imprinted Genes in the Brain. Dev. Cell 2010, 18, 191–202. [Google Scholar] [CrossRef]
- Valle-García, D.; Qadeer, Z.A.; McHugh, D.S.; Ghiraldini, F.G.; Chowdhury, A.H.; Hasson, D.; Dyer, M.A.; Recillas-Targa, F.; Bernstein, E. ATRX binds to atypical chromatin domains at the 3′ exons of zinc finger genes to preserve H3K9me3 enrichment. Epigenetics 2016, 11, 398–414. [Google Scholar] [CrossRef]
- Goldberg, A.D.; Banaszynski, L.A.; Noh, K.M.; Lewis, P.W.; Elsaesser, S.J.; Stadler, S.; Dewell, S.; Law, M.; Guo, X.; Li, X.; et al. Distinct Factors Control Histone Variant H3.3 Localization at Specific Genomic Regions. Cell 2010, 140, 678–691. [Google Scholar] [CrossRef]
- Law, M.J.; Lower, K.M.; Voon, H.P.J.; Hughes, J.R.; Garrick, D.; Viprakasit, V.; Mitson, M.; De Gobbi, M.; Marra, M.; Morris, A.; et al. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell 2010, 143, 367–378. [Google Scholar] [CrossRef]
- Voon, H.P.J.; Hughes, J.R.; Rode, C.; DeLaRosa-Velázquez, I.A.; Jenuwein, T.; Feil, R.; Higgs, D.R.; Gibbons, R.J. ATRX Plays a Key Role in Maintaining Silencing at Interstitial Heterochromatic Loci and Imprinted Genes. Cell Rep. 2015, 11, 405–418. [Google Scholar] [CrossRef]
- Thiriet, C.; Hayes, J.J. Histone dynamics during transcription: Exchange of H2A/H2B dimers and H3/H4 tetramers during pol II elongation. Results Probl. Cell Differ. 2006, 41, 77–90. [Google Scholar]
- Hamiche, A.; Shuaib, M. Chaperoning the histone H3 family. Biochim. Biophys. Acta Gene Regul. Mech. 2012, 1819, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Sauer, P.V.; Gu, Y.; Liu, W.H.; Mattiroli, F.; Panne, D.; Luger, K.; Churchill, M.E.A. Mechanistic insights into histone deposition and nucleosome assembly by the chromatin assembly factor-1. Nucleic Acids Res. 2018, 46, 9907–9917. [Google Scholar] [CrossRef] [PubMed]
- Drané, P.; Ouararhni, K.; Depaux, A.; Shuaib, M.; Hamiche, A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010, 24, 1253–1265. [Google Scholar] [CrossRef] [PubMed]
- Lewis, P.W.; Elsaesser, S.J.; Noh, K.M.; Stadler, S.C.; Allis, C.D. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. USA 2010, 107, 14075–14080. [Google Scholar] [CrossRef]
- Eustermann, S.; Yang, J.C.; Law, M.J.; Amos, R.; Chapman, L.M.; Jelinska, C.; Garrick, D.; Clynes, D.; Gibbons, R.J.; Rhodes, D.; et al. Combinatorial readout of histone H3 modifications specifies localization of ATRX to heterochromatin. Nat. Struct. Mol. Biol. 2011, 18, 777–782. [Google Scholar] [CrossRef] [PubMed]
- Baumann, C.; Viveiros, M.M.; De La Fuente, R. Loss of maternal ATRX results in centromere instability and aneuploidy in the mammalian oocyte and Pre- implantation embryo. PLoS Genet. 2010, 6. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.H.; McGhie, J.D.; Sim, M.; Anderson, M.A.; Ahn, S.; Hannan, R.D.; George, A.J.; Morgan, K.A.; Mann, J.R.; Choo, K.H.A. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010, 20, 351–360. [Google Scholar] [CrossRef]
- Ahmad, K.; Henikoff, S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 2002, 9, 1191–1200. [Google Scholar] [CrossRef]
- Tagami, H.; Ray-Gallet, D.; Almouzni, G.; Nakatani, Y. Histone H3.1 and H3.3 Complexes Mediate Nucleosome Assembly Pathways Dependent or Independent of DNA Synthesis. Cell 2004, 116, 51–61. [Google Scholar] [CrossRef]
- Maul, G.G.; Negorev, D.; Bell, P.; Ishov, A.M. Review: Properties and assembly mechanisms of ND10, PML2 bodies, or PODs. J. Struct. Biol. 2000, 129, 278–287. [Google Scholar] [CrossRef]
- Bernardi, R.; Pandolfi, P.P. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 2007, 8, 1006–1016. [Google Scholar] [CrossRef]
- De Thé, H.; Le Bras, M.; Lallemand-Breitenbach, V. Acute promyelocytic leukemia, arsenic, and PML bodies. J. Cell Biol. 2012, 198, 11–21. [Google Scholar] [CrossRef]
- Delbarre, E.; Ivanauskiene, K.; Küntziger, T.; Collas, P. DAXX-dependent supply of soluble (H3.3-H4) dimers to PML bodies pending deposition into chromatin. Genome Res. 2013, 23, 440–451. [Google Scholar] [CrossRef]
- Ishov, A.M.; Sotnikov, A.G.; Negorev, D.; Vladimirova, O.V.; Neff, N.; Kamitani, T.; Yeh, E.T.H.; Strauss, J.F.; Maul, G.G. PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 1999, 147, 221–233. [Google Scholar] [CrossRef]
- Wang, J.; Shiels, C.; Sasieni, P.; Wu, P.J.; Islam, S.A.; Freemont, P.S.; Sheer, D. Promyelocytic leukemia nuclear bodies associate with transcriptionally active genomic regions. J. Cell Biol. 2004, 164, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Tavalai, N.; Stamminger, T. New insights into the role of the subnuclear structure ND10 for viral infection. Biochim. Biophys. Acta Mol. Cell Res. 2008, 1783, 2207–2221. [Google Scholar] [CrossRef] [PubMed]
- Di Masi, A.; Cilli, D.; Berardinelli, F.; Talarico, A.; Pallavicini, I.; Pennisi, R.; Leone, S.; Antoccia, A.; Noguera, N.I.; Lo-Coco, F.; et al. PML nuclear body disruption impairs DNA double-strand break sensing and repair in APL. Cell Death Dis. 2016, 7, e2308. [Google Scholar] [CrossRef] [PubMed]
- Niwa-Kawakita, M.; Ferhi, O.; Soilihi, H.; Le Bras, M.; Lallemand-Breitenbach, V.; de Thé, H. PML is a ROS sensor activating p53 upon oxidative stress. J. Exp. Med. 2017, 214, 3197–3206. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.R.; Munkhjargal, A.; Kim, M.J.; Park, S.Y.; Jung, E.; Ryu, J.H.; Yang, Y.; Lim, J.S.; Kim, Y. The functional roles of PML nuclear bodies in genome maintenance. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2018, 809, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.H.; Ren, H.; Williams, E.; McGhie, J.; Ann, S.; Sim, M.; Tam, A.; Earle, E.; Anderson, M.A.; Mann, J.; et al. Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome Res. 2009, 19, 404–414. [Google Scholar] [CrossRef]
- Cheng, X.; Guo, S.; Liu, Y.; Chu, H.; Hakimi, P.; Berger, N.A.; Hanson, R.W.; Kao, H.Y. Ablation of promyelocytic leukemia protein (PML) re-patterns energy balance and protects mice from obesity induced by a western diet. J. Biol. Chem. 2013, 288, 29746–29759. [Google Scholar] [CrossRef]
- Delbarre, E.; Ivanauskiene, K.; Spirkoski, J.; Shah, A.; Vekterud, K.; Moskaug, J.; Bøe, S.O.; Wong, L.H.; Küntziger, T.; Collas, P. PML protein organizes heterochromatin domains where it regulates histone H3.3 deposition by ATRX/DAXX. Genome Res. 2017, 27, 913–921. [Google Scholar] [CrossRef]
- Danussi, C.; Bose, P.; Parthasarathy, P.T.; Silberman, P.C.; Van Arnam, J.S.; Vitucci, M.; Tang, O.Y.; Heguy, A.; Wang, Y.; Chan, T.A.; et al. Atrx inactivation drives disease-defining phenotypes in glioma cells of origin through global epigenomic remodeling. Nat. Commun. 2018, 9, 1057. [Google Scholar] [CrossRef]
- Han, B.; Meng, X.; Wu, P.; Li, Z.; Li, S.; Zhang, Y.; Zha, C.; Ye, Q.; Jiang, C.; Cai, J.; et al. ATRX/EZH2 complex epigenetically regulates FADD/PARP1 axis, contributing to TMZ resistance in glioma. Theranostics 2020, 10, 3351–3365. [Google Scholar] [CrossRef]
- Oppel, F.; Tao, T.; Shi, H.; Ross, K.N.; Zimmerman, M.W.; He, S.; Tong, G.; Aster, J.C.; Look, A.T. Loss of atrx cooperates with p53-deficiency to promote the development of sarcomas and other malignancies. PLoS Genet. 2019, 15, e1008039. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Liu, H.; Li, G.; Qian, J.; Gao, R.; Zhou, Y.; Wang, X. Global changes in chromatin accessibility and transcription following ATRX inactivation in human cancer cells. FEBS Lett. 2020, 594, 67–78. [Google Scholar] [CrossRef]
- De La Fuente, R.; Baumann, C.; Viveiros, M.M. ATRX contributes to epigenetic asymmetry and silencing of major satellite transcripts in the maternal genome of the mouse embryo. Development 2015, 142, 1806–1817. [Google Scholar] [CrossRef] [PubMed]
- Flynn, R.L.; Cox, K.E.; Jeitany, M.; Wakimoto, H.; Bryll, A.R.; Ganem, N.J.; Bersani, F.; Pineda, J.R.; Suvà, M.L.; Benes, C.H.; et al. Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science 2015, 347, 273–277. [Google Scholar] [CrossRef]
- Voon, H.P.J.; Wong, L.H. New players in heterochromatin silencing: Histone variant H3.3 and the ATRX/DAXX chaperone. Nucleic Acids Res. 2016, 44, 1496–1501. [Google Scholar] [CrossRef]
- Fioriniello, S.; Marano, D.; Fiorillo, F.; D’esposito, M.; Ragione, F. Della Epigenetic factors that control pericentric heterochromatin organization in mammals. Genes 2020, 11, 595. [Google Scholar] [CrossRef]
- Peters, A.H.F.M.; Kubicek, S.; Mechtler, K.; O’Sullivan, R.J.; Derijck, A.A.H.A.; Perez-Burgos, L.; Kohlmaier, A.; Opravil, S.; Tachibana, M.; Shinkai, Y.; et al. Partitioning and Plasticity of Repressive Histone Methylation States in Mammalian Chromatin. Mol. Cell 2003, 12, 1577–1589. [Google Scholar] [CrossRef]
- Rao, V.K.; Pal, A.; Taneja, R. A drive in SUVs: From development to disease. Epigenetics 2017, 12, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Müller-Ott, K.; Erdel, F.; Matveeva, A.; Mallm, J.; Rademacher, A.; Hahn, M.; Bauer, C.; Zhang, Q.; Kaltofen, S.; Schotta, G.; et al. Specificity, propagation, and memory of pericentric heterochromatin. Mol. Syst. Biol. 2014, 10, 746. [Google Scholar] [CrossRef]
- Probst, A.V.; Almouzni, G. Pericentric heterochromatin: Dynamic Organization during early Development in Mammals. Differentiation 2008, 76, 15–23. [Google Scholar] [CrossRef]
- Almouzni, G.; Probst, A.V. Heterochromatin maintenance and establishment: Lessons from the mouse pericentromere. Nucleus 2011, 2. [Google Scholar] [CrossRef]
- Yamamoto, K.; Sonoda, M. Self-interaction of heterochromatin protein 1 is required for direct binding to histone methyltransferase, SUV39H1. Biochem. Biophys. Res. Commun. 2003, 301, 287–292. [Google Scholar] [CrossRef]
- Maison, C.; Almouzni, G. HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell Biol. 2004, 5, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Maison, C.; Bailly, D.; Roche, D.; De Oca, R.M.; Probst, A.V.; Vassias, I.; Dingli, F.; Lombard, B.; Loew, D.; Quivy, J.P.; et al. SUMOylation promotes de novo targeting of HP1 ± to pericentric heterochromatin. Nat. Genet. 2011, 43, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Saksouk, N.; Barth, T.K.; Ziegler-Birling, C.; Olova, N.; Nowak, A.; Rey, E.; Mateos-Langerak, J.; Urbach, S.; Reik, W.; Torres-Padilla, M.E.; et al. Redundant Mechanisms to Form Silent Chromatin at Pericentromeric Regions Rely on BEND3 and DNA Methylation. Mol. Cell 2014, 56, 580–594. [Google Scholar] [CrossRef]
- Marano, D.; Fioriniello, S.; Fiorillo, F.; Gibbons, R.J.; D’esposito, M.; Ragione, F. Della ATRX contributes to MECP2-mediated pericentric heterochromatin organization during neural differentiation. Int. J. Mol. Sci. 2019, 20, 5371. [Google Scholar] [CrossRef] [PubMed]
- Bérubé, N.G.; Smeenk, C.A.; Picketts, D.J. Cell cycle-dependent phosphorylation of the ATRX protein correlates with changes in nuclear matrix and chromatin association. Hum. Mol. Genet. 2000, 9, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Lechner, M.S.; Schultz, D.C.; Negorev, D.; Maul, G.G.; Rauscher, F.J. The mammalian heterochromatin protein 1 binds diverse nuclear proteins through a common motif that targets the chromoshadow domain. Biochem. Biophys. Res. Commun. 2005, 331, 929–937. [Google Scholar] [CrossRef]
- McDowell, T.L.; Gibbons, R.J.; Sutherland, H.; O’Rourke, D.M.; Bickmore, W.A.; Pombo, A.; Turley, H.; Gatter, K.; Picketts, D.J.; Buckle, V.J.; et al. Localization of a putative transcriptional regulator (ATRX) at pericentromeric heterochromatin and the short arms of acrocentric chromosomes. Proc. Natl. Acad. Sci. USA 1999, 96, 13983–13988. [Google Scholar] [CrossRef] [PubMed]
- García-Cao, M.; O’Sullivan, R.; Peters, A.H.F.M.; Jenuwein, T.; Blasco, M.A. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat. Genet. 2003, 36, 94–99. [Google Scholar] [CrossRef]
- Gonzalo, S.; García-Cao, M.; Fraga, M.F.; Schotta, G.; Peters, A.H.F.M.; Cotter, S.E.; Eguía, R.; Dean, D.C.; Esteller, M.; Jenuwein, T.; et al. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat. Cell Biol. 2005, 7, 420–428. [Google Scholar] [CrossRef]
- Gonzalo, S.; Jaco, I.; Fraga, M.F.; Chen, T.; Li, E.; Esteller, M.; Blasco, M.A. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat. Cell Biol. 2006, 8, 416–424. [Google Scholar] [CrossRef] [PubMed]
- Blasco, M.A. The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet. 2007, 8, 299–309. [Google Scholar] [CrossRef]
- Gauchier, M.; Kan, S.; Barral, A.; Sauzet, S.; Agirre, E.; Bonnell, E.; Saksouk, N.; Barth, T.K.; Ide, S.; Urbach, S.; et al. SETDB1-dependent heterochromatin stimulates alternative lengthening of telomeres. Sci. Adv. 2019, 5. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Hills, M.; Conomos, D.; Stutz, M.D.; Dagg, R.A.; Lau, L.M.S.; Reddel, R.R.; Pickett, H.A. Telomere extension by telomerase and ALT generates variant repeats by mechanistically distinct processes. Nucleic Acids Res. 2014, 42, 1733–1746. [Google Scholar] [CrossRef] [PubMed]
- Udroiu, I.; Sgura, A. Alternative lengthening of telomeres and chromatin status. Genes 2020, 11, 45. [Google Scholar] [CrossRef] [PubMed]
- Schultz, D.C.; Ayyanathan, K.; Negorev, D.; Maul, G.G.; Rauscher, F.J. SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002, 16, 919–932. [Google Scholar] [CrossRef]
- Negishi, Y.; Kawaji, H.; Minoda, A.; Usui, K. Identification of chromatin marks at TERRA promoter and encoding region. Biochem. Biophys. Res. Commun. 2015, 467, 1052–1057. [Google Scholar] [CrossRef]
- Bandaria, J.N.; Qin, P.; Berk, V.; Chu, S.; Yildiz, A. Shelterin protects chromosome ends by compacting telomeric chromatin. Cell 2016, 164, 735–746. [Google Scholar] [CrossRef]
- Cubiles, M.D.; Barroso, S.; Vaquero-Sedas, M.I.; Enguix, A.; Aguilera, A.; Vega-Palas, M.A. Epigenetic features of human telomeres. Nucleic Acids Res. 2018, 46, 2347–2355. [Google Scholar] [CrossRef]
- Benetti, R.; Gonzalo, S.; Jaco, I.; Schotta, G.; Klatt, P.; Jenuwein, T.; Blasco, M.A. Suv4-20h deficiency results in telomere elongation and derepression of telomere recombination. J. Cell Biol. 2007, 178, 925–936. [Google Scholar] [CrossRef] [PubMed]
- Benetti, R.; Gonzalo, S.; Jaco, I.; Muñoz, P.; Gonzalez, S.; Schoeftner, S.; Murchison, E.; Andl, T.; Chen, T.; Klatt, P.; et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat. Struct. Mol. Biol. 2008, 15, 268–279. [Google Scholar] [CrossRef] [PubMed]
- Episkopou, H.; Draskovic, I.; Van Beneden, A.; Tilman, G.; Mattiussi, M.; Gobin, M.; Arnoult, N.; Londoño-Vallejo, A.; Decottignies, A. Alternative Lengthening of Telomeres is characterized by reduced compaction of telomeric chromatin. Nucleic Acids Res. 2014, 42, 4391–4405. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Deng, Z.; Zhang, L.; Wu, C.; Jin, Y.; Hwang, I.; Vladimirova, O.; Xu, L.; Yang, L.; Lu, B.; et al. ATRX loss induces telomere dysfunction and necessitates induction of alternative lengthening of telomeres during human cell immortalization. EMBO J. 2019, 38, e96659. [Google Scholar] [CrossRef] [PubMed]
- Montero, J.J.; López-Silanes, I.; Megías, D.; Fraga, M.; Castells-García, Á.; Blasco, M.A. TERRA recruitment of polycomb to telomeres is essential for histone trymethylation marks at telomeric heterochromatin. Nat. Commun. 2018, 9, 1–14. [Google Scholar] [CrossRef]
- Nguyen, D.T.; Voon, H.P.J.; Xella, B.; Scott, C.; Clynes, D.; Babbs, C.; Ayyub, H.; Kerry, J.; Sharpe, J.A.; Sloane-Stanley, J.A.; et al. The chromatin remodelling factor ATRX suppresses R-loops in transcribed telomeric repeats. EMBO Rep. 2017, 18, 914–928. [Google Scholar] [CrossRef]
- Clynes, D.; Jelinska, C.; Xella, B.; Ayyub, H.; Taylor, S.; Mitson, M.; Bachrati, C.Z.; Higgs, D.R.; Gibbons, R.J. ATRX Dysfunction Induces Replication Defects in Primary Mouse Cells. PLoS ONE 2014, 9, e92915. [Google Scholar] [CrossRef]
- Schlacher, K.; Wu, H.; Jasin, M. A Distinct Replication Fork Protection Pathway Connects Fanconi Anemia Tumor Suppressors to RAD51-BRCA1/2. Cancer Cell 2012, 22, 106–116. [Google Scholar] [CrossRef]
- Chaudhury, I.; Sareen, A.; Raghunandan, M.; Sobeck, A. FANCD2 regulates BLM complex functions independently of FANCI to promote replication fork recovery. Nucleic Acids Res. 2013, 41, 6444–6459. [Google Scholar] [CrossRef]
- Sato, K.; Shimomuki, M.; Katsuki, Y.; Takahashi, D.; Kobayashi, W.; Ishiai, M.; Miyoshi, H.; Takata, M.; Kurumizaka, H. FANCI-FANCD2 stabilizes the RAD51-DNA complex by binding RAD51 and protects the 5′-DNA end. Nucleic Acids Res. 2016, 44, 10758–10771. [Google Scholar] [CrossRef]
- Elbakry, A.; Juhász, S.; Chan, K.C.; Löbrich, M. ATRX and RECQ5 define distinct homologous recombination subpathways. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Heyer, W.D.; Ehmsen, K.T.; Liu, J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010, 44, 113–139. [Google Scholar] [CrossRef]
- Mehta, A.; Haber, J.E. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Chun, J.; Powell, S.N. BRCA1 and BRCA2: Different roles in a common pathway of genome protection. Nat. Rev. Cancer 2012, 12, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Sullivan-Reed, K.; Bolton-Gillespie, E.; Dasgupta, Y.; Langer, S.; Siciliano, M.; Nieborowska-Skorska, M.; Hanamshet, K.; Belyaeva, E.A.; Bernhardy, A.J.; Lee, J.; et al. Simultaneous Targeting of PARP1 and RAD52 Triggers Dual Synthetic Lethality in BRCA-Deficient Tumor Cells. Cell Rep. 2018, 23, 3127–3136. [Google Scholar] [CrossRef] [PubMed]
- Paeschke, K.; Capra, J.A.; Zakian, V.A. DNA Replication through G-Quadruplex Motifs Is Promoted by the Saccharomyces cerevisiae Pif1 DNA Helicase. Cell 2011, 145, 678–691. [Google Scholar] [CrossRef]
- Sarkies, P.; Reams, C.; Simpson, L.J.; Sale, J.E. Epigenetic Instability due to Defective Replication of Structured DNA. Mol. Cell 2010, 40, 703–713. [Google Scholar] [CrossRef]
- Clynes, D.; Higgs, D.R.; Gibbons, R.J. The chromatin remodeller ATRX: A repeat offender in human disease. Trends Biochem. Sci. 2013, 38, 461–466. [Google Scholar] [CrossRef]
- Lewis, P.W.; Müller, M.M.; Koletsky, M.S.; Cordero, F.; Lin, S.; Banaszynski, L.A.; Garcia, B.A.; Muir, T.W.; Becher, O.J.; Allis, C.D. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 2013, 340, 857–861. [Google Scholar] [CrossRef]
- Levy, M.A.; Kernohan, K.D.; Jiang, Y.; Bérubé, N.G. ATRX promotes gene expression by facilitating transcriptional elongation through guanine-rich coding regions. Hum. Mol. Genet. 2014, 24, 1824–1835. [Google Scholar] [CrossRef]
- Özer, Ö.; Hickson, I.D. Pathways for maintenance of telomeres and common fragile sites during DNA replication stress. Open Biol. 2018, 8, 180018. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, A.; García-Muse, T. R Loops: From Transcription Byproducts to Threats to Genome Stability. Mol. Cell 2012, 46, 115–124. [Google Scholar] [CrossRef]
- Duquette, M.L.; Handa, P.; Vincent, J.A.; Taylor, A.F.; Maizels, N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004, 18, 1618–1629. [Google Scholar] [CrossRef]
- Sarkies, P.; Murat, P.; Phillips, L.G.; Patel, K.J.; Balasubramanian, S.; Sale, J.E. FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DNA. Nucleic Acids Res. 2012, 40, 1485–1498. [Google Scholar] [CrossRef]
- Schwab, R.A.; Nieminuszczy, J.; Shah, F.; Langton, J.; Lopez Martinez, D.; Liang, C.C.; Cohn, M.A.; Gibbons, R.J.; Deans, A.J.; Niedzwiedz, W. The Fanconi Anemia Pathway Maintains Genome Stability by Coordinating Replication and Transcription. Mol. Cell 2015, 60, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Arora, R.; Lee, Y.; Wischnewski, H.; Brun, C.M.; Schwarz, T.; Azzalin, C.M. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Clynes, D.; Jelinska, C.; Xella, B.; Ayyub, H.; Scott, C.; Mitson, M.; Taylor, S.; Higgs, D.R.; Gibbons, R.J. Suppression of the alternative lengthening of telomere pathway by the chromatin remodelling factor ATRX. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef]
- Min, J.; Wright, W.E.; Shay, J.W. Alternative Lengthening of Telomeres Mediated by Mitotic DNA Synthesis Engages Break-Induced Replication Processes. Mol. Cell. Biol. 2017, 37. [Google Scholar] [CrossRef] [PubMed]
- Amato, R.; Valenzuela, M.; Berardinelli, F.; Salvati, E.; Maresca, C.; Leone, S.; Antoccia, A.; Sgura, A. G-quadruplex stabilization fuels the ALT pathway in ALT-positive osteosarcoma cells. Genes 2020, 11, 304. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Elledge, S.J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003, 300, 1542–1548. [Google Scholar] [CrossRef]
- Flynn, R.L.; Centore, R.C.; O’Sullivan, R.J.; Rai, R.; Tse, A.; Songyang, Z.; Chang, S.; Karlseder, J.; Zou, L. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature 2011, 471, 532–538. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, R.J.; Arnoult, N.; Lackner, D.H.; Oganesian, L.; Haggblom, C.; Corpet, A.; Almouzni, G.; Karlseder, J. Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nat. Struct. Mol. Biol. 2014, 21, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Dunham, M.A.; Neumann, A.A.; Fasching, C.L.; Reddel, R.R. Telomere maintenance by recombination in human cells. Nat. Genet. 2000, 26, 447–450. [Google Scholar] [CrossRef] [PubMed]
- Sjögren, C.; Nasmyth, K. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr. Biol. 2001, 11, 991–995. [Google Scholar] [CrossRef]
- Canudas, S.; Houghtaling, B.R.; Kim, J.Y.; Dynek, J.N.; Chang, W.G.; Smith, S. Protein requirements for sister telomere association in human cells. EMBO J. 2007, 26, 4867–4878. [Google Scholar] [CrossRef]
- Canudas, S.; Smith, S. Differential regulation of telomere and centromere cohesion by the Scc3 homologues SA1 and SA2, respectively, in human cells. J. Cell Biol. 2009, 187, 165–173. [Google Scholar] [CrossRef]
- Remeseiro, S.; Cuadrado, A.; Carretero, M.; Martínez, P.; Drosopoulos, W.C.; Cañamero, M.; Schildkraut, C.L.; Blasco, M.A.; Losada, A. Cohesin-SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. EMBO J. 2012, 31, 2076–2089. [Google Scholar] [CrossRef]
- Dynek, J.N.; Smith, S. Resolution of Sister Telomere Association Is Required for Progression Through Mitosis. Science 2004, 304, 97–100. [Google Scholar] [CrossRef]
- Smith, S.; Giriat, I.; Schmitt, A.; De Lange, T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 1998, 282, 1484–1487. [Google Scholar] [CrossRef]
- Kustatscher, G.; Hothorn, M.; Pugieux, C.; Scheffzek, K.; Ladurner, A.G. Splicing regulates NAD metabolite binding to histone macroH2A. Nat. Struct. Mol. Biol. 2005, 12, 624–625. [Google Scholar] [CrossRef]
- Timinszky, G.; Till, S.; Hassa, P.O.; Hothorn, M.; Kustatscher, G.; Nijmeijer, B.; Colombelli, J.; Altmeyer, M.; Stelzer, E.H.K.; Scheffzek, K.; et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat. Struct. Mol. Biol. 2009, 16, 923–929. [Google Scholar] [CrossRef]
- Lovejoy, C.A.; Takai, K.; Huh, M.S.; Picketts, D.J.; de Lange, T. ATRX affects the repair of telomeric DSBs by promoting cohesion and a DAXX-dependent activity. PLoS Biol. 2020, 18, e3000594. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Sun, C.; Tran, A.D.; Chin, P.J.; Ruiz, P.D.; Wang, K.; Gibbons, R.J.; Gamble, M.J.; Liu, Y.; Oberdoerffer, P. The macroH2A1.2 histone variant links ATRX loss to alternative telomere lengthening. Nat. Struct. Mol. Biol. 2019, 26, 213–219. [Google Scholar] [CrossRef]
- Khurana, S.; Kruhlak, M.J.; Kim, J.; Tran, A.D.; Liu, J.; Nyswaner, K.; Shi, L.; Jailwala, P.; Sung, M.H.; Hakim, O.; et al. A macrohistone variant links dynamic chromatin compaction to BRCA1-dependent genome maintenance. Cell Rep. 2014, 8, 1049–1062. [Google Scholar] [CrossRef] [PubMed]
- Bouwman, P.; Aly, A.; Escandell, J.M.; Pieterse, M.; Bartkova, J.; Van Der Gulden, H.; Hiddingh, S.; Thanasoula, M.; Kulkarni, A.; Yang, Q.; et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 2010, 17, 688–695. [Google Scholar] [CrossRef]
- Xu, Y.; Ning, S.; Wei, Z.; Xu, R.; Xu, X.; Xing, M.; Guo, R.; Xu, D. 53BP1 and BRCA1 control pathway choice for stalled replication restart. eLife 2017, 6. [Google Scholar] [CrossRef]
- Baumann, C.; Schmidtmann, A.; Muegge, K.; De La Fuente, R. Association of ATRX with pericentric heterochromatin and the Y chromosome of neonatal mouse spermatogonia. BMC Mol. Biol. 2008, 9, 29. [Google Scholar] [CrossRef]
- Clemson, C.M.; McNeil, J.A.; Willard, H.F.; Lawrence, J.B. XIST RNA paints the inactive X chromosome at interphase: Evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell Biol. 1996, 132, 259–275. [Google Scholar] [CrossRef] [PubMed]
- Disteche, C.; Berletch, J.B. X-chromosome inactivation and escape. J. Genet. 2015, 94, 591–599. [Google Scholar] [CrossRef]
- Müller, J.; Verrijzer, P. Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr. Opin. Genet. Dev. 2009, 19, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.A.; Kingston, R.E. Mechanisms of Polycomb gene silencing: Knowns and unknowns. Nat. Rev. Mol. Cell Biol. 2009, 10, 697–708. [Google Scholar] [CrossRef] [PubMed]
- Costanzi, C.; Pehrson, J.R. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature 1998, 393, 599–601. [Google Scholar] [CrossRef]
- Mermoud, J.E.; Costanzi, C.; Pehrson, J.R.; Brockdorff, N. Histone macroH2A1.2 relocates to the inactive X chromosome after initiation and propagation of X-inactivation. J. Cell Biol. 1999, 147, 1399–1408. [Google Scholar] [CrossRef] [PubMed]
- Changolkar, L.N.; Costanzi, C.; Leu, N.A.; Chen, D.; McLaughlin, K.J.; Pehrson, J.R. Developmental Changes in Histone macroH2A1-Mediated Gene Regulation. Mol. Cell. Biol. 2007, 27, 2758–2764. [Google Scholar] [CrossRef] [PubMed]
- Tanasijevic, B.; Rasmussen, T.P. X Chromosome Inactivation and Differentiation Occur Readily in ES Cells Doubly-Deficient for MacroH2A1 and MacroH2A2. PLoS ONE 2011, 6, e21512. [Google Scholar] [CrossRef] [PubMed]
- Pehrson, J.R.; Changolkar, L.N.; Costanzi, C.; Leu, N.A. Mice without MacroH2A Histone Variants. Mol. Cell. Biol. 2014, 34, 4523–4533. [Google Scholar] [CrossRef]
- Buschbeck, M.; Uribesalgo, I.; Wibowo, I.; Rué, P.; Martin, D.; Gutierrez, A.; Morey, L.; Guigó, R.; López-Schier, H.; Di Croce, L. The histone variant macroH2A is an epigenetic regulator of key developmental genes. Nat. Struct. Mol. Biol. 2009, 16, 1074–1079. [Google Scholar] [CrossRef] [PubMed]
- Gamble, M.J.; Frizzell, K.M.; Yang, C.; Krishnakumar, R.; Kraus, W.L. The histone variant macroH2A1 marks repressed autosomal chromatin, but protects a subset of its target genes from silencing. Genes Dev. 2010, 24, 21–32. [Google Scholar] [CrossRef]
- Dunford, A.; Weinstock, D.M.; Savova, V.; Schumacher, S.E.; Cleary, J.P.; Yoda, A.; Sullivan, T.J.; Hess, J.M.; Gimelbrant, A.A.; Beroukhim, R.; et al. Tumor-suppressor genes that escape from X-inactivation contribute to cancer sex bias. Nat. Genet. 2017, 49, 10–16. [Google Scholar] [CrossRef]
- Elsässer, S.J.; Noh, K.M.; Diaz, N.; Allis, C.D.; Banaszynski, L.A. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature 2015, 522, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Rowe, H.M.; Jakobsson, J.; Mesnard, D.; Rougemont, J.; Reynard, S.; Aktas, T.; Maillard, P.V.; Layard-Liesching, H.; Verp, S.; Marquis, J.; et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 2010, 463, 237–240. [Google Scholar] [CrossRef] [PubMed]
- Sadic, D.; Schmidt, K.; Groh, S.; Kondofersky, I.; Ellwart, J.; Fuchs, C.; Theis, F.J.; Schotta, G. Atrx promotes heterochromatin formation at retrotransposons. EMBO Rep. 2015, 16, 836–850. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.C.; Wong, C.W.; Liang, P.P.; Shi, M.; Cao, Y.; Rao, S.T.; Tsui, S.K.W.; Waye, M.M.Y.; Zhang, Q.; Fu, W.M.; et al. Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019, 20, 84. [Google Scholar] [CrossRef] [PubMed]
- Symer, D.E.; Connelly, C.; Szak, S.T.; Caputo, E.M.; Cost, G.J.; Parmigiani, G.; Boeke, J.D. Human L1 retrotransposition is associated with genetic instability in vivo. Cell 2002, 110, 327–338. [Google Scholar] [CrossRef]
- Catez, F.; Picard, C.; Held, K.; Gross, S.; Rousseau, A.; Theil, D.; Sawtell, N.; Labetoulle, M.; Lomonte, P. HSV-1 Genome Subnuclear Positioning and Associations with Host-Cell PML-NBs and Centromeres Regulate LAT Locus Transcription during Latency in Neurons. PLoS Pathog. 2012, 8, e1002852. [Google Scholar] [CrossRef] [PubMed]
- Everett, R.D. Interactions between DNA viruses, ND10 and the DNA damage response. Cell. Microbiol. 2006, 8, 365–374. [Google Scholar] [CrossRef]
- Everett, R.D.; Parada, C.; Gripon, P.; Sirma, H.; Orr, A. Replication of ICP0-Null Mutant Herpes Simplex Virus Type 1 Is Restricted by both PML and Sp100. J. Virol. 2008, 82, 2661–2672. [Google Scholar] [CrossRef]
- Lukashchuk, V.; Everett, R.D. Regulation of ICP0-Null Mutant Herpes Simplex Virus Type 1 Infection by ND10 Components ATRX and hDaxx. J. Virol. 2010, 84, 4026–4040. [Google Scholar] [CrossRef]
- Glass, M.; Everett, R.D. Components of Promyelocytic Leukemia Nuclear Bodies (ND10) Act Cooperatively To Repress Herpesvirus Infection. J. Virol. 2013, 87, 2174–2185. [Google Scholar] [CrossRef]
- Cohen, C.; Corpet, A.; Roubille, S.; Maroui, M.A.; Poccardi, N.; Rousseau, A.; Kleijwegt, C.; Binda, O.; Texier, P.; Sawtell, N.; et al. Promyelocytic leukemia (PML) nuclear bodies (NBs) induce latent/quiescent HSV-1 genomes chromatinization through a PML NB/Histone H3.3/H3.3 Chaperone Axis. PLoS Pathog. 2018, 14. [Google Scholar] [CrossRef]
- Cabral, J.M.; Oh, H.S.; Knipe, D.M. ATRX promotes maintenance of herpes simplex virus heterochromatin during chromatin stress. eLife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Tsai, K.; Thikmyanova, N.; Wojcechowskyj, J.A.; Delecluse, H.-J.; Lieberman, P.M. EBV Tegument Protein BNRF1 Disrupts DAXX-ATRX to Activate Viral Early Gene Transcription. PLoS Pathog. 2011, 7, e1002376. [Google Scholar] [CrossRef] [PubMed]
- Jurak, I.; Silverstein, L.B.; Sharma, M.; Coen, D.M. Herpes Simplex Virus Is Equipped with RNA- and Protein-Based Mechanisms To Repress Expression of ATRX, an Effector of Intrinsic Immunity. J. Virol. 2012, 86, 10093–10102. [Google Scholar] [CrossRef] [PubMed]
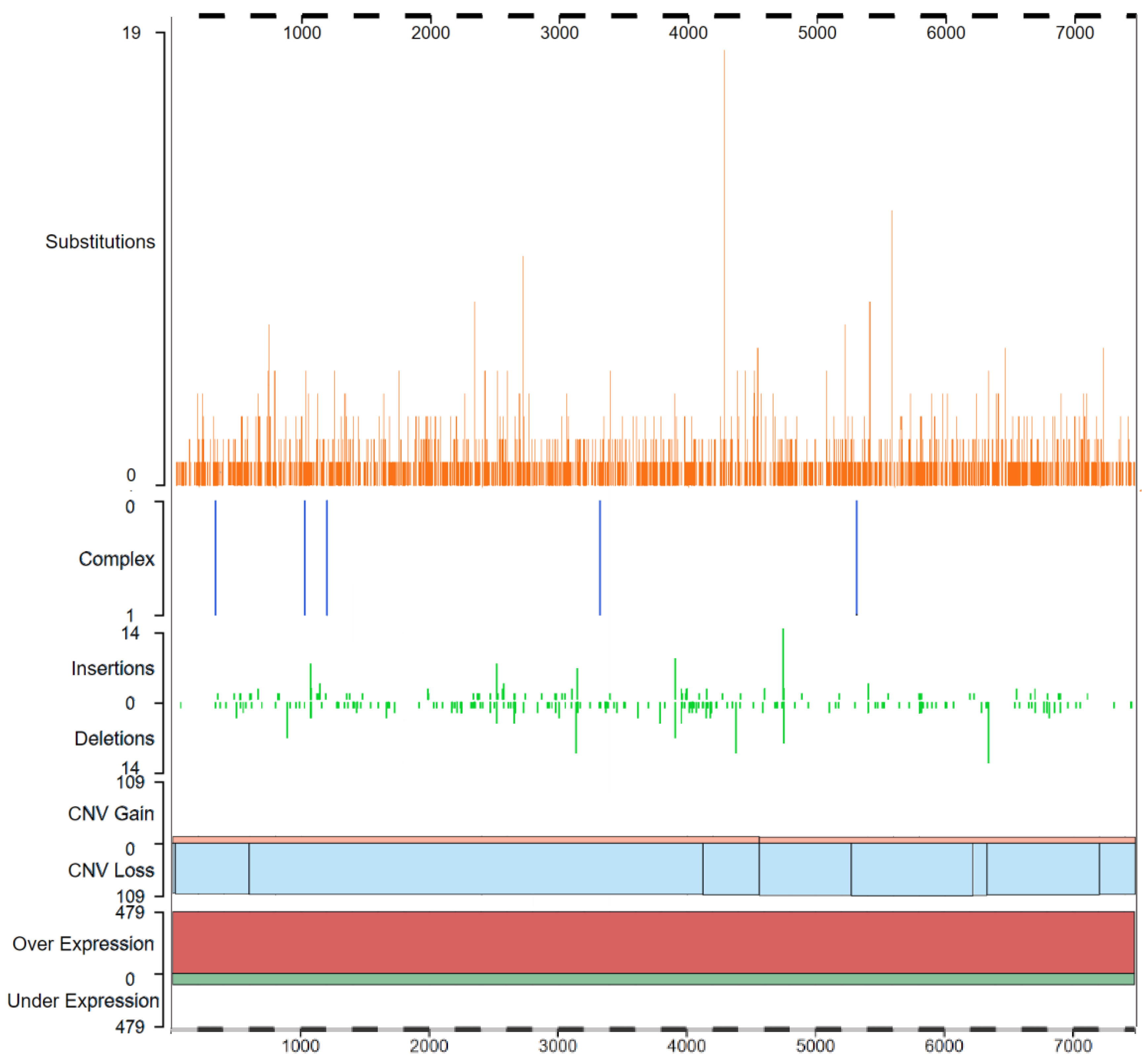
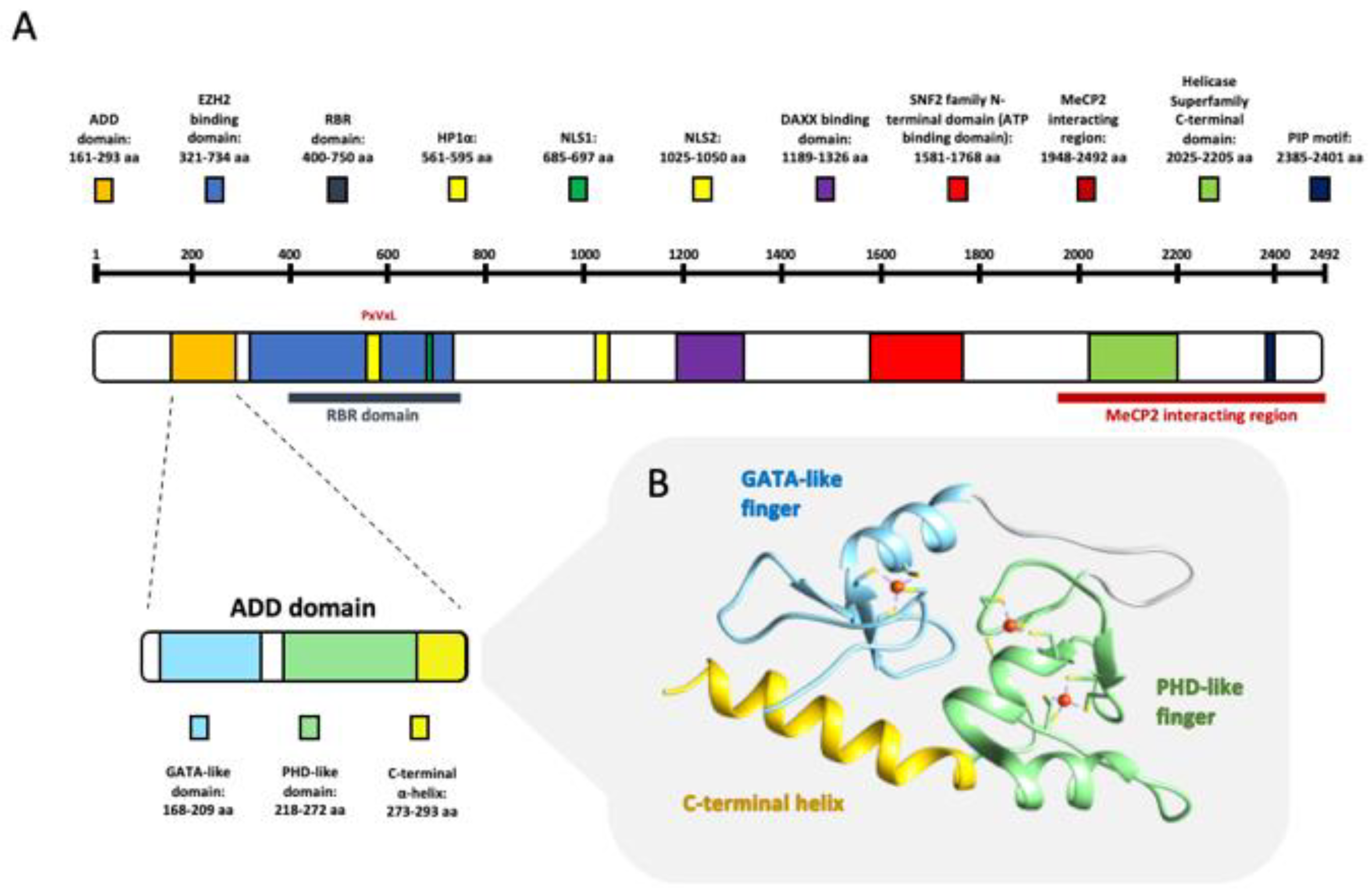
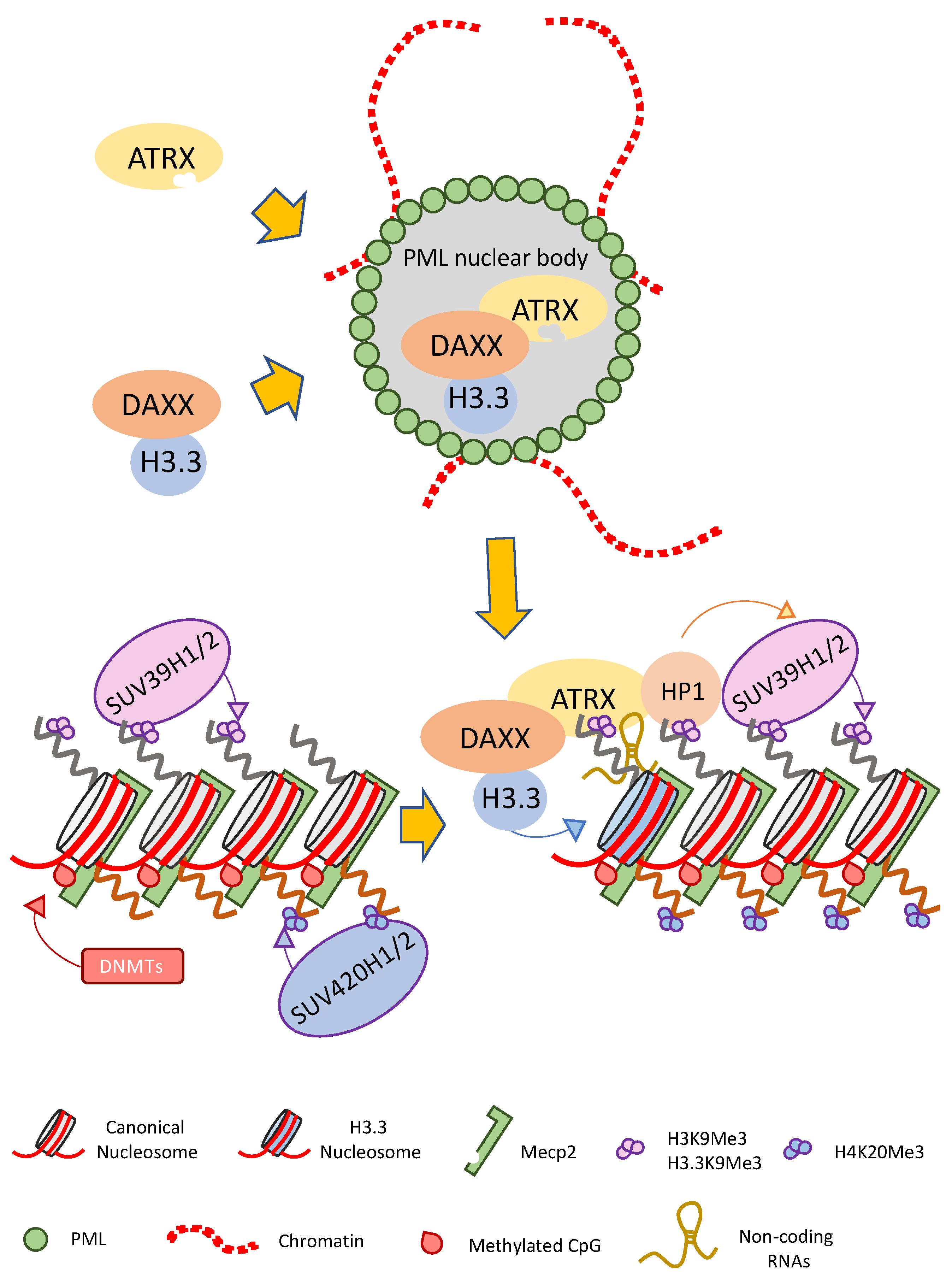
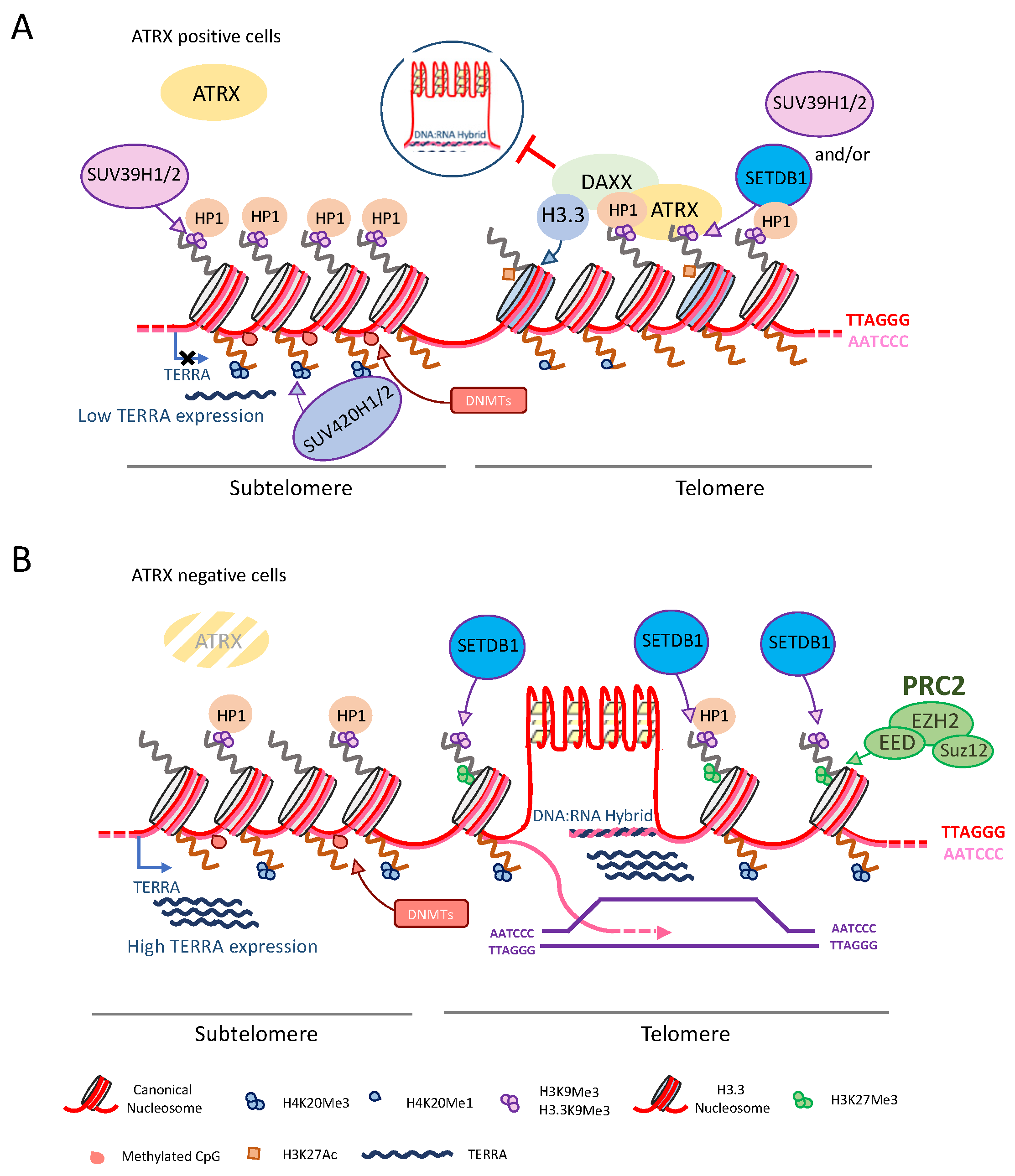
| PROTEIN | INTERACTION DOMAIN | FUNCTION | REF. |
|---|---|---|---|
| DAXX | DAXX binding domain | H3.3 deposition | [37] |
| EZH2 | EZH2 binding domain | DNA methylation | [38] |
| HP1α | HP1α binding domain | DNA methylation and PCH formation | [39] |
| MeCP2 | Mecp2 interacting region | DNA methylation and PCH formation | [40] |
| FANCD2 | Unknown | HR | [41] |
| MRE11 | Unknown | DNA DBS Repair, Replicative stress | [42] |
| NBN | Unknown | DNA DBS Repair, Replicative stress | [43] |
| RAD50 | Unknown | DNA DBS Repair, Replicative stress | [43] |
| CtIP | Unknown | Replicative stress | [41] |
| RFC1 | Unknown | HR | [44] |
| PCNA | Unknown | HR | [44] |
| MacroH2A1.1 | 322–841 aa | Telomere cohesion | [45,46] |
| MacroH2A1.2 | 322–841 aa * | X chromosome inactivation | [45,47] |
| MacroH2A2 | 322–841 aa * | X chromosome inactivation | [45] |
| XIST | RBR domain | X-chromosome inactivation | [48] |
| TERRA | Probably RBR domain | Telomeric maintenance | [49] |
| CHRO1 | Probably RBR domain | Heterochromatin organization | [50] |
| H3K9me3 | ADD domain | Heterochromatin organization | [51] |
| H3K4me0 | ADD domain | Heterochromatin organization | [52] |
| SMC1 | Unknown | Regulation of mitotic chromosome cohesion | [53] |
| SMC3 | Unknown | Regulation of mitotic chromosome cohesion | [53] |
| ZNF274 | Unknown | Heterochromatin maintenance | [54] |
| TRIM28 | Unknown | Heterochromatin maintenance | [54] |
| SETDB1 | Unknown | Heterochromatin maintenance | [54] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valenzuela, M.; Amato, R.; Sgura, A.; Antoccia, A.; Berardinelli, F. The Multiple Facets of ATRX Protein. Cancers 2021, 13, 2211. https://doi.org/10.3390/cancers13092211
Valenzuela M, Amato R, Sgura A, Antoccia A, Berardinelli F. The Multiple Facets of ATRX Protein. Cancers. 2021; 13(9):2211. https://doi.org/10.3390/cancers13092211
Chicago/Turabian StyleValenzuela, Martina, Roberta Amato, Antonella Sgura, Antonio Antoccia, and Francesco Berardinelli. 2021. "The Multiple Facets of ATRX Protein" Cancers 13, no. 9: 2211. https://doi.org/10.3390/cancers13092211
APA StyleValenzuela, M., Amato, R., Sgura, A., Antoccia, A., & Berardinelli, F. (2021). The Multiple Facets of ATRX Protein. Cancers, 13(9), 2211. https://doi.org/10.3390/cancers13092211








