Simple Summary
Stomach cancer is one of the most common cancers in the world, with over one million new cases diagnosed in 2020. Despite recent advances in cancer treatments, gastric cancer remains a serious clinical problem. This disease is tightly linked to gastric infections with Helicobacter pylori bacterium, Epstein–Barr virus, and some other less known pathogens. Here, we discuss how gastric pathogens induce tumorigenic changes in the stomach.
Abstract
Gastric cancer (GC) is one of the deadliest malignancies worldwide. In contrast to many other tumor types, gastric carcinogenesis is tightly linked to infectious events. Infections with Helicobacter pylori (H. pylori) bacterium and Epstein–Barr virus (EBV) are the two most investigated risk factors for GC. These pathogens infect more than half of the world’s population. Fortunately, only a small fraction of infected individuals develops GC, suggesting high complexity of tumorigenic processes in the human stomach. Recent studies suggest that the multifaceted interplay between microbial, environmental, and host genetic factors underlies gastric tumorigenesis. Many aspects of these interactions still remain unclear. In this review, we update on recent discoveries, focusing on the roles of various gastric pathogens and gastric microbiome in tumorigenesis.
1. Introduction
Approximately 13–15% of human cancers worldwide can be attributed to infectious agents [1]. One demonstrative example is gastric cancer (GC), which is strongly associated with infections caused by Helicobacter pylori (H. pylori) bacteria and other pathogens. Despite all efforts, gastric cancer remains a serious clinical problem. Over seven hundred thousands of deaths related to GC have been reported in 2020, ranking the fourth most-deadliest tumor in the World [2]. The incidence of GC is characterized by complex dynamics and geographical variation. Its occurrence slowly declines in North America and most Western European countries, but its burden remains very high in Asia, Latin America, and Eastern Europe [3]. Multiple histological and anatomical classifications of GC have been proposed over time. For more than half a century, the characterization of GC was largely based on Lauren’s criteria, in which GC was divided into intestinal, diffuse and, undetermined types [4,5]. In 2010, the World Health Organization (WHO) expanded this classification by identifying papillary, tubular, mucinous and poorly cohesive (including signet ring cell carcinoma and other variants), and unusual histological variants [6].
Another approach to GC classification is based on the molecular profiling using gene expression and DNA sequencing analyses. A comprehensive study by the Cancer Genome Atlas consortium (TCGA) proposed four molecular subtypes of GC: (1) tumors positive for Epstein–Barr virus (EBV), (2) microsatellite unstable tumors (MSI), (3) genomically stable tumors (GS), and (4) tumors with chromosomal instability (CIN) [7]. More clinically relevant molecular classification has been presented by the Asian Cancer Research Group (ACRG). This study used gene expression data to describe molecular subtypes linked to distinct patterns of molecular alterations and disease progression and prognosis. Based on these criteria, GC was separated into four groups: MSI-high, microsatellite-stable/p53 inactive (MSS/TP53−), microsatellite-stable/p53 active (MSS/TP53+), and microsatellite-stable/epithelial-to-mesenchymal transition (MSS/EMT) subtypes [8]. Additional classifications have also been proposed [9].
In this review, we discuss various bacterial and viral pathogens associated with tumorigenic alterations in the stomach.
2. H. pylori and Gastric Cancer
H. pylori is a spiral-shaped gram-negative microaerophilic bacterium that, in the process of evolution, adapted to survive and thrive in the human stomach. Since the seminal discovery of H. pylori and its role in gastritis and peptic ulcer disease by Robin Warren and Barry Marshall [10], studies of this pathogen have been continuing for more than three decades. Among many important findings during this period of time, discovery of the relationship between H. pylori and noncardia gastric cancer, and the characterization of gastric tumorigenesis as a stepwise inflammatory process, initiated by H. pylori, played key roles [11]. The stepwise model that emphasizes the role of chronic inflammation and consecutive pathological changes has been proposed by Dr. Pelayo Correa, and has stood the test of time [12]. According to this model, intestinal-type GC is the end result of lengthy progressive changes in the gastric mucosa that start with chronic gastritis, followed by atrophic gastritis, intestinal metaplasia (IM), dysplasia and invasive tumor. In 1994, H. pylori was recognized as a type I carcinogen by the International Agency for Research on Cancer [13]. The clinicopathological role of H. pylori was further highlighted by studies showing that H. pylori eradication reduces gastric inflammation and decreases the risk of premalignant and malignant lesions in the stomach [14]. Several effective anti–H. pylori treatment regiments have been developed and successfully used in clinic [15,16,17].
Besides IM, another type of metaplasia, called spasmolytic polypeptide-expressing metaplasia (SPEM), is also associated with chronic H. pylori infection and gastric adenocarcinoma [18]. It develops as a results of transdifferentiation of chief cells following persistent stomach injury and loss of parietal cells in the gastric oxyntic mucosa [18,19].
H. pylori typically infects humans at an early age, leading to decades-long chronic infection and mucosal injury that may progress to GC at older age. H. pylori is responsible for almost 90% of all noncardia gastric cancers [11,20]. Although infection with H. pylori is very common worldwide, only a small fraction of infected individuals develops GC, indicating complexity of tumorigenic interactions between bacteria and host cells. Among H. pylori virulence factors cytotoxin-associated gene A (CagA) protein and vacuolating cytotoxin A (VacA) are the most studied determinants associated with gastric carcinogenesis (Figure 1) [21,22,23,24].
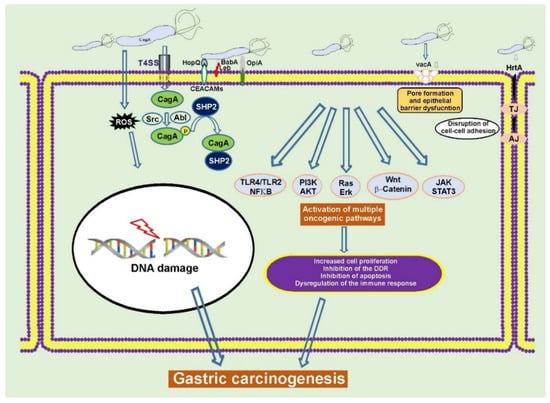
Figure 1.
H. pylori alters cellular homeostasis during infection. H. pylori colonization of the human stomach is responsible for aberrant activation of multiple oncogenic pathways, induction of DNA damage, disruption of the epithelial barrier, and modulation of the host immune response. CagA, VacA, and other virulence factors play a key role in these processes [22,25,26].
2.1. The cag Pathogenicity Island (cag PAI) and CagA Protein
The cagA gene, which encodes CagA protein, is located at the 3′ end of the cag pathogenicity island, a 40-kilobase bacterial genomic DNA fragment that is thought to be acquired by horizontal transfer of genetic material. The products of the cag PAI form highly organized type IV secretion system (T4SS) pili that functions as a sophisticated molecular machine delivering CagA inside gastric epithelial cells. There are also evidences that bacterial lipopolysaccharides, peptidoglycans, and DNA can be delivered by the T4SS [22,26,27,28,29]. After translocation, CagA is phosphorylated by host tyrosine kinases belonging to the SRC and ABL families at the EPIYA (Glu-Pro-Ile-Tyr-Ala) repeatable motifs located at the carboxy-terminal end of the CagA molecule. The EPIYA motifs are responsible for binding of CagA to multiple host proteins and dysregulation of their functions. Currently, four distinct EPIYA types (-A, -B, -C, and -D) have been identified based on surrounding amino acid sequences. The EPIYA motifs are commonly assembled in the A-B-C(D) arrangements, where the EPIYA-C and rarely EPIYA-D fragments can be present in multiple copies. H. pylori strains carrying the EPIYA-C and EPIYA-D motifs have different geographical distribution. The EPIYA-C motif is typically found outside East Asia, whereas the East Asian strains predominantly carry the EPIYA-D motif [30].
This phenomenon has clinicopathological significance. Systematic review and meta-analysis of published research have shown that the presence of EPIYA-D and multiple EPIYA-C motifs are significantly associated with an increased risk of gastric cancer in the United States/Europe and Asia [31,32].
In addition to the EPIYA motif, the C-terminus of the CagA protein contains another repeatable sequence named the CagA-multimerization motif (CM) [33]. The CM motif comprises 16 amino acid residues and is responsible for homodimerization of CagA and interaction with PAR1b/MARK kinase, playing a critical role in the epithelial cell polarity. [33]. East Asian CagA usually has a single copy of East Asian type of the CM motif, while Western CagA retains multiple copies of Western type of the CM motifs. The polymorphism of the CM and EPIYA motifs explains differences in molecular weight of CagA protein that can vary from 120 to 145 kDa between H. pylori variants [33].
Based on current understanding, CagA is the most significant single factor defining gastric tumorigenesis. Multiple human studies have found considerable associations between infections with CagA-positive H. pylori bacteria and an increased risk of gastric cancer [21,34,35,36]. There are also multiple experimental evidences showing that CagA functions as an oncoprotein. CagA transgenic mice, in which effects of other virulence factors were excluded, developed gastric epithelial hyperplasia and hematopoietic and gastrointestinal malignancies, including gastric adenocarcinoma [37]. Similarly, transgenic expression of CagA in zebrafish causes intestinal epithelial hyperplasia and, in combination with loss of p53, produces intestinal small cell carcinomas and adenocarcinomas [38]. CagA also enhances growth and invasion of tumors generated by expression of oncogenic Ras in Drosophila [39].
Oncogenic pursuit of CagA is mediated by aberrant activation of multiple signaling cascades that are known to be altered in gastric cancer (RAS/ERK, WNT/β-catenin, JAK/STAT, PI3K/AKT, and others) and inhibition of tumor suppressors. CagA is the first bacterial protein that has been shown to induce degradation of p53 tumor suppressor, activating the PI3K/AKT/MDM2/ARF-BP1 and ERK/MDM2-pathways [40,41] (Figure 2). Previously, only viral proteins, such as HPV E6, were known to degrade p53 [30]. CagA is responsible for altering expression of N-terminally truncated p53 isoforms: ∆133p53 and ∆160p53 [42]. Interestingly, the dysregulation of p53 occurs in a strain-specific manner, with tumorigenic H. pylori strains having a stronger ability to affect p53 [40,43]. Tumorigenic H. pylori strains also decrease activity of other tumor suppressors: p14ARF, SIVA1, and p27(KIP1) [43,44,45,46].
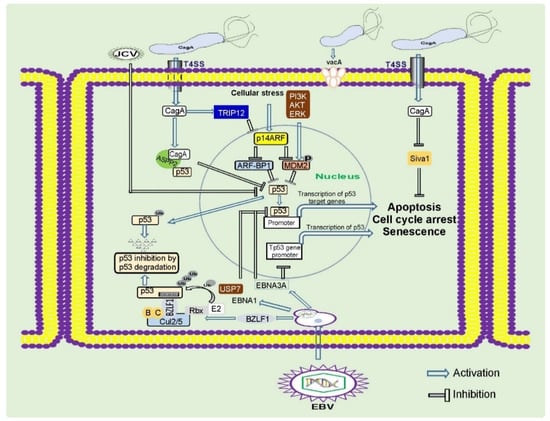
Figure 2.
Regulation of tumor suppressor proteins by gastric pathogens. Gastric pathogens: H. pylori and oncogenic viruses inhibit key tumor suppressors proteins p53, p14ARF, and others. These events result in inhibition of the DNA damage and oncogenic stress responses, two key mechanisms important for prevention of gastric carcinogenesis.
Interaction of H. pylori with gastric cells increases the levels of reactive oxygen and nitrogen species and induces oxidative stress and DNA damage in a CagA-dependent and -independent manner [47,48,49,50]. Although the entire spectrum of H. pylori–induced DNA damage is currently unknown, the formation of oxidized nitrated DNA lesions and single- and double-strand DNA breaks has been shown. Double-strand breaks in DNA are particularly detrimental, as these lesions are extremely difficult to repair resulting in highly cytotoxic and mutagenic effects [47,49,51,52,53,54,55,56]. H. pylori can also induce damage of mitochondrial DNA likely contributing to cellular senescence and gastric cancer initiation [57].
Induction of DNA damage by H. pylori is exacerbated by inhibition of p53 and multiple DNA repair pathways that are important for proper activation of the DNA damage response [40,42,49,52,58,59,60,61].
CagA is known to function as an anti-apoptotic protein. Multiple prosurvival factors and pathways have been shown to be induced by CagA, and among them are kinases AKT and ERK; antiapoptotic members of the B-cell lymphoma 2 (BCL-2) protein family MCL-1, BCL-2, and BCL-Xl; and others [62,63,64,65]. CagA is responsible for the suppression of proapoptotic factors such as SIVA1, BIM, and BAD; downregulation of autophagy; and induction of inflammation [46,62]. Human infections with CagA-positive H. pylori strains are characterized by strong inflammation and severe damage of gastric tissues [66,67,68,69,70].
It has been reported that CagA protein has a profound impact on various cellular functions, including epithelial cell barrier, cell polarity, proliferation, apoptosis, EMT, autophagy, miRNA biogenesis, inflammatory and DNA damage responses, and others. It affects activities of multiple kinases and cell signaling pathways. A partial list includes the following: EGFR, c-MET, SRC, cABL, CSC, aPKC, PAR1, PI3K, AKT, FAK, GSK-3, JAK, PAK, MAP, MDM2, p53, p14ARF, p27, RAS, β-catenin, NFκB, and multiple NFκB-related pathways [43,44,46,71,72,73,74,75,76,77,78]. It is not completely clear how one bacterial protein produces so pleiotropic effect. One plausible explanation is that CagA acts as a scaffolding protein that interacts with a large number of the host regulatory proteins, tethering them into aberrant enzymatic complexes and altering their normal functions [79].
2.2. VacA and Other Virulence Factors
VacA toxin is another virulence factor that plays a major role in tumorigenesis, associated with H. pylori infection. Its name originates from the ability to cause cell vacuolation in cultured eukaryotic cells. VacA has been classified as a pore-forming toxin. Although many toxins can form pores, the amino acid sequence of VacA is not closely resembled sequences of other known bacterial toxins [22,80,81]. The biosynthesis of VacA includes several sequential steps. Following protein translation, the VacA precursor undergoes complex proteolytic cleavage that produces 88 kDa active toxin that either secreted into the extracellular space or retained on the bacterial surface. The secreted VacA protein binds to target cellular membranes, forming an anion-selective membrane channel [82,83].
Multiple functions have been found to be associated with VacA activity, including disruption of the gastric epithelial barrier, interference with antigen presentation, suppression of autophagy and phagocytosis, inhibition of T cells and B cells that are thought to help bacteria to establish persistent infection [84,85,86,87,88]. The ability of VacA to inhibit autophagy and lysosomal degradation facilitates the accumulation of oncogenic protein CagA in gastric epithelial cells [89].
There are considerable variations in VacA sequences. Three main regions of diversity have been recognized in VacA: the signal sequence region (or “s”), the intermediate region (or “i”), and the middle region (or “m”). Based on sequence heterogeneity, the s region was subdivided into s1 (further subdivided into s1a, s1b, and s1c) and s2 types, the i region was subdivided into i1 and i2 types, and the m region was subdivided into m1 and m2 (further subdivided into m2a and m2b) types [85,90]. The incidence of GC has been found to be higher in populations infected with H. pylori variants containing type s1/i1/m1 of vacA, compared to populations infected with H. pylori type s2/i2/m2 of vacA [36,84,85]. Bacterial strains carrying type s1 and m1 vacA alleles have been associated with epithelial damage, increased gastric inflammation, and duodenal ulceration [91,92,93].
Besides CagA and VacA, H. pylori expresses a number of other cancer-associated virulence determinants. Outer-membrane proteins (OMPs) are among them. These proteins are important for bacterial adherence, colonization, survival, and persistence [94]. These factors also promote gastric diseases by affecting the signaling pathways in the host cells, enhancing activity of the T4SS, and altering immune responses [94]. H. pylori expresses a large repertoire of OMPs divided into five major families based on their sequence similarities [95]. The largest and the most studied family is the Family 1, which comprises the Hop (for H. pylori OMP) and Hor (for Hop related) proteins. The two most studied H. pylori adhesins in the Hop subgroup are BabA(HopS) and SabA(HopP), which have been originally identified to interact with the fucosylated-Lewis B (LeB) and the sialylated-Lewis X (sLeX) blood group antigens, respectively, mediating binding of H. pylori to extracellular matrix and gastric epithelial cells [96,97]. BabA potentiates activity of the T4SS [98] and is involved in induction of double-strand breaks in host cells [49]. SabA increases the colonization density and inflammation in human stomach [97,99]. Several studies analyzed associations of BabA and SabA expression with clinical outcome. The BabA status of infecting bacteria has been found to be associated with the presence of intestinal metaplasia, gastric adenocarcinoma, and MALT (Mucosa-Associated Lymphoid Tissue) lymphoma [96,100,101,102,103,104]. Similarly, the SabA status was correlated with an increased risk of premalignant lesions and gastric cancer [105,106]; however, some studies produced contradictory results [99,107].
Other OMPs, such as OipA(HopH), HopQ, and HomB, have also been implicated in gastric tumorigenesis [102,108,109,110,111,112,113,114,115]. Further studies are needed to better characterize properties of OMPs and their roles in gastric tumorigenesis.
3. Gastric Microbiota
The stomach is not a sterile organ, despite its high acidity. It is populated by complex gastric microbial communities that affect tumorigenic processes and are important for the maintenance of human health.
The composition of normal gastric microbiota is diverse and highly dynamic with the most abundant phyla: Proteobacteria, Firmicutes, Bacteroidetes, Fusobacteria, Actinobacteria, and others [116,117]. On the other hand, H. pylori has been found to be the most prevalent bacteria in the stomach of H. pylori-infected individuals [116,118]. H. pylori can induce profound changes in the composition of gastric microbiota [117,119,120,121,122,123,124,125]. Analyses of gastric microbiota in specific pathogen-free (SPF) mice revealed that H. pylori infection decreases abundance of normal gastric flora, such as Lactobacilli, and increases the presence of Clostridia, Ruminococcus spp., Eubacterium spp., Bacteroides/Prevotella spp., and others [122]. Similar phenomenon was observed in Mongolian gerbils [124,125,126]. These alterations can be explained, at least in part, by physiological changes caused by persistent H. pylori infection [127]. Induction of chronic inflammation and suppression of acid production can facilitate growth of various non–H. pylori bacterial species [127,128,129]. Many aspects of these interactions still remain controversial. Some studies did not find significant differences in the microbial composition between H. pylori–positive and –negative individuals [116,130,131]. It is likely that multiple confounding factors, such as level and type of inflammation, drug treatment (such as treatment with proton pump inhibitors), and the presence of precancerous and cancerous lesions, have to be taken into consideration during analyses of gastric microbiota.
Phylogenetic diversity of the stomach microbiome is changed during progression from gastritis to intestinal metaplasia and GC in human patients [120,132,133,134]. H. pylori colonization of the human stomach is frequently decreased in patients with advanced premalignant and malignant lesions, while abundance of Streptococcus, Lactobacillus, Veillonella, Clostridia, and others is increased [132,133,134,135,136]. Decline in Porphyromonas, Neisseria, and S. sinensis species and concomitant increase in Lactobacillus coleohominis and Lachnospiraceae were found to correlate with progression from gastritis to GC [133]. Changes in the gastric microbiota were also observed after surgical treatment of GC patients [137].
Synergetic interactions of bacteria with H. pylori to promote gastric neoplasia have been convincingly demonstrated by using transgenic insulin–gastrin (INS–GAS) mice [138,139]. It was found that H. pylori infection causes less severe gastric lesions and delayed onset of gastric intraepithelial neoplasms (GINs) in germ-free INS–GAS mice compared to mice with complex gastric microbiota [140]. In another study, infection of INS–GAS mice with restricted Altered Schaedler flora (rASF), containing Clostridium, Lactobacillus, and Bacteroides species, was sufficient to develop gastric dysplasia [141]. Infection with H. pylori further accelerated the onset of gastric lesions in rASF-infected mice [142]. Notably, antimicrobial therapies delayed onset of GIN not only in INS–GAS mice infected with H. pylori, but also in animals without H. pylori infection, thus indicating that non–H. pylori bacteria, including those considered as commensals, may represent an additional GC risk, particularly in H. pylori–infected susceptible individuals. [140,142,143,144].
It is not completely clear how non–H. pylori microbiota synergizes with H. pylori to induce GC. One plausible explanation includes overgrowth of bacteria, converting nitrogen compounds into potentially carcinogenic N-nitroso compounds [129]. It was shown that reduction of gastric acidity causes growth of nitrate-reducing bacteria, which produce carcinogenic N-nitrosamine [128,145,146]. It is also possible that various non–H. pylori bacteria promote sustained inflammation that contributes to development of GC.
Interactions within the gastric microbiome are complex and may result in various outcomes. Colonization of C57BL/6 mice with the enterohepatic Helicobacter species, H. bilis or H. muridarum, before challenge with H. pylori, was found to reduce H. pylori–induced gastric injury [147,148]. Similarly, oral Lactobacillus strains were shown to suppress H. pylori– and H. felis–induced inflammation in both mice and gerbils [125,149,150,151,152]. Consistent with rodent models, certain Lactobacilli were also found to suppress H. pylori growth and gastric mucosal inflammation in human individuals [153,154].
One interesting aspect of complex biological interactions in the stomach is the influence of helminthiasis. Parasitic worms are known to be involved in the development of various human malignancies [155]. However, certain types of helminths can decrease the risk of GC [156,157]. Infection of mice with enteric helminth (Heligmosomoides polygyrus) has been found to attenuate progression of premalignant gastric lesions induced by H. pylori and H. felis [156,158]. It was also suggested that helminths may decrease the incidence of H. pylori-associated GC in certain world populations due to their immunomodulating effects [157,159].
4. EBV and Gastric Carcinogenesis
In addition to pathogenic bacteria, compelling evidences point to significant contribution of viral infections to gastric carcinogenesis [1,160,161,162]. EBV is the most characterized gastric oncogenic virus. EBV is a member of the herpes virus family (Herpesviridae) and one of the eight known herpesviruses infecting humans. EBV is classified as the human gamma herpesviruses 4 (HHV-4). It was discovered in cultured tumor cells derived from African Burkitt’s lymphoma in 1964 [163]. Successive studies revealed that EBV infects more than 90% of the world’s population [164]. Infection with EBV typically occurs at a young age. Virus is often transmitted through saliva, infecting oral epithelial cells and multiple types of immune cells. EBV infections are usually asymptomatic, but some patients develop the clinical syndrome of infectious mononucleosis that predominantly affects adolescents and young adults. The occurrence of EBV-associated gastritis has also been reported, especially in cases of co-infection with H. pylori [165].
The most severe consequence of EBV infection is the development of tumors. Infection with EBV is strongly associated with the various types of lymphomas, and non-lymphoid malignancies, such as leiomyosarcoma and gastric and nasopharyngeal carcinomas. EBV contributes to approximately 1.5% of all cases of human cancers worldwide. Similar to H. pylori, it has been classified as a group I carcinogen by the International Agency for Research on Cancer [1,166,167,168]. Interestingly, EBV-associated GC (EBVaGC) has been found to be correlated with a higher survival rate than other GC subtypes [169]; but some studies produced contradictory results [170]. A number of antiviral treatments against EBV have been found to efficiently inhibit viral replication in laboratory testings [171,172]. However, they have had limited success in clinic and are not currently approved for treatment of EBV infection [173]. At the same time, promising results were reported in treatment of EBVaGC based on its distinct pathological characteristics [174].
Based on differences in sequences of the EBNA latency genes, EBV strains have been classified as type 1 (T1) and type 2 (T2) [175]. T1 EBV infections are more common worldwide and primarily found in Europe, Asia, North and South America, whereas T2 strains are prevalent in Africa and New Guinea [176,177]. The role of specific EBV types in the etiology of different cancers is not clear, however accumulating data suggest that additional cofactors (co-infections, comorbidities, etc.) significantly contribute to development of EBV-associated malignances [178,179,180,181].
The association of EBV with lymphoepithelial carcinoma of the stomach was initially reported by Burke et al. the in 1990 [182]. Two years later, Shibata and Weiss, detected EBV genetic material in 16% of gastric adenocarcinomas. The EBV has been specifically found within GC cells and adjusted dysplastic epithelium, but not in surrounding lymphocytes and normal gastric mucosa [183]. EBV is primarily localized in the proximal stomach and manifest itself as adenocarcinoma or as rare lymphoepithelioma-like carcinoma. The comprehensive genomic analysis conducted by the TCGA revealed that approximately 9% of human gastric cancers are positive for EBV. Multiple molecular abnormalities have been detected in EBVaGC: extensive DNA hypermethylation; high-frequency mutations of PIK3CA, ARID1A, and BCOR; and amplification of JAK2, PD-L1, and PD-L2 [7,184]. A closely followed study by the ACRG found 6.5% of EPV-positive GC patients [8,185]. An even higher positivity rate has been reported, when EBV was detected by RNA-Seq instead of traditional EBER1/2 in situ hybridization [186].
The EBV entry into epithelial cells is relatively well-studied. It involves multiple interactions between viral and host proteins. Among host proteins, ephrin receptor A2, integrins (αVβ5, αVβ6, and αVβ8), neuropilin 1, complement receptor type 2 (CR2), and nonmuscle myosin heavy chain IIA help virus entry [187]. It is not completely clear how EBV infects gastric epithelium, owing to the hostile acidic environment of the stomach. Most plausibly, EBV is delivered by infected B lymphocytes, attracted to the stomach by inflammatory processes that precede gastric tumor development [188]. It was found that membrane vesicular products secreted by epithelial cells can activate virus in latent EBV-infected B lymphocytes, resulting in virus production and infection [189]. Another possibility is that EBV is delivered in contaminated saliva, which is constantly ingested, and, in certain circumstances, the virus may survive in the harsh environment of the stomach and infect gastric mucosal cells [167].
One prominent feature of EBV is its ability to establish chronic infections alternating lytic and latent virus cycles. Following the virus entry into host cells, double-stranded viral DNA is maintained as a multiple-copy episome, establishing the base for latent infection that is characterized by remarkable variation and plasticity in viral transcription and replication. Depending on the pattern of viral gene expression, primary EBV infection can be categorized into three latency types (latency types I, II, and III). The latent pattern of EBVaGC corresponds to the unique latency I/II with expression of EBV-determined nuclear antigen 1 (EBNA1), noncoding RNAs (EBER1, EBER2), and BamHI-A rightward transcripts (BART miRNAs) that is characteristic for the latency type I. In addition, latent membrane protein 2 (LMP2A), which is characteristic for latency type II, is expressed in approximately 50% of EBVaGCs [190,191]. Expression of both latent and lytic genes play an important role in gastric carcinogenesis [192].
Several latency factors have been shown to have oncogenic properties. Among them is EBV-encoded latent membrane protein 2A (LMP2A) that activates the PI3K/AKT proliferation pathway, increases survival of infected cells via upregulation of survivin gene expression, inhibits TGFβ1-induced apoptosis, and promotes cellular migration through targeting the Notch signaling pathway [193,194,195]. LMP2A is also responsible for activation of DNA methyltransferase 1 (DNMT1) via STAT3 pathway, leading to the promoter methylation of PTEN tumor suppressor [196].
Another latency factor, EBNA 1 protein that is responsible for viral DNA replication and EBV persistence, induces protein degradation of promyelocytic leukemia (PML) tumor suppressor, inhibiting the formation of nuclear bodies (PML NBs). This inhibition results in impairment of the DNA damage response and increase in cell survival [197].
Not only viral proteins are responsible for tumorigenic alterations. EBV produces multiple viral noncoding RNAs (ncRNAs) during the acute and latent stages of infection that regulate expression of viral and host genes. Among them are EBV-encoded RNAs (EBERs), BamHI-A rightward transcripts (BARTs), viral small nucleolar RNA1 (v-snoRNA1), and viral microRNAs (miRNAs). They play a critical role in host cell proliferation, survival, apoptosis, immune escape, and regulation of host ncRNAs [198]. The list of newly discovered EBV ncRNAs keeps growing.
EBV causes multiple epigenetic abnormalities in host cells. It has been found that Tet Methylcytosine Dioxygenase 2 (TET2), a regulator of DNA methylation, is downregulated during EBV infection, contributing to an abnormal DNA methylation profile [199]. As a result, many tumor-suppressor genes, such as p16, p14, APC, and TP73, become methylated [200,201,202,203,204,205].
Similar to H. pylori, EBV inhibits activity of p53 tumor suppressor. Interestingly, multiple viral proteins are involved in this process. EBNA1 and EBNA3C repress p53-dependent transcription and augment its ubiquitination and degradation [206]. The immediate-early protein BZLF1 (BamHI Z fragment leftward open reading frame 1) also inhibits p53-dependent transcription [207,208]. This protein functions as an adaptor component of the ECS ubiquitin ligase complex (Elongin B/C-Cul2/5-SOCS-box protein) that facilitates p53 degradation [209] (Figure 2). Direct viral inhibition of p53 might explain why EBVaGC rarely harbors mutations in the TP53 gene [7].
5. Other Carcinogenic Viruses
In addition to EBV, a number of viruses have been found to contribute to tumorigenic alterations in the stomach. Among them is human polyomavirus 2, commonly referred as the John Cunningham virus (JCV). JCV is a small DNA virus and etiological agent of progressive multifocal leukoencephalopathy (PML), a debilitating and frequently fatal central nervous system disease. JCV infection typically occurs during childhood and persists for the lifetime of the host [210,211]. Immune system of healthy individuals prevents JCV replication suppressing the virus and keeping it in the latent state, but JCV can be reactivated as a result of various immunodeficiencies.
Several studies reported the presence of JCV in gastric, esophageal, and colorectal cancer tissues [212,213,214,215]. Expression of JCV T-antigen (T-Ag) in gastric tumor tissues was found to be in the range of 26–86% [214,215,216]. It was shown the correlation of JCV infection with activation of β-catenin, absence of p53 mutations, allelic losses and aberrant methylation of multiple genes, including tumor suppressors p14 and p16 [214,215,216,217]. The mechanism of JCV-induced gastric tumorigenesis is not well understood, but is likely mediated by large and small T-antigens that can inhibit retinoblastoma protein (pRb) and p53 tumor suppressor, activate β-catenin, and dysregulate cell cycle in infected cells [218,219,220].
Another virus that promotes gastric tumorigenesis is the human cytomegalovirus (HCMV), also termed human herpesvirus 5 (HHV-5). It is a member of the Herpesviridae family that, similar to EBV, is widely distributed around the World, with a prevalence of about 80% in industrialized countries and almost 100% in developing countries [221]. After initial infection, HCMV commonly remains in the latent state, but it can be activated in immunocompromised individuals. Several studies reported that HCMV infection is associated with an increased risk of GC [222,223,224]. The HCMV load was found to be significantly higher in GC epithelium compared to non-tumorous tissues. HCMV infection was also correlated with early onset of GC, lymphatic metastasis, and inhibition of negative regulator of the Wnt signaling pathway CTNNBIP1 [223,225,226].
Chronic infections with hepatitis B (HBV) or C (HCV) viruses are known risk factors for hepatocellular carcinoma. However, discovery of viral antigens and DNA in gastric tissues suggested a possible involvement of these viruses in gastric tumorigenesis. Histological examinations have shown that viral markers, HBV X protein (HBx) and core antibody (HBcAb), were expressed at higher levels in gastric tumor tissues than in normal counterparts [227]. Some case-control and population-based studies reported correlations between gastric precancerous and cancerous lesions and the presence of HBV surface antigen (HBsAg) or HCV antibody (HCV Ab) [228,229,230,231,232]. A recent meta-analytical study was consistent with these findings and reported association between HBV infection and an increased risk of GC [227]. Another recent large-scale study found that eradication of HCV reduces the risk of gastric cancer, particularly among younger individuals [233].
Infections with human retroviruses HIV (human immunodeficiency virus) or HTLV (human T-cell lymphotropic virus) increase the risk of developing various tumors, and can also indirectly affect gastric tumorigenesis [234,235]. HIV infection, if left untreated, leads to acquired immunodeficiency syndrome, a disease characterized by immune suppression and loss of immune-mediated control against diverse opportunistic pathogens. The analyses of HIV-positive patients found a significantly higher load of EBV DNA than in uninfected individuals [236,237]. Sixty percent of HIV-infected patients were positive for antibodies to EBV early antigen (EA) compared to 12% of uninfected individuals [237]. GC precursor lesions, including intestinal metaplasia and atrophic gastritis, were also common in patients infected with HIV [234]. In contrast to HIV, a large population-based study of HTLV-1-infected subjects found a reduced risk of GC development [238]. A recent meta-analysis of epidemiological studies further confirmed this correlation [235].
Currently, involvement of human papillomaviruses (HPV) in gastric tumorigenesis remains to be controversial and requires further investigation. While some studies suggested a potential link between HPV infection and gastric tumor, and also reported the presence of high-risk carcinogenic HPVs in GC and gastritis tissues [239,240], other studies did not found such associations [241,242].
6. Conclusions
Gastric cancer is a heterogeneous disease developed as a result of multifactorial interactions between infectious agents, the gastric microbiome, and host genetic and environmental factors (Figure 3). Significant progress has been made on the way toward appreciation complexity of host–pathogen interactions. These findings open up new exciting avenues for future research that will certainly lead to a better understanding of gastric tumorigenesis and novel, more effective GC therapies.
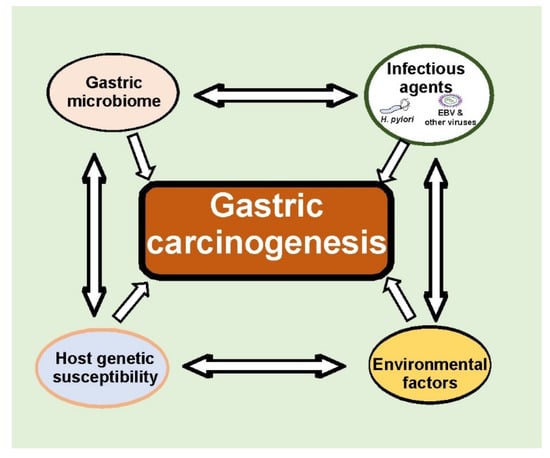
Figure 3.
Multifaceted interactions between infectious agents, gastric microbiomes, and host genetic and environmental factors define tumorigenic processes in the stomach. Many aspects of these interactions remain currently unknown.
Author Contributions
M.P. and E.Z. wrote the manuscript. W.E.-R. and J.Q. discussed various aspect of the manuscript. A.I.Z. coordinated the project, and corrected the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the National Cancer Institute R01 138833, R01 206564, Department of Veterans Affairs BX002115, and the Sylvester Comprehensive Cancer Center P30CA24013. The contents of this work are solely the responsibility of the authors and do not necessarily represent the official views of the Department of Veterans Affairs, National Institutes of Health or University of Miami.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors thank the Sylvester Comprehensive Cancer Center and University of Miami Miller School of Medicine, Miami, FL, USA, for their help and support.
Conflicts of Interest
The authors have declared that no conflict of interest exists.
References
- De Martel, C.; Georges, D.; Bray, F.; Ferlay, J.; Clifford, G.M. Global burden of cancer attributable to infections in 2018: A worldwide incidence analysis. Lancet Glob. Health 2020, 8, e180–e190. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Lauren, P. The two histological main types of gastric carcinoma: Diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol. Microbiol. Scand. 1965, 64, 31–49. [Google Scholar] [CrossRef]
- Van Cutsem, E.; Sagaert, X.; Topal, B.; Haustermans, K.; Prenen, H. Gastric cancer. Lancet 2016, 388, 2654–2664. [Google Scholar] [CrossRef]
- Bosman, F.T.; Carneiro, F.; Hruban, R.H.; Theise, N.D. WHO Classification of Tumours of the Digestive System, 4th ed.; IARC: Lyon, France, 2010. [Google Scholar]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Cristescu, R.; Lee, J.; Nebozhyn, M.; Kim, K.M.; Ting, J.C.; Wong, S.S.; Liu, J.; Yue, Y.G.; Wang, J.; Yu, K.; et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 2015, 21, 449–456. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, G.; Hu, C. Molecular classification of gastric adenocarcinoma. Gastroenterol. Res. 2019, 12, 275–282. [Google Scholar] [CrossRef]
- Warren, J.R.; Marshall, B. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet 1983, 1, 1273–1275. [Google Scholar]
- Moss, S.F. The clinical evidence linking Helicobacter pylori to gastric cancer. Cell. Mol. Gastroenterol. Hepatol. 2017, 3, 183–191. [Google Scholar] [CrossRef]
- Correa, P. A human model of gastric carcinogenesis. Cancer Res. 1988, 48, 3554–3560. [Google Scholar] [PubMed]
- IARC working group on the evaluation of carcinogenic risks to humans. Schistosomes, liver flukes and Helicobacter pylori. Lyon, (FR): International Agency for Research on Cancer. IARC Monogr. Eval. Carcinog. Risks Hum. 1994, 61, 1–241.
- Lee, Y.C.; Chiang, T.H.; Chou, C.K.; Tu, Y.K.; Liao, W.C.; Wu, M.S.; Graham, D.Y. Association between Helicobacter pylori eradication and gastric cancer incidence: A systematic review and meta-analysis. Gastroenterology 2016, 150, 1113–1124.e5. [Google Scholar] [CrossRef]
- Doorakkers, E.; Lagergren, J.; Engstrand, L.; Brusselaers, N. Helicobacter pylori eradication treatment and the risk of gastric adenocarcinoma in a Western population. Gut 2018, 67, 2092–2096. [Google Scholar] [CrossRef] [PubMed]
- Wong, B.C.; Lam, S.K.; Wong, W.M.; Chen, J.S.; Zheng, T.T.; Feng, R.E.; Lai, K.C.; Hu, W.H.; Yuen, S.T.; Leung, S.Y.; et al. Helicobacter pylori eradication to prevent gastric cancer in a high-risk region of China: A randomized controlled trial. JAMA 2004, 291, 187–194. [Google Scholar] [CrossRef]
- Zhang, W.; Chen, Q.; Liang, X.; Liu, W.; Xiao, S.; Graham, D.Y.; Lu, H. Bismuth, lansoprazole, amoxicillin and metronidazole or clarithromycin as first-line Helicobacter pylori therapy. Gut 2015, 64, 1715–1720. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, P.H.; Lee, J.R.; Joshi, V.; Playford, R.J.; Poulsom, R.; Wright, N.A.; Goldenring, J.R. Identification of a metaplastic cell lineage associated with human gastric adenocarcinoma. Lab. Investig. 1999, 79, 639–646. [Google Scholar] [PubMed]
- Saenz, J.B.; Vargas, N.; Mills, J.C. Tropism for spasmolytic polypeptide-expressing metaplasia allows Helicobacter pylori to expand its intragastric niche. Gastroenterology 2019, 156, 160–174.e7. [Google Scholar] [CrossRef] [PubMed]
- Plummer, M.; Franceschi, S.; Vignat, J.; Forman, D.; de Martel, C. Global burden of gastric cancer attributable to Helicobacter pylori. Int. J. Cancer 2015, 136, 487–490. [Google Scholar] [CrossRef]
- Blaser, M.J.; Perez-Perez, G.I.; Kleanthous, H.; Cover, T.L.; Peek, R.M.; Chyou, P.H.; Stemmermann, G.N.; Nomura, A. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 1995, 55, 2111–2115. [Google Scholar]
- Naumann, M.; Sokolova, O.; Tegtmeyer, N.; Backert, S. Helicobacter pylori: A paradigm pathogen for subverting host cell signal transmission. Trends Microbiol. 2017, 25, 316–328. [Google Scholar] [CrossRef]
- Abadi, A.T.; Lee, Y.Y. Helicobacter pylori vacA as marker for gastric cancer and gastroduodenal diseases: One but not the only factor. J. Clin. Microbiol. 2014, 52, 4451. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Vidal, Y.; Ponce-de-Leon, S.; Castillo-Rojas, G.; Barreto-Zuniga, R.; Torre-Delgadillo, A. High diversity of vacA and cagA Helicobacter pylori genotypes in patients with and without gastric cancer. PLoS ONE 2008, 3, e3849. [Google Scholar] [CrossRef] [PubMed]
- Wilson, K.T.; Crabtree, J.E. Immunology of Helicobacter pylori: Insights into the failure of the immune response and perspectives on vaccine studies. Gastroenterology 2007, 133, 288–308. [Google Scholar] [CrossRef]
- Zaika, A.I.; Wei, J.; Noto, J.M.; Peek, R.M. Microbial regulation of p53 tumor suppressor. PLoS Pathog. 2015, 11, e1005099. [Google Scholar] [CrossRef]
- Stein, S.C.; Faber, E.; Bats, S.H.; Murillo, T.; Speidel, Y.; Coombs, N.; Josenhans, C. Helicobacter pylori modulates host cell responses by CagT4SS-dependent translocation of an intermediate metabolite of LPS inner core heptose biosynthesis. PLoS Pathog. 2017, 13, e1006514. [Google Scholar] [CrossRef]
- Varga, M.G.; Shaffer, C.L.; Sierra, J.C.; Suarez, G.; Piazuelo, M.B.; Whitaker, M.E.; Romero-Gallo, J.; Krishna, U.S.; Delgado, A.; Gomez, M.A.; et al. Pathogenic Helicobacter pylori strains translocate DNA and activate TLR9 via the cancer-associated cag type IV secretion system. Oncogene 2016, 35, 6262–6269. [Google Scholar] [CrossRef] [PubMed]
- Viala, J.; Chaput, C.; Boneca, I.G.; Cardona, A.; Girardin, S.E.; Moran, A.P.; Athman, R.; Memet, S.; Huerre, M.R.; Coyle, A.J.; et al. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat. Immunol. 2004, 5, 1166–1174. [Google Scholar] [CrossRef]
- Naito, M.; Yamazaki, T.; Tsutsumi, R.; Higashi, H.; Onoe, K.; Yamazaki, S.; Azuma, T.; Hatakeyama, M. Influence of EPIYA-repeat polymorphism on the phosphorylation-dependent biological activity of Helicobacter pylori CagA. Gastroenterology 2006, 130, 1181–1190. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Liu, J.; Gong, Y.; Yuan, Y. Association of CagA EPIYA-D or EPIYA-C phosphorylation sites with peptic ulcer and gastric cancer risks: A meta-analysis. Medicine 2017, 96, e6620. [Google Scholar] [CrossRef]
- Hatakeyama, M. Malignant Helicobacter pylori-associated diseases: Gastric cancer and MALT lymphoma. Adv. Exp. Med. Biol. 2019, 1149, 135–149. [Google Scholar] [CrossRef]
- Ren, S.; Higashi, H.; Lu, H.; Azuma, T.; Hatakeyama, M. Structural basis and functional consequence of Helicobacter pylori CagA multimerization in cells. J. Biol. Chem. 2006, 281, 32344–32352. [Google Scholar] [CrossRef]
- Parsonnet, J.; Friedman, G.D.; Orentreich, N.; Vogelman, H. Risk for gastric cancer in people with CagA positive or CagA negative Helicobacter pylori infection. Gut 1997, 40, 297–301. [Google Scholar] [CrossRef]
- Huang, J.Q.; Zheng, G.F.; Sumanac, K.; Irvine, E.J.; Hunt, R.H. Meta-analysis of the relationship between cagA seropositivity and gastric cancer. Gastroenterology 2003, 125, 1636–1644. [Google Scholar] [CrossRef] [PubMed]
- Matos, J.I.; de Sousa, H.A.; Marcos-Pinto, R.; Dinis-Ribeiro, M. Helicobacter pylori CagA and VacA genotypes and gastric phenotype: A meta-analysis. Eur. J. Gastroenterol. Hepatol. 2013, 25, 1431–1441. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, N.; Yuasa, H.; Tanaka, S.; Sawa, H.; Miura, M.; Matsui, A.; Higashi, H.; Musashi, M.; Iwabuchi, K.; Suzuki, M.; et al. Transgenic expression of Helicobacter pylori CagA induces gastrointestinal and hematopoietic neoplasms in mouse. Proc. Natl. Acad. Sci. USA 2008, 105, 1003–1008. [Google Scholar] [CrossRef] [PubMed]
- Neal, J.T.; Peterson, T.S.; Kent, M.L.; Guillemin, K.H. pylori virulence factor CagA increases intestinal cell proliferation by Wnt pathway activation in a transgenic zebrafish model. Dis. Model. Mech. 2013, 6, 802–810. [Google Scholar] [CrossRef]
- Wandler, A.M.; Guillemin, K. Transgenic expression of the Helicobacter pylori virulence factor CagA promotes apoptosis or tumorigenesis through JNK activation in Drosophila. PLoS Pathog. 2012, 8, e1002939. [Google Scholar] [CrossRef]
- Wei, J.; Nagy, T.A.; Vilgelm, A.; Zaika, E.; Ogden, S.R.; Romero-Gallo, J.; Piazuelo, M.B.; Correa, P.; Washington, M.K.; El-Rifai, W.; et al. Regulation of p53 tumor suppressor by Helicobacter pylori in gastric epithelial cells. Gastroenterology 2010, 139, 1333–1343.e4. [Google Scholar] [CrossRef]
- Bhardwaj, V.; Noto, J.M.; Wei, J.; Andl, C.; El-Rifai, W.; Peek, R.M.; Zaika, A.I. Helicobacter pylori bacteria alter the p53 stress response via ERK-HDM2 pathway. Oncotarget 2015, 6, 1531–1543. [Google Scholar] [CrossRef]
- Wei, J.; Noto, J.; Zaika, E.; Romero-Gallo, J.; Correa, P.; El-Rifai, W.; Peek, R.M.; Zaika, A. Pathogenic bacterium Helicobacter pylori alters the expression profile of p53 protein isoforms and p53 response to cellular stresses. Proc. Natl. Acad. Sci. USA 2012, 109, E2543–E2550. [Google Scholar] [CrossRef]
- Wei, J.; Noto, J.M.; Zaika, E.; Romero-Gallo, J.; Piazuelo, M.B.; Schneider, B.; El-Rifai, W.; Correa, P.; Peek, R.M.; Zaika, A.I. Bacterial CagA protein induces degradation of p53 protein in a p14ARF-dependent manner. Gut 2015, 64, 1040–1048. [Google Scholar] [CrossRef]
- Horvat, A.; Noto, J.M.; Ramatchandirin, B.; Zaika, E.; Palrasu, M.; Wei, J.; Schneider, B.G.; El-Rifai, W.; Peek, R.M., Jr.; Zaika, A.I. Helicobacter pylori pathogen regulates p14ARF tumor suppressor and autophagy in gastric epithelial cells. Oncogene 2018, 37, 5054–5065. [Google Scholar] [CrossRef] [PubMed]
- Eguchi, H.; Herschenhous, N.; Kuzushita, N.; Moss, S.F. Helicobacter pylori increases proteasome-mediated degradation of p27(kip1) in gastric epithelial cells. Cancer Res. 2003, 63, 4739–4746. [Google Scholar] [PubMed]
- Palrasu, M.; Zaika, E.; El-Rifai, W.; Garcia-Buitrago, M.; Piazuelo, M.B.; Wilson, K.T.; Peek, R.M., Jr.; Zaika, A.I. Bacterial CagA protein compromises tumor suppressor mechanisms in gastric epithelial cells. J. Clin. Investig. 2020, 130, 2422–2434. [Google Scholar] [CrossRef] [PubMed]
- Koeppel, M.; Garcia-Alcalde, F.; Glowinski, F.; Schlaermann, P.; Meyer, T.F. Helicobacter pylori infection causes characteristic DNA damage patterns in human cells. Cell Rep. 2015, 11, 1703–1713. [Google Scholar] [CrossRef] [PubMed]
- Papa, A.; Danese, S.; Sgambato, A.; Ardito, R.; Zannoni, G.; Rinelli, A.; Vecchio, F.M.; Gentiloni-Silveri, N.; Cittadini, A.; Gasbarrini, G.; et al. Role of Helicobacter pylori CagA+ infection in determining oxidative DNA damage in gastric mucosa. Scand. J. Gastroenterol. 2002, 37, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Toller, I.M.; Neelsen, K.J.; Steger, M.; Hartung, M.L.; Hottiger, M.O.; Stucki, M.; Kalali, B.; Gerhard, M.; Sartori, A.A.; Lopes, M.; et al. Carcinogenic bacterial pathogen Helicobacter pylori triggers DNA double-strand breaks and a DNA damage response in its host cells. Proc. Natl. Acad. Sci. USA 2011, 108, 14944–14949. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Suzuki, H.; Saya, H.; Hatakeyama, M.; Hirayama, T.; Hirata, K.; Nagano, O.; Matsuzaki, J.; Hibi, T. Reactive oxygen species-induced autophagic degradation of Helicobacter pylori CagA is specifically suppressed in cancer stem-like cells. Cell Host Microbe 2012, 12, 764–777. [Google Scholar] [CrossRef] [PubMed]
- Hanada, K.; Uchida, T.; Tsukamoto, Y.; Watada, M.; Yamaguchi, N.; Yamamoto, K.; Shiota, S.; Moriyama, M.; Graham, D.Y.; Yamaoka, Y. Helicobacter pylori infection introduces DNA double-strand breaks in host cells. Infect. Immun. 2014, 82, 4182–4189. [Google Scholar] [CrossRef] [PubMed]
- Hartung, M.L.; Gruber, D.C.; Koch, K.N.; Gruter, L.; Rehrauer, H.; Tegtmeyer, N.; Backert, S.; Muller, A.H. pylori-Induced DNA strand breaks are introduced by nucleotide excision repair endonucleases and promote NF-kappaB target gene expression. Cell Rep. 2015, 13, 70–79. [Google Scholar] [CrossRef]
- Raza, Y.; Khan, A.; Farooqui, A.; Mubarak, M.; Facista, A.; Akhtar, S.S.; Khan, S.; Kazi, J.I.; Bernstein, C.; Kazmi, S.U. Oxidative DNA damage as a potential early biomarker of Helicobacter pylori associated carcinogenesis. Pathol. Oncol. Res. 2014, 20, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Uehara, T.; Ma, D.; Yao, Y.; Lynch, J.P.; Morales, K.; Ziober, A.; Feldman, M.; Ota, H.; Sepulveda, A.R. H. pylori infection is associated with DNA damage of Lgr5-positive epithelial stem cells in the stomach of patients with gastric cancer. Dig. Dis. Sci. 2013, 58, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Xu, L.Y.; Yang, Z.; Cao, X.M.; Li, W.; Lu, N.H. Expression of gammaH2AX in various gastric pathologies and its association with Helicobacter pylori infection. Oncol. Lett. 2014, 7, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Zamperone, A.; Cohen, D.; Stein, M.; Viard, C.; Musch, A. Inhibition of polarity-regulating kinase PAR1b contributes to Helicobacter pylori inflicted DNA Double Strand Breaks in gastric cells. Cell Cycle 2019, 18, 299–311. [Google Scholar] [CrossRef]
- Machado, A.M.; Desler, C.; Boggild, S.; Strickertsson, J.A.; Friis-Hansen, L.; Figueiredo, C.; Seruca, R.; Rasmussen, L.J. Helicobacter pylori infection affects mitochondrial function and DNA repair, thus, mediating genetic instability in gastric cells. Mech. Ageing Dev. 2013, 134, 460–466. [Google Scholar] [CrossRef]
- Kim, J.J.; Tao, H.; Carloni, E.; Leung, W.K.; Graham, D.Y.; Sepulveda, A.R. Helicobacter pylori impairs DNA mismatch repair in gastric epithelial cells. Gastroenterology 2002, 123, 542–553. [Google Scholar] [CrossRef]
- Machado, A.M.; Figueiredo, C.; Touati, E.; Maximo, V.; Sousa, S.; Michel, V.; Carneiro, F.; Nielsen, F.C.; Seruca, R.; Rasmussen, L.J. Helicobacter pylori infection induces genetic instability of nuclear and mitochondrial DNA in gastric cells. Clin. Cancer Res. 2009, 15, 2995–3002. [Google Scholar] [CrossRef]
- Park, D.I.; Park, S.H.; Kim, S.H.; Kim, J.W.; Cho, Y.K.; Kim, H.J.; Sohn, C.I.; Jeon, W.K.; Kim, B.I.; Cho, E.Y.; et al. Effect of Helicobacter pylori infection on the expression of DNA mismatch repair protein. Helicobacter 2005, 10, 179–184. [Google Scholar] [CrossRef]
- Strickertsson, J.A.; Desler, C.; Rasmussen, L.J. Impact of bacterial infections on aging and cancer: Impairment of DNA repair and mitochondrial function of host cells. Exp. Gerontol. 2014, 56, 164–174. [Google Scholar] [CrossRef]
- Vallejo-Flores, G.; Torres, J.; Sandoval-Montes, C.; Arevalo-Romero, H.; Meza, I.; Camorlinga-Ponce, M.; Torres-Morales, J.; Chavez-Rueda, A.K.; Legorreta-Haquet, M.V.; Fuentes-Panana, E.M. Helicobacter pylori CagA suppresses apoptosis through activation of AKT in a nontransformed epithelial cell model of glandular acini formation. BioMed Res. Int. 2015, 2015, 761501. [Google Scholar] [CrossRef]
- Lin, W.C.; Tsai, H.F.; Kuo, S.H.; Wu, M.S.; Lin, C.W.; Hsu, P.I.; Cheng, A.L.; Hsu, P.N. Translocation of Helicobacter pylori CagA into Human B lymphocytes, the origin of mucosa-associated lymphoid tissue lymphoma. Cancer Res. 2010, 70, 5740–5748. [Google Scholar] [CrossRef]
- Mimuro, H.; Suzuki, T.; Nagai, S.; Rieder, G.; Suzuki, M.; Nagai, T.; Fujita, Y.; Nagamatsu, K.; Ishijima, N.; Koyasu, S.; et al. Helicobacter pylori dampens gut epithelial self-renewal by inhibiting apoptosis, a bacterial strategy to enhance colonization of the stomach. Cell Host Microbe 2007, 2, 250–263. [Google Scholar] [CrossRef] [PubMed]
- Noto, J.M.; Piazuelo, M.B.; Chaturvedi, R.; Bartel, C.A.; Thatcher, E.J.; Delgado, A.; Romero-Gallo, J.; Wilson, K.T.; Correa, P.; Patton, J.G.; et al. Strain-specific suppression of microRNA-320 by carcinogenic Helicobacter pylori promotes expression of the antiapoptotic protein Mcl-1. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 305, G786–G796. [Google Scholar] [CrossRef]
- Abu-Taleb, A.M.F.; Abdelattef, R.S.; Abdel-Hady, A.A.; Omran, F.H.; El-Korashi, L.A.; Abdel-Aziz El-Hady, H.; El-Gebaly, A.M. Prevalence of Helicobacter pylori cagA and iceA genes and their association with gastrointestinal diseases. Int. J. Microbiol. 2018, 2018, 4809093. [Google Scholar] [CrossRef]
- Peek, R.M., Jr.; Blaser, M.J. Pathophysiology of Helicobacter pylori-induced gastritis and peptic ulcer disease. Am. J. Med. 1997, 102, 200–207. [Google Scholar] [CrossRef]
- Figura, N.; Vindigni, C.; Covacci, A.; Presenti, L.; Burroni, D.; Vernillo, R.; Banducci, T.; Roviello, F.; Marrelli, D.; Biscontri, M.; et al. cagA positive and negative Helicobacter pylori strains are simultaneously present in the stomach of most patients with non-ulcer dyspepsia: Relevance to histological damage. Gut 1998, 42, 772–778. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Tang, B.; Jia, Y.P.; Zhu, P.; Zhuang, Y.; Fang, Y.; Li, Q.; Wang, K.; Zhang, W.J.; Guo, G.; et al. Helicobacter pylori CagA protein negatively regulates autophagy and promotes inflammatory response via c-Met-PI3K/Akt-mTOR signaling pathway. Front. Cell. Infect. Microbiol. 2017, 7, 417. [Google Scholar] [CrossRef]
- Yamaoka, Y.; Kita, M.; Kodama, T.; Sawai, N.; Kashima, K.; Imanishi, J. Induction of various cytokines and development of severe mucosal inflammation by cagA gene positive Helicobacter pylori strains. Gut 1997, 41, 442–451. [Google Scholar] [CrossRef]
- Wei, J.; O’Brien, D.; Vilgelm, A.; Piazuelo, M.B.; Correa, P.; Washington, M.K.; El-Rifai, W.; Peek, R.M.; Zaika, A. Interaction of Helicobacter pylori with gastric epithelial cells is mediated by the p53 protein family. Gastroenterology 2008, 134, 1412–1423. [Google Scholar] [CrossRef]
- Bauer, B.; Pang, E.; Holland, C.; Kessler, M.; Bartfeld, S.; Meyer, T.F. The Helicobacter pylori virulence effector CagA abrogates human beta-defensin 3 expression via inactivation of EGFR signaling. Cell Host Microbe 2012, 11, 576–586. [Google Scholar] [CrossRef] [PubMed]
- Higashi, H.; Nakaya, A.; Tsutsumi, R.; Yokoyama, K.; Fujii, Y.; Ishikawa, S.; Higuchi, M.; Takahashi, A.; Kurashima, Y.; Teishikata, Y.; et al. Helicobacter pylori CagA induces Ras-independent morphogenetic response through SHP-2 recruitment and activation. J. Biol. Chem. 2004, 279, 17205–17216. [Google Scholar] [CrossRef]
- Mueller, D.; Tegtmeyer, N.; Brandt, S.; Yamaoka, Y.; De Poire, E.; Sgouras, D.; Wessler, S.; Torres, J.; Smolka, A.; Backert, S. c-Src and c-Abl kinases control hierarchic phosphorylation and function of the CagA effector protein in Western and East Asian Helicobacter pylori strains. J. Clin. Investig. 2012, 122, 1553–1566. [Google Scholar] [CrossRef]
- Tabassam, F.H.; Graham, D.Y.; Yamaoka, Y. Helicobacter pylori activate epidermal growth factor receptor- and phosphatidylinositol 3-OH kinase-dependent Akt and glycogen synthase kinase 3beta phosphorylation. Cell. Microbiol. 2009, 11, 70–82. [Google Scholar] [CrossRef]
- Tegtmeyer, N.; Hartig, R.; Delahay, R.M.; Rohde, M.; Brandt, S.; Conradi, J.; Takahashi, S.; Smolka, A.J.; Sewald, N.; Backert, S. A small fibronectin-mimicking protein from bacteria induces cell spreading and focal adhesion formation. J. Biol. Chem. 2010, 285, 23515–23526. [Google Scholar] [CrossRef]
- Tegtmeyer, N.; Wittelsberger, R.; Hartig, R.; Wessler, S.; Martinez-Quiles, N.; Backert, S. Serine phosphorylation of cortactin controls focal adhesion kinase activity and cell scattering induced by Helicobacter pylori. Cell Host Microbe 2011, 9, 520–531. [Google Scholar] [CrossRef]
- Tsutsumi, R.; Takahashi, A.; Azuma, T.; Higashi, H.; Hatakeyama, M. Focal adhesion kinase is a substrate and downstream effector of SHP-2 complexed with Helicobacter pylori CagA. Mol. Cell. Biol. 2006, 26, 261–276. [Google Scholar] [CrossRef]
- Hatakeyama, M. Structure and function of Helicobacter pylori CagA, the first-identified bacterial protein involved in human cancer. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2017, 93, 196–219. [Google Scholar] [CrossRef]
- Torres, V.J.; Ivie, S.E.; McClain, M.S.; Cover, T.L. Functional properties of the p33 and p55 domains of the Helicobacter pylori vacuolating cytotoxin. J. Biol. Chem. 2005, 280, 21107–21114. [Google Scholar] [CrossRef]
- Wang, W.C.; Wang, H.J.; Kuo, C.H. Two distinctive cell binding patterns by vacuolating toxin fused with glutathione S-transferase: One high-affinity m1-specific binding and the other lower-affinity binding for variant m forms. Biochemistry 2001, 40, 11887–11896. [Google Scholar] [CrossRef]
- Czajkowsky, D.M.; Iwamoto, H.; Cover, T.L.; Shao, Z. The vacuolating toxin from Helicobacter pylori forms hexameric pores in lipid bilayers at low pH. Proc. Natl. Acad. Sci. USA 1999, 96, 2001–2006. [Google Scholar] [CrossRef]
- Tombola, F.; Carlesso, C.; Szabo, I.; de Bernard, M.; Reyrat, J.M.; Telford, J.L.; Rappuoli, R.; Montecucco, C.; Papini, E.; Zoratti, M. Helicobacter pylori vacuolating toxin forms anion-selective channels in planar lipid bilayers: Possible implications for the mechanism of cellular vacuolation. Biophys. J. 1999, 76, 1401–1409. [Google Scholar] [CrossRef]
- Pyburn, T.M.; Foegeding, N.J.; Gonzalez-Rivera, C.; McDonald, N.A.; Gould, K.L.; Cover, T.L.; Ohi, M.D. Structural organization of membrane-inserted hexamers formed by Helicobacter pylori VacA toxin. Mol. Microbiol. 2016, 102, 22–36. [Google Scholar] [CrossRef]
- Atherton, J.C.; Cao, P.; Peek, R.M., Jr.; Tummuru, M.K.; Blaser, M.J.; Cover, T.L. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J. Biol. Chem. 1995, 270, 17771–17777. [Google Scholar] [CrossRef] [PubMed]
- Boncristiano, M.; Paccani, S.R.; Barone, S.; Ulivieri, C.; Patrussi, L.; Ilver, D.; Amedei, A.; D’Elios, M.M.; Telford, J.L.; Baldari, C.T. The Helicobacter pylori vacuolating toxin inhibits T cell activation by two independent mechanisms. J. Exp. Med. 2003, 198, 1887–1897. [Google Scholar] [CrossRef]
- Gebert, B.; Fischer, W.; Weiss, E.; Hoffmann, R.; Haas, R. Helicobacter pylori vacuolating cytotoxin inhibits T lymphocyte activation. Science 2003, 301, 1099–1102. [Google Scholar] [CrossRef] [PubMed]
- Sundrud, M.S.; Torres, V.J.; Unutmaz, D.; Cover, T.L. Inhibition of primary human T cell proliferation by Helicobacter pylori vacuolating toxin (VacA) is independent of VacA effects on IL-2 secretion. Proc. Natl. Acad. Sci. USA 2004, 101, 7727–7732. [Google Scholar] [CrossRef]
- Abdullah, M.; Greenfield, L.K.; Bronte-Tinkew, D.; Capurro, M.I.; Rizzuti, D.; Jones, N.L. VacA promotes CagA accumulation in gastric epithelial cells during Helicobacter pylori infection. Sci. Rep. 2019, 9, 38. [Google Scholar] [CrossRef]
- Van Doorn, L.J.; Figueiredo, C.; Sanna, R.; Pena, S.; Midolo, P.; Ng, E.K.; Atherton, J.C.; Blaser, M.J.; Quint, W.G. Expanding allelic diversity of Helicobacter pylori vacA. J. Clin. Microbiol. 1998, 36, 2597–2603. [Google Scholar] [CrossRef]
- Atherton, J.C.; Peek, R.M., Jr.; Tham, K.T.; Cover, T.L.; Blaser, M.J. Clinical and pathological importance of heterogeneity in vacA, the vacuolating cytotoxin gene of Helicobacter pylori. Gastroenterology 1997, 112, 92–99. [Google Scholar] [CrossRef]
- Sinnett, C.G.; Letley, D.P.; Narayanan, G.L.; Patel, S.R.; Hussein, N.R.; Zaitoun, A.M.; Robinson, K.; Atherton, J.C. Helicobacter pylori vacA transcription is genetically-determined and stratifies the level of human gastric inflammation and atrophy. J. Clin. Pathol. 2016, 69, 968–973. [Google Scholar] [CrossRef] [PubMed]
- Van Doorn, L.J.; Figueiredo, C.; Sanna, R.; Plaisier, A.; Schneeberger, P.; de Boer, W.; Quint, W. Clinical relevance of the cagA, vacA, and iceA status of Helicobacter pylori. Gastroenterology 1998, 115, 58–66. [Google Scholar] [CrossRef]
- Kao, C.Y.; Sheu, B.S.; Wu, J.J. Helicobacter pylori infection: An overview of bacterial virulence factors and pathogenesis. Biomed. J. 2016, 39, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Alm, R.A.; Bina, J.; Andrews, B.M.; Doig, P.; Hancock, R.E.; Trust, T.J. Comparative genomics of Helicobacter pylori: Analysis of the outer membrane protein families. Infect. Immun. 2000, 68, 4155–4168. [Google Scholar] [CrossRef]
- Fujimoto, S.; Olaniyi Ojo, O.; Arnqvist, A.; Wu, J.Y.; Odenbreit, S.; Haas, R.; Graham, D.Y.; Yamaoka, Y. Helicobacter pylori BabA expression, gastric mucosal injury, and clinical outcome. Clin. Gastroenterol. Hepatol. 2007, 5, 49–58. [Google Scholar] [CrossRef]
- Mahdavi, J.; Sonden, B.; Hurtig, M.; Olfat, F.O.; Forsberg, L.; Roche, N.; Angstrom, J.; Larsson, T.; Teneberg, S.; Karlsson, K.A.; et al. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science 2002, 297, 573–578. [Google Scholar] [CrossRef]
- Ishijima, N.; Suzuki, M.; Ashida, H.; Ichikawa, Y.; Kanegae, Y.; Saito, I.; Boren, T.; Haas, R.; Sasakawa, C.; Mimuro, H. BabA-mediated adherence is a potentiator of the Helicobacter pylori type IV secretion system activity. J. Biol. Chem. 2011, 286, 25256–25264. [Google Scholar] [CrossRef] [PubMed]
- Sheu, B.S.; Odenbreit, S.; Hung, K.H.; Liu, C.P.; Sheu, S.M.; Yang, H.B.; Wu, J.J. Interaction between host gastric Sialyl-Lewis X and H. pylori SabA enhances H. pylori density in patients lacking gastric Lewis B antigen. Am. J. Gastroenterol. 2006, 101, 36–44. [Google Scholar] [CrossRef]
- Azevedo, M.; Eriksson, S.; Mendes, N.; Serpa, J.; Figueiredo, C.; Resende, L.P.; Ruvoen-Clouet, N.; Haas, R.; Boren, T.; Le Pendu, J.; et al. Infection by Helicobacter pylori expressing the BabA adhesin is influenced by the secretor phenotype. J. Pathol. 2008, 215, 308–316. [Google Scholar] [CrossRef]
- Gerhard, M.; Lehn, N.; Neumayer, N.; Boren, T.; Rad, R.; Schepp, W.; Miehlke, S.; Classen, M.; Prinz, C. Clinical relevance of the Helicobacter pylori gene for blood-group antigen-binding adhesin. Proc. Natl. Acad. Sci. USA 1999, 96, 12778–12783. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.L.; Huang, H.L.; Huang, B.S.; Chen, P.C.; Chen, C.S.; Wang, H.L.; Lin, P.H.; Chieh, M.S.; Wu, J.J.; Yang, J.C.; et al. Combination of OipA, BabA, and SabA as candidate biomarkers for predicting Helicobacter pylori-related gastric cancer. Sci. Rep. 2016, 6, 36442. [Google Scholar] [CrossRef]
- Yu, J.; Leung, W.K.; Go, M.Y.; Chan, M.C.; To, K.F.; Ng, E.K.; Chan, F.K.; Ling, T.K.; Chung, S.C.; Sung, J.J. Relationship between Helicobacter pylori babA2 status with gastric epithelial cell turnover and premalignant gastric lesions. Gut 2002, 51, 480–484. [Google Scholar] [CrossRef]
- Prinz, C.; Schoniger, M.; Rad, R.; Becker, I.; Keiditsch, E.; Wagenpfeil, S.; Classen, M.; Rosch, T.; Schepp, W.; Gerhard, M. Key importance of the Helicobacter pylori adherence factor blood group antigen binding adhesin during chronic gastric inflammation. Cancer Res. 2001, 61, 1903–1909. [Google Scholar]
- Lehours, P.; Menard, A.; Dupouy, S.; Bergey, B.; Richy, F.; Zerbib, F.; Ruskone-Fourmestraux, A.; Delchier, J.C.; Megraud, F. Evaluation of the association of nine Helicobacter pylori virulence factors with strains involved in low-grade gastric mucosa-associated lymphoid tissue lymphoma. Infect. Immun. 2004, 72, 880–888. [Google Scholar] [CrossRef]
- Yamaoka, Y.; Ojo, O.; Fujimoto, S.; Odenbreit, S.; Haas, R.; Gutierrez, O.; El-Zimaity, H.M.; Reddy, R.; Arnqvist, A.; Graham, D.Y. Helicobacter pylori outer membrane proteins and gastroduodenal disease. Gut 2006, 55, 775–781. [Google Scholar] [CrossRef]
- Chen, M.Y.; He, C.Y.; Meng, X.; Yuan, Y. Association of Helicobacter pylori babA2 with peptic ulcer disease and gastric cancer. World J. Gastroenterol. 2013, 19, 4242–4251. [Google Scholar] [CrossRef]
- Zhao, Q.; Song, C.; Wang, K.; Li, D.; Yang, Y.; Liu, D.; Wang, L.; Zhou, N.; Xie, Y. Prevalence of Helicobacter pylori babA, oipA, sabA, and homB genes in isolates from Chinese patients with different gastroduodenal diseases. Med. Microbiol. Immunol. 2020, 209, 565–577. [Google Scholar] [CrossRef]
- Bartpho, T.S.; Wattanawongdon, W.; Tongtawee, T.; Paoin, C.; Kangwantas, K.; Dechsukhum, C. Precancerous gastric lesions with Helicobacter pylori vacA (+)/babA2(+)/oipA (+) genotype increase the risk of gastric cancer. BioMed Res. Int. 2020, 2020, 7243029. [Google Scholar] [CrossRef]
- Liu, J.; He, C.; Chen, M.; Wang, Z.; Xing, C.; Yuan, Y. Association of presence/absence and on/off patterns of Helicobacter pylori oipA gene with peptic ulcer disease and gastric cancer risks: A meta-analysis. BMC Infect. Dis. 2013, 13, 555. [Google Scholar] [CrossRef]
- Cao, P.; Lee, K.J.; Blaser, M.J.; Cover, T.L. Analysis of hopQ alleles in East Asian and Western strains of Helicobacter pylori. FEMS Microbiol. Lett. 2005, 251, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Leylabadlo, H.E.; Yekani, M.; Ghotaslou, R. Helicobacter pylori hopQ alleles (type I and II) in gastric cancer. Biomed. Rep. 2016, 4, 601–604. [Google Scholar] [CrossRef]
- Jung, S.W.; Sugimoto, M.; Graham, D.Y.; Yamaoka, Y. homB status of Helicobacter pylori as a novel marker to distinguish gastric cancer from duodenal ulcer. J. Clin. Microbiol. 2009, 47, 3241–3245. [Google Scholar] [CrossRef]
- Kang, J.; Jones, K.R.; Jang, S.; Olsen, C.H.; Yoo, Y.J.; Merrell, D.S.; Cha, J.H. The geographic origin of Helicobacter pylori influences the association of the homB gene with gastric cancer. J. Clin. Microbiol. 2012, 50, 1082–1085. [Google Scholar] [CrossRef] [PubMed]
- Abadi, A.T.B.; Rafiei, A.; Ajami, A.; Hosseini, V.; Taghvaei, T.; Jones, K.R.; Merrell, D.S. Helicobacter pylori homB, but not cagA, is associated with gastric cancer in Iran. J. Clin. Microbiol. 2011, 49, 3191–3197. [Google Scholar] [CrossRef]
- Bik, E.M.; Eckburg, P.B.; Gill, S.R.; Nelson, K.E.; Purdom, E.A.; Francois, F.; Perez-Perez, G.; Blaser, M.J.; Relman, D.A. Molecular analysis of the bacterial microbiota in the human stomach. Proc. Natl. Acad. Sci. USA 2006, 103, 732–737. [Google Scholar] [CrossRef] [PubMed]
- Li, X.X.; Wong, G.L.; To, K.F.; Wong, V.W.; Lai, L.H.; Chow, D.K.; Lau, J.Y.; Sung, J.J.; Ding, C. Bacterial microbiota profiling in gastritis without Helicobacter pylori infection or non-steroidal anti-inflammatory drug use. PLoS ONE 2009, 4, e7985. [Google Scholar] [CrossRef] [PubMed]
- Andersson, A.F.; Lindberg, M.; Jakobsson, H.; Backhed, F.; Nyren, P.; Engstrand, L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS ONE 2008, 3, e2836. [Google Scholar] [CrossRef]
- Espinoza, J.L.; Matsumoto, A.; Tanaka, H.; Matsumura, I. Gastric microbiota: An emerging player in Helicobacter pylori-induced gastric malignancies. Cancer Lett. 2018, 414, 147–152. [Google Scholar] [CrossRef]
- Coker, O.O.; Dai, Z.; Nie, Y.; Zhao, G.; Cao, L.; Nakatsu, G.; Wu, W.K.; Wong, S.H.; Chen, Z.; Sung, J.J.Y.; et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut 2018, 67, 1024–1032. [Google Scholar] [CrossRef]
- Chen, X.H.; Wang, A.; Chu, A.N.; Gong, Y.H.; Yuan, Y. Mucosa-associated microbiota in gastric cancer tissues compared with non-cancer tissues. Front. Microbiol. 2019, 10, 1261. [Google Scholar] [CrossRef]
- Aebischer, T.; Fischer, A.; Walduck, A.; Schlotelburg, C.; Lindig, M.; Schreiber, S.; Meyer, T.F.; Bereswill, S.; Gobel, U.B. Vaccination prevents Helicobacter pylori-induced alterations of the gastric flora in mice. FEMS Immunol. Med. Microbiol. 2006, 46, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Maldonado-Contreras, A.; Goldfarb, K.C.; Godoy-Vitorino, F.; Karaoz, U.; Contreras, M.; Blaser, M.J.; Brodie, E.L.; Dominguez-Bello, M.G. Structure of the human gastric bacterial community in relation to Helicobacter pylori status. ISME J. 2011, 5, 574–579. [Google Scholar] [CrossRef]
- Noto, J.M.; Rose, K.L.; Hachey, A.J.; Delgado, A.G.; Romero-Gallo, J.; Wroblewski, L.E.; Schneider, B.G.; Shah, S.C.; Cover, T.L.; Wilson, K.T.; et al. Carcinogenic Helicobacter pylori strains selectively dysregulate the in vivo gastric proteome, which may be associated with stomach cancer progression. Mol. Cell. Proteom. 2019, 18, 352–371. [Google Scholar] [CrossRef] [PubMed]
- Osaki, T.; Matsuki, T.; Asahara, T.; Zaman, C.; Hanawa, T.; Yonezawa, H.; Kurata, S.; Woo, T.D.; Nomoto, K.; Kamiya, S. Comparative analysis of gastric bacterial microbiota in Mongolian gerbils after long-term infection with Helicobacter pylori. Microb. Pathog. 2012, 53, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.Q.; Monstein, H.J.; Nilsson, L.E.; Petersson, F.; Borch, K. Profiling and identification of eubacteria in the stomach of Mongolian gerbils with and without Helicobacter pylori infection. Helicobacter 2003, 8, 149–157. [Google Scholar] [CrossRef]
- Blaser, M.J.; Atherton, J.C. Helicobacter pylori persistence: Biology and disease. J. Clin. Investig. 2004, 113, 321–333. [Google Scholar] [CrossRef]
- Sanduleanu, S.; Jonkers, D.; De Bruine, A.; Hameeteman, W.; Stockbrugger, R.W. Non-Helicobacter pylori bacterial flora during acid-suppressive therapy: Differential findings in gastric juice and gastric mucosa. Aliment. Pharmacol. Ther. 2001, 15, 379–388. [Google Scholar] [CrossRef]
- Mowat, C.; Williams, C.; Gillen, D.; Hossack, M.; Gilmour, D.; Carswell, A.; Wirz, A.; Preston, T.; McColl, K.E. Omeprazole, Helicobacter pylori status, and alterations in the intragastric milieu facilitating bacterial N-nitrosation. Gastroenterology 2000, 119, 339–347. [Google Scholar] [CrossRef]
- Khosravi, Y.; Dieye, Y.; Poh, B.H.; Ng, C.G.; Loke, M.F.; Goh, K.L.; Vadivelu, J. Culturable bacterial microbiota of the stomach of Helicobacter pylori positive and negative gastric disease patients. Sci. World J. 2014, 2014, 610421. [Google Scholar] [CrossRef]
- Tan, M.P.; Kaparakis, M.; Galic, M.; Pedersen, J.; Pearse, M.; Wijburg, O.L.; Janssen, P.H.; Strugnell, R.A. Chronic Helicobacter pylori infection does not significantly alter the microbiota of the murine stomach. Appl. Environ. Microbiol. 2007, 73, 1010–1013. [Google Scholar] [CrossRef]
- Eun, C.S.; Kim, B.K.; Han, D.S.; Kim, S.Y.; Kim, K.M.; Choi, B.Y.; Song, K.S.; Kim, Y.S.; Kim, J.F. Differences in gastric mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing methods. Helicobacter 2014, 19, 407–416. [Google Scholar] [CrossRef]
- Aviles-Jimenez, F.; Vazquez-Jimenez, F.; Medrano-Guzman, R.; Mantilla, A.; Torres, J. Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Sci. Rep. 2014, 4, 4202. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.M.; Pereira-Marques, J.; Pinto-Ribeiro, I.; Costa, J.L.; Carneiro, F.; Machado, J.C.; Figueiredo, C. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut 2018, 67, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Dicksved, J.; Lindberg, M.; Rosenquist, M.; Enroth, H.; Jansson, J.K.; Engstrand, L. Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. J. Med. Microbiol. 2009, 58, 509–516. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Shao, L.; Liu, X.; Ji, F.; Mei, Y.; Cheng, Y.; Liu, F.; Yan, C.; Li, L.; Ling, Z. Alterations of gastric mucosal microbiota across different stomach microhabitats in a cohort of 276 patients with gastric cancer. EBioMedicine 2019, 40, 336–348. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.H.; Lin, J.T.; Ho, H.J.; Lai, Z.L.; Wang, C.B.; Tang, S.L.; Wu, C.Y. Gastric microbiota and predicted gene functions are altered after subtotal gastrectomy in patients with gastric cancer. Sci. Rep. 2016, 6, 20701. [Google Scholar] [CrossRef]
- Fox, J.G.; Li, X.; Cahill, R.J.; Andrutis, K.; Rustgi, A.K.; Odze, R.; Wang, T.C. Hypertrophic gastropathy in Helicobacter felis-infected wild-type C57BL/6 mice and p53 hemizygous transgenic mice. Gastroenterology 1996, 110, 155–166. [Google Scholar] [CrossRef]
- Fox, J.G.; Rogers, A.B.; Ihrig, M.; Taylor, N.S.; Whary, M.T.; Dockray, G.; Varro, A.; Wang, T.C. Helicobacter pylori-associated gastric cancer in INS-GAS mice is gender specific. Cancer Res. 2003, 63, 942–950. [Google Scholar]
- Lofgren, J.L.; Whary, M.T.; Ge, Z.; Muthupalani, S.; Taylor, N.S.; Mobley, M.; Potter, A.; Varro, A.; Eibach, D.; Suerbaum, S.; et al. Lack of commensal flora in Helicobacter pylori-infected INS-GAS mice reduces gastritis and delays intraepithelial neoplasia. Gastroenterology 2011, 140, 210–220.e4. [Google Scholar] [CrossRef]
- Lertpiriyapong, K.; Whary, M.T.; Muthupalani, S.; Lofgren, J.L.; Gamazon, E.R.; Feng, Y.; Ge, Z.; Wang, T.C.; Fox, J.G. Gastric colonisation with a restricted commensal microbiota replicates the promotion of neoplastic lesions by diverse intestinal microbiota in the Helicobacter pylori INS-GAS mouse model of gastric carcinogenesis. Gut 2014, 63, 54–63. [Google Scholar] [CrossRef]
- Lee, C.W.; Rickman, B.; Rogers, A.B.; Muthupalani, S.; Takaishi, S.; Yang, P.; Wang, T.C.; Fox, J.G. Combination of sulindac and antimicrobial eradication of Helicobacter pylori prevents progression of gastric cancer in hypergastrinemic INS-GAS mice. Cancer Res. 2009, 69, 8166–8174. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.W.; Rickman, B.; Rogers, A.B.; Ge, Z.; Wang, T.C.; Fox, J.G. Helicobacter pylori eradication prevents progression of gastric cancer in hypergastrinemic INS-GAS mice. Cancer Res. 2008, 68, 3540–3548. [Google Scholar] [CrossRef] [PubMed]
- Rolig, A.S.; Cech, C.; Ahler, E.; Carter, J.E.; Ottemann, K.M. The degree of Helicobacter pylori-triggered inflammation is manipulated by preinfection host microbiota. Infect. Immun. 2013, 81, 1382–1389. [Google Scholar] [CrossRef]
- Leach, S.A.; Thompson, M.; Hill, M. Bacterially catalysed N-nitrosation reactions and their relative importance in the human stomach. Carcinogenesis 1987, 8, 1907–1912. [Google Scholar] [CrossRef] [PubMed]
- Stockbruegger, R.W. Bacterial overgrowth as a consequence of reduced gastric acidity. Scand. J. Gastroenterol. Suppl. 1985, 111, 7–16. [Google Scholar] [CrossRef]
- Lemke, L.B.; Ge, Z.; Whary, M.T.; Feng, Y.; Rogers, A.B.; Muthupalani, S.; Fox, J.G. Concurrent Helicobacter bilis infection in C57BL/6 mice attenuates proinflammatory H. pylori-induced gastric pathology. Infect. Immun. 2009, 77, 2147–2158. [Google Scholar] [CrossRef]
- Ge, Z.; Feng, Y.; Muthupalani, S.; Eurell, L.L.; Taylor, N.S.; Whary, M.T.; Fox, J.G. Coinfection with Enterohepatic Helicobacter species can ameliorate or promote Helicobacter pylori-induced gastric pathology in C57BL/6 mice. Infect. Immun. 2011, 79, 3861–3871. [Google Scholar] [CrossRef]
- Chen, Y.H.; Tsai, W.H.; Wu, H.Y.; Chen, C.Y.; Yeh, W.L.; Chen, Y.H.; Hsu, H.Y.; Chen, W.W.; Chen, Y.W.; Chang, W.W.; et al. Probiotic Lactobacillus spp. act against Helicobacter pylori-induced inflammation. J. Clin. Med. 2019, 8, 90. [Google Scholar] [CrossRef]
- Kabir, A.M.; Aiba, Y.; Takagi, A.; Kamiya, S.; Miwa, T.; Koga, Y. Prevention of Helicobacter pylori infection by lactobacilli in a gnotobiotic murine model. Gut 1997, 41, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Zaman, C.; Osaki, T.; Hanawa, T.; Yonezawa, H.; Kurata, S.; Kamiya, S. Analysis of the microbial ecology between Helicobacter pylori and the gastric microbiota of Mongolian gerbils. J. Med. Microbiol. 2014, 63, 129–137. [Google Scholar] [CrossRef][Green Version]
- Schmitz, J.M.; Durham, C.G.; Schoeb, T.R.; Soltau, T.D.; Wolf, K.J.; Tanner, S.M.; McCracken, V.J.; Lorenz, R.G. Helicobacter felis—Associated gastric disease in microbiota-restricted mice. J. Histochem. Cytochem. 2011, 59, 826–841. [Google Scholar] [CrossRef]
- Sakamoto, I.; Igarashi, M.; Kimura, K.; Takagi, A.; Miwa, T.; Koga, Y. Suppressive effect of Lactobacillus gasseri OLL 2716 (LG21) on Helicobacter pylori infection in humans. J. Antimicrob. Chemother. 2001, 47, 709–710. [Google Scholar] [CrossRef] [PubMed]
- De Klerk, N.; Maudsdotter, L.; Gebreegziabher, H.; Saroj, S.D.; Eriksson, B.; Eriksson, O.S.; Roos, S.; Linden, S.; Sjolinder, H.; Jonsson, A.B. Lactobacilli reduce Helicobacter pylori attachment to host gastric epithelial cells by inhibiting adhesion gene expression. Infect. Immun. 2016, 84, 1526–1535. [Google Scholar] [CrossRef] [PubMed]
- Scholte, L.L.S.; Pascoal-Xavier, M.A.; Nahum, L.A. Helminths and cancers from the evolutionary perspective. Front. Med. 2018, 5, 90. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.G.; Beck, P.; Dangler, C.A.; Whary, M.T.; Wang, T.C.; Shi, H.N.; Nagler-Anderson, C. Concurrent enteric helminth infection modulates inflammation and gastric immune responses and reduces helicobacter-induced gastric atrophy. Nat. Med. 2000, 6, 536–542. [Google Scholar] [CrossRef] [PubMed]
- Whary, M.T.; Sundina, N.; Bravo, L.E.; Correa, P.; Quinones, F.; Caro, F.; Fox, J.G. Intestinal helminthiasis in Colombian children promotes a Th2 response to Helicobacter pylori: Possible implications for gastric carcinogenesis. Cancer Epidemiol. Biomark. Prev. 2005, 14, 1464–1469. [Google Scholar] [CrossRef] [PubMed]
- Whary, M.T.; Muthupalani, S.; Ge, Z.; Feng, Y.; Lofgren, J.; Shi, H.N.; Taylor, N.S.; Correa, P.; Versalovic, J.; Wang, T.C.; et al. Helminth co-infection in Helicobacter pylori infected INS-GAS mice attenuates gastric premalignant lesions of epithelial dysplasia and glandular atrophy and preserves colonization resistance of the stomach to lower bowel microbiota. Microbes Infect. 2014, 16, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Fuenmayor-Boscan, A.; Hernandez-Rincon, I.; Arismendi-Morillo, G.; Mengual, E.; Rivero, Z.; Romero, G.; Lizarzabal, M.; Alvarez-Mon, M. Changes in the severity of gastric mucosal inflammation associated with Helicobacter pylori in humans coinfected with intestinal helminths. Indian J. Gastroenterol. 2020, 39, 186–195. [Google Scholar] [CrossRef]
- Fattahi, S.; Kosari-Monfared, M.; Ghadami, E.; Golpour, M.; Khodadadi, P.; Ghasemiyan, M.; Akhavan-Niaki, H. Infection-associated epigenetic alterations in gastric cancer: New insight in cancer therapy. J. Cell Physiol. 2018, 233, 9261–9270. [Google Scholar] [CrossRef]
- Plummer, M.; de Martel, C.; Vignat, J.; Ferlay, J.; Bray, F.; Franceschi, S. Global burden of cancers attributable to infections in 2012: A synthetic analysis. Lancet Glob. Health 2016, 4, e609–e616. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, J.; Cheng, A.S.L.; Yu, J.; To, K.F.; Kang, W. Gastric cancer: Genome damaged by bugs. Oncogene 2020, 39, 3427–3442. [Google Scholar] [CrossRef]
- Epstein, M.A.; Achong, B.G.; Barr, Y.M. Virus particles in cultured lymphoblasts from Burkitt’s lymphoma. Lancet 1964, 1, 702–703. [Google Scholar] [CrossRef]
- Lieberman, P.M. Virology. Epstein-Barr virus turns 50. Science 2014, 343, 1323–1325. [Google Scholar] [CrossRef]
- Cardenas-Mondragon, M.G.; Carreon-Talavera, R.; Camorlinga-Ponce, M.; Gomez-Delgado, A.; Torres, J.; Fuentes-Panana, E.M. Epstein Barr virus and Helicobacter pylori co-infection are positively associated with severe gastritis in pediatric patients. PLoS ONE 2013, 8, e62850. [Google Scholar] [CrossRef] [PubMed]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Epstein-Barr Virus and Kaposi’s Sarcoma Herpesvirus/Human Herpesvirus 8; IARC: Lyon, France, 1997; Volume 70. [Google Scholar]
- Farrell, P.J. Epstein-Barr virus and cancer. Annu. Rev. Pathol. Mech. Dis. 2019, 14, 29–53. [Google Scholar] [CrossRef]
- Shannon-Lowe, C.; Rickinson, A. The global landscape of EBV-associated tumors. Front. Oncol. 2019, 9, 713. [Google Scholar] [CrossRef] [PubMed]
- Camargo, M.C.; Kim, W.H.; Chiaravalli, A.M.; Kim, K.M.; Corvalan, A.H.; Matsuo, K.; Yu, J.; Sung, J.J.; Herrera-Goepfert, R.; Meneses-Gonzalez, F.; et al. Improved survival of gastric cancer with tumour Epstein-Barr virus positivity: An international pooled analysis. Gut 2014, 63, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, J.; Iizasa, H.; Yoshiyama, H.; Shimokuri, K.; Kobayashi, Y.; Sasaki, S.; Nakamura, M.; Yanai, H.; Sakai, K.; Suehiro, Y.; et al. Clinical Importance of Epstein(-)Barr Virus-Associated Gastric Cancer. Cancers 2018, 10, 167. [Google Scholar] [CrossRef]
- Neyts, J.; Sadler, R.; De Clercq, E.; Raab-Traub, N.; Pagano, J.S. The antiviral agent cidofovir [(S)-1-(3-hydroxy-2-phosphonyl-methoxypropyl)cytosine] has pronounced activity against nasopharyngeal carcinoma grown in nude mice. Cancer Res. 1998, 58, 384–388. [Google Scholar]
- Yoshizaki, T.; Wakisaka, N.; Kondo, S.; Murono, S.; Shimizu, Y.; Nakashima, M.; Tsuji, A.; Furukawa, M. Treatment of locally recurrent Epstein-Barr virus-associated nasopharyngeal carcinoma using the anti-viral agent cidofovir. J. Med. Virol. 2008, 80, 879–882. [Google Scholar] [CrossRef]
- Andrei, G.; Trompet, E.; Snoeck, R. Novel Therapeutics for Epstein(-)Barr Virus. Molecules 2019, 24, 997. [Google Scholar] [CrossRef]
- Kang, B.W.; Baek, D.W.; Kang, H.; Baek, J.H.; Kim, J.G. Novel Therapeutic Approaches for Epstein-Barr Virus Associated Gastric Cancer. Anticancer Res. 2019, 39, 4003–4010. [Google Scholar] [CrossRef]
- Sample, J.; Young, L.; Martin, B.; Chatman, T.; Kieff, E.; Rickinson, A.; Kieff, E. Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J. Virol. 1990, 64, 4084–4092. [Google Scholar] [CrossRef]
- Gratama, J.W.; Ernberg, I. Molecular epidemiology of Epstein-Barr virus infection. Adv. Cancer Res. 1995, 67, 197–255. [Google Scholar]
- Palser, A.L.; Grayson, N.E.; White, R.E.; Corton, C.; Correia, S.; Ba Abdullah, M.M.; Watson, S.J.; Cotten, M.; Arrand, J.R.; Murray, P.G.; et al. Genome diversity of Epstein-Barr virus from multiple tumor types and normal infection. J. Virol. 2015, 89, 5222–5237. [Google Scholar] [CrossRef]
- Thorley-Lawson, D.; Deitsch, K.W.; Duca, K.A.; Torgbor, C. The link between Plasmodium falciparum malaria and endemic Burkitt’s lymphoma-new insight into a 50-year-old enigma. PLoS Pathog. 2016, 12, e1005331. [Google Scholar] [CrossRef]
- Murphy, G.; Pfeiffer, R.; Camargo, M.C.; Rabkin, C.S. Meta-analysis shows that prevalence of Epstein-Barr virus-positive gastric cancer differs based on sex and anatomic location. Gastroenterology 2009, 137, 824–833. [Google Scholar] [CrossRef]
- Liu, S.; Zhao, Z.; Han, L.; Liu, S.; Luo, B. Epstein-Barr virus infection in gastric remnant carcinoma and recurrent gastric carcinoma in Qingdao of Northern China. PLoS ONE 2016, 11, e0148342. [Google Scholar] [CrossRef]
- Choi, M.G.; Jeong, J.Y.; Kim, K.M.; Bae, J.M.; Noh, J.H.; Sohn, T.S.; Kim, S. Clinical significance of gastritis cystica profunda and its association with Epstein-Barr virus in gastric cancer. Cancer 2012, 118, 5227–5233. [Google Scholar] [CrossRef]
- Burke, A.P.; Yen, T.S.; Shekitka, K.M.; Sobin, L.H. Lymphoepithelial carcinoma of the stomach with Epstein-Barr virus demonstrated by polymerase chain reaction. Mod. Pathol. 1990, 3, 377–380. [Google Scholar]
- Shibata, D.; Weiss, L.M. Epstein-Barr virus-associated gastric adenocarcinoma. Am. J. Pathol. 1992, 140, 769–774. [Google Scholar] [PubMed]
- Dong, M.; Wang, H.Y.; Zhao, X.X.; Chen, J.N.; Zhang, Y.W.; Huang, Y.; Xue, L.; Li, H.G.; Du, H.; Wu, X.Y.; et al. Expression and prognostic roles of PIK3CA, JAK2, PD-L1, and PD-L2 in Epstein-Barr virus-associated gastric carcinoma. Hum. Pathol. 2016, 53, 25–34. [Google Scholar] [CrossRef]
- Camargo, M.C.; Bowlby, R.; Chu, A.; Pedamallu, C.S.; Thorsson, V.; Elmore, S.; Mungall, A.J.; Bass, A.J.; Gulley, M.L.; Rabkin, C.S. Validation and calibration of next-generation sequencing to identify Epstein-Barr virus-positive gastric cancer in The Cancer Genome Atlas. Gastric Cancer 2016, 19, 676–681. [Google Scholar] [CrossRef] [PubMed]
- Sadato, D.; Ogawa, M.; Hirama, C.; Hishima, T.; Horiguchi, S.I.; Harada, Y.; Shimoyama, T.; Itokawa, M.; Ohashi, K.; Oboki, K. Potential prognostic impact of EBV RNA-seq reads in gastric cancer: A reanalysis of The Cancer Genome Atlas cohort. FEBS Open Bio 2020, 10, 455–467. [Google Scholar] [CrossRef]
- Chen, J.; Longnecker, R. Epithelial cell infection by Epstein–Barr virus. FEMS Microbiol. Rev. 2019, 43, 674–683. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.G.; Wang, T.C. Inflammation, atrophy, and gastric cancer. J. Clin. Investig. 2007, 117, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Swan, K.; Zhang, X.; Cao, S.; Brett, Z.; Drury, S.; Strong, M.J.; Fewell, C.; Puetter, A.; Wang, X.; et al. Secreted Oral Epithelial Cell Membrane Vesicles Induce Epstein-Barr Virus Reactivation in Latently Infected B Cells. J. Virol. 2016, 90, 3469–3479. [Google Scholar] [CrossRef]
- Luo, B.; Wang, Y.; Wang, X.F.; Liang, H.; Yan, L.P.; Huang, B.H.; Zhao, P. Expression of Epstein-Barr virus genes in EBV-associated gastric carcinomas. World J. Gastroenterol. 2005, 11, 629–633. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, M.; Imai, S.; Tokunaga, M.; Koizumi, S.; Uchizawa, M.; Okamoto, K.; Osato, T. Transcriptional analysis of Epstein-Barr virus gene expression in EBV-positive gastric carcinoma: Unique viral latency in the tumour cells. Br. J. Cancer 1996, 74, 625–631. [Google Scholar] [CrossRef]
- Borozan, I.; Zapatka, M.; Frappier, L.; Ferretti, V. Analysis of Epstein-Barr virus genomes and expression profiles in gastric adenocarcinoma. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Fukuda, M.; Longnecker, R. Latent membrane protein 2A inhibits transforming growth factor-beta 1-induced apoptosis through the phosphatidylinositol 3-kinase/Akt pathway. J. Virol. 2004, 78, 1697–1705. [Google Scholar] [CrossRef]
- Hino, R.; Uozaki, H.; Inoue, Y.; Shintani, Y.; Ushiku, T.; Sakatani, T.; Takada, K.; Fukayama, M. Survival advantage of EBV-associated gastric carcinoma: Survivin up-regulation by viral latent membrane protein 2A. Cancer Res. 2008, 68, 1427–1435. [Google Scholar] [CrossRef]
- Pal, A.D.; Basak, N.P.; Banerjee, A.S.; Banerjee, S. Epstein-Barr virus latent membrane protein-2A alters mitochondrial dynamics promoting cellular migration mediated by Notch signaling pathway. Carcinogenesis 2014, 35, 1592–1601. [Google Scholar] [CrossRef]
- Hino, R.; Uozaki, H.; Murakami, N.; Ushiku, T.; Shinozaki, A.; Ishikawa, S.; Morikawa, T.; Nakaya, T.; Sakatani, T.; Takada, K.; et al. Activation of DNA methyltransferase 1 by EBV latent membrane protein 2A leads to promoter hypermethylation of PTEN gene in gastric carcinoma. Cancer Res. 2009, 69, 2766–2774. [Google Scholar] [CrossRef]
- Sivachandran, N.; Dawson, C.W.; Young, L.S.; Liu, F.F.; Middeldorp, J.; Frappier, L. Contributions of the Epstein-Barr virus EBNA1 protein to gastric carcinoma. J. Virol. 2012, 86, 60–68. [Google Scholar] [CrossRef]
- Chavez-Calvillo, G.; Martin, S.; Hamm, C.; Sztuba-Solinska, J. The structure-to-function relationships of gammaherpesvirus-encoded long non-coding RNAs and their contributions to viral pathogenesis. Noncoding RNA 2018, 4, 24. [Google Scholar] [CrossRef]
- Namba-Fukuyo, H.; Funata, S.; Matsusaka, K.; Fukuyo, M.; Rahmutulla, B.; Mano, Y.; Fukayama, M.; Aburatani, H.; Kaneda, A. TET2 functions as a resistance factor against DNA methylation acquisition during Epstein-Barr virus infection. Oncotarget 2016, 7, 81512–81526. [Google Scholar] [CrossRef]
- Wang, J.; Liu, W.; Zhang, X.; Zhang, Y.; Xiao, H.; Luo, B. LMP2A induces DNA methylation and expression repression of AQP3 in EBV-associated gastric carcinoma. Virology 2019, 534, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Geddert, H.; zur Hausen, A.; Gabbert, H.E.; Sarbia, M. EBV-infection in cardiac and non-cardiac gastric adenocarcinomas is associated with promoter methylation of p16, p14 and APC, but not hMLH1. Cell. Oncol. 2011, 34, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Liang, Q.; Cheung, K.F.; Kang, W.; Dong, Y.; Lung, R.W.; Tong, J.H.; To, K.F.; Sung, J.J.; Yu, J. Somatostatin receptor 1, a novel EBV-associated CpG hypermethylated gene, contributes to the pathogenesis of EBV-associated gastric cancer. Br. J. Cancer 2013, 108, 2557–2564. [Google Scholar] [CrossRef]
- He, D.; Zhang, Y.W.; Zhang, N.N.; Zhou, L.; Chen, J.N.; Jiang, Y.; Shao, C.K. Aberrant gene promoter methylation of p16, FHIT, CRBP1, WWOX, and DLC-1 in Epstein-Barr virus-associated gastric carcinomas. Med. Oncol. 2015, 32, 92. [Google Scholar] [CrossRef]
- Yu, J.; Liang, Q.; Wang, J.; Wang, K.; Gao, J.; Zhang, J.; Zeng, Y.; Chiu, P.W.; Ng, E.K.; Sung, J.J. REC8 functions as a tumor suppressor and is epigenetically downregulated in gastric cancer, especially in EBV-positive subtype. Oncogene 2017, 36, 182–193. [Google Scholar] [CrossRef]
- Ushiku, T.; Chong, J.M.; Uozaki, H.; Hino, R.; Chang, M.S.; Sudo, M.; Rani, B.R.; Sakuma, K.; Nagai, H.; Fukayama, M. p73 gene promoter methylation in Epstein-Barr virus-associated gastric carcinoma. Int. J. Cancer 2007, 120, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Yi, F.; Saha, A.; Murakami, M.; Kumar, P.; Knight, J.S.; Cai, Q.; Choudhuri, T.; Robertson, E.S. Epstein-Barr virus nuclear antigen 3C targets p53 and modulates its transcriptional and apoptotic activities. Virology 2009, 388, 236–247. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.T.; Chang, Y.T.; Chen, S.C.; Chuang, Y.C.; Chen, Y.R.; Lin, C.S.; Chen, J.Y. Epstein-Barr virus latent membrane protein 1 represses p53-mediated DNA repair and transcriptional activity. Oncogene 2005, 24, 2635–2646. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Gutsch, D.; Kenney, S. Functional and physical interaction between p53 and BZLF1: Implications for Epstein-Barr virus latency. Mol. Cell. Biol. 1994, 14, 1929–1938. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Kamura, T.; Shirata, N.; Murata, T.; Kudoh, A.; Iwahori, S.; Nakayama, S.; Isomura, H.; Nishiyama, Y.; Tsurumi, T. Degradation of phosphorylated p53 by viral protein-ECS E3 ligase complex. PLoS Pathog. 2009, 5, e1000530. [Google Scholar] [CrossRef] [PubMed]
- Gossai, A.; Waterboer, T.; Nelson, H.H.; Michel, A.; Willhauck-Fleckenstein, M.; Farzan, S.F.; Hoen, A.G.; Christensen, B.C.; Kelsey, K.T.; Marsit, C.J.; et al. Seroepidemiology of human polyomaviruses in a US population. Am. J. Epidemiol. 2016, 183, 61–69. [Google Scholar] [CrossRef]
- Lundstig, A.; Dillner, J. Serological diagnosis of human polyomavirus infection. Adv. Exp. Med. Biol. 2006, 577, 96–101. [Google Scholar] [CrossRef]
- Del Valle, L.; White, M.K.; Enam, S.; Pina Oviedo, S.; Bromer, M.Q.; Thomas, R.M.; Parkman, H.P.; Khalili, K. Detection of JC virus DNA sequences and expression of viral T antigen and agnoprotein in esophageal carcinoma. Cancer 2005, 103, 516–527. [Google Scholar] [CrossRef]
- Hori, R.; Murai, Y.; Tsuneyama, K.; Abdel-Aziz, H.O.; Nomoto, K.; Takahashi, H.; Cheng, C.M.; Kuchina, T.; Harman, B.V.; Takano, Y. Detection of JC virus DNA sequences in colorectal cancers in Japan. Virchows Arch. 2005, 447, 723–730. [Google Scholar] [CrossRef]
- Murai, Y.; Zheng, H.C.; Abdel Aziz, H.O.; Mei, H.; Kutsuna, T.; Nakanishi, Y.; Tsuneyama, K.; Takano, Y. High JC virus load in gastric cancer and adjacent non-cancerous mucosa. Cancer Sci. 2007, 98, 25–31. [Google Scholar] [CrossRef]
- Shin, S.K.; Li, M.S.; Fuerst, F.; Hotchkiss, E.; Meyer, R.; Kim, I.T.; Goel, A.; Boland, C.R. Oncogenic T-antigen of JC virus is present frequently in human gastric cancers. Cancer 2006, 107, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Ksiaa, F.; Ziadi, S.; Mokni, M.; Korbi, S.; Trimeche, M. The presence of JC virus in gastric carcinomas correlates with patient’s age, intestinal histological type and aberrant methylation of tumor suppressor genes. Mod. Pathol. 2010, 23, 522–530. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka, S.; Yamamoto, H.; Nosho, K.; Taniguchi, H.; Adachi, Y.; Sasaki, S.; Arimura, Y.; Imai, K.; Shinomura, Y. Genetic and epigenetic characteristics of gastric cancers with JC virus T-antigen. World J. Gastroenterol. 2009, 15, 5579–5585. [Google Scholar] [CrossRef] [PubMed]
- Baez, C.F.; Brandao Varella, R.; Villani, S.; Delbue, S. Human Polyomaviruses: The battle of large and small tumor antigens. Virol. Res. Treat. 2017, 8. [Google Scholar] [CrossRef]
- Caracciolo, V.; Macaluso, M.; D’Agostino, L.; Montanari, M.; Scheff, J.; Reiss, K.; Khalili, K.; Giordano, A. Cross-talk between T-Ag presence and pRb family and p53/p73 signaling in mouse and human medulloblastoma. J. Cell. Biochem. 2010, 110, 182–190. [Google Scholar] [CrossRef]
- Enam, S.; Del Valle, L.; Lara, C.; Gan, D.D.; Ortiz-Hidalgo, C.; Palazzo, J.P.; Khalili, K. Association of human polyomavirus JCV with colon cancer: Evidence for interaction of viral T-antigen and beta-catenin. Cancer Res. 2002, 62, 7093–7101. [Google Scholar]
- Al Mana, H.; Yassine, H.M.; Younes, N.N.; Al-Mohannadi, A.; Al-Sadeq, D.W.; Alhababi, D.; Nasser, E.A.; Nasrallah, G.K. The current status of Cytomegalovirus (CMV) prevalence in the MENA region: A systematic review. Pathogens 2019, 8, 213. [Google Scholar] [CrossRef]
- Lv, Y.L.; Han, F.F.; An, Z.L.; Jia, Y.; Xuan, L.L.; Gong, L.L.; Zhang, W.; Ren, L.L.; Yang, S.; Liu, H.; et al. Cytomegalovirus infection is a risk factor in gastrointestinal cancer: A cross-sectional and meta-analysis study. Intervirology 2020, 63, 10–16. [Google Scholar] [CrossRef]
- Zhang, L.; Guo, G.; Xu, J.; Sun, X.; Chen, W.; Jin, J.; Hu, C.; Zhang, P.; Shen, X.; Xue, X. Human cytomegalovirus detection in gastric cancer and its possible association with lymphatic metastasis. Diagn. Microbiol. Infect. Dis. 2017, 88, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Hu, C.; Wang, P.; Chen, J.; Wu, T.; Chen, W.; Ye, L.; Zhu, G.; Zhang, L.; Xue, X.; et al. Latent infection of human cytomegalovirus is associated with the development of gastric cancer. Oncol. Lett. 2014, 8, 898–904. [Google Scholar] [CrossRef] [PubMed]
- Fattahi, S.; Nikbakhsh, N.; Taheri, H.; Ghadami, E.; Kosari-Monfared, M.; Amirbozorgi, G.; Asouri, M.; Pilehchian-Langroudi, M.; Ranaee, M.; Samadani, A.A.; et al. Prevalence of multiple infections and the risk of gastric adenocarcinoma development at earlier age. Diagn. Microbiol. Infect. Dis. 2018, 92, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Kosari-Monfared, M.; Nikbakhsh, N.; Fattahi, S.; Ghadami, E.; Ranaei, M.; Taheri, H.; Amjadi-Moheb, F.; Godazandeh, G.A.; Shafaei, S.; Pilehchian-Langroudi, M.; et al. CTNNBIP1 downregulation is associated with tumor grade and viral infections in gastric adenocarcinoma. J. Cell. Physiol. 2019, 234, 2895–2904. [Google Scholar] [CrossRef]
- Cui, H.; Jin, Y.; Chen, F.; Ni, H.; Hu, C.; Xu, Y.; Xuan, H.; Hu, D.; Deng, W.; Zhang, Y.; et al. Clinicopathological evidence of hepatitis B virus infection in the development of gastric adenocarcinoma. J. Med. Virol. 2020, 92, 71–77. [Google Scholar] [CrossRef]
- Baghbanian, M.; Hoseini Mousa, S.A.; Doosti, M.; Moghimi, M. Association between gastric pathology and Hepatitis B virus infection in patients with or without Helicobacter pylori. Asian Pac. J. Cancer Prev. 2019, 20, 2177–2180. [Google Scholar] [CrossRef]
- Chen, C.W.; Cheng, J.S.; Chen, T.D.; Le, P.H.; Ku, H.P.; Chang, M.L. The irreversible HCV-associated risk of gastric cancer following interferon-based therapy: A joint study of hospital-based cases and nationwide population-based cohorts. Ther. Adv. Gastroenterol. 2019, 12, 1756284819855732. [Google Scholar] [CrossRef]
- Kai, K.; Miyoshi, A.; Kitahara, K.; Masuda, M.; Takase, Y.; Miyazaki, K.; Noshiro, H.; Tokunaga, O. Analysis of extrahepatic multiple primary malignancies in patients with hepatocellular carcinoma according to viral infection status. Int. J. Hepatol. 2012, 2012, 495950. [Google Scholar] [CrossRef]
- Kocoglu, H.; Karaca, M.; Tural, D.; Hocaoglu, E.; Okuturlar, Y.; Fetullahoglu, Z.; Gunaldi, M.; Ciftci, R.; Tuna, S.; Yucil, O.K.; et al. Hepatitis B and C rates are significantly increased in certain solid tumors: A large retrospective study. J. Cancer Res. Ther. 2018, 14, S774–S778. [Google Scholar] [CrossRef]
- Song, C.; Lv, J.; Liu, Y.; Chen, J.G.; Ge, Z.; Zhu, J.; Dai, J.; Du, L.B.; Yu, C.; Guo, Y.; et al. Associations between Hepatitis B virus infection and risk of all cancer types. JAMA Netw. Open 2019, 2, e195718. [Google Scholar] [CrossRef]
- Huang, C.F.; Lai, H.C.; Chen, C.Y.; Tseng, K.C.; Kuo, H.T.; Hung, C.H.; Wang, J.H.; Chen, J.J.; Lee, P.L.; Chien, R.N.; et al. Extrahepatic malignancy among patients with chronic Hepatitis C after antiviral therapy: A real-world nationwide Study on Taiwanese chronic Hepatitis C cohort (T-COACH). Am. J. Gastroenterol. 2020. [Google Scholar] [CrossRef]
- Kang, J.S.; Lee, S.H.; Lee, S.; Kim, G.H.; Park, Y.J.; Han, I.S.; Lee, J.E.; Lee, S.O.; Moon, C. Role of upper gastrointestinal endoscopy in patients with human immunodeficiency virus infection in the era of combination antiretroviral therapy. Infect. Chemother. 2019, 51, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Schierhout, G.; McGregor, S.; Gessain, A.; Einsiedel, L.; Martinello, M.; Kaldor, J. Association between HTLV-1 infection and adverse health outcomes: A systematic review and meta-analysis of epidemiological studies. Lancet Infect. Dis. 2020, 20, 133–143. [Google Scholar] [CrossRef]
- Falasca, F.; Maida, P.; Gaeta, A.; Verzaro, S.; Mezzaroma, I.; Fantauzzi, A.; Donato, G.; Bonci, E.; Castilletti, C.; Antonelli, G.; et al. Detection and quantification of EBV, HHV-6 and CMV DNA in the gastrointestinal tract of HIV-positive patients. Infection 2014, 42, 1033–1037. [Google Scholar] [CrossRef]
- Kayamba, V.; Monze, M.; Asombang, A.W.; Zyambo, K.; Kelly, P. Serological response to Epstein-Barr virus early antigen is associated with gastric cancer and human immunodeficiency virus infection in Zambian adults: A case-control study. Pan Afr. Med. J. 2016, 23, 45. [Google Scholar] [CrossRef]
- Matsumoto, S.; Yamasaki, K.; Tsuji, K.; Shirahama, S. Human T lymphotropic virus type 1 infection and gastric cancer development in Japan. J. Infect. Dis. 2008, 198, 10–15. [Google Scholar] [CrossRef]
- Bozdayi, G.; Dinc, B.; Avcikucuk, H.; Turhan, N.; Altay-Kocak, A.; Ozkan, S.; Ozin, Y.; Bostanci, B. Is human papillomavirus and Helicobacter pylori related in gastric lesions? Clin. Lab. 2019, 65. [Google Scholar] [CrossRef]
- Zeng, Z.M.; Luo, F.F.; Zou, L.X.; He, R.Q.; Pan, D.H.; Chen, X.; Xie, T.T.; Li, Y.Q.; Peng, Z.G.; Chen, G. Human papillomavirus as a potential risk factor for gastric cancer: A meta-analysis of 1917 cases. OncoTargets Ther. 2016, 9, 7105–7114. [Google Scholar] [CrossRef]
- De Souza, C.R.T.; Almeida, M.C.A.; Khayat, A.S.; da Silva, E.L.; Soares, P.C.; Chaves, L.C.; Burbano, R.M.R. Association between Helicobacter pylori, Epstein-Barr virus, human papillomavirus and gastric adenocarcinomas. World J. Gastroenterol. 2018, 24, 4928–4938. [Google Scholar] [CrossRef]
- Snietura, M.; Waniczek, D.; Piglowski, W.; Kopec, A.; Nowakowska-Zajdel, E.; Lorenc, Z.; Muc-Wierzgon, M. Potential role of human papilloma virus in the pathogenesis of gastric cancer. World J. Gastroenterol. 2014, 20, 6632–6637. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).