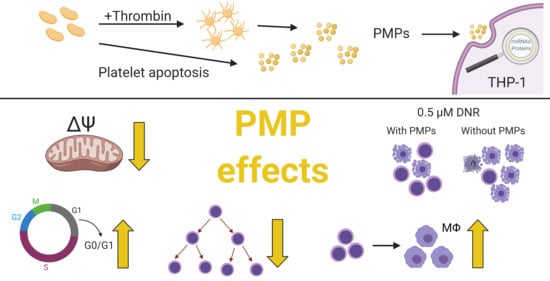
Platelet Microparticles Protect Acute Myelogenous Leukemia Cells against Daunorubicin-Induced Apoptosis
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
2.1. Platelet-Associated microRNAs Are Increased in THP-1 Cells after PMP Co-Incubation
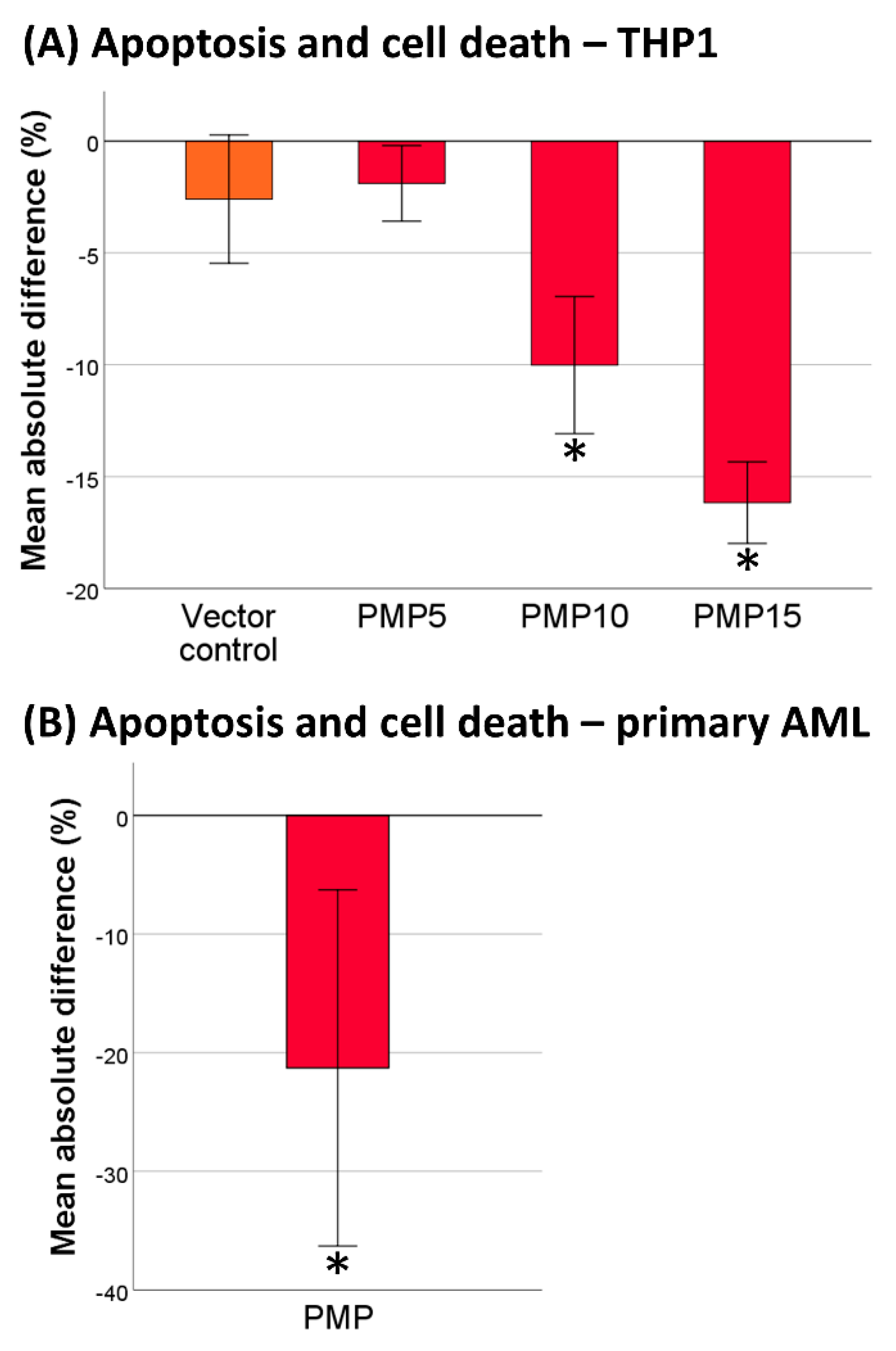
2.2. PMPs Lead to Increased Resistance of THP-1 Cells to DNR
2.3. Co-Incubation of Primary AML Cells with PMPs Also Increased Resistance to DNR
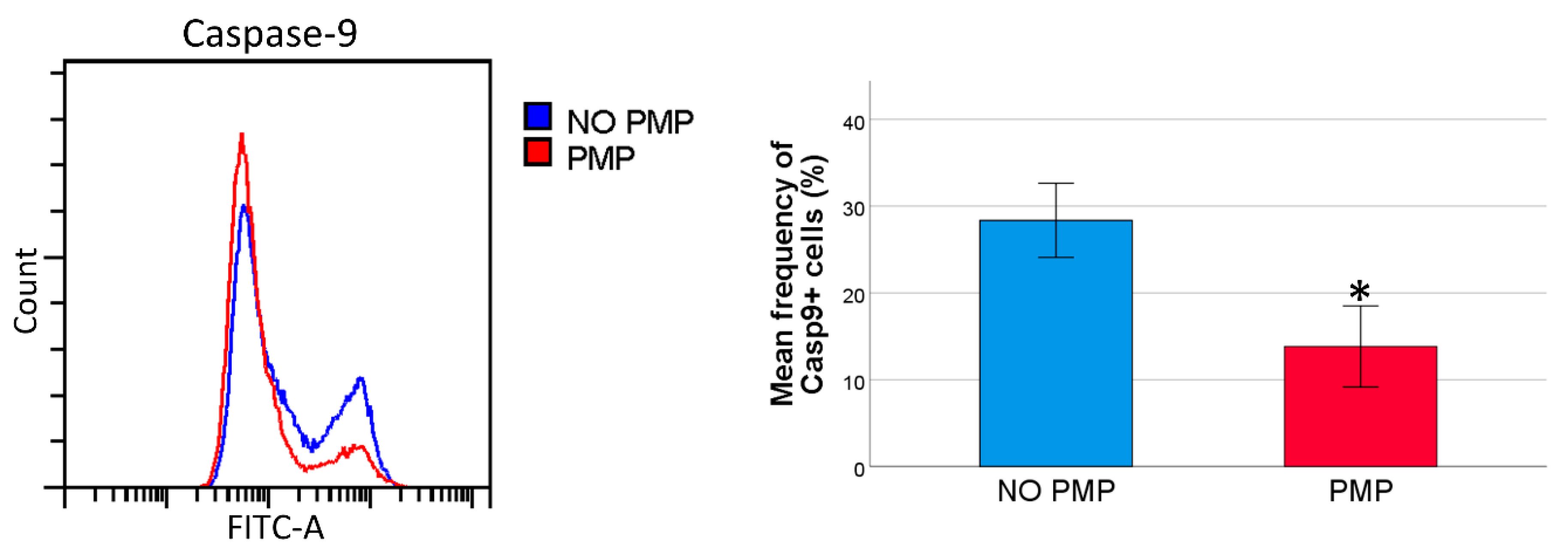
2.4. THP-1 Cells Co-Incubated with PMPs Had Lower Caspase-9 Activity Following DNR-Treatment
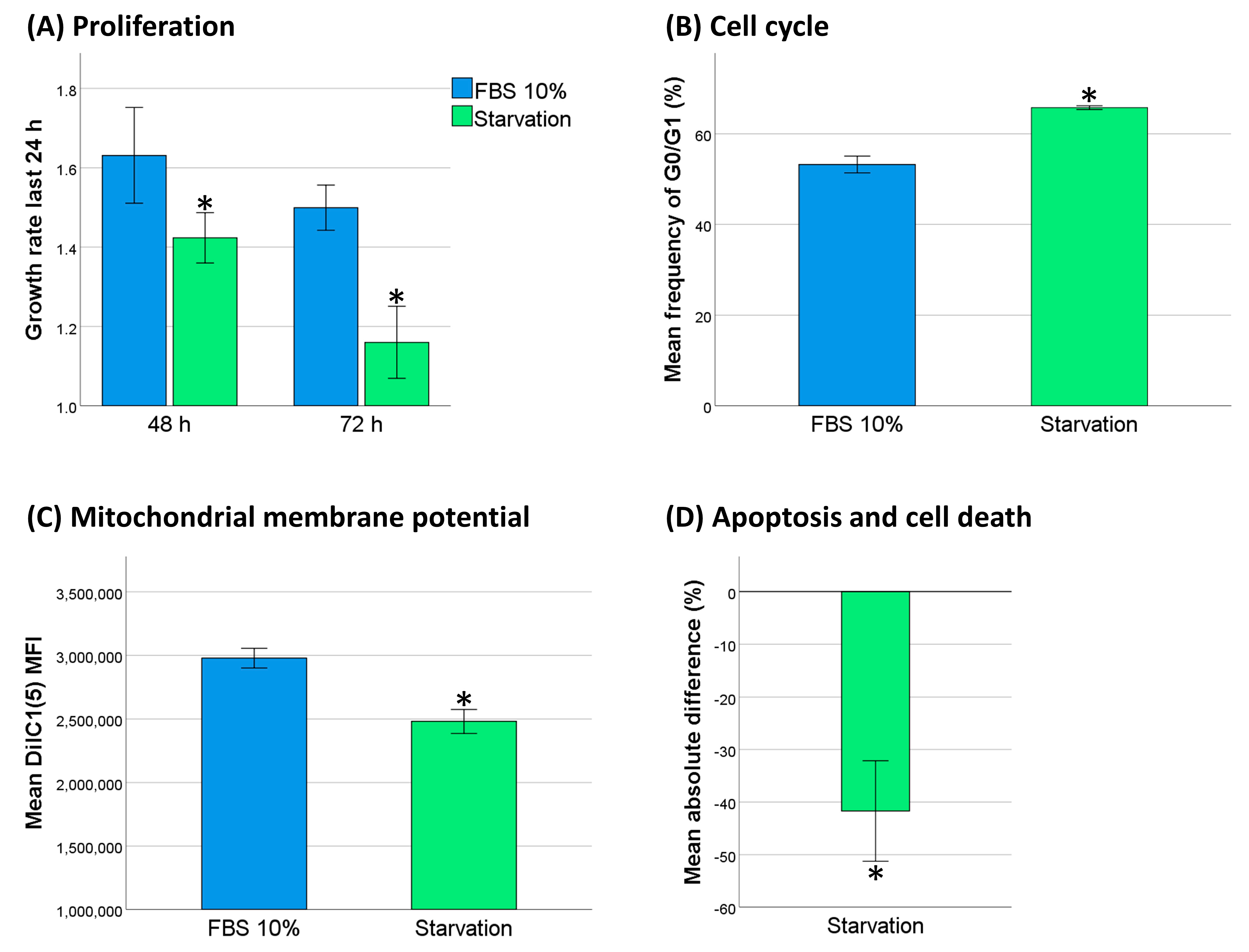
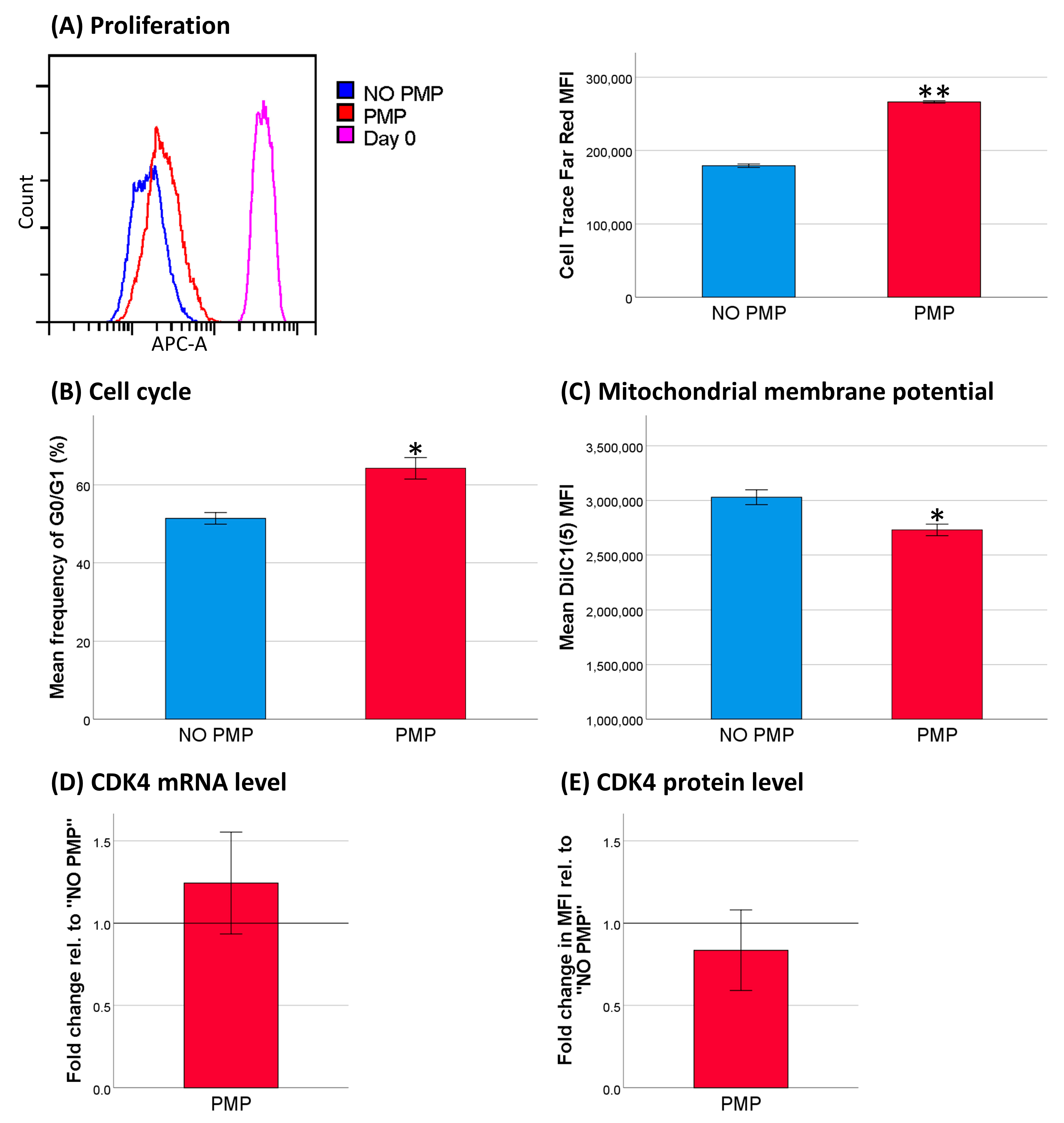
2.5. Decreased Cell Activity Protected THP-1 Cells against DNR
2.6. PMP Co-Incubation Increased Differentiation of THP-1 Cells toward Macrophages
3. Discussion
4. Materials and Methods
4.1. Cell Line
4.2. Primary AML Cells
4.3. Platelet Concentrate
4.4. Platelet Releasate
4.5. Platelet Microparticles Isolation, Co-Culture, and Measurement
4.6. Acridine Orange Staining of Platelet Microparticles
4.7. mRNA and microRNA Analysis
4.8. Daunorubicin Apoptosis Assay
4.9. Caspase-9 Activity
4.10. Mitochondrial Membrane Potential
4.11. Cell Cycle Analysis
4.12. Flow Cytometry Proliferation Analysis
4.13. Flow Cytometry Immunophenotyping
4.14. Measurement of CDK4 by Indirect Intracellular Flow Cytomtery
4.15. Measurement of Cell Cross-Sectional Area
4.16. Serum Starvation
4.17. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sarry, J.-E.; Murphy, K.; Perry, R.; Sanchez, P.V.; Secreto, A.; Keefer, C.; Swider, C.R.; Strzelecki, A.-C.; Cavelier, C.; Récher, C.; et al. Human acute myelogenous leukemia stem cells are rare and heterogeneous when assayed in nod/scid/il2rγc-deficient mice. J. Clin. Investig. 2011, 121, 384–395. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.W.K.; Mitchell, A.; Kennedy, J.A.; Chen, W.C.; McLeod, J.; Ibrahimova, N.; Arruda, A.; Popescu, A.; Gupta, V.; Schimmer, A.D.; et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature 2016, 540, 433–437. [Google Scholar] [CrossRef]
- Krivtsov, A.V.; Twomey, D.; Feng, Z.; Stubbs, M.C.; Wang, Y.; Faber, J.; Levine, J.E.; Wang, J.; Hahn, W.C.; Gilliland, D.G.; et al. Transformation from committed progenitor to leukaemia stem cell initiated by mll-af9. Nature 2006, 442, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Kantarjian, H. Acute myeloid leukemia--major progress over four decades and glimpses into the future. Am. J. Hematol. 2016, 91, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Herold, T.; Rothenberg-Thurley, M.; Grunwald, V.V.; Janke, H.; Goerlich, D.; Sauerland, M.C.; Konstandin, N.P.; Dufour, A.; Schneider, S.; Neusser, M.; et al. Validation and refinement of the revised 2017 european leukemianet genetic risk stratification of acute myeloid leukemia. Leukemia 2020, 34, 3161–3172. [Google Scholar] [CrossRef] [PubMed]
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and management of aml in adults: 2017 eln recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef]
- Zhang, Q.; Hu, H.; Liu, H.; Jin, J.; Zhu, P.; Wang, S.; Shen, K.; Hu, Y.; Li, Z.; Zhan, P.; et al. Rna sequencing enables systematic identification of platelet transcriptomic alterations in nsclc patients. Biomed. Pharmacother. 2018, 105, 204–214. [Google Scholar] [CrossRef]
- Nieswandt, B.; Hafner, M.; Echtenacher, B.; Männel, D.N. Lysis of tumor cells by natural killer cells in mice is impeded by platelets. Cancer Res. 1999, 59, 1295–1300. [Google Scholar]
- Kopp, H.G.; Placke, T.; Salih, H.R. Platelet-derived transforming growth factor-beta down-regulates nkg2d thereby inhibiting natural killer cell antitumor reactivity. Cancer Res. 2009, 69, 7775–7783. [Google Scholar] [CrossRef]
- Yu, M.; Bardia, A.; Wittner, B.S.; Stott, S.L.; Smas, M.E.; Ting, D.T.; Isakoff, S.J.; Ciciliano, J.C.; Wells, M.N.; Shah, A.M.; et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013, 339, 580–584. [Google Scholar] [CrossRef]
- Takemoto, A.; Okitaka, M.; Takagi, S.; Takami, M.; Sato, S.; Nishio, M.; Okumura, S.; Fujita, N. A critical role of platelet tgf-β release in podoplanin-mediated tumour invasion and metastasis. Sci. Rep. 2017, 7, 42186. [Google Scholar] [CrossRef] [PubMed]
- Palacios-Acedo, A.L.; Mège, D.; Crescence, L.; Dignat-George, F.; Dubois, C.; Panicot-Dubois, L. Platelets, thrombo-inflammation, and cancer: Collaborating with the enemy. Front. Immunol. 2019, 10, 1805. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Jurasz, P. The role of platelets in the tumor microenvironment: From solid tumors to leukemia. Biochim. Biophys. Acta 2016, 1863, 392–400. [Google Scholar] [CrossRef] [PubMed]
- De Paoli, S.H.; Tegegn, T.Z.; Elhelu, O.K.; Strader, M.B.; Patel, M.; Diduch, L.L.; Tarandovskiy, I.D.; Wu, Y.; Zheng, J.; Ovanesov, M.V.; et al. Dissecting the biochemical architecture and morphological release pathways of the human platelet extracellular vesiculome. Cell. Mol. Life Sci. Cmls 2018, 75, 3781–3801. [Google Scholar] [CrossRef]
- Black, A.; Orsó, E.; Kelsch, R.; Pereira, M.; Kamhieh-Milz, J.; Salama, A.; Fischer, M.B.; Meyer, E.; Frey, B.M.; Schmitz, G. Analysis of platelet-derived extracellular vesicles in plateletpheresis concentrates: A multicenter study. Transfusion 2017, 57, 1459–1469. [Google Scholar] [CrossRef]
- Vasina, E.M.; Cauwenberghs, S.; Staudt, M.; Feijge, M.A.H.; Weber, C.; Koenen, R.R.; Heemskerk, J.W.M. Aging- and activation-induced platelet microparticles suppress apoptosis in monocytic cells and differentially signal to proinflammatory mediator release. Am. J. Blood Res. 2013, 3, 107–123. [Google Scholar]
- Laffont, B.; Corduan, A.; Ple, H.; Duchez, A.C.; Cloutier, N.; Boilard, E.; Provost, P. Activated platelets can deliver mrna regulatory ago2*microrna complexes to endothelial cells via microparticles. Blood 2013, 122, 253–261. [Google Scholar] [CrossRef]
- Risitano, A.; Beaulieu, L.M.; Vitseva, O.; Freedman, J.E. Platelets and platelet-like particles mediate intercellular rna transfer. Blood 2012, 119, 6288–6295. [Google Scholar] [CrossRef]
- Anene, C.; Graham, A.M.; Boyne, J.; Roberts, W. Platelet microparticle delivered microrna-let-7a promotes the angiogenic switch. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 2633–2643. [Google Scholar] [CrossRef]
- Maynard, D.M.; Heijnen, H.F.; Horne, M.K.; White, J.G.; Gahl, W.A. Proteomic analysis of platelet alpha-granules using mass spectrometry. J. Thromb. Haemost. 2007, 5, 1945–1955. [Google Scholar] [CrossRef]
- Parsons, M.E.M.; Szklanna, P.B.; Guererro, J.A.; Wynne, K.; Dervin, F.; O’Connell, K.; Allen, S.; Egan, K.; Bennett, C.; McGuigan, C.; et al. Platelet releasate proteome profiling reveals a core set of proteins with low variance between healthy adults. Proteomics 2018, 18, e1800219. [Google Scholar] [CrossRef] [PubMed]
- Pienimaeki-Roemer, A.; Konovalova, T.; Musri, M.M.; Sigruener, A.; Boettcher, A.; Meister, G.; Schmitz, G. Transcriptomic profiling of platelet senescence and platelet extracellular vesicles. Transfusion 2017, 57, 144–156. [Google Scholar] [CrossRef] [PubMed]
- Ambrose, A.R.; Alsahli, M.A.; Kurmani, S.A.; Goodall, A.H. Comparison of the release of micrornas and extracellular vesicles from platelets in response to different agonists. Platelets 2018, 29, 446–454. [Google Scholar] [CrossRef]
- Liang, H.; Yan, X.; Pan, Y.; Wang, Y.; Wang, N.; Li, L.; Liu, Y.; Chen, X.; Zhang, C.Y.; Gu, H.; et al. Microrna-223 delivered by platelet-derived microvesicles promotes lung cancer cell invasion via targeting tumor suppressor epb41l3. Mol. Cancer 2015, 14, 58. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Jiang, L.; Lin, Y.; Wu, X.; Wang, K.; He, Q.; Wang, X.; Li, W. Platelet microparticle-mediated transfer of mir-939 to epithelial ovarian cancer cells promotes epithelial to mesenchymal transition. Oncotarget 2017, 8, 97464–97475. [Google Scholar] [CrossRef] [PubMed]
- Michael, J.V.; Wurtzel, J.G.T.; Mao, G.F.; Rao, A.K.; Kolpakov, M.A.; Sabri, A.; Hoffman, N.E.; Rajan, S.; Tomar, D.; Madesh, M.; et al. Platelet microparticles infiltrating solid tumors transfer mirnas that suppress tumor growth. Blood 2017, 130, 567–580. [Google Scholar] [CrossRef]
- Cassier, P.A.; Castets, M.; Belhabri, A.; Vey, N. Targeting apoptosis in acute myeloid leukaemia. Br. J. Cancer 2017, 117, 1089–1098. [Google Scholar] [CrossRef]
- Ashkenazi, A.; Fairbrother, W.J.; Leverson, J.D.; Souers, A.J. From basic apoptosis discoveries to advanced selective bcl-2 family inhibitors. Nat. Rev. Drug Discov. 2017, 16, 273–284. [Google Scholar] [CrossRef]
- Velez, J.; Enciso, L.J.; Suarez, M.; Fiegl, M.; Grismaldo, A.; López, C.; Barreto, A.; Cardozo, C.; Palacios, P.; Morales, L.; et al. Platelets promote mitochondrial uncoupling and resistance to apoptosis in leukemia cells: A novel paradigm for the bone marrow microenvironment. Cancer Microenviron. 2014, 7, 79–90. [Google Scholar] [CrossRef]
- Bai, H.; Zhou, L.; Wang, C.; Xu, X.; Jiang, J.; Qin, Y.; Wang, X.; Zhao, C.; Shao, S. Involvement of mir-125a in resistance to daunorubicin by inhibiting apoptosis in leukemia cell lines. Tumour Biol. 2017, 39, 1010428317695964. [Google Scholar] [CrossRef]
- Zhou, L.; Bai, H.; Wang, C.; Wei, D.; Qin, Y.; Xu, X. Microrna125b promotes leukemia cell resistance to daunorubicin by inhibiting apoptosis. Mol. Med. Rep. 2014, 9, 1909–1916. [Google Scholar] [CrossRef] [PubMed]
- Gabra, M.M.; Salmena, L. Micrornas and acute myeloid leukemia chemoresistance: A mechanistic overview. Front. Oncol. 2017, 7, 255. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Volinia, S.; Liu, C.G.; Fernandez-Cymering, C.; Palumbo, T.; Pichiorri, F.; Fabbri, M.; Coombes, K.; Alder, H.; Nakamura, T.; et al. Microrna signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood 2008, 111, 3183–3189. [Google Scholar] [CrossRef]
- Lim, E.L.; Trinh, D.L.; Ries, R.E.; Wang, J.; Gerbing, R.B.; Ma, Y.; Topham, J.; Hughes, M.; Pleasance, E.; Mungall, A.J.; et al. Microrna expression-based model indicates event-free survival in pediatric acute myeloid leukemia. J. Clin. Oncol. 2017, 35, 3964–3977. [Google Scholar] [CrossRef] [PubMed]
- Bruserud, O.; Gjertsen, B.T.; Foss, B.; Huang, T.S. New strategies in the treatment of acute myelogenous leukemia (aml): In vitro culture of aml cells—The present use in experimental studies and the possible importance for future therapeutic approaches. Stem Cells (Dayt. Ohio) 2001, 19, 1–11. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. Mirdb: An online database for prediction of functional microrna targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microrna target sites in mammalian mrnas. Elife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Youle, R.J.; Strasser, A. The bcl-2 protein family: Opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 2008, 9, 47–59. [Google Scholar] [CrossRef]
- Trotter, E.W.; Hagan, I.M. Release from cell cycle arrest with cdk4/6 inhibitors generates highly synchronized cell cycle progression in human cell culture. Open Biol. 2020, 10, 200200. [Google Scholar] [CrossRef]
- Skowron, M.A.; Vermeulen, M.; Winkelhausen, A.; Becker, T.K.; Bremmer, F.; Petzsch, P.; Schönberger, S.; Calaminus, G.; Köhrer, K.; Albers, P.; et al. Cdk4/6 inhibition presents as a therapeutic option for paediatric and adult germ cell tumours and induces cell cycle arrest and apoptosis via canonical and non-canonical mechanisms. Br. J. Cancer 2020, 123, 378–391. [Google Scholar] [CrossRef]
- Daigneault, M.; Preston, J.A.; Marriott, H.M.; Whyte, M.K.; Dockrell, D.H. The identification of markers of macrophage differentiation in pma-stimulated thp-1 cells and monocyte-derived macrophages. PLoS ONE 2010, 5, e8668. [Google Scholar] [CrossRef]
- Bogason, A.; Quartino, A.L.; Lafolie, P.; Masquelier, M.; Karlsson, M.O.; Paul, C.; Gruber, A.; Vitols, S. Inverse relationship between leukaemic cell burden and plasma concentrations of daunorubicin in patients with acute myeloid leukaemia. Br. J. Clin. Pharmacol. 2011, 71, 514–521. [Google Scholar] [CrossRef]
- Burnett, A.K.; Russell, N.H.; Hills, R.K.; Kell, J.; Cavenagh, J.; Kjeldsen, L.; McMullin, M.F.; Cahalin, P.; Dennis, M.; Friis, L.; et al. A randomized comparison of daunorubicin 90 mg/m2 vs. 60 mg/m2 in aml induction: Results from the uk ncri aml17 trial in 1206 patients. Blood 2015, 125, 3878–3885. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, K.L.; Huang, K.; Zhang, J.; Chen, Y.; Luo, X. Inactivation of prosurvival bcl-2 proteins activates bax/bak through the outer mitochondrial membrane. Genes Dev. 2016, 30, 973–988. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Yu, M.C.; Gao, G.L.; Liang, H.W.; Zhou, X.Y.; Zhu, Z.T.; Zhang, C.Y.; Wang, Y.B.; Chen, X. Mir-125a-5p functions as a tumour suppressor in breast cancer by downregulating bap1. J. Cell. Biochem. 2018, 119, 8773–8783. [Google Scholar] [CrossRef]
- Tong, Z.; Liu, N.; Lin, L.; Guo, X.; Yang, D.; Zhang, Q. Mir-125a-5p inhibits cell proliferation and induces apoptosis in colon cancer via targeting bcl2, bcl2l12 and mcl1. Biomed. Pharmacother. 2015, 75, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Marinello, J.; Delcuratolo, M.; Capranico, G. Anthracyclines as topoisomerase ii poisons: From early studies to new perspectives. Int. J. Mol. Sci. 2018, 19, 3480. [Google Scholar] [CrossRef]
- Al-Aamri, H.M.; Ku, H.; Irving, H.R.; Tucci, J.; Meehan-Andrews, T.; Bradley, C. Time dependent response of daunorubicin on cytotoxicity, cell cycle and DNA repair in acute lymphoblastic leukaemia. Bmc Cancer 2019, 19, 179. [Google Scholar] [CrossRef] [PubMed]
- Gažová, I.; Lefevre, L.; Bush, S.J.; Clohisey, S.; Arner, E.; de Hoon, M.; Severin, J.; van Duin, L.; Andersson, R.; Lengeling, A.; et al. The transcriptional network that controls growth arrest and macrophage differentiation in the human myeloid leukemia cell line thp-1. Front. Cell Dev. Biol. 2020, 8, 498. [Google Scholar] [CrossRef]
- Bernardi, M.; Agostini, F.; Chieregato, K.; Amati, E.; Durante, C.; Rassu, M.; Ruggeri, M.; Sella, S.; Lombardi, E.; Mazzucato, M.; et al. The production method affects the efficacy of platelet derivatives to expand mesenchymal stromal cells in vitro. J. Transl. Med. 2017, 15, 90. [Google Scholar] [CrossRef]
- Agostini, F.; Polesel, J.; Battiston, M.; Lombardi, E.; Zanolin, S.; Da Ponte, A.; Astori, G.; Durante, C.; Mazzucato, M. Standardization of platelet releasate products for clinical applications in cell therapy: A mathematical approach. J. Transl. Med. 2017, 15, 107. [Google Scholar] [CrossRef]
- Plantureux, L.; Mège, D.; Crescence, L.; Carminita, E.; Robert, S.; Cointe, S.; Brouilly, N.; Ezzedine, W.; Dignat-George, F.; Dubois, C.; et al. The interaction of platelets with colorectal cancer cells inhibits tumor growth but promotes metastasis. Cancer Res. 2020, 80, 291–303. [Google Scholar] [CrossRef]
- Żmigrodzka, M.; Witkowska-Piłaszewicz, O.; Winnicka, A. Platelets extracellular vesicles as regulators of cancer progression-an updated perspective. Int. J. Mol. Sci. 2020, 21, 5195. [Google Scholar] [CrossRef]
- Giacomazzi, A.; Degan, M.; Calabria, S.; Meneguzzi, A.; Minuz, P. Antiplatelet agents inhibit the generation of platelet-derived microparticles. Front. Pharmacol. 2016, 7, 314. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.T.; Ogino, S.; Fuchs, C.S. Aspirin use and survival after diagnosis of colorectal cancer. JAMA 2009, 302, 649–658. [Google Scholar] [CrossRef]
- Bains, S.J.; Mahic, M.; Myklebust, T.A.; Smastuen, M.C.; Yaqub, S.; Dorum, L.M.; Bjornbeth, B.A.; Moller, B.; Brudvik, K.W.; Tasken, K. Aspirin as secondary prevention in patients with colorectal cancer: An unselected population-based study. J. Clin. Oncol. 2016, 34, 2501–2508. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Nishihara, R.; Wu, K.; Wang, M.; Ogino, S.; Willett, W.C.; Spiegelman, D.; Fuchs, C.S.; Giovannucci, E.L.; Chan, A.T. Population-wide impact of long-term use of aspirin and the risk for cancer. JAMA Oncol. 2016, 2, 762–769. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Yang, T.; Gan, Y.; Li, W.; Wang, C.; Gong, Y.; Lu, Z. Associations between aspirin use and the risk of cancers: A meta-analysis of observational studies. Bmc Cancer 2018, 18, 288. [Google Scholar] [CrossRef]
- Poncelet, P.; Robert, S.; Bouriche, T.; Bez, J.; Lacroix, R.; Dignat-George, F. Standardized counting of circulating platelet microparticles using currently available flow cytometers and scatter-based triggering: Forward or side scatter? Cytom. Part A 2016, 89, 148–158. [Google Scholar] [CrossRef]
- Ludwig, L.M.; Maxcy, K.L.; LaBelle, J.L. Flow cytometry-based detection and analysis of bcl-2 family proteins and mitochondrial outer membrane permeabilization (momp). Methods Mol. Biol. (Cliftonn. J.) 2019, 1877, 77–91. [Google Scholar]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. Nih image to imagej: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]


| # | Sex | Age | Prev. Myeloid Disease | FAB | Cytogenetics | FLT3 | NPM1 | CEBPA | CD11b | CD14 | CD33 | CD34 | CD45 | CD64 | CD117 | HLA-DR | L-MPO |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 22 | No | M5 | del(9) | ITD (low ratio) | wt | neg | neg | pos | pos | dim | pos | pos | dim | ||
| 2 | F | 60 | PV | del(7) | wt | wt | wt | neg | neg | pos | dim | pos | pos | ||||
| 3 | F | 56 | No | M4 | inv(16) | wt | wt | neg | neg | pos | pos | dim | neg | pos | pos | pos | |
| 4 | M | 44 | No | M4 | inv(16) | wt | wt | neg | neg | neg | neg | dim | dim | neg | pos | ||
| 5 | F | 92 | No | M1 | neg | neg | dim | neg | dim | dim | pos | pos | pos | ||||
| 6 | M | 49 | No | M4 | 45, XY | wt | ins | wt | neg | neg | neg | dim | dim | pos | pos | ||
| 7 | M | 76 | No | M5 | Normal | wt | ins | wt | hetero | hetero | neg | hetero | pos | hetero | pos | ||
| 8 | F | 95 | No | M4 | Normal | wt | wt | wt | dim | dim | dim | ||||||
| 9 | M | 29 | No | M4 | Normal | ITD (high ratio) | wt | wt | neg | neg | neg | pos | dim | neg | neg | pos | pos |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cacic, D.; Reikvam, H.; Nordgård, O.; Meyer, P.; Hervig, T. Platelet Microparticles Protect Acute Myelogenous Leukemia Cells against Daunorubicin-Induced Apoptosis. Cancers 2021, 13, 1870. https://doi.org/10.3390/cancers13081870
Cacic D, Reikvam H, Nordgård O, Meyer P, Hervig T. Platelet Microparticles Protect Acute Myelogenous Leukemia Cells against Daunorubicin-Induced Apoptosis. Cancers. 2021; 13(8):1870. https://doi.org/10.3390/cancers13081870
Chicago/Turabian StyleCacic, Daniel, Håkon Reikvam, Oddmund Nordgård, Peter Meyer, and Tor Hervig. 2021. "Platelet Microparticles Protect Acute Myelogenous Leukemia Cells against Daunorubicin-Induced Apoptosis" Cancers 13, no. 8: 1870. https://doi.org/10.3390/cancers13081870
APA StyleCacic, D., Reikvam, H., Nordgård, O., Meyer, P., & Hervig, T. (2021). Platelet Microparticles Protect Acute Myelogenous Leukemia Cells against Daunorubicin-Induced Apoptosis. Cancers, 13(8), 1870. https://doi.org/10.3390/cancers13081870