Ropporin-1 and 1B Are Widely Expressed in Human Melanoma and Evoke Strong Humoral Immune Responses
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
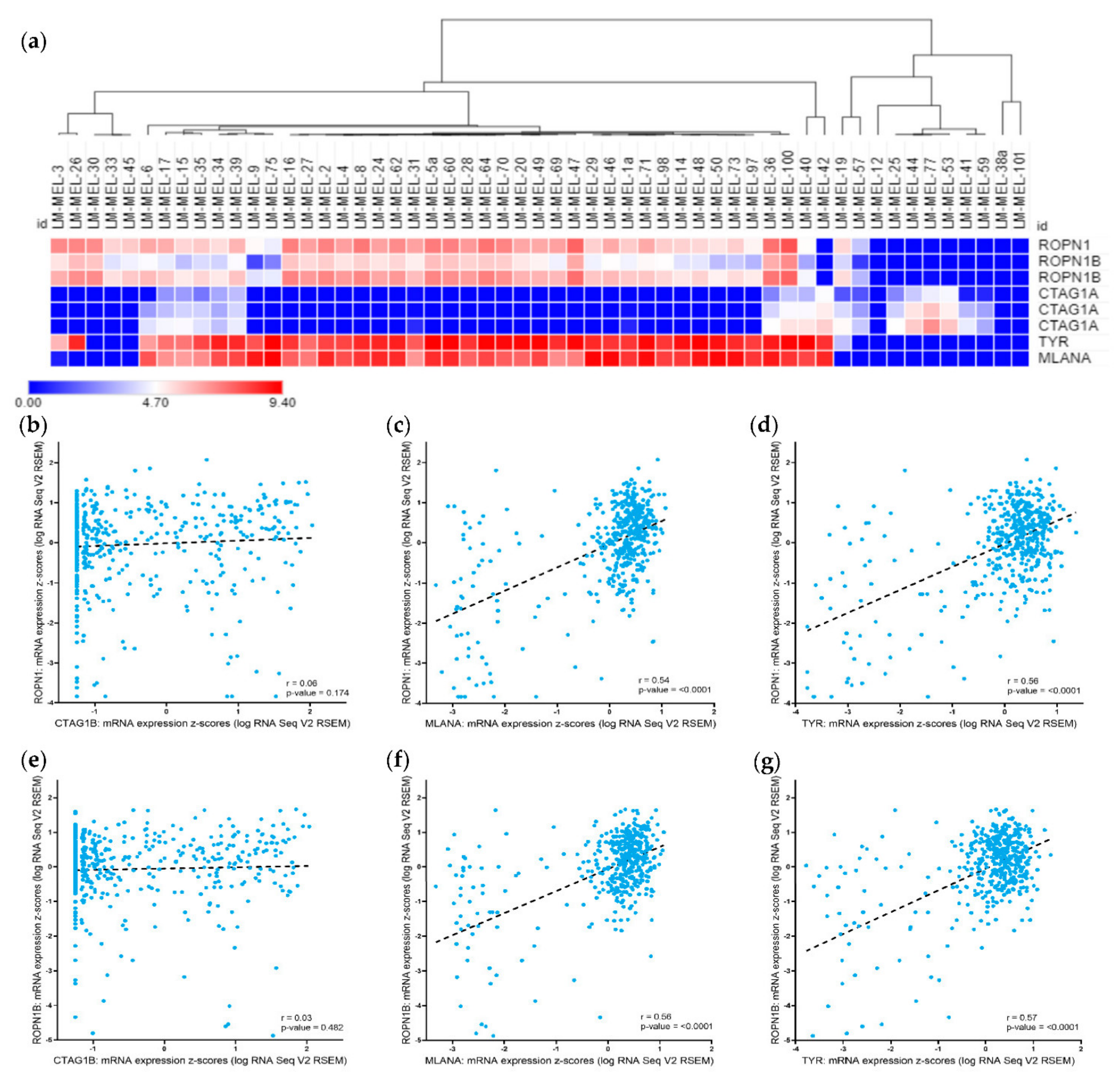
2.1. Ropporin-1 (ROPN1) and Ropporin-1B (ROPN1B) Genes Are Expressed in Melanoma Samples and Correlated with Melanoma Differentiation Antigens
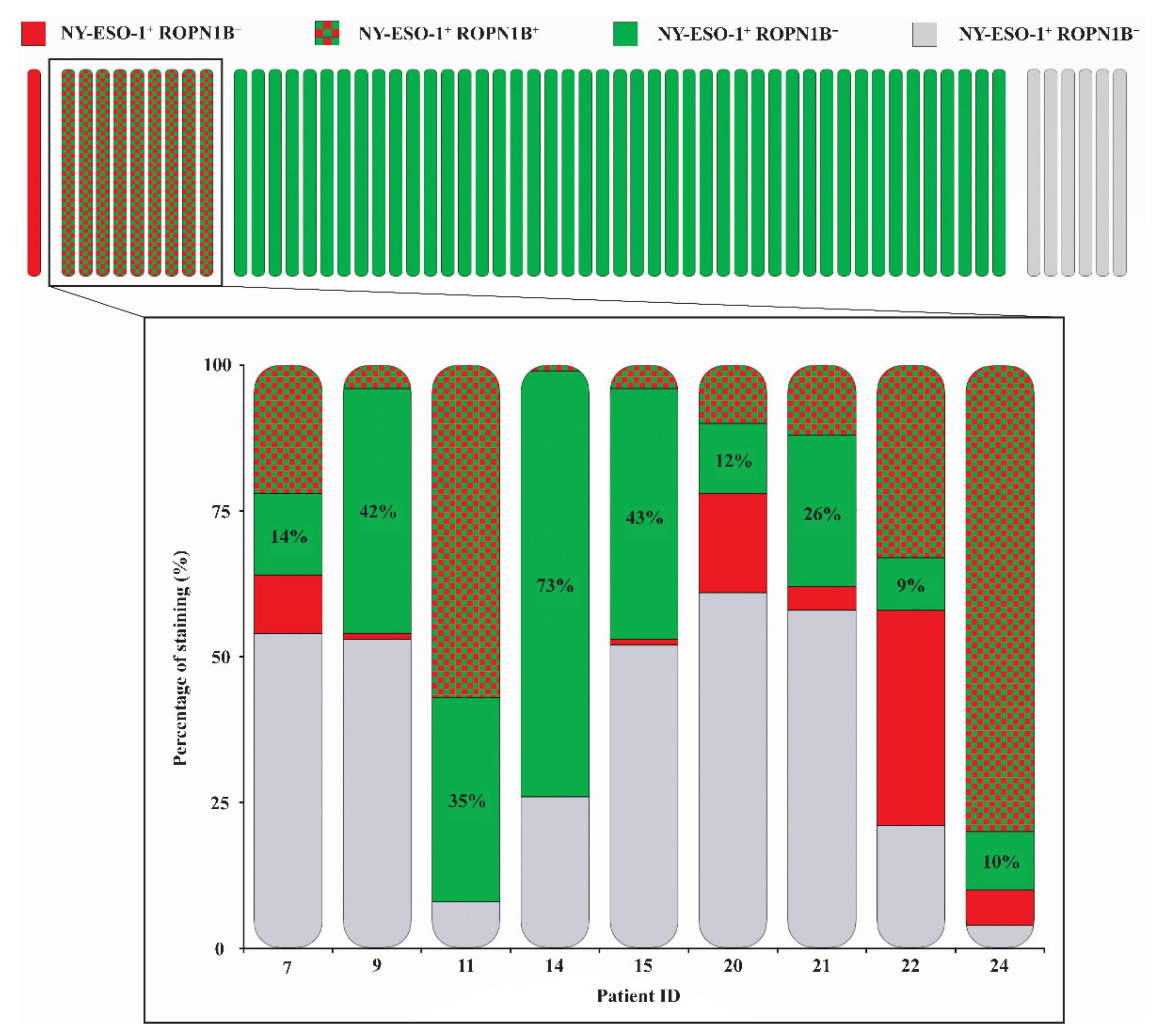
2.2. Ropporin-1B (ROPN1B) Protein Is Expressed in Melanoma Tumours
2.3. Ropporin-1A (ROPN1A) and Ropporin-1B (ROPN1B) Are Immunogenic Antigens in Melanoma
3. Discussion
4. Materials and Methods
4.1. Human Ethics Approval
4.2. Melanoma Cell Lines and Cell Culture
4.3. Cell Pellet DNA Extraction
4.4. Gene Expression—Cell Lines
4.5. Gene Expression—The Cancer Genome Atlas (TCGA)
4.6. Multispectral Immunohistochemistry
4.7. Antibody Profiling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Larkin, J.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.J.; Rutkowski, P.; Lao, C.D.; Cowey, C.L.; Schadendorf, D.; Wagstaff, J.; Dummer, R.; et al. Five-year survival with combined nivolumab and ipilimumab in advanced melanoma. N. Engl. J. Med. 2019, 381, 1535–1546. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, N.A.; Hellmann, M.D.; Snyder, A.; Kvistborg, P.; Makarov, V.; Havel, J.J.; Lee, W.; Yuan, J.; Wong, P.; Ho, T.S.; et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015, 348, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Malaker, S.A.; Penny, S.A.; Steadman, L.G.; Myers, P.T.; Loke, J.C.; Raghavan, M.; Bai, D.L.; Shabanowitz, J.; Hunt, D.F.; Cobbold, M. Identification of Glycopeptides as Posttranslationally Modified Neoantigens in Leukemia. Cancer Immunol. Res. 2017, 5, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Ott, P.A.; Hu, Z.; Keskin, D.B.; Shukla, S.A.; Sun, J.; Bozym, D.J.; Zhang, W.; Luoma, A.; Giobbie-Hurder, A.; Peter, L.; et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 2017, 547, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.J.R.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Børresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef]
- Chalmers, Z.R.; Connelly, C.F.; Fabrizio, D.; Gay, L.; Ali, S.M.; Ennis, R.; Schrock, A.; Campbell, B.; Shlien, A.; Chmielecki, J.; et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Fourcade, J.; Sun, Z.; Pagliano, O.; Chauvin, J.-M.; Sander, C.; Janjic, B.; Tarhini, A.A.; Tawbi, H.A.; Kirkwood, J.M.; Moschos, S.; et al. PD-1 and Tim-3 regulate the expansion of tumor antigen-specific CD8+ T cells induced by melanoma vaccines. Cancer Res. 2014, 74, 1045–1055. [Google Scholar] [CrossRef] [PubMed]
- Mahvi, D.A.; Meyers, J.V.; Tatar, A.J.; Contreras, A.; Suresh, M.; Leverson, G.E.; Sen, S.; Cho, C.S. Ctla-4 blockade plus adoptive T-cell transfer promotes optimal melanoma immunity in mice. J. Immunother. 2015, 38, 54–61. [Google Scholar] [CrossRef]
- Kuramitsu, S.; Yamamichi, A.; Ohka, F.; Motomura, K.; Hara, M.; Natsume, A. Adoptive immunotherapy for the treatment of glioblastoma: Progress and possibilities. Immunotherapy 2016, 8, 1393–1404. [Google Scholar] [CrossRef]
- Davis, I.D.; Chen, W.; Jackson, H.; Parente, P.; Shackleton, M.; Hopkins, W.; Chen, Q.; Dimopoulos, N.; Luke, T.; Murphy, R.; et al. Recombinant NY-ESO-1 protein with ISCOMATRIX adjuvant induces broad integrated antibody and CD4(+) and CD8(+) T cell responses in humans. Proc. Natl. Acad. Sci. USA 2004, 101, 10697–10702. [Google Scholar] [CrossRef]
- Maraskovsky, E. NY-ESO-1 Protein Formulated in ISCOMATRIX Adjuvant Is a Potent Anticancer Vaccine Inducing Both Humoral and CD8+ T-Cell-Mediated Immunity and Protection against NY-ESO-1+ Tumors. Clin. Cancer Res. 2004, 10, 2879–2890. [Google Scholar] [CrossRef][Green Version]
- Phan, G.Q.; Rosenberg, S.A. Adoptive cell transfer for patients with metastatic melanoma: The potential and promise of cancer immunotherapy. Cancer Control 2013, 20, 289–297. [Google Scholar] [CrossRef]
- Geldmacher, A.; Freier, A.; Losch, F.O.; Walden, P. Therapeutic vaccination for cancer immunotherapy: Antigen selection and clinical responses. Hum. Vaccin. 2011, 7, 115–119. [Google Scholar] [CrossRef]
- Jackson, H.; Dimopoulos, N.; Mifsud, N.A.; Tai, T.Y.; Chen, Q.; Svobodova, S.; Browning, J.; Luescher, I.; Stockert, L.; Old, L.J.; et al. Striking immunodominance hierarchy of naturally occurring CD8+ and CD4+ T cell responses to tumor antigen NY-ESO-1. J. Immunol. 2006, 176, 5908–5917. [Google Scholar] [CrossRef]
- Stockert, E.; Jäger, E.; Chen, Y.T.; Scanlan, M.J.; Gout, I.; Karbach, J.; Arand, M.; Knuth, A.; Old, L.J. A survey of the humoral immune response of cancer patients to a panel of human tumor antigens. J. Exp. Med. 1998, 187, 1349–1354. [Google Scholar] [CrossRef]
- Knights, A.J.; Nuber, N.; Thomson, C.W.; De La Rosa, O.; Jäger, E.; Tiercy, J.M.; Van Den Broek, M.; Pascolo, S.; Knuth, A.; Zippelius, A. Modified tumour antigen-encoding mRNA facilitates the analysis of naturally occurring and vaccine-induced CD4 and CD8 T cells in cancer patients. Cancer Immunol. Immunother. 2009, 58, 325–338. [Google Scholar] [CrossRef]
- Barrow, C.; Browning, J.; MacGregor, D.; Davis, I.D.; Sturrock, S.; Jungbluth, A.A.; Cebon, J. Tumor antigen expression in melanoma varies according to antigen and stage. Clin. Cancer Res. 2006, 12, 764–771. [Google Scholar] [CrossRef]
- Filipazzi, P.; Pilla, L.; Mariani, L.; Patuzzo, R.; Castelli, C.; Camisaschi, C.; Maurichi, A.; Cova, A.; Rigamonti, G.; Giardino, F.; et al. Limited induction of tumor cross-reactive T cells without a measurable clinical benefit in early melanoma patients vaccinated with human leukocyte antigen class I-modified peptides. Clin. Cancer Res. 2012, 18, 6485–6496. [Google Scholar] [CrossRef]
- Slingluff, C.L. The present and future of peptide vaccines for cancer: Single or multiple, long or short, alone or in combination? Cancer J. 2011, 17, 343–350. [Google Scholar] [CrossRef]
- Slingluff, C.L.; Petroni, G.R.; Yamshchikov, G.V.; Hibbitts, S.; Grosh, W.W.; Chianese-Bullock, K.A.; Bissonette, E.A.; Barnd, D.L.; Deacon, D.H.; Patterson, J.W.; et al. Immunologic and clinical outcomes of vaccination with a multiepitope melanoma peptide vaccine plus low-dose interleukin-2 administered either concurrently or on a delayed schedule. J. Clin. Oncol. 2004, 22, 4474–4485. [Google Scholar] [CrossRef]
- Kumai, T.; Fan, A.; Harabuchi, Y.; Celis, E. Cancer immunotherapy: Moving forward with peptide T cell vaccines. Curr. Opin. Immunol. 2017, 47, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Hersey, P.; Gallagher, S.J.; Kirkwood, J.M.; Cebon, J. Melanoma Vaccines. In Cutaneous Melanoma; Balch, C.M., Atkins, M.B., Garbe, C., Gershenwald, J.E., Halpern, A.C., Kirkwood, J.M., McArthur, G.A., Thompson, J.F., Sober, A.J., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 1243–1265. ISBN 978-3-030-05070-2. [Google Scholar]
- Grenier, J.M.; Yeung, S.T.; Qiu, Z.; Jellison, E.R. Combining Adoptive Cell Therapy with Cytomegalovirus-Based Vaccine Is Protective against Solid Skin Tumors. Front. Immunol. 2017, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, O.; Caballero, O.L.; Stevenson, B.J.; Chen, Y.-T.Y.; Cohen, T.; Chua, R.; Maher, C.A.; Panji, S.; Schaefer, U.; Kruger, A.; et al. Genome-wide analysis of cancer/testis gene expression. Proc. Natl. Acad. Sci. USA 2008, 105, 20422–20427. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Zhu, W.; Xue, S.; Han, D. Testicular defense systems: Immune privilege and innate immunity. Cell. Mol. Immunol. 2014, 11, 428–437. [Google Scholar] [CrossRef]
- Fujita, A.; Nakamura, K.I.; Kato, T.; Watanabe, N.; Ishizaki, T.; Kimura, K.; Mizoguchi, A.; Narumiya, S. Ropporin, a sperm-specific binding protein of rhophilin, that is localized in the fibrous sheath of sperm flagella. J. Cell Sci. 2000, 113, 103–112. [Google Scholar]
- Carr, D.W.; Fujita, A.; Stentz, C.L.; Liberty, G.A.; Olson, G.E.; Narumiya, S. Identification of Sperm-specific Proteins that Interact with A-kinase Anchoring Proteins in a Manner Similar to the Type II Regulatory Subunit of PKA. J. Biol. Chem. 2001, 276, 17332–17338. [Google Scholar] [CrossRef]
- Chen, J.; Wang, Y.; Wei, B.; Lai, Y.; Yan, Q.; Gui, Y.; Cai, Z. Functional expression of ropporin in human testis and ejaculated spermatozoa. J. Androl. 2011, 32, 26–32. [Google Scholar] [CrossRef]
- Li, Z.; Li, W.; Meklat, F.; Wang, Z.; Zhang, J.; Zhang, Y.; Lim, S.H. A yeast two-hybrid system using Sp17 identified Ropporin as a novel cancer-testis antigen in hematologic malignancies. Int. J. Cancer 2007, 121, 1507–1511. [Google Scholar] [CrossRef]
- Liu, Q.; Huang, X.; Li, Q.; He, L.; Li, S.; Chen, X.; Ouyang, Y.; Wang, X.; Lin, C. Rhophilin-associated tail protein 1 promotes migration and metastasis in triple negative breast cancer via activation of RhoA. FASEB J. 2020, 34, 9959–9971. [Google Scholar] [CrossRef]
- Chiriva-Internati, M.; Mirandola, L.; Yu, Y.; Jenkins, M.R.; Gornati, R.; Bernardini, G.; Gioia, M.; Chiaramonte, R.; Cannon, M.J.; Kast, W.M.; et al. Cancer testis antigen, ropporin, is a potential target for multiple myeloma immunotherapy. J. Immunother. 2011, 34, 490–499. [Google Scholar] [CrossRef]
- Behren, A.; Anaka, M.; Lo, P.-H.; Vella, L.J.; Davis, I.D.; Catimel, J.; Cardwell, T.; Gedye, C.; Hudson, C.; Stan, R.; et al. The Ludwig institute for cancer research Melbourne melanoma cell line panel. Pigment. Cell Melanoma Res. 2013, 26, 597–600. [Google Scholar] [CrossRef]
- Tarhini, A.A.; Leng, S.; Moschos, S.J.; Yin, Y.; Sander, C.; Lin, Y.; Gooding, W.E.; Kirkwood, J.M. Safety and immunogenicity of vaccination with MART-1 (26-35, 27L), gp100 (209-217, 210M), and tyrosinase (368-376, 370D) in adjuvant with PF-3512676 and GM-CSF in metastatic melanoma. J. Immunother. 2012, 35, 359–366. [Google Scholar] [CrossRef]
- Schumacher, T.N.; Schreiber, R.D. Realising the Promise: Neoantigens in cancer immunotherapy. Sci. Mag. 2015, 348. [Google Scholar]
- Roesch, A. Tumor heterogeneity and plasticity as elusive drivers for resistance to MAPK pathway inhibition in melanoma. Oncogene 2015, 34, 2951–2957. [Google Scholar] [CrossRef]
- Monjazeb, A.M.; Zamora, A.E.; Grossenbacher, S.K.; Mirsoian, A.; Sckisel, G.D.; Murphy, W.J. Immunoediting and antigen loss: Overcoming the Achilles heel of immunotherapy with antigen non-specific therapies. Front. Oncol. 2013, 3, 1–10. [Google Scholar] [CrossRef]
- Landsberg, J.; Kohlmeyer, J.; Renn, M.; Bald, T.; Rogava, M.; Cron, M.; Fatho, M.; Lennerz, V.; Wölfel, T.; Hölzel, M.; et al. Melanomas resist T-cell therapy through inflammation-induced reversible dedifferentiation. Nature 2012, 490, 412–416. [Google Scholar] [CrossRef]
- Woods, K.; Pasam, A.; Jayachandran, A.; Andrews, M.C.; Cebon, J. Effects of epithelial to mesenchymal transition on t cell targeting of melanoma cells. Front. Oncol. 2014, 4, 1–7. [Google Scholar] [CrossRef]
- Cebon, J.; Caballero, O.; John, T.; Klein, O. Cancer Testis Antigens. In Tumor-Associated Antigens; Gires, O., Seliger, B., Eds.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2009; pp. 161–177. ISBN 9783527320844. [Google Scholar]
- Scanlan, M.J.; Gure, A.O.; Jungbluth, A.A.; Old, L.J.; Chen, Y. Cancer/testis antigens: An expanding family of targets for cancer immunotherapy. Immunol. Rev. 2002, 188, 22–32. [Google Scholar] [CrossRef]
- Cebon, J.S.; Gore, M.; Thompson, J.F.; Davis, I.D.; McArthur, G.A.; Walpole, E.; Smithers, M.; Cerundolo, V.; Dunbar, P.R.; MacGregor, D.; et al. Results of a randomized, double-blind phase II clinical trial of NY-ESO-1 vaccine with ISCOMATRIX adjuvant versus ISCOMATRIX alone in participants with high-risk resected melanoma. J. Immunother. Cancer 2020, 8, 1–11. [Google Scholar] [CrossRef]
- Maire, C.; Vercambre-Darras, S.; Devos, P.; D’Herbomez, M.; Dubucquoi, S.; Mortier, L. Metastatic melanoma: Spontaneous occurrence of auto antibodies is a good prognosis factor in a prospective cohort. J. Eur. Acad. Dermatol. Venereol. 2013, 27, 92–96. [Google Scholar] [CrossRef]
- Kupsch, J.M.; Tidman, N.H.; Kang, N.V.; Truman, H.; Hamilton, S.; Patel, N.; Bishop, J.A.N.; Leigh, I.M.; Crowe, J.S. Isolation of human tumor-specific antibodies by selection of an antibody phage library on melanoma cells. Clin. Cancer Res. 1999, 5, 925–931. [Google Scholar]
- Ohue, Y.; Wada, H.; Oka, M.; Nakayama, E. Antibody response to cancer/testis (CT) antigens: A prognostic marker in cancer patients. Oncoimmunology 2014, 3, e970032-1–e970032-3. [Google Scholar] [CrossRef]
- Casadevall, A.; Pirofski, L. Antibody-mediated regulation of cellular immunity and the inflammatory response. Trends Immunol. 2003, 24, 474–478. [Google Scholar] [CrossRef]
- Jäger, E.; Chen, Y.T.; Drijfhout, J.W.; Karbach, J.; Ringhoffer, M.; Jäger, D.; Arand, M.; Wada, H.; Noguchi, Y.; Stockert, E.; et al. Simultaneous humoral and cellular immune response against cancer-testis antigen NY-ESO-1: Definition of human histocompatibility leukocyte antigen (HLA)-A2-binding peptide epitopes. J. Exp. Med. 1998, 187, 265–270. [Google Scholar] [CrossRef]
- Beeton-Kempen, N.; Duarte, J.; Shoko, A.; Serufuri, J.-M.; John, T.; Cebon, J.; Blackburn, J. Development of a novel, quantitative protein microarray platform for the multiplexed serological analysis of autoantibodies to cancer-testis antigens. Int. J. Cancer 2014, 135, 1842–1851. [Google Scholar] [CrossRef]
- Gnjatic, S.; Atanackovic, D.; Jäger, E.; Matsuo, M.; Selvakumar, A.; Altorki, N.K.; Maki, R.G.; Dupont, B.; Ritter, G.; Chen, Y.-T.; et al. Survey of naturally occurring CD4+ T cell responses against NY-ESO-1 in cancer patients: Correlation with antibody responses. Proc. Natl. Acad. Sci. USA 2003, 100, 8862–8867. [Google Scholar] [CrossRef]
- Svobodová, S.; Browning, J.; MacGregor, D.; Pollara, G.; Scolyer, R.A.; Murali, R.; Thompson, J.F.; Deb, S.; Azad, A.; Davis, I.D.; et al. Cancer-testis antigen expression in primary cutaneous melanoma has independent prognostic value comparable to that of Breslow thickness, ulceration and mitotic rate. Eur. J. Cancer 2011, 47, 460–469. [Google Scholar] [CrossRef]
- Mirandola, L.; Wade, R.; Verma, R.; Pena, C.; Hosiriluck, N.; Figueroa, J.A.; Cobos, E.; Jenkins, M.R.; Chiriva-Internati, M. Sex-Driven Differences in Immunological Responses: Challenges and Opportunities for the Immunotherapies of the Third Millennium. Int. Rev. Immunol. 2015, 34, 134–142. [Google Scholar] [CrossRef]
- Joosse, A.; Collette, S.; Suciu, S.; Nijsten, T.; Lejeune, F.; Kleeberg, U.R.; Coebergh, J.W.W.; Eggermont, A.M.M.; de Vries, E. Superior outcome of women with stage I/II cutaneous melanoma: Pooled analysis of four European Organisation for Research and Treatment of Cancer phase III trials. J. Clin. Oncol. 2012, 30, 2240–2247. [Google Scholar] [CrossRef]
- Joosse, A.; Collette, S.; Suciu, S.; Nijsten, T.; Patel, P.M.; Keilholz, U.; Eggermont, A.M.M.; Coebergh, J.W.W.; de Vries, E. Sex is an independent prognostic indicator for survival and relapse/progression-free survival in metastasized stage III to IV melanoma: A pooled analysis of five European organisation for research and treatment of cancer randomized controlled trials. J. Clin. Oncol. 2013, 31, 2337–2346. [Google Scholar] [CrossRef]
- Mittal, D.; Gubin, M.M.; Schreiber, R.D.; Smyth, M.J. New insights into cancer immunoediting and its three component phases--elimination, equilibrium and escape. Curr. Opin. Immunol. 2014, 27, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Ott, P.A.; Hodi, F.S.; Kaufman, H.L.; Wigginton, J.M.; Wolchok, J.D. Combination immunotherapy: A road map. J. Immunother. Cancer 2017, 5, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Gjerstorff, M.F.; Andersen, M.H.; Ditzel, H.J. Oncogenic cancer/testis antigens: Prime candidates for immunotherapy. Oncotarget 2015, 6, 15772–15787. [Google Scholar] [CrossRef] [PubMed]
- Anaka, M.; Freyer, C.; Gedye, C.; Caballero, O.; Davis, I.D.; Behren, A.; Cebon, J. Stem cell media culture of melanoma results in the induction of a nonrepresentative neural expression profile. Stem Cells 2012, 30, 336–343. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Da Gama Duarte, J.; Goosen, R.W.; Lawry, P.J.; Blackburn, J.M. PMA: Protein Microarray Analyser, a user-friendly tool for data processing and normalization. BMC Res. Notes 2018, 11, 156. [Google Scholar] [CrossRef]


| Cohort 1 (n = 52) | Cohort 2 (n = 469) | Cohort 3 (n = 61) | Cohort 4 (n = 104) | |
|---|---|---|---|---|
| Sample Type | Cell lines | Tumours | Tumours | Serum or Plasma |
| Used to Determine | Gene Expression | Gene Expression | Protein Expression | Circulating Antibodies |
| Age–yr | ||||
| Median | 56 | 58 | 53 | 54 |
| Range | 25–83 | 15–90 | 21–88 | 21–87 |
| Gender–no. (%) | ||||
| Unknown | 0 (0) | 0 (0) | 0 (0) | 1 (0.9) |
| Male | 33 (63.5) | 289 (61.6) | 35 (57.4) | 63 (60.6) |
| Female | 19 (36.5) | 180 (38.4) | 26 (42.6) | 40 (38.5) |
| Stage–no. (%) | ||||
| Unknown | 9 (17.3) | 59 (12.6) | 6 (9.8) | 7 (6.7) |
| I | 0 (0) | 77 (16.4) | 5 (8.2) | 2 (1.9) |
| II | 1 (1.9) | 140 (29.9) | 21 (34.4) | 9 (8.7) |
| III | 22 (42.3) | 170 (36.2) | 4 (6.6) | 35 (33.7) |
| IV | 20 (38.5) | 23 (4.9) | 25 (41.0) | 51 (49.0) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Da Gama Duarte, J.; Woods, K.; Quigley, L.T.; Deceneux, C.; Tutuka, C.; Witkowski, T.; Ostrouska, S.; Hudson, C.; Tsao, S.C.-H.; Pasam, A.; et al. Ropporin-1 and 1B Are Widely Expressed in Human Melanoma and Evoke Strong Humoral Immune Responses. Cancers 2021, 13, 1805. https://doi.org/10.3390/cancers13081805
Da Gama Duarte J, Woods K, Quigley LT, Deceneux C, Tutuka C, Witkowski T, Ostrouska S, Hudson C, Tsao SC-H, Pasam A, et al. Ropporin-1 and 1B Are Widely Expressed in Human Melanoma and Evoke Strong Humoral Immune Responses. Cancers. 2021; 13(8):1805. https://doi.org/10.3390/cancers13081805
Chicago/Turabian StyleDa Gama Duarte, Jessica, Katherine Woods, Luke T. Quigley, Cyril Deceneux, Candani Tutuka, Tom Witkowski, Simone Ostrouska, Chris Hudson, Simon Chang-Hao Tsao, Anupama Pasam, and et al. 2021. "Ropporin-1 and 1B Are Widely Expressed in Human Melanoma and Evoke Strong Humoral Immune Responses" Cancers 13, no. 8: 1805. https://doi.org/10.3390/cancers13081805
APA StyleDa Gama Duarte, J., Woods, K., Quigley, L. T., Deceneux, C., Tutuka, C., Witkowski, T., Ostrouska, S., Hudson, C., Tsao, S. C.-H., Pasam, A., Dobrovic, A., Blackburn, J. M., Cebon, J., & Behren, A. (2021). Ropporin-1 and 1B Are Widely Expressed in Human Melanoma and Evoke Strong Humoral Immune Responses. Cancers, 13(8), 1805. https://doi.org/10.3390/cancers13081805








