GSK-3α Inhibition in Drug-Resistant CML Cells Promotes Susceptibility to NK Cell-Mediated Lysis in an NKG2D- and NKp30-Dependent Manner
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Reagents
2.2. NK Cell Expansion
2.3. Antibodies
2.4. RT-PCR
2.5. RNA Interference
2.6. NK Cell Degranulation Assay
2.7. NK Cell Intracellular IFN-γ Production Assay
2.8. NK Cell Cytotoxicity Assay
2.9. Immunoblot Analysis
2.10. In Vivo Leukemia Clearance Assay
2.11. Statistical Analysis
3. Results
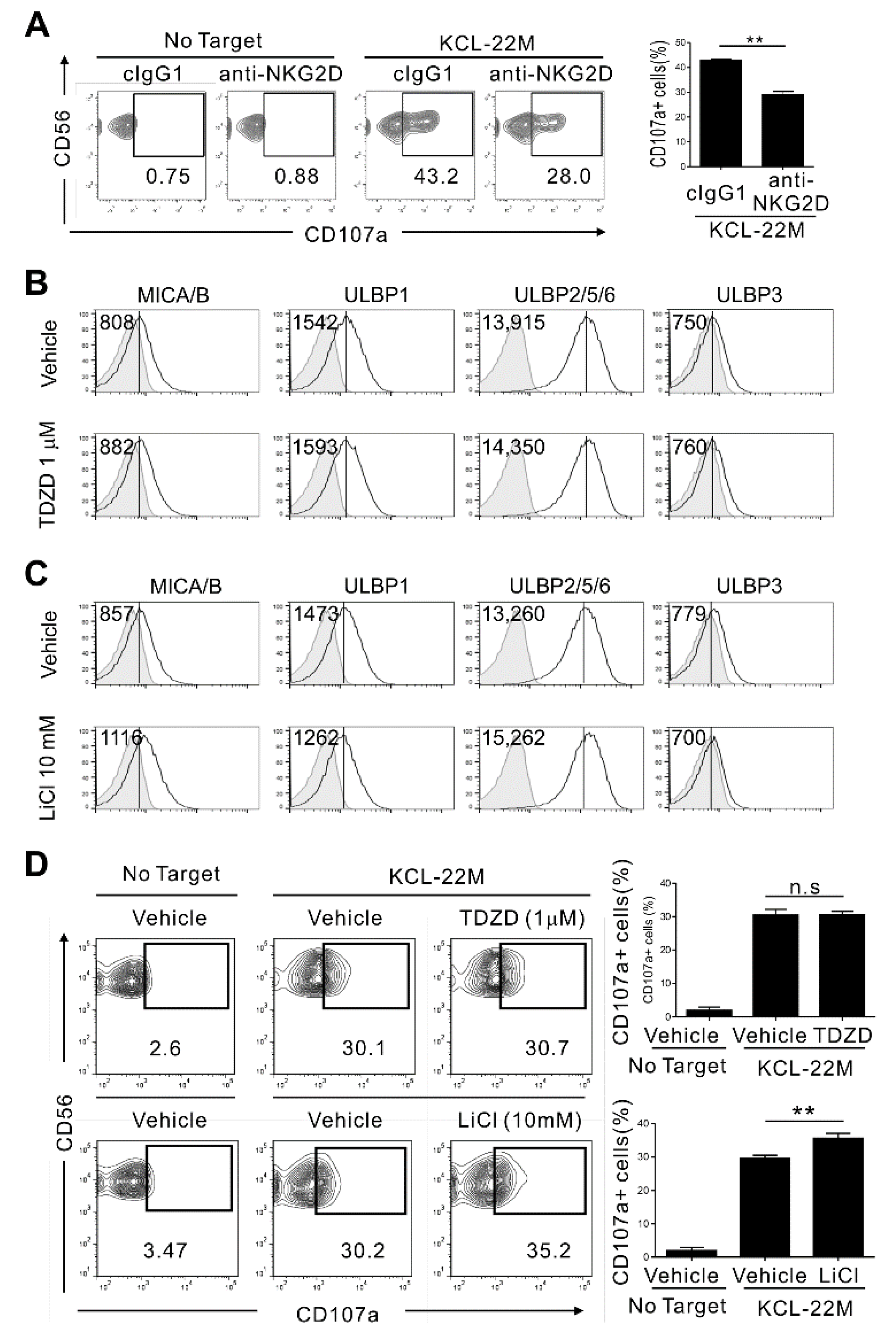
3.1. Pharmacologic Inhibition of GSK-3 in TKI-Resistant CML Cells Enhances Their Expression of NKG2D Ligands and Susceptibility to NK Cells
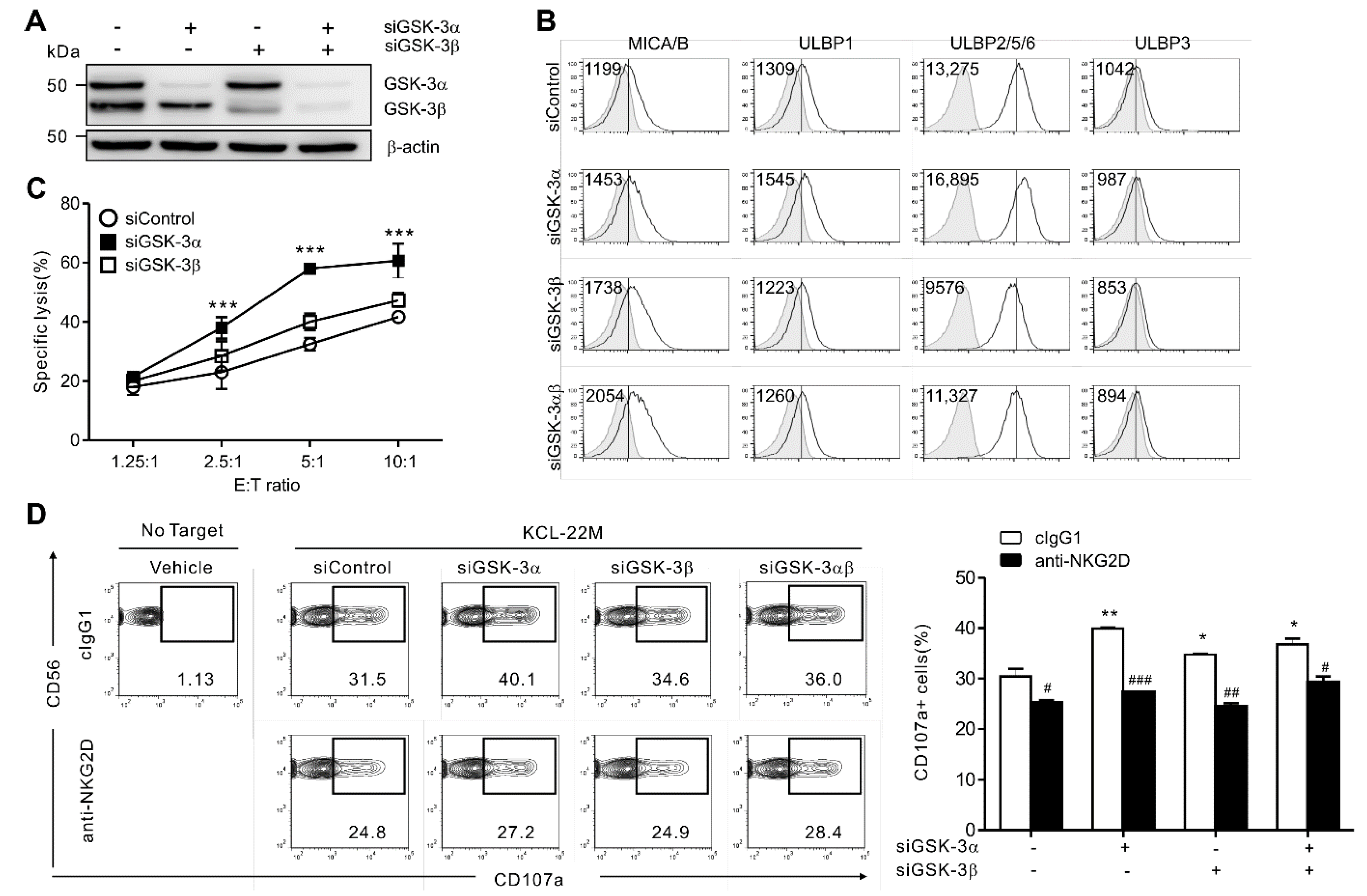
3.2. GSK-3α Selectively Regulates NKG2DL Expression and Cytolysis by NK Cells
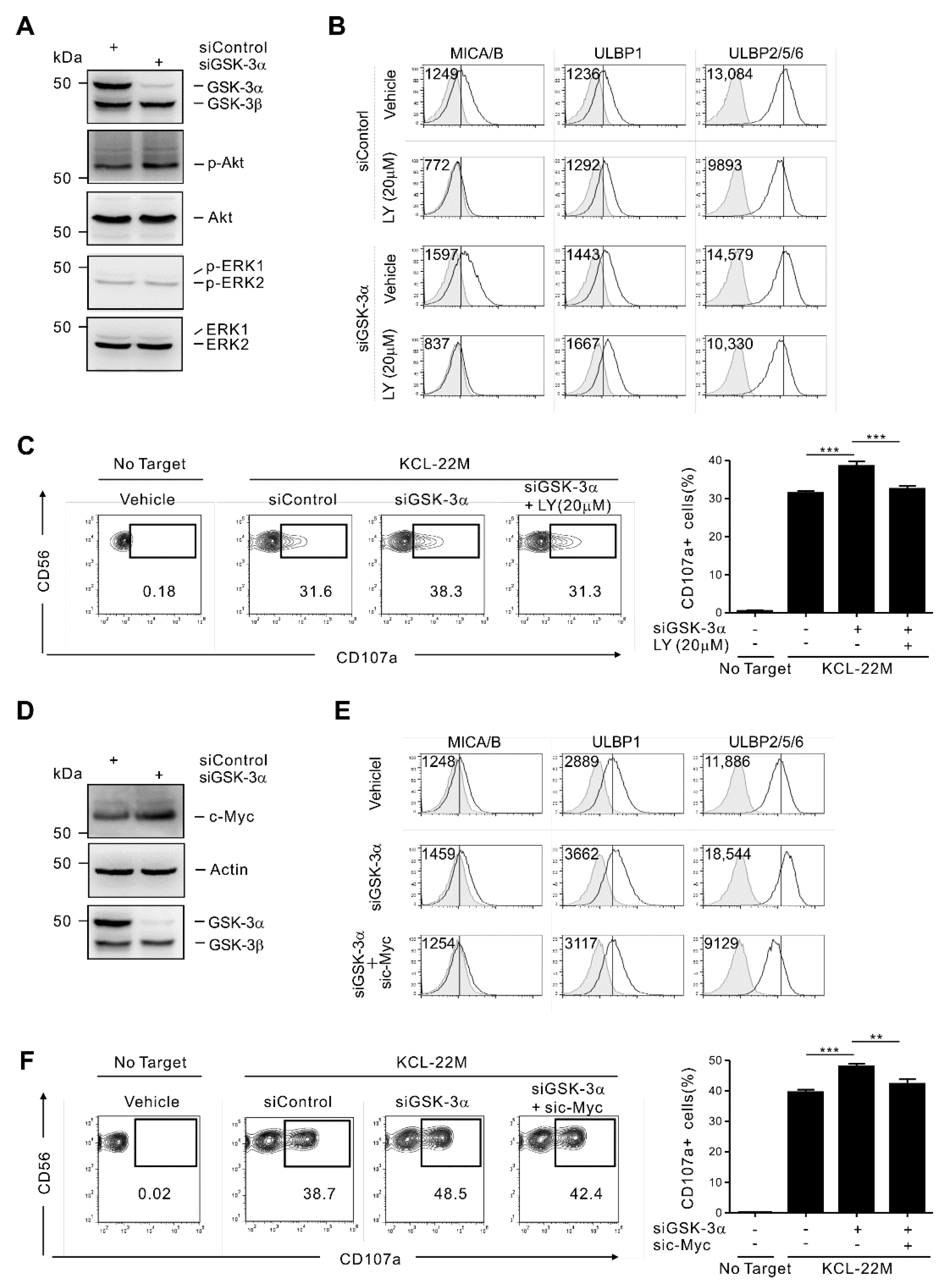
3.3. GSK-3α Regulates ULBP2/5/6 by PI3K/Akt- and c-Myc-Dependent Pathways
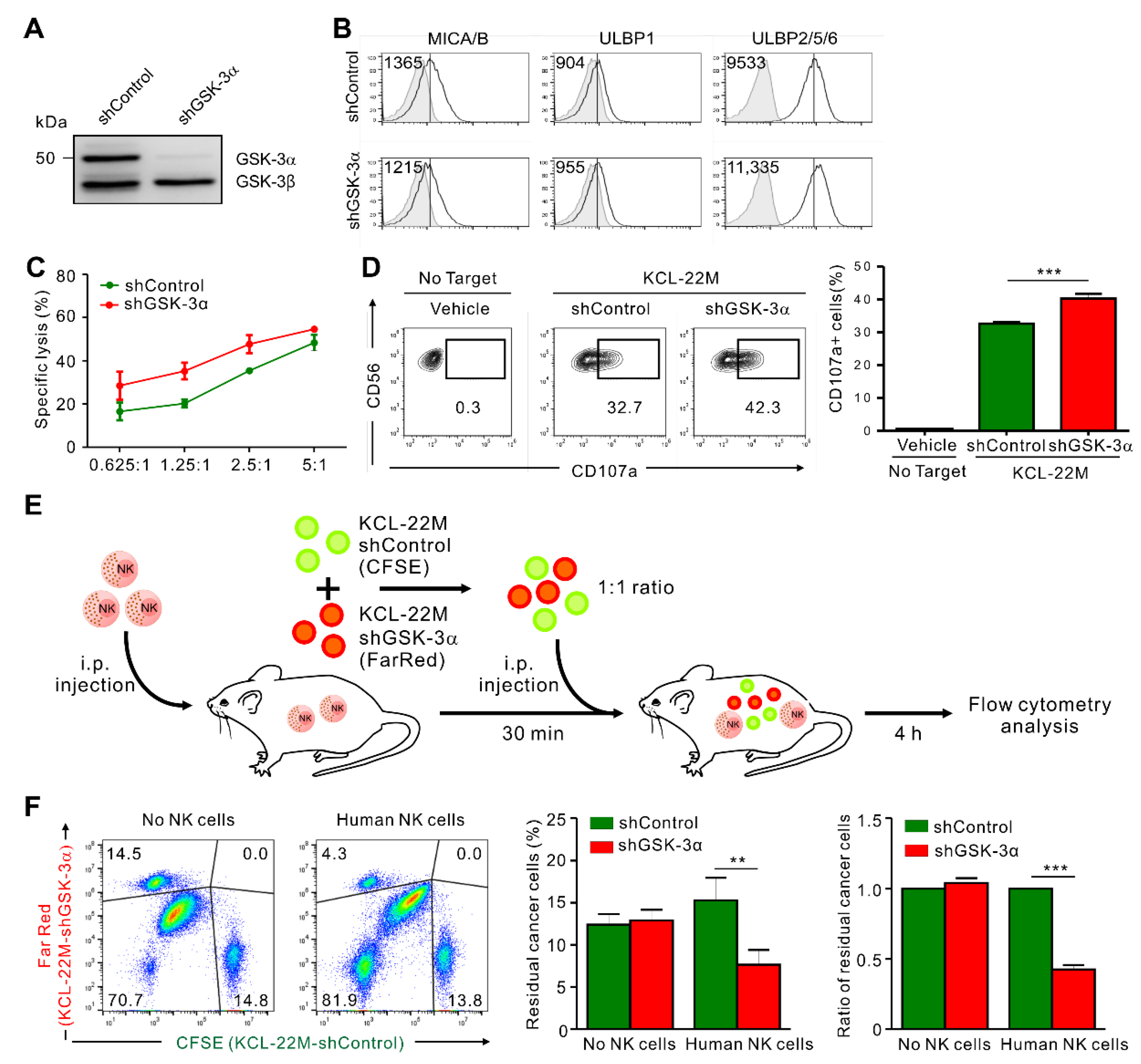
3.4. GSK-3α Knockdown Enhances the Susceptibility of KCL-22M Cells to NK Cells In Vivo
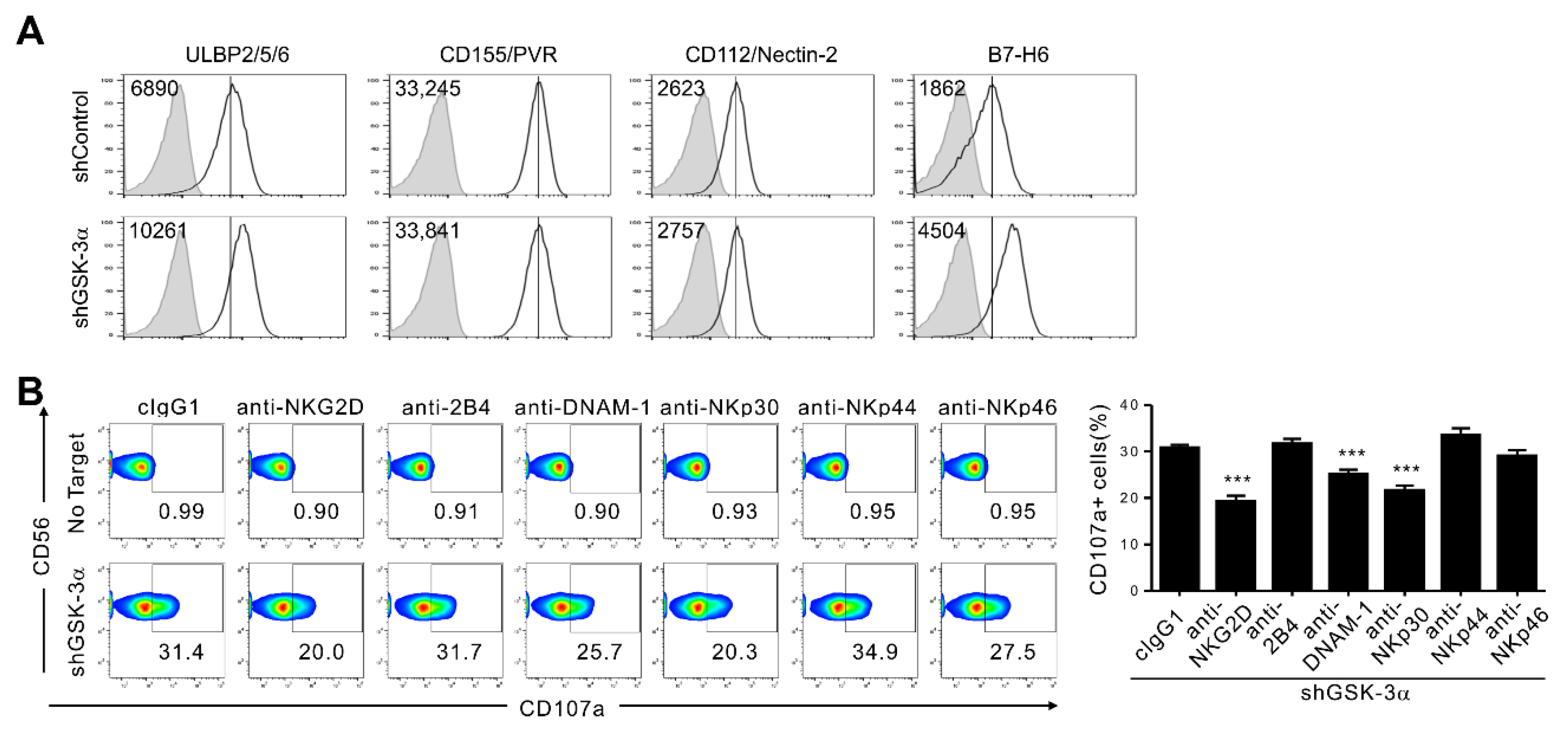
3.5. GSK-3α Knockdown Upregulates NKp30L (B7-H6) in Addition to NKG2DL
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Long, E.O.; Sik Kim, H.; Liu, D.; Peterson, M.E.; Rajagopalan, S. Controlling natural killer cell responses: Integration of signals for activation and inhibition. Annu. Rev. Immunol. 2013, 31, 227–258. [Google Scholar] [CrossRef]
- Morvan, M.G.; Lanier, L.L. NK cells and cancer: You can teach innate cells new tricks. Nat. Rev. Cancer 2016, 16, 7–19. [Google Scholar] [CrossRef] [PubMed]
- Carlsten, M.; Jaras, M. Natural Killer Cells in Myeloid Malignancies: Immune Surveillance, NK Cell Dysfunction, and Pharmacological Opportunities to Bolster the Endogenous NK Cells. Front. Immunol. 2019, 10, 2357. [Google Scholar] [CrossRef] [PubMed]
- Guillerey, C.; Huntington, N.D.; Smyth, M.J. Targeting natural killer cells in cancer immunotherapy. Nat. Immunol. 2016, 17, 1025–1036. [Google Scholar] [CrossRef]
- Lopez-Soto, A.; Gonzalez, S.; Smyth, M.J.; Galluzzi, L. Control of Metastasis by NK Cells. Cancer Cell 2017, 32, 135–154. [Google Scholar] [CrossRef] [PubMed]
- Shimasaki, N.; Jain, A.; Campana, D. NK cells for cancer immunotherapy. Nat. Rev. Drug Discov. 2020, 19, 200–218. [Google Scholar] [CrossRef]
- Beurel, E.; Michalek, S.M.; Jope, R.S. Innate and adaptive immune responses regulated by glycogen synthase kinase-3 (GSK3). Trends Immunol. 2010, 31, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Jope, R.S.; Johnson, G.V. The glamour and gloom of glycogen synthase kinase-3. Trends Biochem. Sci. 2004, 29, 95–102. [Google Scholar] [CrossRef]
- Augello, G.; Emma, M.R.; Cusimano, A.; Azzolina, A.; Montalto, G.; McCubrey, J.A.; Cervello, M. The Role of GSK-3 in Cancer Immunotherapy: GSK-3 Inhibitors as a New Frontier in Cancer Treatment. Cells 2020, 9, 1427. [Google Scholar] [CrossRef]
- Pandey, M.K.; DeGrado, T.R. Glycogen Synthase Kinase-3 (GSK-3)-Targeted Therapy and Imaging. Theranostics 2016, 6, 571–593. [Google Scholar] [CrossRef]
- Woodgett, J.R. Molecular cloning and expression of glycogen synthase kinase-3/factor A. EMBO J. 1990, 9, 2431–2438. [Google Scholar] [CrossRef]
- Doble, B.W.; Patel, S.; Wood, G.A.; Kockeritz, L.K.; Woodgett, J.R. Functional redundancy of GSK-3alpha and GSK-3beta in Wnt/beta-catenin signaling shown by using an allelic series of embryonic stem cell lines. Dev. Cell 2007, 12, 957–971. [Google Scholar] [CrossRef] [PubMed]
- Hoeflich, K.P.; Luo, J.; Rubie, E.A.; Tsao, M.S.; Jin, O.; Woodgett, J.R. Requirement for glycogen synthase kinase-3beta in cell survival and NF-kappaB activation. Nature 2000, 406, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Kerkela, R.; Kockeritz, L.; Macaulay, K.; Zhou, J.; Doble, B.W.; Beahm, C.; Greytak, S.; Woulfe, K.; Trivedi, C.M.; Woodgett, J.R.; et al. Deletion of GSK-3beta in mice leads to hypertrophic cardiomyopathy secondary to cardiomyoblast hyperproliferation. J. Clin. Investig. 2008, 118, 3609–3618. [Google Scholar] [CrossRef] [PubMed]
- MacAulay, K.; Doble, B.W.; Patel, S.; Hansotia, T.; Sinclair, E.M.; Drucker, D.J.; Nagy, A.; Woodgett, J.R. Glycogen synthase kinase 3alpha-specific regulation of murine hepatic glycogen metabolism. Cell Metab. 2007, 6, 329–337. [Google Scholar] [CrossRef]
- Phiel, C.J.; Wilson, C.A.; Lee, V.M.; Klein, P.S. GSK-3alpha regulates production of Alzheimer’s disease amyloid-beta peptides. Nature 2003, 423, 435–439. [Google Scholar] [CrossRef]
- Linding, R.; Jensen, L.J.; Ostheimer, G.J.; van Vugt, M.A.; Jorgensen, C.; Miron, I.M.; Diella, F.; Colwill, K.; Taylor, L.; Elder, K.; et al. Systematic discovery of in vivo phosphorylation networks. Cell 2007, 129, 1415–1426. [Google Scholar] [CrossRef]
- Sutherland, C. What Are the bona fide GSK3 Substrates? Int. J. Alzheimers Dis. 2011, 2011, 505607. [Google Scholar] [CrossRef]
- Patel, P.; Woodgett, J.R. Glycogen Synthase Kinase 3: A Kinase for All Pathways? Curr. Top. Dev. Biol. 2017, 123, 277–302. [Google Scholar] [CrossRef]
- Beurel, E.; Grieco, S.F.; Jope, R.S. Glycogen synthase kinase-3 (GSK3): Regulation, actions, and diseases. Pharmacol. Ther. 2015, 148, 114–131. [Google Scholar] [CrossRef]
- McCubrey, J.A.; Steelman, L.S.; Bertrand, F.E.; Davis, N.M.; Abrams, S.L.; Montalto, G.; D’Assoro, A.B.; Libra, M.; Nicoletti, F.; Maestro, R.; et al. Multifaceted roles of GSK-3 and Wnt/beta-catenin in hematopoiesis and leukemogenesis: Opportunities for therapeutic intervention. Leukemia 2014, 28, 15–33. [Google Scholar] [CrossRef]
- Luo, J. Glycogen synthase kinase 3beta (GSK3beta) in tumorigenesis and cancer chemotherapy. Cancer Lett. 2009, 273, 194–200. [Google Scholar] [CrossRef]
- Wang, Z.; Smith, K.S.; Murphy, M.; Piloto, O.; Somervaille, T.C.; Cleary, M.L. Glycogen synthase kinase 3 in MLL leukaemia maintenance and targeted therapy. Nature 2008, 455, 1205–1209. [Google Scholar] [CrossRef]
- Wu, X.; Stenson, M.; Abeykoon, J.; Nowakowski, K.; Zhang, L.; Lawson, J.; Wellik, L.; Li, Y.; Krull, J.; Wenzl, K.; et al. Targeting glycogen synthase kinase 3 for therapeutic benefit in lymphoma. Blood 2019, 134, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Gunn, W.G.; Krause, U.; Lee, N.; Gregory, C.A. Pharmaceutical inhibition of glycogen synthetase kinase-3beta reduces multiple myeloma-induced bone disease in a novel murine plasmacytoma xenograft model. Blood 2011, 117, 1641–1651. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.J.; Kwon, S.J.; Lee, H.; Park, H.R.; Choi, G.E.; Kang, S.W.; Kwon, S.W.; Kim, N.; Lee, S.Y.; Ryu, S.; et al. NK cell function triggered by multiple activating receptors is negatively regulated by glycogen synthase kinase-3beta. Cell Signal. 2015, 27, 1731–1741. [Google Scholar] [CrossRef]
- Parameswaran, R.; Ramakrishnan, P.; Moreton, S.A.; Xia, Z.; Hou, Y.; Lee, D.A.; Gupta, K.; deLima, M.; Beck, R.C.; Wald, D.N. Repression of GSK3 restores NK cell cytotoxicity in AML patients. Nat. Commun. 2016, 7, 11154. [Google Scholar] [CrossRef] [PubMed]
- Fionda, C.; Malgarini, G.; Soriani, A.; Zingoni, A.; Cecere, F.; Iannitto, M.L.; Ricciardi, M.R.; Federico, V.; Petrucci, M.T.; Santoni, A.; et al. Inhibition of glycogen synthase kinase-3 increases NKG2D ligand MICA expression and sensitivity to NK cell-mediated cytotoxicity in multiple myeloma cells: Role of STAT3. J. Immunol. 2013, 190, 6662–6672. [Google Scholar] [CrossRef]
- Kim, N.; Lee, H.H.; Lee, H.J.; Choi, W.S.; Lee, J.; Kim, H.S. Natural killer cells as a promising therapeutic target for cancer immunotherapy. Arch. Pharm. Res. 2019, 42, 591–606. [Google Scholar] [CrossRef]
- Raulet, D.H.; Gasser, S.; Gowen, B.G.; Deng, W.; Jung, H. Regulation of ligands for the NKG2D activating receptor. Annu. Rev. Immunol. 2013, 31, 413–441. [Google Scholar] [CrossRef]
- Brandt, C.S.; Baratin, M.; Yi, E.C.; Kennedy, J.; Gao, Z.; Fox, B.; Haldeman, B.; Ostrander, C.D.; Kaifu, T.; Chabannon, C.; et al. The B7 family member B7-H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. J. Exp. Med. 2009, 206, 1495–1503. [Google Scholar] [CrossRef] [PubMed]
- Fiegler, N.; Textor, S.; Arnold, A.; Rolle, A.; Oehme, I.; Breuhahn, K.; Moldenhauer, G.; Witzens-Harig, M.; Cerwenka, A. Downregulation of the activating NKp30 ligand B7-H6 by HDAC inhibitors impairs tumor cell recognition by NK cells. Blood 2013, 122, 684–693. [Google Scholar] [CrossRef]
- Yuan, H.; Wang, Z.; Gao, C.; Chen, W.; Huang, Q.; Yee, J.K.; Bhatia, R.; Chen, W. BCR-ABL gene expression is required for its mutations in a novel KCL-22 cell culture model for acquired resistance of chronic myelogenous leukemia. J. Biol. Chem. 2010, 285, 5085–5096. [Google Scholar] [CrossRef]
- Kwon, H.J.; Choi, G.E.; Ryu, S.; Kwon, S.J.; Kim, S.C.; Booth, C.; Nichols, K.E.; Kim, H.S. Stepwise phosphorylation of p65 promotes NF-kappaB activation and NK cell responses during target cell recognition. Nat. Commun. 2016, 7, 11686. [Google Scholar] [CrossRef]
- Jun, E.; Song, A.Y.; Choi, J.W.; Lee, H.H.; Kim, M.Y.; Ko, D.H.; Kang, H.J.; Kim, S.W.; Bryceson, Y.; Kim, S.C.; et al. Progressive Impairment of NK Cell Cytotoxic Degranulation Is Associated With TGF-beta1 Deregulation and Disease Progression in Pancreatic Cancer. Front. Immunol. 2019, 10, 1354. [Google Scholar] [CrossRef]
- Hyun, Y.M.; Seo, S.U.; Choi, W.S.; Kwon, H.J.; Kim, D.Y.; Jeong, S.; Kang, G.Y.; Yi, E.; Kim, M.; Ryu, H.J.; et al. Endogenous DEL-1 restrains melanoma lung metastasis by limiting myeloid cell-associated lung inflammation. Sci. Adv. 2020, 6, eabc4882. [Google Scholar] [CrossRef]
- Kim, N.; Kim, M.Y.; Cho, Y.U.; Chen, W.; Lee, K.H.; Kim, H.S. PVR and ICAM-1 on Blast Crisis CML Stem and Progenitor Cells with TKI Resistance Confer Susceptibility to NK Cells. Cancers 2020, 12, 1923. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.D.; Noh, J.Y.; Shin, J.H.; Lin, J.J.; Lee, S.Y. PTEN regulation by the Akt/GSK-3beta axis during RANKL signaling. Bone 2013, 55, 126–131. [Google Scholar] [CrossRef]
- Wang, Q.; Zhou, Y.; Wang, X.; Evers, B.M. Glycogen synthase kinase-3 is a negative regulator of extracellular signal-regulated kinase. Oncogene 2006, 25, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Raulet, D.H.; Marcus, A.; Coscoy, L. Dysregulated cellular functions and cell stress pathways provide critical cues for activating and targeting natural killer cells to transformed and infected cells. Immunol. Rev. 2017, 280, 93–101. [Google Scholar] [CrossRef]
- Liu, X.V.; Ho, S.S.; Tan, J.J.; Kamran, N.; Gasser, S. Ras activation induces expression of Raet1 family NK receptor ligands. J. Immunol. 2012, 189, 1826–1834. [Google Scholar] [CrossRef]
- Borchers, M.T.; Harris, N.L.; Wesselkamper, S.C.; Vitucci, M.; Cosman, D. NKG2D ligands are expressed on stressed human airway epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2006, 291, L222–L231. [Google Scholar] [CrossRef]
- Kauffmann-Zeh, A.; Rodriguez-Viciana, P.; Ulrich, E.; Gilbert, C.; Coffer, P.; Downward, J.; Evan, G. Suppression of c-Myc-induced apoptosis by Ras signalling through PI(3)K and PKB. Nature 1997, 385, 544–548. [Google Scholar] [CrossRef]
- Skorski, T.; Bellacosa, A.; Nieborowska-Skorska, M.; Majewski, M.; Martinez, R.; Choi, J.K.; Trotta, R.; Wlodarski, P.; Perrotti, D.; Chan, T.O.; et al. Transformation of hematopoietic cells by BCR/ABL requires activation of a PI-3k/Akt-dependent pathway. EMBO J. 1997, 16, 6151–6161. [Google Scholar] [CrossRef]
- Nanbakhsh, A.; Pochon, C.; Mallavialle, A.; Amsellem, S.; Bourhis, J.H.; Chouaib, S. c-Myc regulates expression of NKG2D ligands ULBP1/2/3 in AML and modulates their susceptibility to NK-mediated lysis. Blood 2014, 123, 3585–3595. [Google Scholar] [CrossRef]
- Sears, R.; Nuckolls, F.; Haura, E.; Taya, Y.; Tamai, K.; Nevins, J.R. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000, 14, 2501–2514. [Google Scholar] [CrossRef]
- Gregory, M.A.; Qi, Y.; Hann, S.R. Phosphorylation by glycogen synthase kinase-3 controls c-myc proteolysis and subnuclear localization. J. Biol. Chem. 2003, 278, 51606–51612. [Google Scholar] [CrossRef] [PubMed]
- Doble, B.W.; Woodgett, J.R. GSK-3: Tricks of the trade for a multi-tasking kinase. J. Cell Sci. 2003, 116, 1175–1186. [Google Scholar] [CrossRef] [PubMed]
- Force, T.; Woodgett, J.R. Unique and overlapping functions of GSK-3 isoforms in cell differentiation and proliferation and cardiovascular development. J. Biol. Chem. 2009, 284, 9643–9647. [Google Scholar] [CrossRef] [PubMed]
- Shi, P.; Yin, T.; Zhou, F.; Cui, P.; Gou, S.; Wang, C. Valproic acid sensitizes pancreatic cancer cells to natural killer cell-mediated lysis by upregulating MICA and MICB via the PI3K/Akt signaling pathway. BMC Cancer 2014, 14, 370. [Google Scholar] [CrossRef]
- Xia, C.; He, Z.; Cai, Y.; Liang, S. Vorinostat upregulates MICA via the PI3K/Akt pathway to enhance the ability of natural killer cells to kill tumor cells. Eur. J. Pharmacol. 2020, 875, 173057. [Google Scholar] [CrossRef]
- Cohen, P.; Frame, S. The renaissance of GSK3. Nat. Rev. Mol. Cell Biol. 2001, 2, 769–776. [Google Scholar] [CrossRef]
- Felsher, D.W.; Bishop, J.M. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol. Cell 1999, 4, 199–207. [Google Scholar] [CrossRef]
- Vita, M.; Henriksson, M. The Myc oncoprotein as a therapeutic target for human cancer. Semin. Cancer Biol. 2006, 16, 318–330. [Google Scholar] [CrossRef]
- Unni, A.M.; Bondar, T.; Medzhitov, R. Intrinsic sensor of oncogenic transformation induces a signal for innate immunosurveillance. Proc. Natl. Acad. Sci. USA 2008, 105, 1686–1691. [Google Scholar] [CrossRef]
- Kazi, A.; Xiang, S.; Yang, H.; Delitto, D.; Trevino, J.; Jiang, R.H.Y.; Ayaz, M.; Lawrence, H.R.; Kennedy, P.; Sebti, S.M. GSK3 suppression upregulates beta-catenin and c-Myc to abrogate KRas-dependent tumors. Nat. Commun. 2018, 9, 5154. [Google Scholar] [CrossRef]
- Textor, S.; Bossler, F.; Henrich, K.O.; Gartlgruber, M.; Pollmann, J.; Fiegler, N.; Arnold, A.; Westermann, F.; Waldburger, N.; Breuhahn, K.; et al. The proto-oncogene Myc drives expression of the NK cell-activating NKp30 ligand B7-H6 in tumor cells. Oncoimmunology 2016, 5, e1116674. [Google Scholar] [CrossRef]
- Paczulla, A.M.; Rothfelder, K.; Raffel, S.; Konantz, M.; Steinbacher, J.; Wang, H.; Tandler, C.; Mbarga, M.; Schaefer, T.; Falcone, M.; et al. Absence of NKG2D ligands defines leukaemia stem cells and mediates their immune evasion. Nature 2019, 572, 254–259. [Google Scholar] [CrossRef]
- Sconocchia, G.; Lau, M.; Provenzano, M.; Rezvani, K.; Wongsena, W.; Fujiwara, H.; Hensel, N.; Melenhorst, J.; Li, J.; Ferrone, S.; et al. The antileukemia effect of HLA-matched NK and NK-T cells in chronic myelogenous leukemia involves NKG2D-target-cell interactions. Blood 2005, 106, 3666–3672. [Google Scholar] [CrossRef] [PubMed]
- Banerji, V.; Frumm, S.M.; Ross, K.N.; Li, L.S.; Schinzel, A.C.; Hahn, C.K.; Kakoza, R.M.; Chow, K.T.; Ross, L.; Alexe, G.; et al. The intersection of genetic and chemical genomic screens identifies GSK-3alpha as a target in human acute myeloid leukemia. J. Clin. Investig. 2012, 122, 935–947. [Google Scholar] [CrossRef] [PubMed]
- Ougolkov, A.V.; Bone, N.D.; Fernandez-Zapico, M.E.; Kay, N.E.; Billadeau, D.D. Inhibition of glycogen synthase kinase-3 activity leads to epigenetic silencing of nuclear factor kappaB target genes and induction of apoptosis in chronic lymphocytic leukemia B cells. Blood 2007, 110, 735–742. [Google Scholar] [CrossRef]
- Mirlashari, M.R.; Randen, I.; Kjeldsen-Kragh, J. Glycogen synthase kinase-3 (GSK-3) inhibition induces apoptosis in leukemic cells through mitochondria-dependent pathway. Leuk. Res. 2012, 36, 499–508. [Google Scholar] [CrossRef]
- Wagner, F.F.; Benajiba, L.; Campbell, A.J.; Weiwer, M.; Sacher, J.R.; Gale, J.P.; Ross, L.; Puissant, A.; Alexe, G.; Conway, A.; et al. Exploiting an Asp-Glu “switch” in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia. Sci. Transl. Med. 2018, 10, eaam8460. [Google Scholar] [CrossRef] [PubMed]
- Guezguez, B.; Almakadi, M.; Benoit, Y.D.; Shapovalova, Z.; Rahmig, S.; Fiebig-Comyn, A.; Casado, F.L.; Tanasijevic, B.; Bresolin, S.; Masetti, R.; et al. GSK3 Deficiencies in Hematopoietic Stem Cells Initiate Pre-neoplastic State that Is Predictive of Clinical Outcomes of Human Acute Leukemia. Cancer Cell 2016, 29, 61–74. [Google Scholar] [CrossRef] [PubMed]
- O’Hare, T.; Zabriskie, M.S.; Eiring, A.M.; Deininger, M.W. Pushing the limits of targeted therapy in chronic myeloid leukaemia. Nat. Rev. Cancer 2012, 12, 513–526. [Google Scholar] [CrossRef] [PubMed]
- Soverini, S.; Mancini, M.; Bavaro, L.; Cavo, M.; Martinelli, G. Chronic myeloid leukemia: The paradigm of targeting oncogenic tyrosine kinase signaling and counteracting resistance for successful cancer therapy. Mol. Cancer 2018, 17, 49. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, N.; Kim, M.Y.; Choi, W.S.; Yi, E.; Lee, H.J.; Kim, H.S. GSK-3α Inhibition in Drug-Resistant CML Cells Promotes Susceptibility to NK Cell-Mediated Lysis in an NKG2D- and NKp30-Dependent Manner. Cancers 2021, 13, 1802. https://doi.org/10.3390/cancers13081802
Kim N, Kim MY, Choi WS, Yi E, Lee HJ, Kim HS. GSK-3α Inhibition in Drug-Resistant CML Cells Promotes Susceptibility to NK Cell-Mediated Lysis in an NKG2D- and NKp30-Dependent Manner. Cancers. 2021; 13(8):1802. https://doi.org/10.3390/cancers13081802
Chicago/Turabian StyleKim, Nayoung, Mi Yeon Kim, Woo Seon Choi, Eunbi Yi, Hyo Jung Lee, and Hun Sik Kim. 2021. "GSK-3α Inhibition in Drug-Resistant CML Cells Promotes Susceptibility to NK Cell-Mediated Lysis in an NKG2D- and NKp30-Dependent Manner" Cancers 13, no. 8: 1802. https://doi.org/10.3390/cancers13081802
APA StyleKim, N., Kim, M. Y., Choi, W. S., Yi, E., Lee, H. J., & Kim, H. S. (2021). GSK-3α Inhibition in Drug-Resistant CML Cells Promotes Susceptibility to NK Cell-Mediated Lysis in an NKG2D- and NKp30-Dependent Manner. Cancers, 13(8), 1802. https://doi.org/10.3390/cancers13081802





