Integrative RNA-Seq and H3 Trimethylation ChIP-Seq Analysis of Human Lung Cancer Cells Isolated by Laser-Microdissection
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue Samples and Laser-Microdissection Microscopy (LMD) Isolation
2.2. RNA-Sequencing
2.3. RNA-Seq Analysis
2.4. Chromatin Immunoprecipitation Sequencing (ChIP-Seq)
2.5. ChIP-Seq Read Mapping and Peaks Calling
2.6. Transcription Factor Binding Sites (TFBSs) Analysis (ENCODE)
3. Results
3.1. Transcriptomic Profiles of Non-Small Cell Lung Cancer (NSCLC) Were Successfully Captured Using LMD-Isolated Samples
3.2. RNA-Seq Analysis Showed Subtype-Specific Differential Expressed Genes (DEGs) and Enriched Pathways in NSCLC
3.3. RNA-Seq Results from LMD-Isolated Tumor Samples Were Concordant with the Cancer Genome Atlas (TCGA) Reference Dataset While Excluding Stroma Parts
3.4. Somatically Altered H3K4me3-Marked Promoters in NSCLC Were Successfully Identified Using LDM-Isolated Samples
3.5. Proximal Tumor-Altered H3K4me3 Regions Were Found at Cancer-Related DEGs
3.6. Altered H3K4me3-Marked Promoters Distant from TSSs Were Annotated with Enhancer Activity of Cancer Regulatory Genes
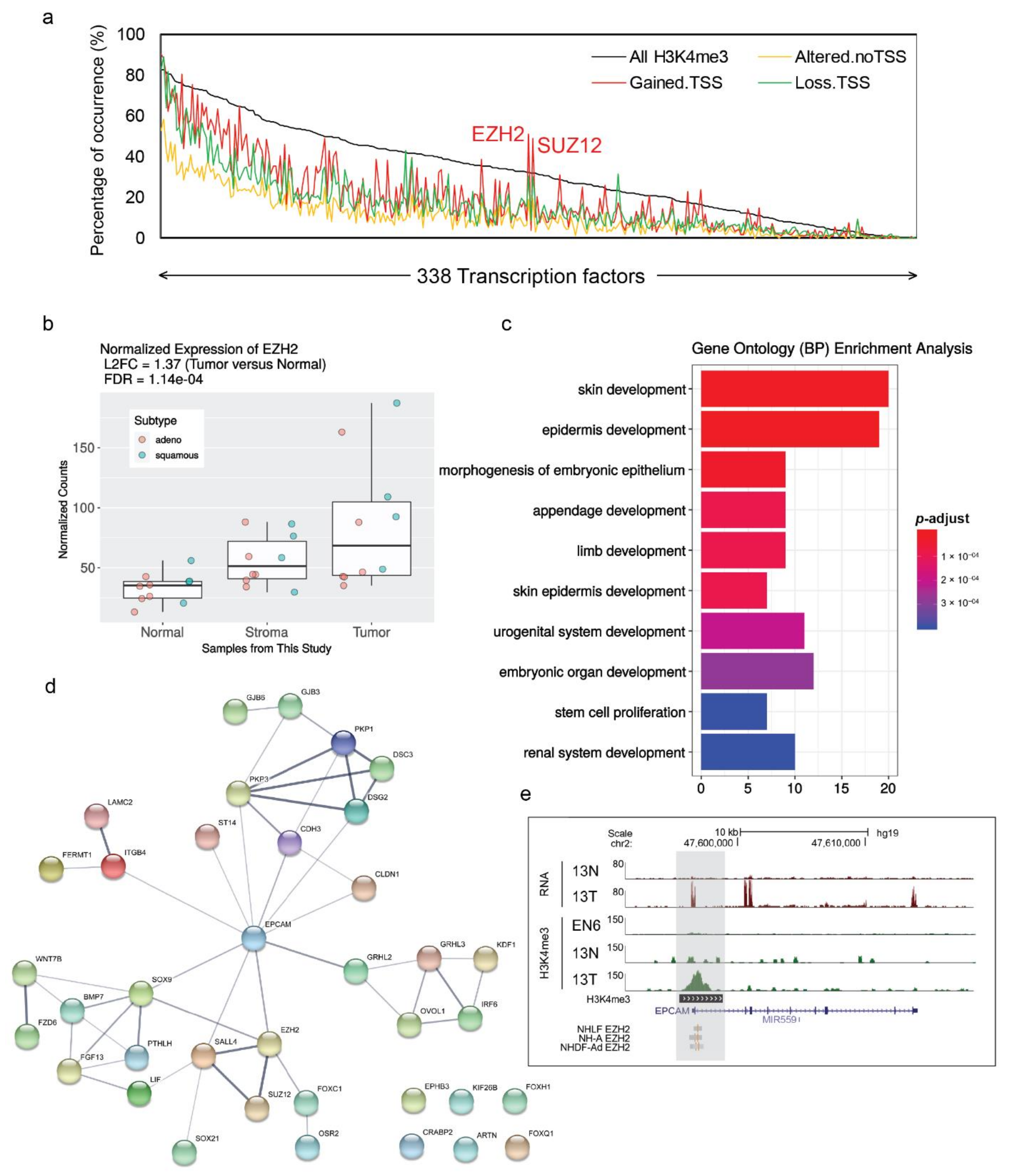
3.7. Proximal Tumor-Gained Promoters Associated with EZH2 and SUZ12 Occupancies and Enriched for Developmental Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stratton, M.R.; Campbell, P.J.; Futreal, P.A. The cancer genome. Nature 2009, 458, 719–724. [Google Scholar] [CrossRef] [PubMed]
- Easwaran, H.; Tsai, H.C.; Baylin, S.B. Cancer Epigenetics: Tumor Heterogeneity, Plasticity of Stem-like States, and Drug Resistance. Mol. Cell 2014, 54, 716–727. [Google Scholar] [CrossRef] [PubMed]
- Muratani, M.; Deng, N.; Ooi, W.F.; Lin, S.J.; Xing, M.; Xu, C.; Qamra, A.; Tay, S.T.; Malik, S.; Wu, J.; et al. Nanoscale chromatin profiling of gastric adenocarcinoma reveals cancer-associated cryptic promoters and somatically acquired regulatory elements. Nat. Commun. 2014, 5, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Rabien, A.; Kristiansen, G. Tissue microdissection. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2016; Volume 1381, pp. 39–52. [Google Scholar] [CrossRef]
- Molina, J.R.; Yang, P.; Cassivi, S.D.; Schild, S.E.; Adjei, A.A. Non-small cell lung cancer: Epidemiology, risk factors, treatment, and survivorship. Mayo Clin. Proc. 2008, 83, 584–594. [Google Scholar] [CrossRef]
- Lu, T.; Yang, X.; Huang, Y.; Zhao, M.; Li, M.; Ma, K.; Yin, J.; Zhan, C.; Wang, Q. Trends in the incidence, treatment, and survival of patients with lung cancer in the last four decades. Cancer Manag. Res. 2019, 11, 943–953. [Google Scholar] [CrossRef] [PubMed]
- Lambrechts, D.; Wauters, E.; Boeckx, B.; Aibar, S.; Nittner, D.; Burton, O.; Bassez, A.; Decaluwé, H.; Pircher, A.; Van den Eynde, K.; et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 2018, 24, 1277–1289. [Google Scholar] [CrossRef]
- Altorki, N.K.; Markowitz, G.J.; Gao, D.; Port, J.L.; Saxena, A.; Stiles, B.; McGraw, T.; Mittal, V. The lung microenvironment: An important regulator of tumour growth and metastasis. Nat. Rev. Cancer 2019, 19, 9–31. [Google Scholar] [CrossRef]
- Ling, J.Q.; Hoffman, A.R. Epigenetics of long-range chromatin interactions. Pediatr. Res. 2007, 61, 11–16. [Google Scholar] [CrossRef]
- Kim, S.K.; Jang, H.R.; Kim, J.H.; Noh, S.M.; Song, K.S.; Kim, M.R.; Kim, S.Y.; Yeom, Y.I.; Kim, N.S.; Yoo, H.S.; et al. The epigenetic silencing of LIMS2 in gastric cancer and its inhibitory effect on cell migration. Biochem. Biophys. Res. Commun. 2006, 349, 1032–1040. [Google Scholar] [CrossRef]
- Travis, W.D.; Brambilla, E.; Burke, A.P.; Marx, A.; Nicholson, A.G. Introduction to the 2015 World Health Organization Classification of Tumors of the Lung, Pleura, Thymus, and Heart. J. Thorac. Oncol. 2015, 10, 1240–1242. [Google Scholar] [CrossRef]
- Husni, R.E.; Shiba-Ishii, A.; Nakagawa, T.; Dai, T.; Kim, Y.; Hong, J.; Sakashita, S.; Sakamoto, N.; Sato, Y.; Noguchi, M. DNA hypomethylation-related overexpression of SFN, GORASP2 and ZYG11A is a novel prognostic biomarker for early stage lung adenocarcinoma. Oncotarget 2019, 10, 1625–1636. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. Available online: https://arxiv.org/abs/1303.3997 (accessed on 8 May 2018).
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Ng, J.H.; Kumar, V.; Muratani, M.; Kraus, P.; Yeo, J.C.; Yaw, L.P.; Xue, K.; Lufkin, T.; Prabhakar, S.; Ng, H.H. In Vivo Epigenomic Profiling of Germ Cells Reveals Germ Cell Molecular Signatures. Dev. Cell 2013, 24, 324–333. [Google Scholar] [CrossRef]
- ChIP dilution buffer. Cold Spring Harb. Protoc. 2006, 2006. [CrossRef]
- Xu, H.; Handoko, L.; Wei, X.; Ye, C.; Sheng, J.; Wei, C.L.; Lin, F.; Sung, W.K. A signal-noise model for significance analysis of ChIP-seq with negative control. Bioinformatics 2010, 26, 1199–1204. [Google Scholar] [CrossRef]
- Amemiya, H.M.; Kundaje, A.; Boyle, A.P. The ENCODE Blacklist: Identification of Problematic Regions of the Genome. Sci. Rep. 2019, 9, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, S.; Arakawa, T.; Fukuda, S.; Furuno, M.; Hasegawa, A.; Hori, F.; Ishikawa-Kato, S.; Kaida, K.; Kaiho, A.; Kanamori-Katayama, M.; et al. FANTOM5 CAGE profiles of human and mouse samples. Sci. Data 2017, 4, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The Human Genome Browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhuang, J.; Iyer, S.; Lin, X.Y.; Greven, M.C.; Kim, B.H.; Moore, J.; Pierce, B.G.; Dong, X.; Virgil, D.; et al. Factorbook.org: A Wiki-based database for transcription factor-binding data generated by the ENCODE consortium. Nucleic Acids Res. 2013, 41, D171. [Google Scholar] [CrossRef] [PubMed]
- Ikari, A.; Sato, T.; Watanabe, R.; Yamazaki, Y.; Sugatani, J. Increase in claudin-2 expression by an EGFR/MEK/ERK/c-Fos pathway in lung adenocarcinoma A549 cells. Biochim. Biophys. Acta Mol. Cell Res. 2012, 1823, 1110–1118. [Google Scholar] [CrossRef] [PubMed]
- Tachihara-Yoshikawa, M.; Ishida, T.; Watanabe, K.; Sugawara, A.; Kanazawa, K.; Kanno, R.; Suzuki, T.; Niimi, T.; Kimura, S.; Munakata, M. Expression of secretoglobin3A2 (SCGB3A2) in primary pulmonary carcinomas. Fukushima J. Med. Sci. 2008, 54, 61–72. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yoshimoto, T.; Matsubara, D.; Soda, M.; Ueno, T.; Amano, Y.; Kihara, A.; Sakatani, T.; Nakano, T.; Shibano, T.; Endo, S.; et al. Mucin 21 is a key molecule involved in the incohesive growth pattern in lung adenocarcinoma. Cancer Sci. 2019, 110, 3006–3011. [Google Scholar] [CrossRef]
- Hawthorn, L.; Stein, L.; Panzarella, J.; Loewen, G.M.; Baumann, H. Characterization of cell-type specific profiles in tissues and isolated cells from squamous cell carcinomas of the lung. Lung Cancer 2006, 53, 129–142. [Google Scholar] [CrossRef]
- Fujii, T.; Dracheva, T.; Player, A.; Chacko, S.; Clifford, R.; Strausberg, R.L.; Buetow, K.; Azumi, N.; Travis, W.D.; Jen, J. A Preliminary Transcriptome Map of Non-Small Cell Lung Cancer. Cancer Res. 2002, 62, 3340–3346. [Google Scholar]
- Chang, H.-H.; Dreyfuss, J.M.; Ramoni, M.F. A transcriptional network signature characterizes lung cancer subtypes. Cancer 2011, 117, 353–360. [Google Scholar] [CrossRef]
- Habib Dakir, E.L.; Feigenbaum, L.; Linnoila, R.I. Constitutive expression of human keratin 14 gene in mouse lung induces premalignant lesions and squamous differentiation. Carcinogenesis 2008, 29, 2377–2384. [Google Scholar] [CrossRef]
- Landt, S.G.; Marinov, G.K.; Kundaje, A.; Kheradpour, P.; Pauli, F.; Batzoglou, S.; Bernstein, B.E.; Bickel, P.; Brown, J.B.; Cayting, P.; et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012, 22, 1813–1831. [Google Scholar] [CrossRef] [PubMed]
- Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.Y.; Schones, D.E.; Wang, Z.; Wei, G.; Chepelev, I.; Zhao, K. High-Resolution Profiling of Histone Methylations in the Human Genome. Cell 2007, 129, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Guenther, M.G.; Levine, S.S.; Boyer, L.A.; Jaenisch, R.; Young, R.A. A Chromatin Landmark and Transcription Initiation at Most Promoters in Human Cells. Cell 2007, 130, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Kok-Sin, T.; Mokhtar, N.M.; Hassan, N.Z.A.; Sagap, I.; Rose, I.M.; Harun, R.; Jamal, R. Identification of diagnostic markers in colorectal cancer via integrative epigenomics and genomics data. Oncol. Rep. 2015, 34, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Sasagawa, T.; Jinno-Oue, A.; Nagamatsu, T.; Morita, K.; Tsuruga, T.; Mori-Uchino, M.; Fujii, T.; Shibuya, M. Production of an anti-angiogenic factor sFLT1 is suppressed via promoter hypermethylation of FLT1 gene in choriocarcinoma cells. BMC Cancer 2020, 20, 112. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Yu, H.; Martin, T.A.; Zhang, Y.; Chen, G.; Jiang, W.G. Effect of junctional adhesion molecule-2 expression on cell growth, invasion and migration in human colorectal cancer. Int. J. Oncol. 2016, 48, 929–936. [Google Scholar] [CrossRef]
- Carboni, G.L.; Gao, B.; Nishizaki, M.; Xu, K.; Minna, J.D.; Roth, J.A.; Ji, L. CACNA2D2-mediated apoptosis in NSCLC cells is associated with alterations of the intracellular calcium signaling and disruption of mitochondria membrane integrity. Oncogene 2003, 22, 615–626. [Google Scholar] [CrossRef]
- Wang, Y.; Marino-Enriquez, A.; Bennett, R.R.; Zhu, M.; Shen, Y.; Eilers, G.; Lee, J.C.; Henze, J.; Fletcher, B.S.; Gu, Z.; et al. Dystrophin is a tumor suppressor in human cancers with myogenic programs. Nat. Genet. 2014, 46, 601–606. [Google Scholar] [CrossRef]
- Sakamoto, K.; Imai, K.; Higashi, T.; Taki, K.; Nakagawa, S.; Okabe, H.; Nitta, H.; Hayashi, H.; Chikamoto, A.; Ishiko, T.; et al. Significance of P-cadherin overexpression and possible mechanism of its regulation in intrahepatic cholangiocarcinoma and pancreatic cancer. Cancer Sci. 2015, 106, 1153–1162. [Google Scholar] [CrossRef]
- Wu, X.; Xiao, J.; Zhao, C.; Zhao, C.; Han, Z.; Wang, F.; Yang, Y.; Jiang, Y.; Fang, F. Claudin1 promotes the proliferation, invasion and migration of nasopharyngeal carcinoma cells by upregulating the expression and nuclear entry of β-catenin. Exp. Ther. Med. 2018, 16, 3445–3451. [Google Scholar] [CrossRef]
- Qin, S.; Liao, Y.; Du, Q.; Wang, W.; Huang, J.; Liu, P.; Shang, C.; Liu, T.; Xia, M.; Yao, S. DSG2 expression is correlated with poor prognosis and promotes early-stage cervical cancer. Cancer Cell Int. 2020, 20. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Maldonado, L.; Tiana, M.; Roche, O.; Prado-Cabrero, A.; Jensen, L.; Fernandez-Barral, A.; Guijarro-Muñoz, I.; Favaro, E.; Moreno-Bueno, G.; Sanz, L.; et al. EFNA3 long noncoding RNAs induced by hypoxia promote metastatic dissemination. Oncogene 2015, 34, 2609–2620. [Google Scholar] [CrossRef]
- Ma, B.; Zhang, L.; Zou, Y.; He, R.; Wu, Q.; Han, C.; Zhang, B. Reciprocal regulation of integrin β4 and KLF4 promotes gliomagenesis through maintaining cancer stem cell traits. J. Exp. Clin. Cancer Res. 2019, 38, 23. [Google Scholar] [CrossRef]
- He, J.; Liu, Y.; Zhang, L.; Zhang, H. Integrin subunit beta 8 (ITGB8) upregulation is an independent predictor of unfavorable survival of high-grade serous ovarian carcinoma patients. Med. Sci. Monit. 2018, 24, 8933–8940. [Google Scholar] [CrossRef]
- Huang, D.; Du, C.; Ji, D.; Xi, J.; Gu, J. Overexpression of LAMC2 predicts poor prognosis in colorectal cancer patients and promotes cancer cell proliferation, migration, and invasion. Tumor Biol. 2017, 39, 101042831770584. [Google Scholar] [CrossRef]
- Li, Q.L.; Wang, D.Y.; Ju, L.G.; Yao, J.; Gao, C.; Lei, P.J.; Li, L.Y.; Zhao, X.L.; Wu, M. The hyper-activation of transcriptional enhancers in breast cancer. Clin. Epigenet. 2019, 11, 48. [Google Scholar] [CrossRef]
- Fishilevich, S.; Nudel, R.; Rappaport, N.; Hadar, R.; Plaschkes, I.; Iny Stein, T.; Rosen, N.; Kohn, A.; Twik, M.; Safran, M.; et al. GeneHancer: Genome-wide integration of enhancers and target genes in GeneCards. Database 2017, 2017. [Google Scholar] [CrossRef]
- Zhang, S.; Mo, Q.; Wang, X. Oncological role of HMGA2 (Review). Int. J. Oncol. 2019, 55, 775–778. [Google Scholar] [CrossRef]
- Wuebben, E.L.; Rizzino, A. The dark side of SOX2: Cancer—A comprehensive overview. Oncotarget 2017, 8, 44917–44943. [Google Scholar] [CrossRef]
- Fu, X.; Pereira, R.; De Angelis, C.; Veeraraghavan, J.; Nanda, S.; Qin, L.; Cataldo, M.L.; Sethunath, V.; Mehravaran, S.; Gutierrez, C.; et al. FOXA1 upregulation promotes enhancer and transcriptional reprogramming in endocrine-resistant breast cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 26823–26834. [Google Scholar] [CrossRef]
- Cassandri, M.; Butera, A.; Amelio, I.; Lena, A.M.; Montanaro, M.; Mauriello, A.; Anemona, L.; Candi, E.; Knight, R.A.; Agostini, M.; et al. ZNF750 represses breast cancer invasion via epigenetic control of prometastatic genes. Oncogene 2020, 39, 4331–4343. [Google Scholar] [CrossRef]
- Yamauchi, T.; Danis, E.; Zhang, X.; Riedel, S.; Huang, H.; Bernt, K.M.; Neff, T. The Role of Gata2 in Murine Models of Acute Myeloid Leukemia. Blood 2016, 128, 1516. [Google Scholar] [CrossRef]
- Fabian, J.; Lodrini, M.; Oehme, I.; Schier, M.C.; Thole, T.M.; Hielscher, T.; Kopp-Schneider, A.; Opitz, L.; Capper, D.; Von Deimling, A.; et al. GRHL1 acts as tumor suppressor in neuroblastoma and is negatively regulated by MYCN and HDAC3. Cancer Res. 2014, 74, 2604–2616. [Google Scholar] [CrossRef]
- Lee, T.I.; Jenner, R.G.; Boyer, L.A.; Guenther, M.G.; Levine, S.S.; Kumar, R.M.; Chevalier, B.; Johnstone, S.E.; Cole, M.F.; Isono, K.; et al. Control of Developmental Regulators by Polycomb in Human Embryonic Stem Cells. Cell 2006, 125, 301–313. [Google Scholar] [CrossRef]
- Qamra, A.; Xing, M.; Padmanabhan, N.; Kwok, J.J.T.; Zhang, S.; Xu, C.; Leong, Y.S.; Lim, A.P.L.; Tang, Q.; Ooi, W.F.; et al. Epigenomic promoter alterations amplify gene isoform and immunogenic diversity in gastric adenocarcinoma. Cancer Discov. 2017, 7, 630–651. [Google Scholar] [CrossRef]
- Comet, I.; Riising, E.M.; Leblanc, B.; Helin, K. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat. Rev. Cancer 2016, 16, 803–810. [Google Scholar] [CrossRef] [PubMed]
- Dimou, A.; Dincman, T.; Evanno, E.; Gemmill, R.M.; Roche, J.; Drabkin, H.A. Epigenetics during EMT in lung cancer: EZH2 as a potential therapeutic target. Cancer Treat. Res. Commun. 2017, 12, 40–48. [Google Scholar] [CrossRef]
- Liu, C.; Shi, X.; Wang, L.; Wu, Y.; Jin, F.; Bai, C.; Song, Y. SUZ12 is involved in progression of non-small cell lung cancer by promoting cell proliferation and metastasis. Tumor Biol. 2014, 35, 6073–6082. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Ikari, A.; Sato, T.; Takiguchi, A.; Atomi, K.; Yamazaki, Y.; Sugatani, J. Claudin-2 knockdown decreases matrix metalloproteinase-9 activity and cell migration via suppression of nuclear Sp1 in A549 cells. Life Sci. 2011, 88, 628–633. [Google Scholar] [CrossRef]
- Karantza, V. Keratins in health and cancer: More than mere epithelial cell markers. Oncogene 2011, 30, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Lu, X.; Chen, X.; Zou, Y.; Liu, A.; Li, W.; He, B.; He, S.; Chen, Q. Eight potential biomarkers for distinguishing between lung adenocarcinoma and squamous cell carcinoma. Oncotarget 2017, 8, 71759–71771. [Google Scholar] [CrossRef] [PubMed]
- Lieu, E.L.; Nguyen, T.; Rhyne, S.; Kim, J. Amino acids in cancer. Exp. Mol. Med. 2020, 52, 15–30. [Google Scholar] [CrossRef]
- Asai, A.; Konno, M.; Koseki, J.; Taniguchi, M.; Vecchione, A.; Ishii, H. One-carbon metabolism for cancer diagnostic and therapeutic approaches. Cancer Lett. 2020, 470, 141–148. [Google Scholar] [CrossRef]
- Janiszewska, M.; Primi, M.C.; Izard, T. Cell adhesion in cancer: Beyond the migration of single cells. J. Biol. Chem. 2020, 295, 2495–2505. [Google Scholar] [CrossRef]
- López-Ferrer, A.; Barranco, C.; de Bolós, C. Differences in the O-Glycosylation Patterns Between Lung Squamous Cell Carcinoma and Adenocarcinoma. Am. J. Clin. Pathol. 2002, 118, 749–755. [Google Scholar] [CrossRef]
- Shi, Y.; Li, Y.; Yan, C.; Su, H.; Ying, K. Identification of key genes and evaluation of clinical outcomes in lung squamous cell carcinoma using integrated bioinformatics analysis. Oncol. Lett. 2019, 18, 5859–5870. [Google Scholar] [CrossRef]
- Steinberg, J. Collagen turnover and the growth state in 3T6 fibroblast cultures. Lab. Investig. 1978, 39, 491–496. [Google Scholar]
- Steinberg, J. The turnover of collagen in fibroblast cultures. J. Cell Sci. 1973, 12, 217–234. [Google Scholar]
- Kauppila, S.; Stenbäck, F.; Risteli, J.; Jukkola, A.; Risteli, L. Aberrant type I and type III collagen gene expression in human breast cancer in vivo. J. Pathol. 1998, 186, 262–268. [Google Scholar] [CrossRef]
- Cheng, Z.; Gao, W.; Fan, X.; Chen, X.; Mei, H.; Liu, J.; Luo, X.; Hu, Y. Extracellular signal-regulated kinase 5 associates with casein kinase II to regulate GPIb-IX-mediated platelet activation via the PTEN/PI3K/Akt pathway. J. Thromb. Haemost. 2017, 15, 1679–1688. [Google Scholar] [CrossRef]
- Kiefer, J.A.; Farach-Carson, M.C. Type I collagen-mediated proliferation of PC3 prostate carcinoma cell line: Implications for enhanced growth in the bone microenvironment. Matrix Biol. 2001, 20, 429–437. [Google Scholar] [CrossRef]
- Imamichi, Y.; König, A.; Gress, T.; Menke, A. Collagen type I-induced Smad-interacting protein 1 expression downregulates E-cadherin in pancreatic cancer. Oncogene 2007, 26, 2381–2385. [Google Scholar] [CrossRef] [PubMed]
- Koenig, A.; Mueller, C.; Hasel, C.; Adler, G.; Menke, A. Collagen type I induces disruption of E-cadherin-mediated cell-cell contacts and promotes proliferation of pancreatic carcinoma cells. Cancer Res. 2006, 66, 4662–4671. [Google Scholar] [CrossRef] [PubMed]
- You, J.S.; Jones, P.A. Cancer Genetics and Epigenetics: Two Sides of the Same Coin? Cancer Cell 2012, 22, 9–20. [Google Scholar] [CrossRef]
- Zhao, X.D.; Han, X.; Chew, J.L.; Liu, J.; Chiu, K.P.; Choo, A.; Orlov, Y.L.; Sung, W.K.; Shahab, A.; Kuznetsov, V.A.; et al. Whole-Genome Mapping of Histone H3 Lys4 and 27 Trimethylations Reveals Distinct Genomic Compartments in Human Embryonic Stem Cells. Cell Stem Cell 2007, 1, 286–298. [Google Scholar] [CrossRef]
- Murray, S.; Lorenz, P.; Howe, F.; Wouters, M.; Brown, T.; Xi, S.; Fischl, H.; Khushaim, W.; Rayappu, J.R.; Angel, A.; et al. H3K4me3 is neither instructive for, nor informed by, transcription. bioRxiv 2019, 709014. [Google Scholar] [CrossRef]
- Madsen, R.R. PI3K in stemness regulation: From development to cancer. Biochem. Soc. Trans. 2020, 48, 301–315. [Google Scholar] [CrossRef]
- Lu, P.; Weaver, V.M.; Werb, Z. The extracellular matrix: A dynamic niche in cancer progression. J. Cell Biol. 2012, 196, 395–406. [Google Scholar] [CrossRef]
- Lim, S.B.; Tan, S.J.; Lim, W.T.; Lim, C.T. An extracellular matrix-related prognostic and predictive indicator for early-stage non-small cell lung cancer. Nat. Commun. 2017, 8, 1–11. [Google Scholar] [CrossRef]
- Hu, D.; Gao, X.; Cao, K.; Morgan, M.A.; Mas, G.; Smith, E.R.; Volk, A.G.; Bartom, E.T.; Crispino, J.D.; Di Croce, L.; et al. Not All H3K4 Methylations Are Created Equal: Mll2/COMPASS Dependency in Primordial Germ Cell Specification. Mol. Cell 2017, 65, 460–475.e6. [Google Scholar] [CrossRef]
- Shen, H.; Xu, W.; Guo, R.; Rong, B.; Gu, L.; Wang, Z.; He, C.; Zheng, L.; Hu, X.; Hu, Z.; et al. Suppression of Enhancer Overactivation by a RACK7-Histone Demethylase Complex. Cell 2016, 165, 331–342. [Google Scholar] [CrossRef] [PubMed]
- Shi, B.; Liang, J.; Yang, X.; Wang, Y.; Zhao, Y.; Wu, H.; Sun, L.; Zhang, Y.; Chen, Y.; Li, R.; et al. Integration of Estrogen and Wnt Signaling Circuits by the Polycomb Group Protein EZH2 in Breast Cancer Cells. Mol. Cell. Biol. 2007, 27, 5105–5119. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.Y.; Jun, S.; Lee, M.; Kim, H.C.; Wang, X.; Ji, H.; McCrea, P.D.; Park, J. Il PAF and EZH2 induce wnt/β-catenin signaling hyperactivation. Mol. Cell 2013, 52, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Went, P.T.; Lugli, A.; Meier, S.; Bundi, M.; Mirlacher, M.; Sauter, G.; Dirnhofer, S. Frequent EpCam Protein Expression in Human Carcinomas. Hum. Pathol. 2004, 35, 122–128. [Google Scholar] [CrossRef]
- Keller, L.; Werner, S.; Pantel, K. Biology and clinical relevance of EpCAM. Cell Stress 2019, 3, 165–180. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ong, Q.; Sakashita, S.; Hanawa, E.; Kaneko, N.; Noguchi, M.; Muratani, M. Integrative RNA-Seq and H3 Trimethylation ChIP-Seq Analysis of Human Lung Cancer Cells Isolated by Laser-Microdissection. Cancers 2021, 13, 1719. https://doi.org/10.3390/cancers13071719
Ong Q, Sakashita S, Hanawa E, Kaneko N, Noguchi M, Muratani M. Integrative RNA-Seq and H3 Trimethylation ChIP-Seq Analysis of Human Lung Cancer Cells Isolated by Laser-Microdissection. Cancers. 2021; 13(7):1719. https://doi.org/10.3390/cancers13071719
Chicago/Turabian StyleOng, Quang, Shingo Sakashita, Emi Hanawa, Naomi Kaneko, Masayuki Noguchi, and Masafumi Muratani. 2021. "Integrative RNA-Seq and H3 Trimethylation ChIP-Seq Analysis of Human Lung Cancer Cells Isolated by Laser-Microdissection" Cancers 13, no. 7: 1719. https://doi.org/10.3390/cancers13071719
APA StyleOng, Q., Sakashita, S., Hanawa, E., Kaneko, N., Noguchi, M., & Muratani, M. (2021). Integrative RNA-Seq and H3 Trimethylation ChIP-Seq Analysis of Human Lung Cancer Cells Isolated by Laser-Microdissection. Cancers, 13(7), 1719. https://doi.org/10.3390/cancers13071719




