Cancer Stem Cells Are Possible Key Players in Regulating Anti-Tumor Immune Responses: The Role of Immunomodulating Molecules and MicroRNAs
Abstract
Simple Summary
Abstract
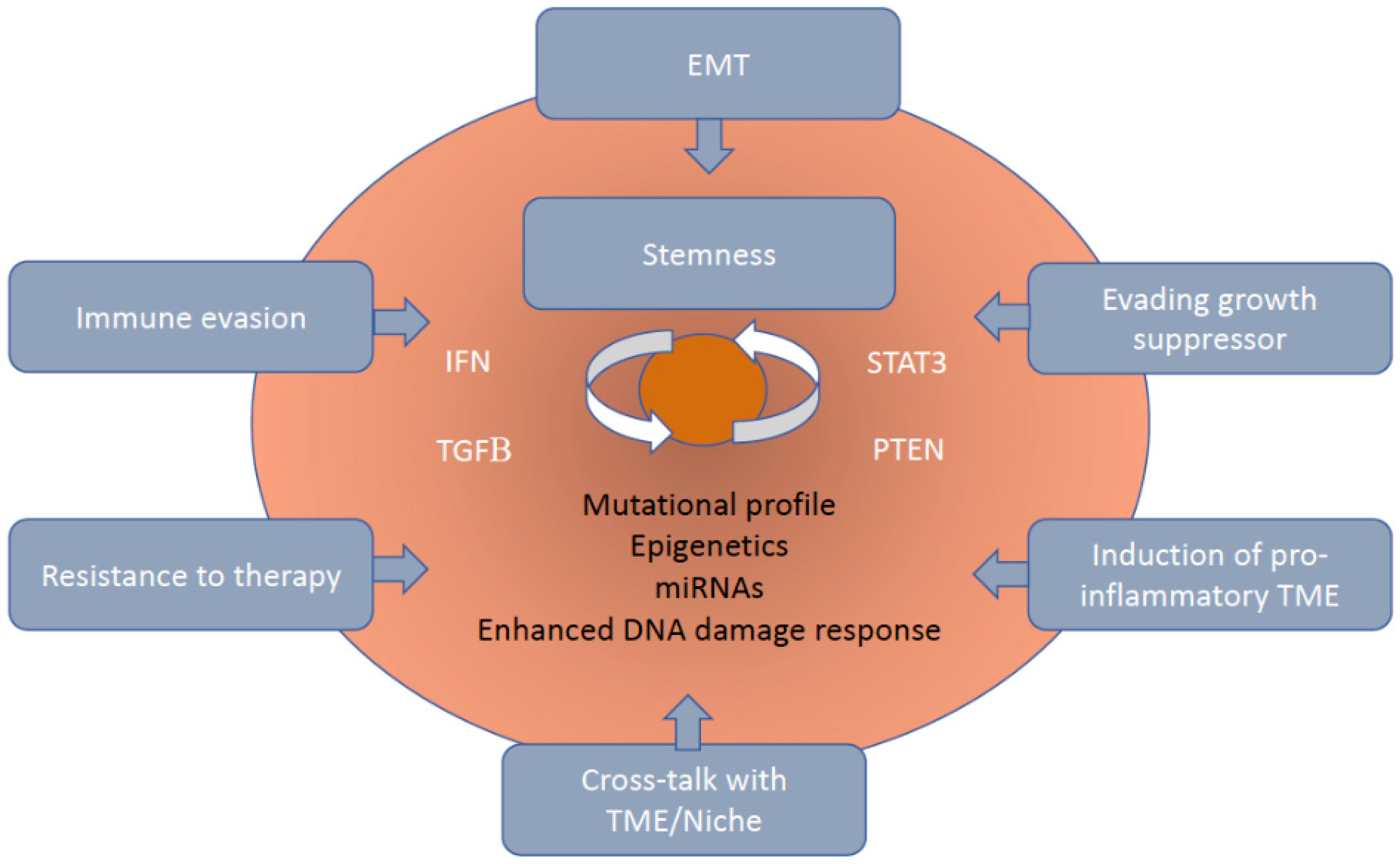
1. Introduction
2. Mechanisms of Immune Resistance of CSCs/CICs to Cell Mediated Immune Responses
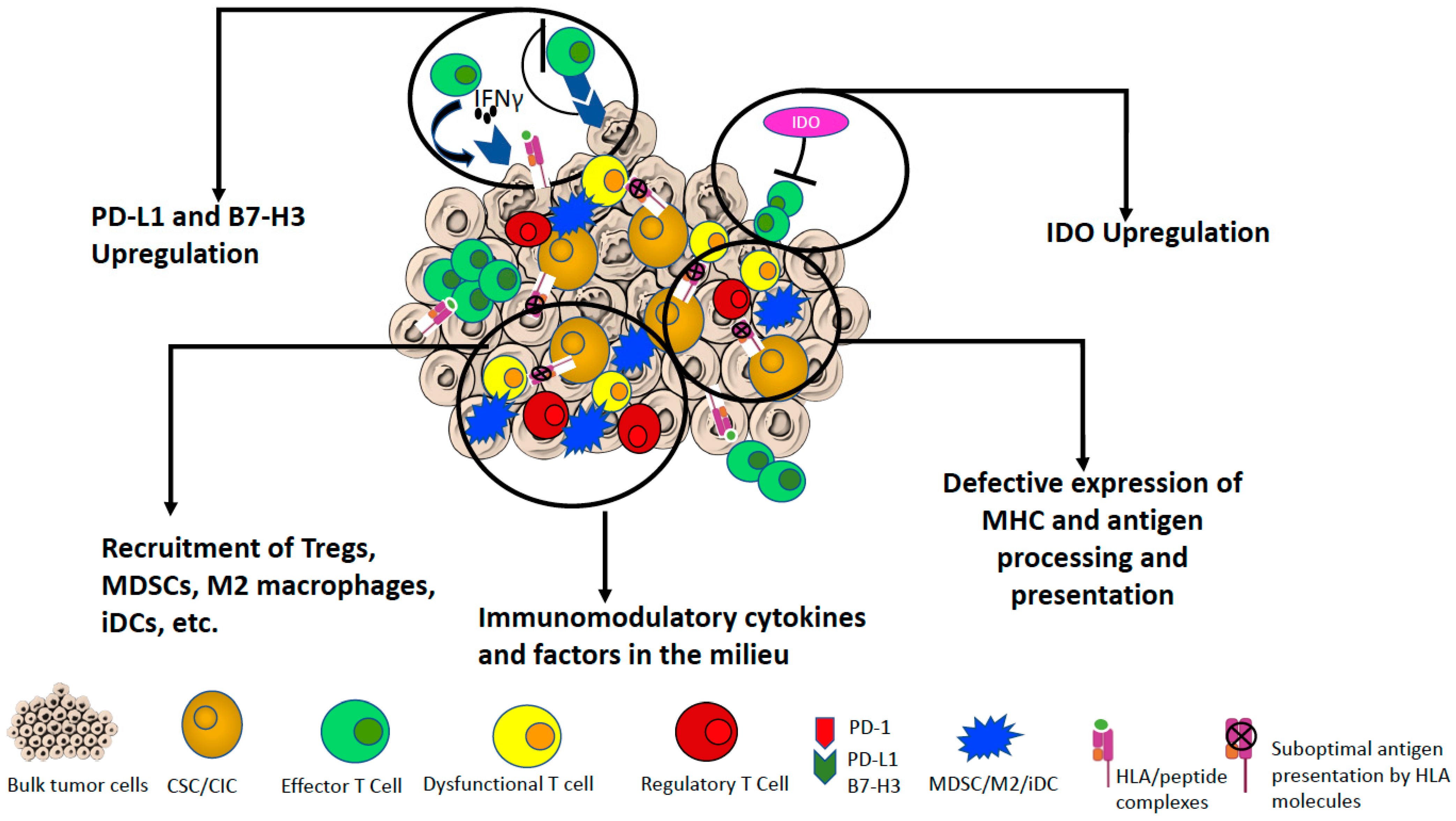
3. Interaction of Immune Cells with CSCs/CICs
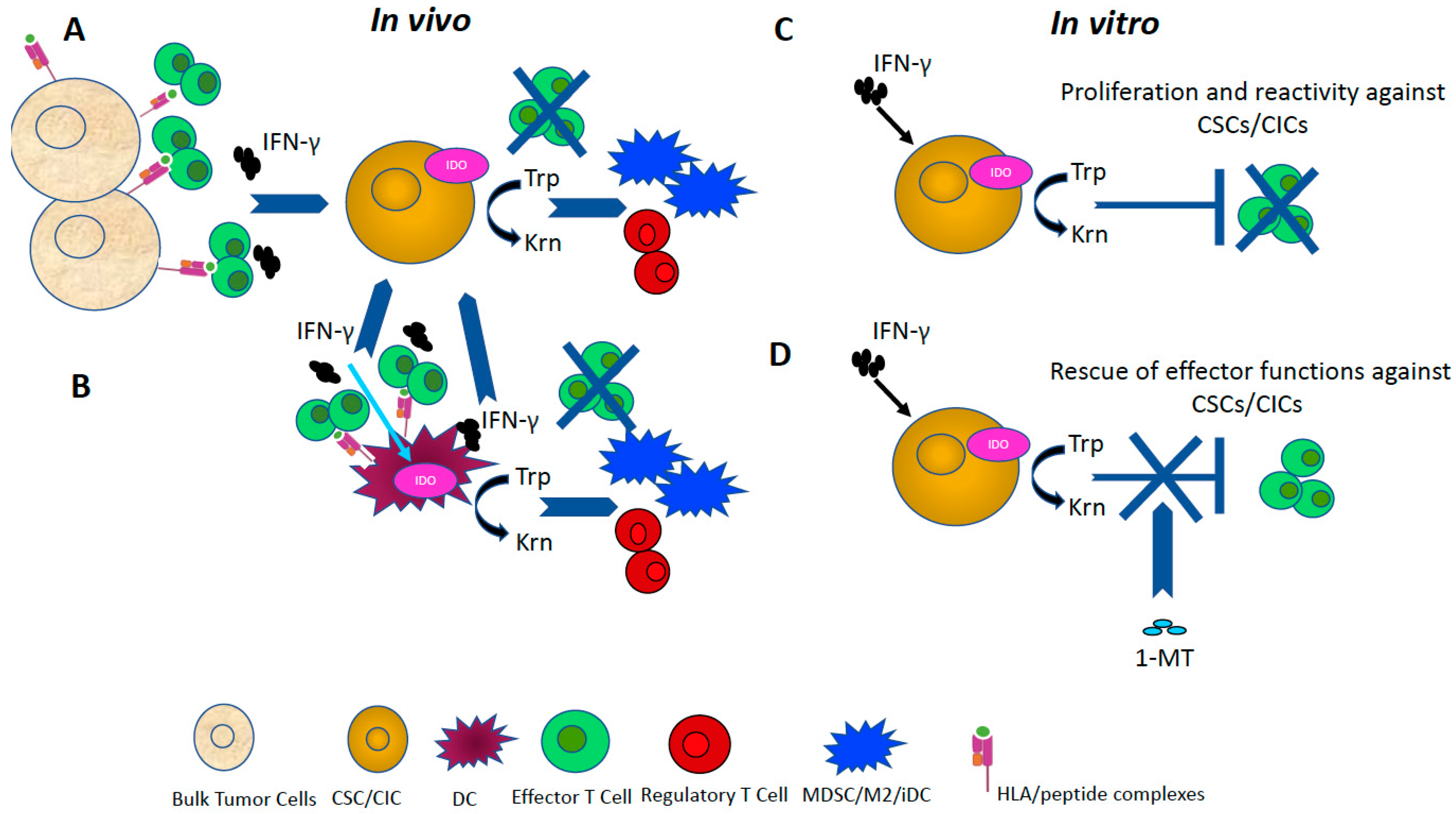
4. Role of IDO in Immunomodulation
5. Immunomodulatory Factors Associated with CSCs/CICs
5.1. Cytokines, Growth Factors and Other Soluble Regulators
5.2. Immune Checkpoints
6. The Dual Role of microRNAs in Orchestrating Pro-Tumoral and Stemness Features and Immunological Functions
6.1. MIRNAs Regulating Tumor and Stemness Properties
6.2. MiRNAs with Immunological Roles
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Li, Y.; Laterra, J. Cancer stem cells: Distinct entities or dynamically regulated phenotypes? Cancer Res. 2012, 72, 576–580. [Google Scholar] [CrossRef] [PubMed]
- Maccalli, C.; De Maria, R. Cancer stem cells: Perspectives for therapeutic targeting. Cancer Immunol. Immunother. 2015, 64, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Shackleton, M.; Quintana, E.; Fearon, E.R.; Morrison, S.J. Heterogeneity in cancer: Cancer stem cells versus clonal evolution. Cell 2009, 138, 822–829. [Google Scholar] [CrossRef] [PubMed]
- Prager, B.C.; Xie, Q.; Bao, S.; Rich, J.N. Cancer Stem Cells: The Architects of the Tumor Ecosystem. Cell Stem Cell 2019, 24, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Adair, J.E.; Kubek, S.P.; Kiem, H.P. Hematopoietic Stem Cell Approaches to Cancer. Hematol. Oncol. Clin. N. Am. 2017, 31, 897–912. [Google Scholar] [CrossRef]
- Clevers, H. The cancer stem cell: Premises, promises and challenges. Nat. Med. 2011, 17, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Kreso, A.; Dick, J.E. Evolution of the cancer stem cell model. Cell Stem Cell 2014, 14, 275–291. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, D.; Dick, J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997, 3, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Ricci-Vitiani, L.; Fabrizi, E.; Palio, E.; De Maria, R. Colon cancer stem cells. J. Mol. Med. 2009, 87, 1097–1104. [Google Scholar] [CrossRef]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [PubMed]
- Morrison, B.J.; Steel, J.C.; Morris, J.C. Reduction of MHC-I expression limits T-lymphocyte-mediated killing of Cancer-initiating cells. BMC Cancer 2018, 18, 469. [Google Scholar] [CrossRef]
- Al-Ejeh, F.; Smart, C.E.; Morrison, B.J.; Chenevix-Trench, G.; Lopez, J.A.; Lakhani, S.R.; Brown, M.P.; Khanna, K.K. Breast cancer stem cells: Treatment resistance and therapeutic opportunities. Carcinogenesis 2011, 32, 650–658. [Google Scholar] [CrossRef] [PubMed]
- Meyerrose, T.E.; Herrbrich, P.; Hess, D.A.; Nolta, J.A. Immune-deficient mouse models for analysis of human stem cells. Biotechniques 2003, 35, 1262–1272. [Google Scholar] [CrossRef] [PubMed]
- Tirino, V.; Desiderio, V.; Paino, F.; Papaccio, G.; De Rosa, M. Methods for cancer stem cell detection and isolation. Methods Mol. Biol. 2012, 879, 513–529. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, K.; Saikawa, Y.; Ohashi, M.; Kumagai, K.; Kitajima, M.; Okano, H.; Matsuzaki, Y.; Kitagawa, Y. Tumor initiating potential of side population cells in human gastric cancer. Int. J. Oncol. 2009, 34, 1201–1207. [Google Scholar] [PubMed]
- Maccalli, C.; Volonte, A.; Cimminiello, C.; Parmiani, G. Immunology of cancer stem cells in solid tumours. A review. Eur. J. Cancer 2014, 50, 649–655. [Google Scholar] [CrossRef] [PubMed]
- Cioffi, M.; D’Alterio, C.; Camerlingo, R.; Tirino, V.; Consales, C.; Riccio, A.; Ierano, C.; Cecere, S.C.; Losito, N.S.; Greggi, S.; et al. Identification of a distinct population of CD133(+)CXCR4(+) cancer stem cells in ovarian cancer. Sci. Rep. 2015, 5, 10357. [Google Scholar] [CrossRef] [PubMed]
- Eramo, A.; Lotti, F.; Sette, G.; Pilozzi, E.; Biffoni, M.; Di Virgilio, A.; Conticello, C.; Ruco, L.; Peschle, C.; De Maria, R. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ 2008, 15, 504–514. [Google Scholar] [CrossRef]
- Ginestier, C.; Hur, M.H.; Charafe-Jauffret, E.; Monville, F.; Dutcher, J.; Brown, M.; Jacquemier, J.; Viens, P.; Kleer, C.G.; Liu, S.; et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 2007, 1, 555–567. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.B.; Fillmore, C.M.; Jiang, G.; Shapira, S.D.; Tao, K.; Kuperwasser, C.; Lander, E.S. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell 2011, 146, 633–644. [Google Scholar] [CrossRef]
- Hadjimichael, C.; Chanoumidou, K.; Papadopoulou, N.; Arampatzi, P.; Papamatheakis, J.; Kretsovali, A. Common stemness regulators of embryonic and cancer stem cells. World J. Stem Cells 2015, 7, 1150–1184. [Google Scholar] [CrossRef] [PubMed]
- Hjelmeland, A.B.; Rich, J.N. The quest for self-identity: Not all cancer stem cells are the same. Clin. Cancer Res. 2012, 18, 3495–3498. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Hawkins, C.; Clarke, I.D.; Squire, J.A.; Bayani, J.; Hide, T.; Henkelman, R.M.; Cusimano, M.D.; Dirks, P.B. Identification of human brain tumour initiating cells. Nature 2004, 432, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Maugeri-Sacca, M.; Vigneri, P.; De Maria, R. Cancer stem cells and chemosensitivity. Clin. Cancer Res. 2011, 17, 4942–4947. [Google Scholar] [CrossRef]
- Bao, S.; Wu, Q.; McLendon, R.E.; Hao, Y.; Shi, Q.; Hjelmeland, A.B.; Dewhirst, M.W.; Bigner, D.D.; Rich, J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006, 444, 756–760. [Google Scholar] [CrossRef]
- Dean, M.; Fojo, T.; Bates, S. Tumour stem cells and drug resistance. Nat. Rev. Cancer 2005, 5, 275–284. [Google Scholar] [CrossRef]
- Diehn, M.; Clarke, M.F. Cancer stem cells and radiotherapy: New insights into tumor radioresistance. J. Natl. Cancer Inst. 2006, 98, 1755–1757. [Google Scholar] [CrossRef]
- Eyler, C.E.; Rich, J.N. Survival of the fittest: Cancer stem cells in therapeutic resistance and angiogenesis. J. Clin. Oncol. 2008, 26, 2839–2845. [Google Scholar] [CrossRef]
- Zheng, X.; Naiditch, J.; Czurylo, M.; Jie, C.; Lautz, T.; Clark, S.; Jafari, N.; Qiu, Y.; Chu, F.; Madonna, M.B. Differential effect of long-term drug selection with doxorubicin and vorinostat on neuroblastoma cells with cancer stem cell characteristics. Cell Death Dis. 2013, 4, e740. [Google Scholar] [CrossRef]
- Xiao, W.; Gao, Z.; Duan, Y.; Yuan, W.; Ke, Y. Notch signaling plays a crucial role in cancer stem-like cells maintaining stemness and mediating chemotaxis in renal cell carcinoma. J. Exp. Clin. Cancer Res. 2017, 36, 41. [Google Scholar] [CrossRef]
- Terry, S.; Savagner, P.; Ortiz-Cuaran, S.; Mahjoubi, L.; Saintigny, P.; Thiery, J.P.; Chouaib, S. New insights into the role of EMT in tumor immune escape. Mol. Oncol. 2017, 11, 824–846. [Google Scholar] [CrossRef] [PubMed]
- Acloque, H.; Adams, M.S.; Fishwick, K.; Bronner-Fraser, M.; Nieto, M.A. Epithelial-mesenchymal transitions: The importance of changing cell state in development and disease. J. Clin. Investig. 2009, 119, 1438–1449. [Google Scholar] [CrossRef]
- Flier, S.N.; Tanjore, H.; Kokkotou, E.G.; Sugimoto, H.; Zeisberg, M.; Kalluri, R. Identification of epithelial to mesenchymal transition as a novel source of fibroblasts in intestinal fibrosis. J. Biol. Chem. 2010, 285, 20202–20212. [Google Scholar] [CrossRef]
- Kalluri, R.; Neilson, E.G. Epithelial-mesenchymal transition and its implications for fibrosis. J. Clin. Investig. 2003, 112, 1776–1784. [Google Scholar] [CrossRef]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [PubMed]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef] [PubMed]
- Scheel, C.; Weinberg, R.A. Cancer stem cells and epithelial-mesenchymal transition: Concepts and molecular links. Semin. Cancer Biol. 2012, 22, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P.; Sleeman, J.P. Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 2006, 7, 131–142. [Google Scholar] [CrossRef]
- Bataille, F.; Rohrmeier, C.; Bates, R.; Weber, A.; Rieder, F.; Brenmoehl, J.; Strauch, U.; Farkas, S.; Furst, A.; Hofstadter, F.; et al. Evidence for a role of epithelial mesenchymal transition during pathogenesis of fistulae in Crohn’s disease. Inflamm. Bowel. Dis. 2008, 14, 1514–1527. [Google Scholar] [CrossRef]
- Dissanayake, S.K.; Wade, M.; Johnson, C.E.; O’Connell, M.P.; Leotlela, P.D.; French, A.D.; Shah, K.V.; Hewitt, K.J.; Rosenthal, D.T.; Indig, F.E.; et al. The Wnt5A/protein kinase C pathway mediates motility in melanoma cells via the inhibition of metastasis suppressors and initiation of an epithelial to mesenchymal transition. J. Biol. Chem. 2007, 282, 17259–17271. [Google Scholar] [CrossRef]
- Jiang, B.; Zhang, T.; Liu, F.; Sun, Z.; Shi, H.; Hua, D.; Yang, C. The co-stimulatory molecule B7-H3 promotes the epithelial-mesenchymal transition in colorectal cancer. Oncotarget 2016, 7, 31755–31771. [Google Scholar] [CrossRef]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Reiman, J.M.; Knutson, K.L.; Radisky, D.C. Immune promotion of epithelial-mesenchymal transition and generation of breast cancer stem cells. Cancer Res. 2010, 70, 3005–3008. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, E.; Tumbar, T.; Guasch, G. Socializing with the neighbors: Stem cells and their niche. Cell 2004, 116, 769–778. [Google Scholar] [CrossRef]
- Brown, Y.; Hua, S.; Tanwar, P.S. Extracellular matrix-mediated regulation of cancer stem cells and chemoresistance. Int. J. Biochem. Cell Biol. 2019, 109, 90–104. [Google Scholar] [CrossRef]
- Cazet, A.S.; Hui, M.N.; Elsworth, B.L.; Wu, S.Z.; Roden, D.; Chan, C.L.; Skhinas, J.N.; Collot, R.; Yang, J.; Harvey, K.; et al. Targeting stromal remodeling and cancer stem cell plasticity overcomes chemoresistance in triple negative breast cancer. Nat. Commun. 2018, 9, 2897. [Google Scholar] [CrossRef] [PubMed]
- Easwaran, H.; Tsai, H.C.; Baylin, S.B. Cancer epigenetics: Tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol. Cell 2014, 54, 716–727. [Google Scholar] [CrossRef]
- Meacham, C.E.; Morrison, S.J. Tumour heterogeneity and cancer cell plasticity. Nature 2013, 501, 328–337. [Google Scholar] [CrossRef] [PubMed]
- Nieto, M.A. Epithelial plasticity: A common theme in embryonic and cancer cells. Science 2013, 342, 1234850. [Google Scholar] [CrossRef]
- Scheel, C.; Weinberg, R.A. Phenotypic plasticity and epithelial-mesenchymal transitions in cancer and normal stem cells? Int. J. Cancer 2011, 129, 2310–2314. [Google Scholar] [CrossRef]
- Prieto-Vila, M.; Takahashi, R.U.; Usuba, W.; Kohama, I.; Ochiya, T. Drug Resistance Driven by Cancer Stem Cells and Their Niche. Int. J. Mol. Sci. 2017, 18, 2574. [Google Scholar] [CrossRef] [PubMed]
- Albini, A.; Bruno, A.; Gallo, C.; Pajardi, G.; Noonan, D.M.; Dallaglio, K. Cancer stem cells and the tumor microenvironment: Interplay in tumor heterogeneity. Connect Tissue Res. 2015, 56, 414–425. [Google Scholar] [CrossRef] [PubMed]
- Codony-Servat, J.; Rosell, R. Cancer stem cells and immunoresistance: Clinical implications and solutions. Transl. Lung Cancer Res. 2015, 4, 689–703. [Google Scholar] [CrossRef]
- Irvin, D.K.; Jouanneau, E.; Duvall, G.; Zhang, X.X.; Zhai, Y.; Sarayba, D.; Seksenyan, A.; Panwar, A.; Black, K.L.; Wheeler, C.J. T cells enhance stem-like properties and conditional malignancy in gliomas. PLoS ONE 2010, 5, e10974. [Google Scholar] [CrossRef] [PubMed]
- Maccalli, C.; Rasul, K.I.; Elawad, M.; Ferrone, S. The role of cancer stem cells in the modulation of anti-tumor immune responses. Semin. Cancer Biol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Maccalli, C.; Parmiani, G.; Ferrone, S. Immunomodulating and Immunoresistance Properties of Cancer-Initiating Cells: Implications for the Clinical Success of Immunotherapy. Immunol. Investig. 2017, 46, 221–238. [Google Scholar] [CrossRef]
- Ravindran, S.; Rasool, S.; Maccalli, C. The Cross Talk between Cancer Stem Cells/Cancer Initiating Cells and Tumor Microenvironment: The Missing Piece of the Puzzle for the Efficient Targeting of these Cells with Immunotherapy. Cancer Microenviron. 2019, 12, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Ljujic, B.; Milovanovic, M.; Volarevic, V.; Murray, B.; Bugarski, D.; Przyborski, S.; Arsenijevic, N.; Lukic, M.L.; Stojkovic, M. Human mesenchymal stem cells creating an immunosuppressive environment and promote breast cancer in mice. Sci. Rep. 2013, 3, 2298. [Google Scholar] [CrossRef] [PubMed]
- Reya, T.; Morrison, S.J.; Clarke, M.F.; Weissman, I.L. Stem cells, cancer, and cancer stem cells. Nature 2001, 414, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Weissman, I.L. Stem cells: Units of development, units of regeneration, and units in evolution. Cell 2000, 100, 157–168. [Google Scholar] [CrossRef]
- Di Tomaso, T.; Mazzoleni, S.; Wang, E.; Sovena, G.; Clavenna, D.; Franzin, A.; Mortini, P.; Ferrone, S.; Doglioni, C.; Marincola, F.M.; et al. Immunobiological characterization of cancer stem cells isolated from glioblastoma patients. Clin. Cancer Res. 2010, 16, 800–813. [Google Scholar] [CrossRef]
- Schatton, T.; Frank, M.H. Antitumor immunity and cancer stem cells. Ann. N. Y. Acad. Sci. 2009, 1176, 154–169. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Tang, D.G.; Rycaj, K. Cancer stem cells: Regulation programs, immunological properties and immunotherapy. Semin. Cancer Biol. 2018, 52, 94–106. [Google Scholar] [CrossRef]
- Zhang, J.; Li, Q.; Chang, A.E. Immunologic Targeting of Cancer Stem Cells. Surg. Oncol. Clin. N. Am. 2019, 28, 431–445. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Barr, J.; Kong, L.Y.; Wang, Y.; Wu, A.; Sharma, A.K.; Gumin, J.; Henry, V.; Colman, H.; Priebe, W.; et al. Glioblastoma cancer-initiating cells inhibit T-cell proliferation and effector responses by the signal transducers and activators of transcription 3 pathway. Mol. Cancer Ther. 2010, 9, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Barr, J.; Kong, L.Y.; Wang, Y.; Wu, A.; Sharma, A.K.; Gumin, J.; Henry, V.; Colman, H.; Sawaya, R.; et al. Glioma-associated cancer-initiating cells induce immunosuppression. Clin. Cancer Res. 2010, 16, 461–473. [Google Scholar] [CrossRef] [PubMed]
- Munn, D.H. Blocking IDO activity to enhance anti-tumor immunity. Front. Biosci. 2012, 4, 734–745. [Google Scholar] [CrossRef]
- Prendergast, G.C. Immune escape as a fundamental trait of cancer: Focus on IDO. Oncogene 2008, 27, 3889–3900. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wu, J.; Shen, H.; Wang, J. The prognostic value of IDO expression in solid tumors: A systematic review and meta-analysis. BMC Cancer 2020, 20, 471. [Google Scholar] [CrossRef] [PubMed]
- Fu, R.; Zhang, Y.W.; Li, H.M.; Lv, W.C.; Zhao, L.; Guo, Q.L.; Lu, T.; Weiss, S.J.; Li, Z.Y.; Wu, Z.Q. LW106, a novel indoleamine 2,3-dioxygenase 1 inhibitor, suppresses tumour progression by limiting stroma-immune crosstalk and cancer stem cell enrichment in tumour micro-environment. Br. J. Pharmacol. 2018, 175, 3034–3049. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Croce, C.M. MicroRNA dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 2012, 4, 143–159. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Z.; Schaffert, S.; Fragoso, R.; Loh, C. Regulation of immune responses and tolerance: The microRNA perspective. Immunol. Rev. 2013, 253, 112–128. [Google Scholar] [CrossRef] [PubMed]
- Huffaker, T.B.; Lee, S.H.; Tang, W.W.; Wallace, J.A.; Alexander, M.; Runtsch, M.C.; Larsen, D.K.; Thompson, J.; Ramstead, A.G.; Voth, W.P.; et al. Antitumor immunity is defective in T cell-specific microRNA-155-deficient mice and is rescued by immune checkpoint blockade. J. Biol. Chem. 2017, 292, 18530–18541. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.M.; Kahn, D.; Gibson, W.S.; Round, J.L.; Scholz, R.L.; Chaudhuri, A.A.; Kahn, M.E.; Rao, D.S.; Baltimore, D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity 2010, 33, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Paladini, L.; Fabris, L.; Bottai, G.; Raschioni, C.; Calin, G.A.; Santarpia, L. Targeting microRNAs as key modulators of tumor immune response. J. Exp. Clin. Cancer Res. 2016, 35, 103. [Google Scholar] [CrossRef] [PubMed]
- Raisch, J.; Darfeuille-Michaud, A.; Nguyen, H.T. Role of microRNAs in the immune system, inflammation and cancer. World J. Gastroenterol. 2013, 19, 2985–2996. [Google Scholar] [CrossRef] [PubMed]
- Tsitsiou, E.; Lindsay, M.A. microRNAs and the immune response. Curr. Opin. Pharmacol. 2009, 9, 514–520. [Google Scholar] [CrossRef]
- Comber, J.D.; Philip, R. MHC class I antigen presentation and implications for developing a new generation of therapeutic vaccines. Ther. Adv. Vaccines 2014, 2, 77–89. [Google Scholar] [CrossRef]
- Schatton, T.; Schutte, U.; Frank, N.Y.; Zhan, Q.; Hoerning, A.; Robles, S.C.; Zhou, J.; Hodi, F.S.; Spagnoli, G.C.; Murphy, G.F.; et al. Modulation of T-cell activation by malignant melanoma initiating cells. Cancer Res. 2010, 70, 697–708. [Google Scholar] [CrossRef] [PubMed]
- Volonte, A.; Di Tomaso, T.; Spinelli, M.; Todaro, M.; Sanvito, F.; Albarello, L.; Bissolati, M.; Ghirardelli, L.; Orsenigo, E.; Ferrone, S.; et al. Cancer-initiating cells from colorectal cancer patients escape from T cell-mediated immunosurveillance in vitro through membrane-bound IL-4. J. Immunol. 2014, 192, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Van Kaer, L. Major histocompatibility complex class I-restricted antigen processing and presentation. Tissue Antigens 2002, 60, 1–9. [Google Scholar] [CrossRef]
- Garbi, N.; Tanaka, S.; van den Broek, M.; Momburg, F.; Hammerling, G.J. Accessory molecules in the assembly of major histocompatibility complex class I/peptide complexes: How essential are they for CD8(+) T-cell immune responses? Immunol. Rev. 2005, 207, 77–88. [Google Scholar] [CrossRef]
- Busse, A.; Letsch, A.; Fusi, A.; Nonnenmacher, A.; Stather, D.; Ochsenreither, S.; Regenbrecht, C.R.; Keilholz, U. Characterization of small spheres derived from various solid tumor cell lines: Are they suitable targets for T cells? Clin. Exp. Metastasis 2013, 30, 781–791. [Google Scholar] [CrossRef]
- Castriconi, R.; Daga, A.; Dondero, A.; Zona, G.; Poliani, P.L.; Melotti, A.; Griffero, F.; Marubbi, D.; Spaziante, R.; Bellora, F.; et al. NK cells recognize and kill human glioblastoma cells with stem cell-like properties. J. Immunol. 2009, 182, 3530–3539. [Google Scholar] [CrossRef]
- Tallerico, R.; Todaro, M.; Di Franco, S.; Maccalli, C.; Garofalo, C.; Sottile, R.; Palmieri, C.; Tirinato, L.; Pangigadde, P.N.; La Rocca, R.; et al. Human NK cells selective targeting of colon cancer-initiating cells: A role for natural cytotoxicity receptors and MHC class I molecules. J. Immunol. 2013, 190, 2381–2390. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Wang, Q.; Wang, Z.; Jiang, J.; Yu, S.C.; Ping, Y.F.; Yang, J.; Xu, S.L.; Ye, X.Z.; Xu, C.; et al. Metastatic consequences of immune escape from NK cell cytotoxicity by human breast cancer stem cells. Cancer Res. 2014, 74, 5746–5757. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.; Wiesner, S.; Xiao, J.; Ericson, K.; Chen, W.; Hall, W.A.; Low, W.C.; Ohlfest, J.R. Expression of MHC I and NK ligands on human CD133+ glioma cells: Possible targets of immunotherapy. J. Neurooncol. 2007, 83, 121–131. [Google Scholar] [CrossRef]
- Carapito, R.; Bahram, S. Genetics, genomics, and evolutionary biology of NKG2D ligands. Immunol. Rev. 2015, 267, 88–116. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xu, H.; Zhu, X.X. Abnormal expression levels of sMICA and NKG2D are correlated with poor prognosis in pancreatic cancer. Ther. Clin. Risk. Manag. 2016, 12, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Maccalli, C.; Scaramuzza, S.; Parmiani, G. TNK cells (NKG2D+ CD8+ or CD4+ T lymphocytes) in the control of human tumors. Cancer Immunol. Immunother. 2009, 58, 801–808. [Google Scholar] [CrossRef]
- Maccalli, C.; Giannarelli, D.; Capocefalo, F.; Pilla, L.; Fonsatti, E.; Di Giacomo, A.M.; Parmiani, G.; Maio, M. Immunological markers and clinical outcome of advanced melanoma patients receiving ipilimumab plus fotemustine in the NIBIT-M1 study. Oncoimmunology 2016, 5, e1071007. [Google Scholar] [CrossRef]
- Maccalli, C.; Giannarelli, D.; Chiarucci, C.; Cutaia, O.; Giacobini, G.; Hendrickx, W.; Amato, G.; Annesi, D.; Bedognetti, D.; Altomonte, M.; et al. Soluble NKG2D ligands are biomarkers associated with the clinical outcome to immune checkpoint blockade therapy of metastatic melanoma patients. Oncoimmunology 2017, 6, e1323618. [Google Scholar] [CrossRef]
- Parmiani, G.; Russo, V.; Maccalli, C.; Parolini, D.; Rizzo, N.; Maio, M. Peptide-based vaccines for cancer therapy. Hum. Vaccin. Immunother. 2014, 10, 3175–3178. [Google Scholar] [CrossRef] [PubMed]
- Hirohashi, Y.; Torigoe, T.; Tsukahara, T.; Kanaseki, T.; Kochin, V.; Sato, N. Immune responses to human cancer stem-like cells/cancer-initiating cells. Cancer Sci. 2016, 107, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Korkaya, H.; Wicha, M.S. HER2 and breast cancer stem cells: More than meets the eye. Cancer Res. 2013, 73, 3489–3493. [Google Scholar] [CrossRef]
- Khandekar, D.; Amara, S.; Tiriveedhi, V. Immunogenicity of Tumor Initiating Stem Cells: Potential Applications in Novel Anticancer Therapy. Front. Oncol. 2019, 9, 315. [Google Scholar] [CrossRef] [PubMed]
- Melero, I.; Gaudernack, G.; Gerritsen, W.; Huber, C.; Parmiani, G.; Scholl, S.; Thatcher, N.; Wagstaff, J.; Zielinski, C.; Faulkner, I.; et al. Therapeutic vaccines for cancer: An overview of clinical trials. Nat. Rev. Clin. Oncol. 2014, 11, 509–524. [Google Scholar] [CrossRef]
- Scanlan, M.J.; Gure, A.O.; Jungbluth, A.A.; Old, L.J.; Chen, Y.T. Cancer/testis antigens: An expanding family of targets for cancer immunotherapy. Immunol. Rev. 2002, 188, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Morita, R.; Hirohashi, Y.; Torigoe, T.; Ito-Inoda, S.; Takahashi, A.; Mariya, T.; Asanuma, H.; Tamura, Y.; Tsukahara, T.; Kanaseki, T.; et al. Olfactory Receptor Family 7 Subfamily C Member 1 Is a Novel Marker of Colon Cancer-Initiating Cells and Is a Potent Target of Immunotherapy. Clin. Cancer Res. 2016, 22, 3298–3309. [Google Scholar] [CrossRef]
- Yamada, R.; Takahashi, A.; Torigoe, T.; Morita, R.; Tamura, Y.; Tsukahara, T.; Kanaseki, T.; Kubo, T.; Watarai, K.; Kondo, T.; et al. Preferential expression of cancer/testis genes in cancer stem-like cells: Proposal of a novel sub-category, cancer/testis/stem gene. Tissue Antigens 2013, 81, 428–434. [Google Scholar] [CrossRef] [PubMed]
- Ilyas, S.; Yang, J.C. Landscape of Tumor Antigens in T Cell Immunotherapy. J. Immunol. 2015, 195, 5117–5122. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.C.; Robbins, P.F. Cancer immunotherapy targeting neoantigens. Semin. Immunol. 2016, 28, 22–27. [Google Scholar] [CrossRef]
- Mennonna, D.; Maccalli, C.; Romano, M.C.; Garavaglia, C.; Capocefalo, F.; Bordoni, R.; Severgnini, M.; De Bellis, G.; Sidney, J.; Sette, A.; et al. T cell neoepitope discovery in colorectal cancer by high throughput profiling of somatic mutations in expressed genes. Gut 2015. [Google Scholar] [CrossRef] [PubMed]
- Dunn, G.P.; Bruce, A.T.; Ikeda, H.; Old, L.J.; Schreiber, R.D. Cancer immunoediting: From immunosurveillance to tumor escape. Nat. Immunol. 2002, 3, 991–998. [Google Scholar] [CrossRef]
- Koebel, C.M.; Vermi, W.; Swann, J.B.; Zerafa, N.; Rodig, S.J.; Old, L.J.; Smyth, M.J.; Schreiber, R.D. Adaptive immunity maintains occult cancer in an equilibrium state. Nature 2007, 450, 903–907. [Google Scholar] [CrossRef] [PubMed]
- Mittal, D.; Gubin, M.M.; Schreiber, R.D.; Smyth, M.J. New insights into cancer immunoediting and its three component phases--elimination, equilibrium and escape. Curr. Opin. Immunol. 2014, 27, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Gajewski, T.F.; Schreiber, H.; Fu, Y.X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013, 14, 1014–1022. [Google Scholar] [CrossRef] [PubMed]
- Gajewski, T.F.; Woo, S.R.; Zha, Y.; Spaapen, R.; Zheng, Y.; Corrales, L.; Spranger, S. Cancer immunotherapy strategies based on overcoming barriers within the tumor microenvironment. Curr. Opin. Immunol. 2013, 25, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Fridman, W.H.; Galon, J. The prognostic impact of anti-cancer immune response: A novel classification of cancer patients. Semin. Immunopathol. 2011, 33, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Fridman, W.H.; Dieu-Nosjean, M.C.; Pages, F.; Cremer, I.; Damotte, D.; Sautes-Fridman, C.; Galon, J. The immune microenvironment of human tumors: General significance and clinical impact. Cancer Microenviron. 2013, 6, 117–122. [Google Scholar] [CrossRef]
- Galon, J.; Costes, A.; Sanchez-Cabo, F.; Kirilovsky, A.; Mlecnik, B.; Lagorce-Pages, C.; Tosolini, M.; Camus, M.; Berger, A.; Wind, P.; et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006, 313, 1960–1964. [Google Scholar] [CrossRef] [PubMed]
- Becht, E.; Giraldo, N.A.; Germain, C.; de Reynies, A.; Laurent-Puig, P.; Zucman-Rossi, J.; Dieu-Nosjean, M.C.; Sautes-Fridman, C.; Fridman, W.H. Immune Contexture, Immunoscore, and Malignant Cell Molecular Subgroups for Prognostic and Theranostic Classifications of Cancers. Adv. Immunol. 2016, 130, 95–190. [Google Scholar] [CrossRef] [PubMed]
- Becht, E.; Giraldo, N.A.; Lacroix, L.; Buttard, B.; Elarouci, N.; Petitprez, F.; Selves, J.; Laurent-Puig, P.; Sautes-Fridman, C.; Fridman, W.H.; et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016, 17, 218. [Google Scholar] [CrossRef]
- Dieu-Nosjean, M.C.; Giraldo, N.A.; Kaplon, H.; Germain, C.; Fridman, W.H.; Sautes-Fridman, C. Tertiary lymphoid structures, drivers of the anti-tumor responses in human cancers. Immunol. Rev. 2016, 271, 260–275. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Pietra, G.; Manzini, C.; Vitale, M.; Balsamo, M.; Ognio, E.; Boitano, M.; Queirolo, P.; Moretta, L.; Mingari, M.C. Natural killer cells kill human melanoma cells with characteristics of cancer stem cells. Int. Immunol. 2009, 21, 793–801. [Google Scholar] [CrossRef]
- Ames, E.; Canter, R.J.; Grossenbacher, S.K.; Mac, S.; Chen, M.; Smith, R.C.; Hagino, T.; Perez-Cunningham, J.; Sckisel, G.D.; Urayama, S.; et al. NK Cells Preferentially Target Tumor Cells with a Cancer Stem Cell Phenotype. J. Immunol. 2015, 195, 4010–4019. [Google Scholar] [CrossRef] [PubMed]
- Jewett, A.; Tseng, H.C.; Arasteh, A.; Saadat, S.; Christensen, R.E.; Cacalano, N.A. Natural killer cells preferentially target cancer stem cells; role of monocytes in protection against NK cell mediated lysis of cancer stem cells. Curr. Drug Deliv. 2012, 9, 5–16. [Google Scholar] [CrossRef]
- Avril, T.; Vauleon, E.; Hamlat, A.; Saikali, S.; Etcheverry, A.; Delmas, C.; Diabira, S.; Mosser, J.; Quillien, V. Human glioblastoma stem-like cells are more sensitive to allogeneic NK and T cell-mediated killing compared with serum-cultured glioblastoma cells. Brain Pathol. 2012, 22, 159–174. [Google Scholar] [CrossRef]
- She, M.; Niu, X.; Chen, X.; Li, J.; Zhou, M.; He, Y.; Le, Y.; Guo, K. Resistance of leukemic stem-like cells in AML cell line KG1a to natural killer cell-mediated cytotoxicity. Cancer Lett. 2012, 318, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Doubrovina, E.S.; Doubrovin, M.M.; Vider, E.; Sisson, R.B.; O’Reilly, R.J.; Dupont, B.; Vyas, Y.M. Evasion from NK cell immunity by MHC class I chain-related molecules expressing colon adenocarcinoma. J. Immunol. 2003, 171, 6891–6899. [Google Scholar] [CrossRef]
- Dudek, A.M.; Martin, S.; Garg, A.D.; Agostinis, P. Immature, Semi-Mature, and Fully Mature Dendritic Cells: Toward a DC-Cancer Cells Interface That Augments Anticancer Immunity. Front. Immunol. 2013, 4, 438. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.L.; Chen, Y.J.; Chang, W.A.; Jian, S.F.; Fan, H.L.; Wang, J.Y.; Kuo, P.L. Interaction between Tumor-Associated Dendritic Cells and Colon Cancer Cells Contributes to Tumor Progression via CXCL1. Int. J. Mol. Sci. 2018, 19, 2427. [Google Scholar] [CrossRef] [PubMed]
- Li, J.K.; Yu, L.; Shen, Y.; Zhou, L.S.; Wang, Y.C.; Zhang, J.H. Inhibition of CXCR4 activity with AMD3100 decreases invasion of human colorectal cancer cells in vitro. World J. Gastroenterol. 2008, 14, 2308–2313. [Google Scholar] [CrossRef] [PubMed]
- Majeti, R.; Chao, M.P.; Alizadeh, A.A.; Pang, W.W.; Jaiswal, S.; Gibbs, K.D., Jr.; van Rooijen, N.; Weissman, I.L. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell 2009, 138, 286–299. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Long, W.; Xing, C.; Chu, J.; Luo, M.; Wang, H.Y.; Liu, Q.; Wang, R.F. Cancer Stem Cells and Immunosuppressive Microenvironment in Glioma. Front. Immunol. 2018, 9, 2924. [Google Scholar] [CrossRef] [PubMed]
- Jinushi, M.; Chiba, S.; Yoshiyama, H.; Masutomi, K.; Kinoshita, I.; Dosaka-Akita, H.; Yagita, H.; Takaoka, A.; Tahara, H. Tumor-associated macrophages regulate tumorigenicity and anticancer drug responses of cancer stem/initiating cells. Proc. Natl. Acad. Sci. USA 2011, 108, 12425–12430. [Google Scholar] [CrossRef]
- Sherry, M.M.; Reeves, A.; Wu, J.K.; Cochran, B.H. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells 2009, 27, 2383–2392. [Google Scholar] [CrossRef]
- Zhou, J.; Wulfkuhle, J.; Zhang, H.; Gu, P.; Yang, Y.; Deng, J.; Margolick, J.B.; Liotta, L.A.; Petricoin, E., 3rd; Zhang, Y. Activation of the PTEN/mTOR/STAT3 pathway in breast cancer stem-like cells is required for viability and maintenance. Proc. Natl. Acad. Sci. USA 2007, 104, 16158–16163. [Google Scholar] [CrossRef]
- Mitchem, J.B.; Brennan, D.J.; Knolhoff, B.L.; Belt, B.A.; Zhu, Y.; Sanford, D.E.; Belaygorod, L.; Carpenter, D.; Collins, L.; Piwnica-Worms, D.; et al. Targeting tumor-infiltrating macrophages decreases tumor-initiating cells, relieves immunosuppression, and improves chemotherapeutic responses. Cancer Res. 2013, 73, 1128–1141. [Google Scholar] [CrossRef]
- Lindau, D.; Gielen, P.; Kroesen, M.; Wesseling, P.; Adema, G.J. The immunosuppressive tumour network: Myeloid-derived suppressor cells, regulatory T cells and natural killer T cells. Immunology 2013, 138, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Panni, R.Z.; Sanford, D.E.; Belt, B.A.; Mitchem, J.B.; Worley, L.A.; Goetz, B.D.; Mukherjee, P.; Wang-Gillam, A.; Link, D.C.; Denardo, D.G.; et al. Tumor-induced STAT3 activation in monocytic myeloid-derived suppressor cells enhances stemness and mesenchymal properties in human pancreatic cancer. Cancer Immunol. Immunother. 2014, 63, 513–528. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Yu, S.; Zhang, X. Hypothesis: Tim-3/galectin-9, a new pathway for leukemia stem cells survival by promoting expansion of myeloid-derived suppressor cells and differentiating into tumor-associated macrophages. Cell Biochem. Biophys. 2014, 70, 273–277. [Google Scholar] [CrossRef]
- Riether, C.; Schurch, C.M.; Ochsenbein, A.F. Regulation of hematopoietic and leukemic stem cells by the immune system. Cell Death Differ. 2015, 22, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Sarvaria, A.; Madrigal, J.A.; Saudemont, A. B cell regulation in cancer and anti-tumor immunity. Cell. Mol. Immunol. 2017, 14, 662–674. [Google Scholar] [CrossRef] [PubMed]
- Smyth, M.J.; Thia, K.Y.; Cretney, E.; Kelly, J.M.; Snook, M.B.; Forbes, C.A.; Scalzo, A.A. Perforin is a major contributor to NK cell control of tumor metastasis. J. Immunol. 1999, 162, 6658–6662. [Google Scholar] [PubMed]
- Cella, M.; Sallusto, F.; Lanzavecchia, A. Origin, maturation and antigen presenting function of dendritic cells. Curr. Opin. Immunol. 1997, 9, 10–16. [Google Scholar] [CrossRef]
- Banchereau, J.; Steinman, R.M. Dendritic cells and the control of immunity. Nature 1998, 392, 245–252. [Google Scholar] [CrossRef]
- Keller, A.M.; Xiao, Y.; Peperzak, V.; Naik, S.H.; Borst, J. Costimulatory ligand CD70 allows induction of CD8+ T-cell immunity by immature dendritic cells in a vaccination setting. Blood 2009, 113, 5167–5175. [Google Scholar] [CrossRef]
- Hanke, N.; Alizadeh, D.; Katsanis, E.; Larmonier, N. Dendritic cell tumor killing activity and its potential applications in cancer immunotherapy. Crit. Rev. Immunol. 2013, 33, 1–21. [Google Scholar] [CrossRef]
- Jonuleit, H.; Schmitt, E.; Stassen, M.; Tuettenberg, A.; Knop, J.; Enk, A.H. Identification and functional characterization of human CD4(+)CD25(+) T cells with regulatory properties isolated from peripheral blood. J. Exp. Med. 2001, 193, 1285–1294. [Google Scholar] [CrossRef]
- Sainz, B., Jr.; Carron, E.; Vallespinos, M.; Machado, H.L. Cancer Stem Cells and Macrophages: Implications in Tumor Biology and Therapeutic Strategies. Med. Inflamm. 2016, 2016, 9012369. [Google Scholar] [CrossRef]
- Filipazzi, P.; Huber, V.; Rivoltini, L. Phenotype, function and clinical implications of myeloid-derived suppressor cells in cancer patients. Cancer Immunol. Immunother. 2012, 61, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Nagaraj, S.; Gabrilovich, D.I. Regulation of suppressive function of myeloid-derived suppressor cells by CD4+ T cells. Semin. Cancer Biol. 2012, 22, 282–288. [Google Scholar] [CrossRef][Green Version]
- Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Bronte, V. Coordinated regulation of myeloid cells by tumours. Nat. Rev. Immunol. 2012, 12, 253–268. [Google Scholar] [CrossRef]
- Taylor, M.W.; Feng, G.S. Relationship between interferon-gamma, indoleamine 2,3-dioxygenase, and tryptophan catabolism. FASEB J. 1991, 5, 2516–2522. [Google Scholar] [CrossRef] [PubMed]
- Ball, H.J.; Sanchez-Perez, A.; Weiser, S.; Austin, C.J.; Astelbauer, F.; Miu, J.; McQuillan, J.A.; Stocker, R.; Jermiin, L.S.; Hunt, N.H. Characterization of an indoleamine 2,3-dioxygenase-like protein found in humans and mice. Gene 2007, 396, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Pan, K.; Wang, H.; Chen, M.S.; Zhang, H.K.; Weng, D.S.; Zhou, J.; Huang, W.; Li, J.J.; Song, H.F.; Xia, J.C. Expression and prognosis role of indoleamine 2,3-dioxygenase in hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 2008, 134, 1247–1253. [Google Scholar] [CrossRef] [PubMed]
- Muller, A.J.; Prendergast, G.C. Marrying immunotherapy with chemotherapy: Why say I DO? Cancer Res. 2005, 65, 8065–8068. [Google Scholar] [CrossRef]
- Munn, D.H.; Mellor, A.L. Indoleamine 2,3-dioxygenase and tumor-induced tolerance. J. Clin. Investig. 2007, 117, 1147–1154. [Google Scholar] [CrossRef] [PubMed]
- Munn, D.H.; Sharma, M.D.; Hou, D.; Baban, B.; Lee, J.R.; Antonia, S.J.; Messina, J.L.; Chandler, P.; Koni, P.A.; Mellor, A.L. Expression of indoleamine 2,3-dioxygenase by plasmacytoid dendritic cells in tumor-draining lymph nodes. J. Clin. Investig. 2004, 114, 280–290. [Google Scholar] [CrossRef] [PubMed]
- Prendergast, G.C.; Malachowski, W.J.; Mondal, A.; Scherle, P.; Muller, A.J. Indoleamine 2,3-Dioxygenase and Its Therapeutic Inhibition in Cancer. Int. Rev. Cell Mol. Biol. 2018, 336, 175–203. [Google Scholar] [CrossRef] [PubMed]
- Prendergast, G.C.; Smith, C.; Thomas, S.; Mandik-Nayak, L.; Laury-Kleintop, L.; Metz, R.; Muller, A.J. Indoleamine 2,3-dioxygenase pathways of pathogenic inflammation and immune escape in cancer. Cancer Immunol. Immunother. 2014, 63, 721–735. [Google Scholar] [CrossRef] [PubMed]
- Platten, M.; Ho, P.P.; Youssef, S.; Fontoura, P.; Garren, H.; Hur, E.M.; Gupta, R.; Lee, L.Y.; Kidd, B.A.; Robinson, W.H.; et al. Treatment of autoimmune neuroinflammation with a synthetic tryptophan metabolite. Science 2005, 310, 850–855. [Google Scholar] [CrossRef] [PubMed]
- Bauer, T.M.; Jiga, L.P.; Chuang, J.J.; Randazzo, M.; Opelz, G.; Terness, P. Studying the immunosuppressive role of indoleamine 2,3-dioxygenase: Tryptophan metabolites suppress rat allogeneic T-cell responses in vitro and in vivo. Transpl. Int. 2005, 18, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Pallotta, M.T.; Orabona, C.; Volpi, C.; Vacca, C.; Belladonna, M.L.; Bianchi, R.; Servillo, G.; Brunacci, C.; Calvitti, M.; Bicciato, S.; et al. Indoleamine 2,3-dioxygenase is a signaling protein in long-term tolerance by dendritic cells. Nat. Immunol. 2011, 12, 870–878. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Jin, W.; Hardegen, N.; Lei, K.J.; Li, L.; Marinos, N.; McGrady, G.; Wahl, S.M. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J. Exp. Med. 2003, 198, 1875–1886. [Google Scholar] [CrossRef]
- Grohmann, U.; Orabona, C.; Fallarino, F.; Vacca, C.; Calcinaro, F.; Falorni, A.; Candeloro, P.; Belladonna, M.L.; Bianchi, R.; Fioretti, M.C.; et al. CTLA-4-Ig regulates tryptophan catabolism in vivo. Nat. Immunol. 2002, 3, 1097–1101. [Google Scholar] [CrossRef]
- Stapelberg, M.; Zobalova, R.; Nguyen, M.N.; Walker, T.; Stantic, M.; Goodwin, J.; Pasdar, E.A.; Thai, T.; Prokopova, K.; Yan, B.; et al. Indoleamine-2,3-dioxygenase elevated in tumor-initiating cells is suppressed by mitocans. Free Radic. Biol. Med. 2014, 67, 41–50. [Google Scholar] [CrossRef]
- Tomei, S.; Volontè, A.; Ravindran, S.; Mazzoleni, S.; Wang, E.; Galli, R. Cristina Maccalli1 MicroRNA profile in glioblastoma stem cells The Journal of Personalized Medicine-Special Issue Role of MicroRNA in Cancer Development and Treatment. 2020; in press. [Google Scholar]
- Munn, D.H.; Mellor, A.L. IDO in the Tumor Microenvironment: Inflammation, Counter-Regulation, and Tolerance. Trends Immunol. 2016, 37, 193–207. [Google Scholar] [CrossRef]
- Bruttel, V.S.; Wischhusen, J. Cancer stem cell immunology: Key to understanding tumorigenesis and tumor immune escape? Front. Immunol. 2014, 5, 360. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Leland, P.; Joshi, B.H.; Puri, R.K. Targeting of IL-4 and IL-13 receptors for cancer therapy. Cytokine 2015, 75, 79–88. [Google Scholar] [CrossRef]
- Chen, Y.; Tan, W.; Wang, C. Tumor-associated macrophage-derived cytokines enhance cancer stem-like characteristics through epithelial-mesenchymal transition. Oncol. Targets Ther. 2018, 11, 3817–3826. [Google Scholar] [CrossRef] [PubMed]
- Cabarcas, S.M.; Mathews, L.A.; Farrar, W.L. The cancer stem cell niche--there goes the neighborhood? Int. J. Cancer 2011, 129, 2315–2327. [Google Scholar] [CrossRef] [PubMed]
- Miyazono, K. Positive and negative regulation of TGF-beta signaling. J. Cell Sci. 2000, 113, 1101–1109. [Google Scholar] [PubMed]
- Wischhusen, J.; Melero, I.; Fridman, W.H. Growth/Differentiation Factor-15 (GDF-15): From Biomarker to Novel Targetable Immune Checkpoint. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Roth, P.; Junker, M.; Tritschler, I.; Mittelbronn, M.; Dombrowski, Y.; Breit, S.N.; Tabatabai, G.; Wick, W.; Weller, M.; Wischhusen, J. GDF-15 contributes to proliferation and immune escape of malignant gliomas. Clin. Cancer Res. 2010, 16, 3851–3859. [Google Scholar] [CrossRef]
- Kuroda, H.; Mabuchi, S.; Yokoi, E.; Komura, N.; Kozasa, K.; Matsumoto, Y.; Kawano, M.; Takahashi, R.; Sasano, T.; Shimura, K.; et al. Prostaglandin E2 produced by myeloid-derived suppressive cells induces cancer stem cells in uterine cervical cancer. Oncotarget 2018, 9, 36317. [Google Scholar] [CrossRef] [PubMed]
- Nangia-Makker, P.; Hogan, V.; Raz, A. Galectin-3 and cancer stemness. Glycobiology 2018, 28, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Ilmer, M.; Mazurek, N.; Gilcrease, M.Z.; Byrd, J.C.; Woodward, W.A.; Buchholz, T.A.; Acklin, K.; Ramirez, K.; Hafley, M.; Alt, E.; et al. Low expression of galectin-3 is associated with poor survival in node-positive breast cancers and mesenchymal phenotype in breast cancer stem cells. Breast Cancer Res. 2016, 18, 97. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.G.; Kim, D.H.; Kim, S.J.; Cho, Y.; Jung, J.; Jang, W.; Chun, K.H. Galectin-3 supports stemness in ovarian cancer stem cells by activation of the Notch1 intracellular domain. Oncotarget 2016, 7, 68229–68241. [Google Scholar] [CrossRef]
- Li, Y.; Xu, X.; Wang, L.; Liu, G.; Li, Y.; Wu, X.; Jing, Y.; Li, H.; Wang, G. Senescent mesenchymal stem cells promote colorectal cancer cells growth via galectin-3 expression. Cell Biosci. 2015, 5, 21. [Google Scholar] [CrossRef] [PubMed]
- Caputo, S.; Grioni, M.; Brambillasca, C.S.; Monno, A.; Brevi, A.; Freschi, M.; Piras, I.S.; Elia, A.R.; Pieri, V.; Baccega, T.; et al. Galectin-3 in Prostate Cancer Stem-Like Cells Is Immunosuppressive and Drives Early Metastasis. Front. Immunol. 2020, 11, 1820. [Google Scholar] [CrossRef]
- Chen, H.Y.; Liu, F.T.; Yang, R.Y. Roles of galectin-3 in immune responses. Arch. Immunol. Ther. Exp. 2005, 53, 497–504. [Google Scholar]
- Dumic, J.; Dabelic, S.; Flogel, M. Galectin-3: An open-ended story. Biochim. Biophys. Acta 2006, 1760, 616–635. [Google Scholar] [CrossRef] [PubMed]
- Hsu, J.M.; Li, C.W.; Lai, Y.J.; Hung, M.C. Posttranslational Modifications of PD-L1 and Their Applications in Cancer Therapy. Cancer Res. 2018, 78, 6349–6353. [Google Scholar] [CrossRef] [PubMed]
- Hsu, J.M.; Xia, W.; Hsu, Y.H.; Chan, L.C.; Yu, W.H.; Cha, J.H.; Chen, C.T.; Liao, H.W.; Kuo, C.W.; Khoo, K.H.; et al. STT3-dependent PD-L1 accumulation on cancer stem cells promotes immune evasion. Nat. Commun. 2018, 9, 1908. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Wang, X.; Jin, K.; Zhu, J.; Wang, Y.; Xiong, S.; Mao, Y.; Zhou, L. B7-H4 is preferentially expressed in non-dividing brain tumor cells and in a subset of brain tumor stem-like cells. J. Neurooncol. 2008, 89, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, W.; Phillips, J.B.; Arora, R.; McClellan, S.; Li, J.; Kim, J.H.; Sobol, R.W.; Tan, M. Immunoregulatory protein B7-H3 regulates cancer stem cell enrichment and drug resistance through MVP-mediated MEK activation. Oncogene 2019, 38, 88–102. [Google Scholar] [CrossRef]
- Bin, Z.; Guangbo, Z.; Yan, G.; Huan, Z.; Desheng, L.; Xueguang, Z. Overexpression of B7-H3 in CD133+ colorectal cancer cells is associated with cancer progression and survival in human patients. J. Surg. Res. 2014, 188, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Buchbinder, E.; Hodi, F.S. Cytotoxic T lymphocyte antigen-4 and immune checkpoint blockade. J. Clin. Investig. 2015, 125, 3377–3383. [Google Scholar] [CrossRef] [PubMed]
- Rubin, K.M.; Olszanski, A.J. Immune Checkpoint Inhibitor-Based Therapy as a Backbone in Cancer Treatment. J. Adv. Pract. Oncol. 2020, 11, 231–233. [Google Scholar] [CrossRef] [PubMed]
- Rotte, A.; Jin, J.Y.; Lemaire, V. Mechanistic overview of immune checkpoints to support the rational design of their combinations in cancer immunotherapy. Ann. Oncol. 2018, 29, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, inflammation, and cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef] [PubMed]
- Osaki, M.; Okada, F.; Ochiya, T. miRNA therapy targeting cancer stem cells: A new paradigm for cancer treatment and prevention of tumor recurrence. Ther. Deliv. 2015, 6, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Kanellopoulou, C.; Muljo, S.A.; Kung, A.L.; Ganesan, S.; Drapkin, R.; Jenuwein, T.; Livingston, D.M.; Rajewsky, K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005, 19, 489–501. [Google Scholar] [CrossRef]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Garg, A.D.; Krysko, D.V.; Verfaillie, T.; Kaczmarek, A.; Ferreira, G.B.; Marysael, T.; Rubio, N.; Firczuk, M.; Mathieu, C.; Roebroek, A.J.; et al. A novel pathway combining calreticulin exposure and ATP secretion in immunogenic cancer cell death. EMBO J. 2012, 31, 1062–1079. [Google Scholar] [CrossRef] [PubMed]
- Agliano, A.; Calvo, A.; Box, C. The challenge of targeting cancer stem cells to halt metastasis. Semin. Cancer Biol. 2017, 44, 25–42. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Jiao, X.; Pestell, T.G.; Fan, C.; Qin, S.; Mirabelli, E.; Ren, H.; Pestell, R.G. MicroRNAs and cancer stem cells: The sword and the shield. Oncogene 2014, 33, 4967–4977. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Kelnar, K.; Vlassov, A.V.; Brown, D.; Wang, J.; Tang, D.G. Distinct microRNA expression profiles in prostate cancer stem/progenitor cells and tumor-suppressive functions of let-7. Cancer Res. 2012, 72, 3393–3404. [Google Scholar] [CrossRef] [PubMed]
- Shimono, Y.; Zabala, M.; Cho, R.W.; Lobo, N.; Dalerba, P.; Qian, D.; Diehn, M.; Liu, H.; Panula, S.P.; Chiao, E.; et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 2009, 138, 592–603. [Google Scholar] [CrossRef] [PubMed]
- Guessous, F.; Alvarado-Velez, M.; Marcinkiewicz, L.; Zhang, Y.; Kim, J.; Heister, S.; Kefas, B.; Godlewski, J.; Schiff, D.; Purow, B.; et al. Oncogenic effects of miR-10b in glioblastoma stem cells. J. Neurooncol. 2013, 112, 153–163. [Google Scholar] [CrossRef]
- Tsukerman, P.; Stern-Ginossar, N.; Gur, C.; Glasner, A.; Nachmani, D.; Bauman, Y.; Yamin, R.; Vitenshtein, A.; Stanietsky, N.; Bar-Mag, T.; et al. MiR-10b downregulates the stress-induced cell surface molecule MICB, a critical ligand for cancer cell recognition by natural killer cells. Cancer Res. 2012, 72, 5463–5472. [Google Scholar] [CrossRef]
- Teplyuk, N.M.; Uhlmann, E.J.; Gabriely, G.; Volfovsky, N.; Wang, Y.; Teng, J.; Karmali, P.; Marcusson, E.; Peter, M.; Mohan, A.; et al. Therapeutic potential of targeting microRNA-10b in established intracranial glioblastoma: First steps toward the clinic. EMBO Mol. Med. 2016, 8, 268–287. [Google Scholar] [CrossRef] [PubMed]
- Wimmer, M.; Zauner, R.; Ablinger, M.; Pinon-Hofbauer, J.; Guttmann-Gruber, C.; Reisenberger, M.; Lettner, T.; Niklas, N.; Proell, J.; Sajinovic, M.; et al. A cancer stem cell-like phenotype is associated with miR-10b expression in aggressive squamous cell carcinomas. Cell Commun. Signal. 2020, 18, 61. [Google Scholar] [CrossRef]
- Shen, J.; Pan, J.; Du, C.; Si, W.; Yao, M.; Xu, L.; Zheng, H.; Xu, M.; Chen, D.; Wang, S.; et al. Silencing NKG2D ligand-targeting miRNAs enhances natural killer cell-mediated cytotoxicity in breast cancer. Cell Death Dis. 2017, 8, e2740. [Google Scholar] [CrossRef]
- Omar, H.A.; El-Serafi, A.T.; Hersi, F.; Arafa, E.A.; Zaher, D.M.; Madkour, M.; Arab, H.H.; Tolba, M.F. Immunomodulatory MicroRNAs in cancer: Targeting immune checkpoints and the tumor microenvironment. FEBS J. 2019, 286, 3540–3557. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Chen, S.; Feng, S.; Li, Y.; Zou, J.; Ling, H.; Zeng, Y.; Zeng, X. Function and mechanisms of microRNA-20a in colorectal cancer. Exp. Ther. Med. 2020, 19, 1605–1616. [Google Scholar] [CrossRef] [PubMed]
- Xi, J.; Huang, Q.; Wang, L.; Ma, X.; Deng, Q.; Kumar, M.; Zhou, Z.; Li, L.; Zeng, Z.; Young, K.H.; et al. miR-21 depletion in macrophages promotes tumoricidal polarization and enhances PD-1 immunotherapy. Oncogene 2018, 37, 3151–3165. [Google Scholar] [CrossRef] [PubMed]
- Pradella, D.; Naro, C.; Sette, C.; Ghigna, C. EMT and stemness: Flexible processes tuned by alternative splicing in development and cancer progression. Mol. Cancer 2017, 16, 8. [Google Scholar] [CrossRef] [PubMed]
- Zottel, A.; Samec, N.; Kump, A.; Raspor Dall’Olio, L.R.; Puzar Dominkus, P.; Romih, R.; Hudoklin, S.; Mlakar, J.; Nikitin, D.; Sorokin, M.; et al. Analysis of miR-9-5p, miR-124-3p, miR-21-5p, miR-138-5p, and miR-1-3p in Glioblastoma Cell Lines and Extracellular Vesicles. Int. J. Mol. Sci. 2020, 21, 8491. [Google Scholar] [CrossRef] [PubMed]
- Aldaz, B.; Sagardoy, A.; Nogueira, L.; Guruceaga, E.; Grande, L.; Huse, J.T.; Aznar, M.A.; Diez-Valle, R.; Tejada-Solis, S.; Alonso, M.M.; et al. Involvement of miRNAs in the differentiation of human glioblastoma multiforme stem-like cells. PLoS ONE 2013, 8, e77098. [Google Scholar] [CrossRef]
- Skafi, N.; Fayyad-Kazan, M.; Badran, B. Immunomodulatory role for MicroRNAs: Regulation of PD-1/PD-L1 and CTLA-4 immune checkpoints expression. Gene 2020, 754, 144888. [Google Scholar] [CrossRef]
- Iliopoulos, D.; Jaeger, S.A.; Hirsch, H.A.; Bulyk, M.L.; Struhl, K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol. Cell 2010, 39, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.Q.; Ahmed, E.I.; Elareer, N.R.; Junejo, K.; Steinhoff, M.; Uddin, S. Role of miRNA-Regulated Cancer Stem Cells in the Pathogenesis of Human Malignancies. Cells 2019, 8, 840. [Google Scholar] [CrossRef]
- Lu, L.F.; Liston, A. MicroRNA in the immune system, microRNA as an immune system. Immunology 2009, 127, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Liu, M.; Xu, Z.; Li, Y.; Guo, H.; Ge, Y.; Liu, Y.; Zheng, D.; Shi, J. A double feedback loop mediated by microRNA-23a/27a/24-2 regulates M1 versus M2 macrophage polarization and thus regulates cancer progression. Oncotarget 2016, 7, 13502–13519. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Xu, J.; Zhao, J.; Bai, J. Knockdown of miR-27a sensitizes colorectal cancer stem cells to TRAIL by promoting the formation of Apaf-1-caspase-9 complex. Oncotarget 2017, 8, 45213–45223. [Google Scholar] [CrossRef]
- Ren, L.; Li, Y.; Zhao, Q.; Fan, L.; Tan, B.; Zang, A.; Yang, H. miR-519 regulates the proliferation of breast cancer cells via targeting human antigen R. Oncol. Lett. 2020, 19, 1567–1576. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Z.; Zhang, C.; Liu, X.; Li, G.; Liu, S.; Sun, L.; Liang, J.; Hu, H.; Liu, Y.; et al. Genetic and clinical characterization of B7-H3 (CD276) expression and epigenetic regulation in diffuse brain glioma. Cancer Sci. 2018, 109, 2697–2705. [Google Scholar] [CrossRef]
- Wang, J.; Chong, K.K.; Nakamura, Y.; Nguyen, L.; Huang, S.K.; Kuo, C.; Zhang, W.; Yu, H.; Morton, D.L.; Hoon, D.S. B7-H3 associated with tumor progression and epigenetic regulatory activity in cutaneous melanoma. J. Investig. Dermatol. 2013, 133, 2050–2058. [Google Scholar] [CrossRef]
- Fan, X.; Chen, X.; Deng, W.; Zhong, G.; Cai, Q.; Lin, T. Up-regulated microRNA-143 in cancer stem cells differentiation promotes prostate cancer cells metastasis by modulating FNDC3B expression. BMC Cancer 2013, 13, 61. [Google Scholar] [CrossRef]
- Wu, Y.; Shi, W.; Tang, T.; Wang, Y.; Yin, X.; Chen, Y.; Zhang, Y.; Xing, Y.; Shen, Y.; Xia, T.; et al. miR-29a contributes to breast cancer cells epithelial-mesenchymal transition, migration, and invasion via down-regulating histone H4K20 trimethylation through directly targeting SUV420H2. Cell Death Dis. 2019, 10, 176. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Kelnar, K.; Liu, B.; Chen, X.; Calhoun-Davis, T.; Li, H.; Patrawala, L.; Yan, H.; Jeter, C.; Honorio, S.; et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat. Med. 2011, 17, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, A.; Zhao, F.; Pechlivanis, S.; Eberle, J.; Steinle, A.; Diederichs, S.; Schadendorf, D.; Paschen, A. Tumor suppressive microRNAs miR-34a/c control cancer cell expression of ULBP2, a stress-induced ligand of the natural killer cell receptor NKG2D. Cancer Res. 2012, 72, 460–471. [Google Scholar] [CrossRef] [PubMed]
- Grenda, A.; Krawczyk, P. New Dancing Couple: PD-L1 and MicroRNA. Scand J. Immunol. 2017, 86, 130–134. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Chen, L.J.; Zhang, G.B.; Jiang, J.T.; Zhu, M.; Tan, Y.; Wang, H.T.; Lu, B.F.; Zhang, X.G. Clinical significance and regulation of the costimulatory molecule B7-H3 in human colorectal carcinoma. Cancer Immunol. Immunother. 2010, 59, 1163–1171. [Google Scholar] [CrossRef]
- Jia, L.; Xi, Q.; Wang, H.; Zhang, Z.; Liu, H.; Cheng, Y.; Guo, X.; Zhang, J.; Zhang, Q.; Zhang, L.; et al. miR-142-5p regulates tumor cell PD-L1 expression and enhances anti-tumor immunity. Biochem. Biophys. Res. Commun. 2017, 488, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Zhang, Y.; Yan, R.; Huang, L.; Mellor, A.L.; Yang, Y.; Chen, X.; Wei, W.; Wu, X.; Yu, L.; et al. Exosome-derived miR-142-5p remodels lymphatic vessels and induces IDO to promote immune privilege in the tumour microenvironment. Cell Death Differ. 2020. [Google Scholar] [CrossRef] [PubMed]
- Troschel, F.M.; Bohly, N.; Borrmann, K.; Braun, T.; Schwickert, A.; Kiesel, L.; Eich, H.T.; Gotte, M.; Greve, B. miR-142-3p attenuates breast cancer stem cell characteristics and decreases radioresistance in vitro. Tumour Biol. 2018, 40. [Google Scholar] [CrossRef] [PubMed]
- Liston, A.; Linterman, M.; Lu, L.F. MicroRNA in the adaptive immune system, in sickness and in health. J. Clin. Immunol. 2010, 30, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, A.; Vigorito, E.; Clare, S.; Warren, M.V.; Couttet, P.; Soond, D.R.; van Dongen, S.; Grocock, R.J.; Das, P.P.; Miska, E.A.; et al. Requirement of bic/microRNA-155 for normal immune function. Science 2007, 316, 608–611. [Google Scholar] [CrossRef]
- Yee, D.; Shah, K.M.; Coles, M.C.; Sharp, T.V.; Lagos, D. MicroRNA-155 induction via TNF-alpha and IFN-gamma suppresses expression of programmed death ligand-1 (PD-L1) in human primary cells. J. Biol. Chem. 2017, 292, 20683–20693. [Google Scholar] [CrossRef]
- Zonari, E.; Pucci, F.; Saini, M.; Mazzieri, R.; Politi, L.S.; Gentner, B.; Naldini, L. A role for miR-155 in enabling tumor-infiltrating innate immune cells to mount effective antitumor responses in mice. Blood 2013, 122, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Zhang, H.W.; Lu, M.H.; He, X.H.; Li, Y.; Gu, H.; Liu, M.F.; Wang, E.D. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 2010, 70, 3119–3127. [Google Scholar] [CrossRef]
- Zhou, X.; Mao, Y.; Zhu, J.; Meng, F.; Chen, Q.; Tao, L.; Li, R.; Fu, F.; Liu, C.; Hu, Y.; et al. TGF-beta1 promotes colorectal cancer immune escape by elevating B7-H3 and B7-H4 via the miR-155/miR-143 axis. Oncotarget 2016, 7, 67196–67211. [Google Scholar] [CrossRef] [PubMed]
- Gasparini, P.; Lovat, F.; Fassan, M.; Casadei, L.; Cascione, L.; Jacob, N.K.; Carasi, S.; Palmieri, D.; Costinean, S.; Shapiro, C.L.; et al. Protective role of miR-155 in breast cancer through RAD51 targeting impairs homologous recombination after irradiation. Proc. Natl. Acad. Sci. USA 2014, 111, 4536–4541. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.M.; Chaudhuri, A.A.; Rao, D.S.; Baltimore, D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc. Natl. Acad. Sci. USA 2009, 106, 7113–7118. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, I.M.; Otero, D.; Kao, E.; Miletic, A.V.; Hother, C.; Ralfkiaer, E.; Rickert, R.C.; Gronbaek, K.; David, M. Onco-miR-155 targets SHIP1 to promote TNFalpha-dependent growth of B cell lymphomas. EMBO Mol. Med. 2009, 1, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Trotta, R.; Chen, L.; Costinean, S.; Josyula, S.; Mundy-Bosse, B.L.; Ciarlariello, D.; Mao, C.; Briercheck, E.L.; McConnell, K.K.; Mishra, A.; et al. Overexpression of miR-155 causes expansion, arrest in terminal differentiation and functional activation of mouse natural killer cells. Blood 2013, 121, 3126–3134. [Google Scholar] [CrossRef]
- Liu, F.; Kong, X.; Lv, L.; Gao, J. TGF-beta1 acts through miR-155 to down-regulate TP53INP1 in promoting epithelial-mesenchymal transition and cancer stem cell phenotypes. Cancer Lett. 2015, 359, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Kong, X.; Lv, L.; Gao, J. MiR-155 targets TP53INP1 to regulate liver cancer stem cell acquisition and self-renewal. FEBS Lett. 2015, 589, 500–506. [Google Scholar] [CrossRef]
- Chen, C.Z.; Li, L.; Lodish, H.F.; Bartel, D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science 2004, 303, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.J.; Chau, J.; Ebert, P.J.; Sylvester, G.; Min, H.; Liu, G.; Braich, R.; Manoharan, M.; Soutschek, J.; Skare, P.; et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell 2007, 129, 147–161. [Google Scholar] [CrossRef]
- Kim, J.; Yao, F.; Xiao, Z.; Sun, Y.; Ma, L. MicroRNAs and metastasis: Small RNAs play big roles. Cancer Metastasis Rev. 2018, 37, 5–15. [Google Scholar] [CrossRef]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef]
- Gao, Y.; Fang, P.; Li, W.J.; Zhang, J.; Wang, G.P.; Jiang, D.F.; Chen, F.P. LncRNA NEAT1 sponges miR-214 to regulate M2 macrophage polarization by regulation of B7-H3 in multiple myeloma. Mol. Immunol. 2020, 117, 20–28. [Google Scholar] [CrossRef]
- Xu, C.X.; Xu, M.; Tan, L.; Yang, H.; Permuth-Wey, J.; Kruk, P.A.; Wenham, R.M.; Nicosia, S.V.; Lancaster, J.M.; Sellers, T.A.; et al. MicroRNA miR-214 regulates ovarian cancer cell stemness by targeting p53/Nanog. J. Biol. Chem. 2012, 287, 34970–34978. [Google Scholar] [CrossRef]
- Pei, X.; Wang, X.; Li, H. LncRNA SNHG1 regulates the differentiation of Treg cells and affects the immune escape of breast cancer via regulating miR-448/IDO. Int. J. Biol. Macromol. 2018, 118, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Ni, K.; Ke, J.; Zhang, W.; Feng, Y.; Mao, Q. miR-448 inhibits the epithelial-mesenchymal transition in breast cancer cells by directly targeting the E-cadherin repressor ZEB1/2. Exp. Biol. Med. 2018, 243, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Bamodu, O.A.; Huang, W.C.; Lee, W.H.; Wu, A.; Wang, L.S.; Hsiao, M.; Yeh, C.T.; Chao, T.Y. Aberrant KDM5B expression promotes aggressive breast cancer through MALAT1 overexpression and downregulation of hsa-miR-448. BMC Cancer 2016, 16, 160. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.C.; Yang, Y.J.; Zhang, J.Q.; Guo, M.; Xiang, L.; Yu, S.F.; Ping, H.; Zhuo, L. microRNA-448 inhibits stemness maintenance and self-renewal of hepatocellular carcinoma stem cells through the MAGEA6-mediated AMPK signaling pathway. J. Cell Physiol. 2019, 234, 23461–23474. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, R.U.; Miyazaki, H.; Ochiya, T. The Roles of MicroRNAs in Breast Cancer. Cancers 2015, 7, 598–616. [Google Scholar] [CrossRef]
- Godlewski, J.; Newton, H.B.; Chiocca, E.A.; Lawler, S.E. MicroRNAs and glioblastoma; the stem cell connection. Cell Death Differ 2010, 17, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Ceppi, P.; Peter, M.E. MicroRNAs regulate both epithelial-to-mesenchymal transition and cancer stem cells. Oncogene 2014, 33, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Toden, S.; Kunitoshi, S.; Cardenas, J.; Gu, J.; Hutchins, E.; Van Keuren-Jensen, K.; Uetake, H.; Toiyama, Y.; Goel, A. Cancer stem cell-associated miRNAs serve as prognostic biomarkers in colorectal cancer. JCI Insight 2019, 4. [Google Scholar] [CrossRef]
- Xia, H.; Hui, K.M. MicroRNAs involved in regulating epithelial-mesenchymal transition and cancer stem cells as molecular targets for cancer therapeutics. Cancer Gene Ther. 2012, 19, 723–730. [Google Scholar] [CrossRef]
- Liu, C.; Tang, D.G. MicroRNA regulation of cancer stem cells. Cancer Res. 2011, 71, 5950–5954. [Google Scholar] [CrossRef] [PubMed]
- Lopez de Andres, J.; Grinan-Lison, C.; Jimenez, G.; Marchal, J.A. Cancer stem cell secretome in the tumor microenvironment: A key point for an effective personalized cancer treatment. J. Hematol. Oncol. 2020, 13, 136. [Google Scholar] [CrossRef] [PubMed]
- Dooley, J.; Linterman, M.A.; Liston, A. MicroRNA regulation of T-cell development. Immunol. Rev. 2013, 253, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Cai, X.; Chen, X.; Liang, H.; Zhang, Y.; Li, J.; Wang, Z.; Chen, X.; Zhang, W.; Yokoyama, S.; et al. Tumor-secreted miR-214 induces regulatory T cells: A major link between immune evasion and tumor growth. Cell Res. 2014, 24, 1164–1180. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, Y.; Fan, H.; Zhao, X.; Liu, D.; Hu, Y.; Kidd, A.R., 3rd; Bao, J.; Hou, Y. MicroRNA-181a regulates local immune balance by inhibiting proliferation and immunosuppressive properties of mesenchymal stem cells. Stem Cells 2012, 30, 1756–1770. [Google Scholar] [CrossRef]
- Adachi, K.; Tamada, K. Immune checkpoint blockade opens an avenue of cancer immunotherapy with a potent clinical efficacy. Cancer Sci. 2015, 106, 945–950. [Google Scholar] [CrossRef]
- Bianchini, G.; Pusztai, L.; Pienkowski, T.; Im, Y.H.; Bianchi, G.V.; Tseng, L.M.; Liu, M.C.; Lluch, A.; Galeota, E.; Magazzù, D.; et al. Immune modulation of pathologic complete response after neoadjuvant HER2-directed therapies in the NeoSphere trial. Ann. Oncol. 2015, 26, 2429–2436. [Google Scholar] [CrossRef]
- Kyi, C.; Postow, M.A. Immune checkpoint inhibitor combinations in solid tumors: Opportunities and challenges. Immunotherapy 2016, 8, 821–837. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Medeiros, L.J.; Young, K.H. Immune checkpoint blockade: Releasing the brake towards hematological malignancies. Blood Rev. 2016, 30, 189–200. [Google Scholar] [CrossRef] [PubMed]
| Immune Responses | Cell Type | Effect of CSCs/CICs | References |
|---|---|---|---|
| Innate | |||
| NK cells | Immune evasion of CSCs/CICs or their killing depending on the expression profile of HLA molecules and ligands of NK activatory receptors. | [84,85,86,116,117,118,119,120,121] | |
| DCs | 1. Differentiation towards iDCs and impairment of anti-tumor (including CSCs/CICs) T cell-mediated responses. 2. Pro-tumoral role by CXCL1 and CXCL12/CXCR4. | [122,123,124,125] | |
| Macrophages | Promote stemness functions through STAT3 signaling pathway. | [126,127,128,129,130] | |
| MDSCs | 1. Promote stemness functions and survival of CSCs/CICs through STAT3 signaling pathway. 2. Impairment of anti-tumor (including CSCs/CICs) immune responses. | [131,132,133,134] | |
| Adaptive | |||
| T lymphocytes | Immune evasion of CSCs/CICs or their killing depending on the levels of the expression of HLA molecules, APM and ligands of T cell co-stimulatory receptors. | [53,55,79,80] | |
| Tregs | Differentiation of Tregs through either the secretion of immunosuppressive factors by CSCs/CICs or their crosstalk with immune suppressive APC (e.g., iDCs, MDSCs, TAMs). | [55,79,126] | |
| Bregs | Establishment of the niche and sustainment of stemness functions. | [126,135] | |
| Molecule | Cancer Types | Functions Mediated by CSCs/CICs | References |
|---|---|---|---|
| Cytokines and growth factors | |||
| IL-4, IL-13 | CRC, GBM, Ovarian, Lung, Breast, Pancreatic and Bladder cancer | Immunoregulatory functions: inhibition of effector functions; differentiation of immunosuppressive cells. | [52,65,79,80,162,163] |
| IL-10 | Multiple | Immunoregulatory functions: inhibition of effector functions; differentiation of Tregs and MDSCs. | [163,164,165] |
| TGF-β | Melanoma, Pancreatic adenocarcinoma, GBM | Growth factor with both immunoregulatory functions and pro-tumoral activity. | [79,166] |
| GDF-15 | Glioma, CRC, Prostate Cancer, Melanoma | Growth factor with immunoregulatory activity. Inhibition of effector immune functions. | [167,168] |
| Soluble regulators | |||
| PGE2 | Uterine Cervical Cancer | Lipid mediator of inflammatory and immune responses. Inhibition of innate immune responses. | [169] |
| Galectin-3 | GBM, CRC, Ovarian and Breast cancer, Leukemia | Regulate the interaction of cells with TME. Inhibition of T cell-mediated responses. | [133,170,171,172,173,174,175,176] |
| IDO | GBM, CRC, Lung, Breast and Pancreatic Cancer | Enzyme regulating tryptophan catabolism: mediating the differentiation of Tregs, inhibitor of effector cell proliferation and cytotoxic activity. | [70,159] |
| Immune checkpoints | |||
| PD-L1/CTLA-4 | Multiple | Inhibitors of cell-mediated immune responses | [55,62,64,79,177,178] |
| B7-H3/B7-H4 | CRC, Melanoma and Brain Tumors | Inhibitors of cell-mediated immune responses | [41,62,179,180,181] |
| miRNA | Tumor Type of CSCs/CICs | Type of miRNA | Functions Associated with Stemness Properties | Target Molecules of miRNAs Involved in Immunological Signaling ∂ | References |
|---|---|---|---|---|---|
| miR-10b | Breast cancer, brain tumors, squamous cell carcinoma | Oncogenic | Proliferation, migration and invasiveness | MICA/B, ULBPs | [195,196,197,198] |
| miR-20a | CRC, breast cancer | Oncogenic | Cell growth, migration, invasiveness, autophagy, PTEN, PI3K/AKT, MAPK/ERK | PTEN, MICA/B, ULBPs | [86,199,200,201] |
| miR-21 | CRC, GBM and other solid tumors | Oncogenic | EMT, stemness by TGF-βR2, PTEN, STAT3 (miR-18 and miR-23) * | IFN-β, TLR (regulated also by miR-125 and let-7), PTEN, STAT3 (miR-18 and miR-23) * | [200,202,203,204,205,206,207] |
| miR-27 | CRC | Oncogenic | Resistance to TNF-related apoptosis | Differentiation of Macrophage and MDSC | [208,209,210,211] |
| miR-29 | Breast, Melanoma, Glioma | Oncogenic | Migration and metastasis due to the upregulation of bFGF | IFN-γ, B7-H3 | [212,213,214,215,216] |
| miR-34 | Breast, prostate and other solid tumors | Tumor suppressor | Stemness by targeting Wnt/β-catenin, suppresses CD44 which inhibits self-renewal and metastasis of CSCs | MICA/B, ULBPs (miR-10) *, PD-L1 (miR-24, miR-30b, miR-1385p, miR-425p and miR-940) * | [76,206,217,218,219,220] |
| miR142 | Breast cancer | Tumor suppressor | Stemness, proliferation, apoptosis | TGF-β, IFN-γ, IDO, PD-L1 | [221,222,223] |
| miR155 | Breast and liver cancer | Oncogenic | Proliferation of BCSCs by activating STAT3 | IFNs, TLR, TGFb1, IDO, B7-H3 and B7-H4, STAT3 | [72,159,208,209,215,224,225,226,227,228,229,230,231,232,233,234,235] |
| miR-181 | Multiple solid tumors | Tumor Suppressor | Drug resistance, PTEN, STAT3/AKT (miR-18 and miR-23) * | PD-L1 (miR-940 and miR-30b) *, PTEN, STAT3 (miR-18 and miR-23) *, IDO (miR-760) * | [72,76,159,207,236,237] |
| miR200 | CRC, breast and prostate cancer | Tumor suppressor | EMT, invasiveness, chemoresistance | PD-L1 | [206,219,238,239] |
| miR214 | Ovarian cancer | Tumor suppressor | EMT, invasiveness and proliferation, JAK2/STAT3 | B7-H3, JAK2/STAT3 | [206,240,241] |
| miR-448 | Brain tumors, breast and hepatic cancer | Tumor suppressor | EMT, TGF-β1 | IDO, TGF-β1 | [222,242,243,244,245] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tomei, S.; Ibnaof, O.; Ravindran, S.; Ferrone, S.; Maccalli, C. Cancer Stem Cells Are Possible Key Players in Regulating Anti-Tumor Immune Responses: The Role of Immunomodulating Molecules and MicroRNAs. Cancers 2021, 13, 1674. https://doi.org/10.3390/cancers13071674
Tomei S, Ibnaof O, Ravindran S, Ferrone S, Maccalli C. Cancer Stem Cells Are Possible Key Players in Regulating Anti-Tumor Immune Responses: The Role of Immunomodulating Molecules and MicroRNAs. Cancers. 2021; 13(7):1674. https://doi.org/10.3390/cancers13071674
Chicago/Turabian StyleTomei, Sara, Ola Ibnaof, Shilpa Ravindran, Soldano Ferrone, and Cristina Maccalli. 2021. "Cancer Stem Cells Are Possible Key Players in Regulating Anti-Tumor Immune Responses: The Role of Immunomodulating Molecules and MicroRNAs" Cancers 13, no. 7: 1674. https://doi.org/10.3390/cancers13071674
APA StyleTomei, S., Ibnaof, O., Ravindran, S., Ferrone, S., & Maccalli, C. (2021). Cancer Stem Cells Are Possible Key Players in Regulating Anti-Tumor Immune Responses: The Role of Immunomodulating Molecules and MicroRNAs. Cancers, 13(7), 1674. https://doi.org/10.3390/cancers13071674








