Oral and Intravenous Iron Therapy Differentially Alter the On- and Off-Tumor Microbiota in Anemic Colorectal Cancer Patients
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. DNA Extraction and 16S rRNA Amplicon Sequencing
2.3. Statistical Analysis
3. Results
3.1. On- and Off-Tumor Bacterial Diversity Following Iron Therapy
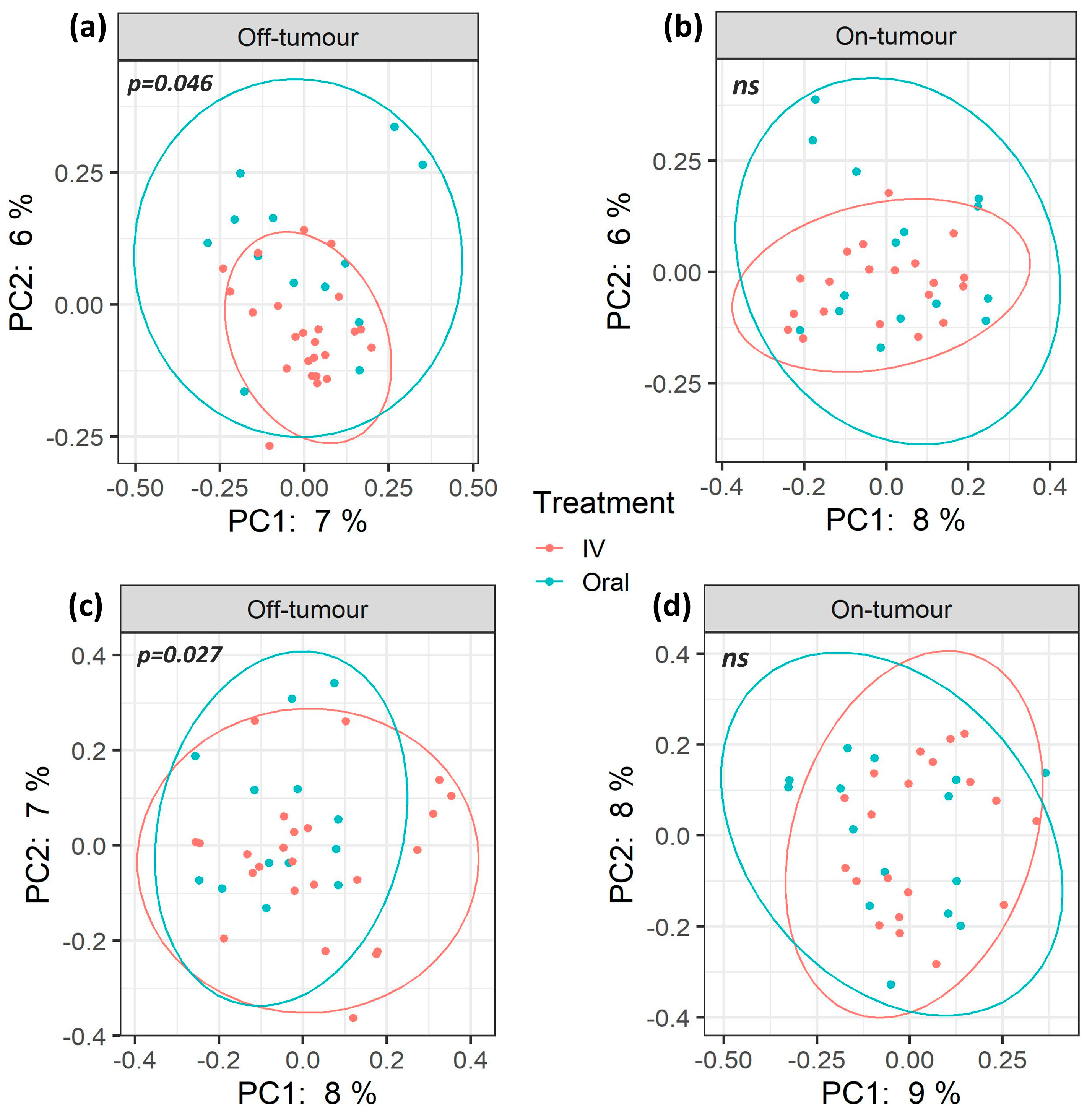
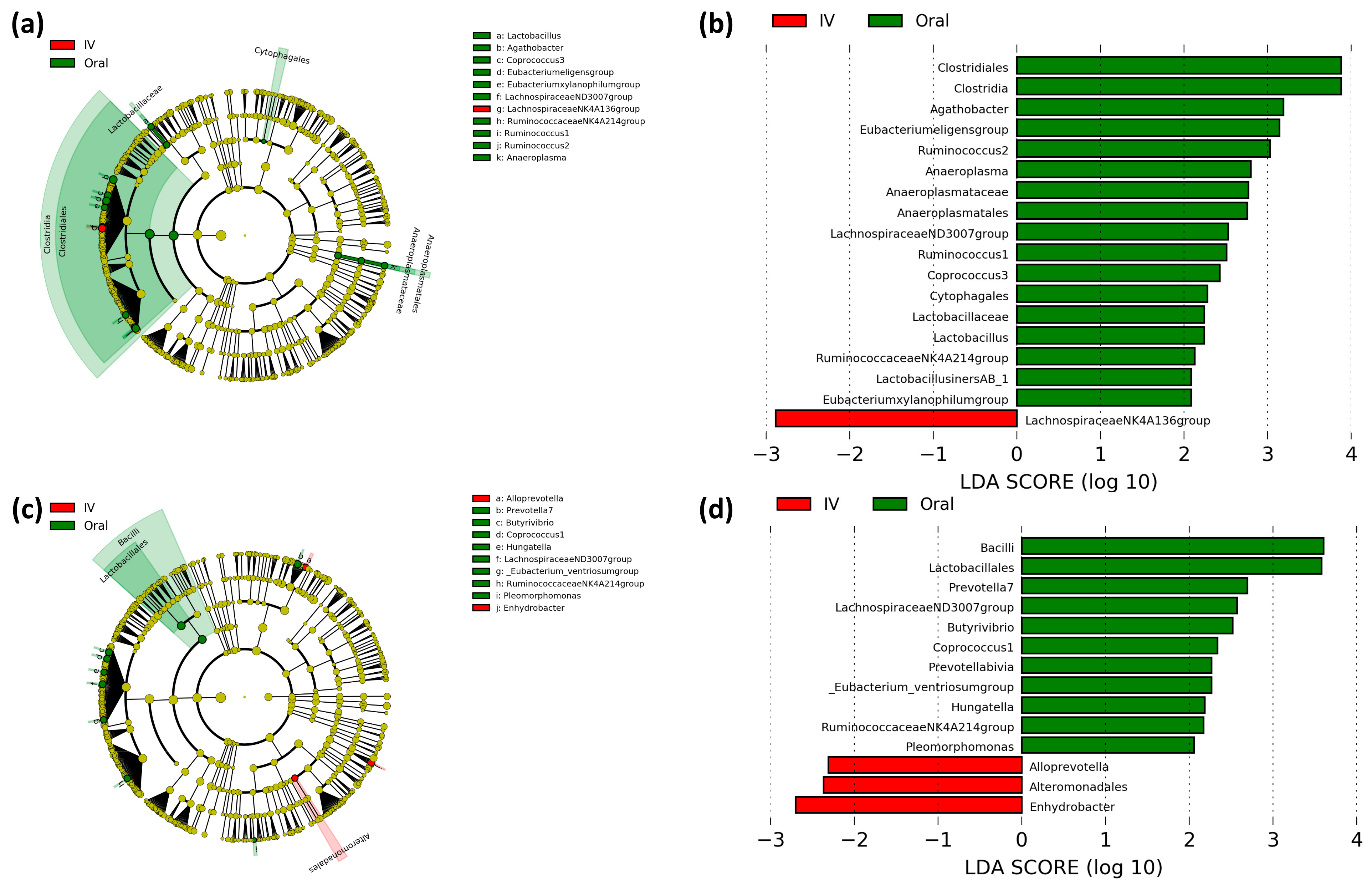
3.2. Oral and Intravenous Iron-Treated Patients Show Differing Bacterial Communities
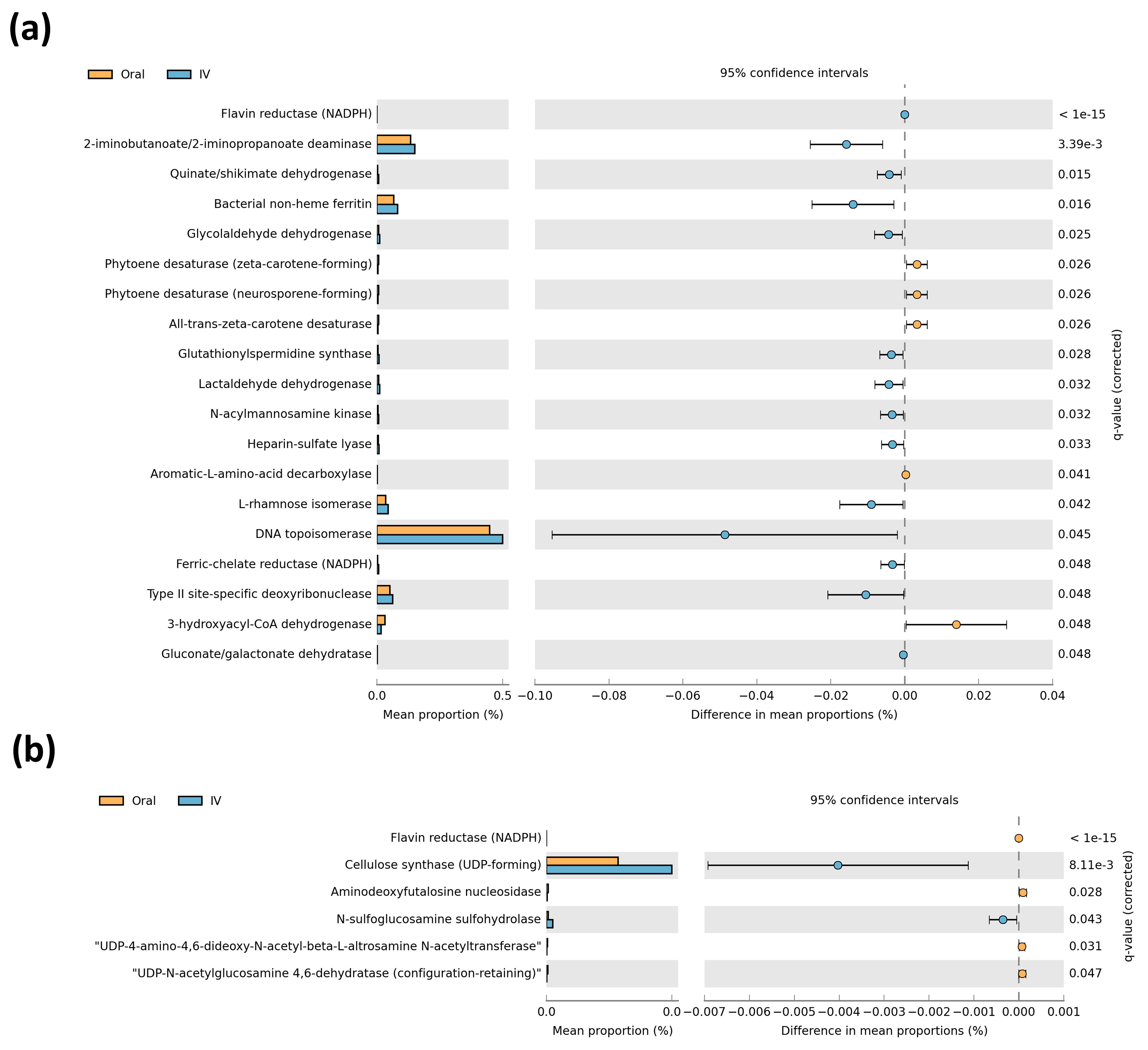
3.3. Differential Predictive Enzymatic Pathways between Iron Treatment Groups
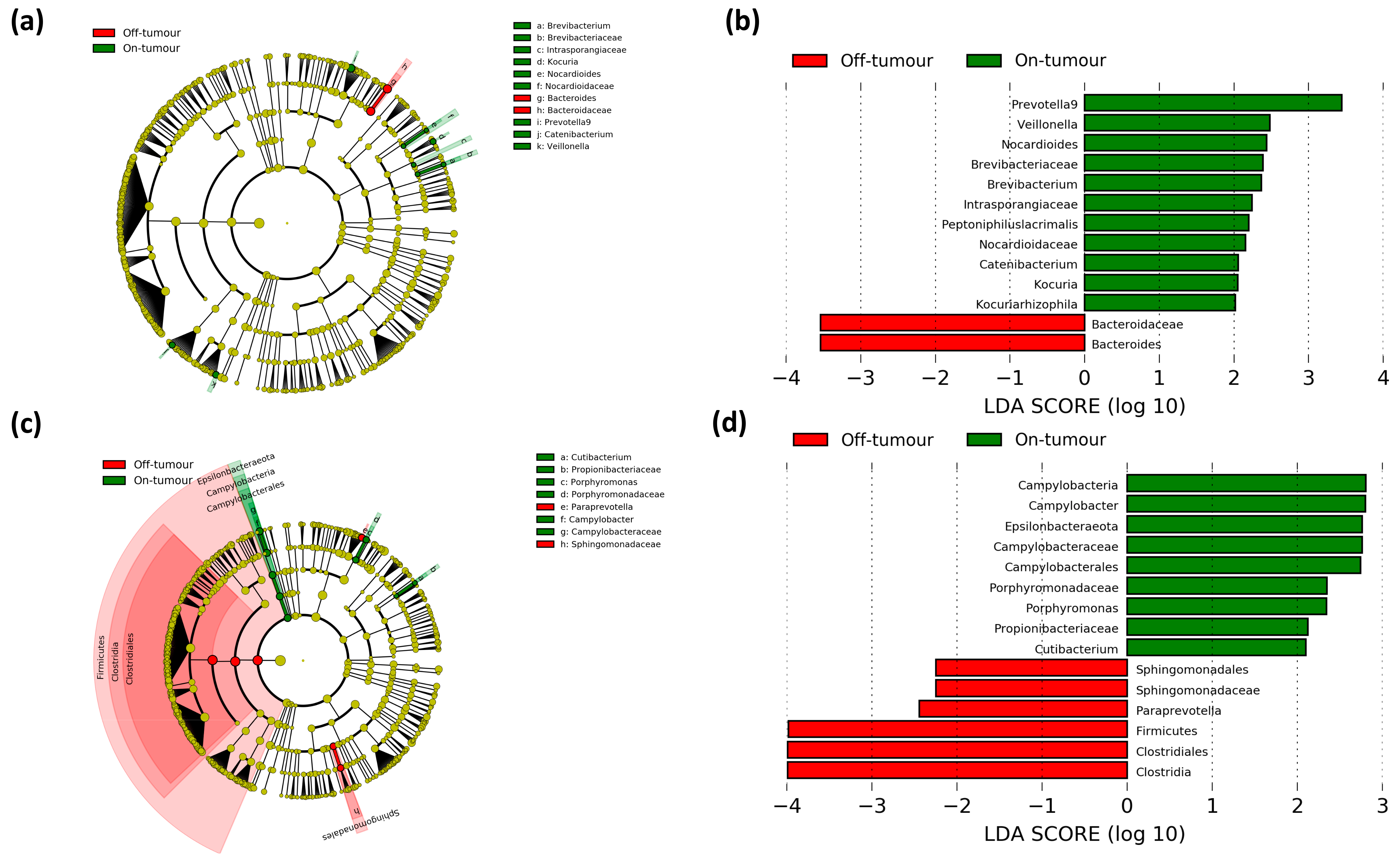
3.4. Paired On- and Off-Tumor Microbiota Show Varying Microbial Communities Following Oral and Intravenous Iron Therapy
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sánchez-Alcoholado, L.; Ramos-Molina, B.; Otero, A.; Laborda-Illanes, A.; Ordóñez, R.; Medina, J.A.; Gómez-Millán, J.; Queipo-Ortuño, M.I. The Role of the Gut Microbiome in Colorectal Cancer Development and Therapy Response. Cancers 2020, 12, 1406. [Google Scholar] [CrossRef]
- Tjalsma, H.; Boleij, A.; Marchesi, J.R.; Dutilh, B.E. A Bacterial Driver–passenger Model for Colorectal Cancer: Beyond the Usual Suspects. Nat. Rev. Microbiol. 2012, 10, 575–582. [Google Scholar] [CrossRef]
- Wirth, U.; Garzetti, D.; Jochum, L.M.; Spriewald, S.; Kühn, F.; Ilmer, M.; Lee, S.M.L.; Niess, H.; Bazhin, A.V.; Andrassy, J.; et al. Microbiome Analysis from Paired Mucosal and Fecal Samples of a Colorectal Cancer Biobank. Cancers 2020, 12, 3702. [Google Scholar] [CrossRef]
- Weersma, R.K.; Zhernakova, A.; Fu, J. Interaction between Drugs and the Gut Microbiome. Gut 2020, 69, 1510–1519. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Chan, A.T.; Sun, J. Influence of the Gut Microbiome, Diet, and Environment on Risk of Colorectal Cancer. Gastroenterology 2020, 158, 322–340. [Google Scholar] [CrossRef] [PubMed]
- Cassat, J.E.; Skaar, E.P. Iron in Infection and Immunity. Cell Host Microbe 2013, 13, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Phipps, O.; Al-Hassi, H.O.; Quraishi, M.N.; Kumar, A.; Brookes, M.J. Influence of Iron on the Gut Microbiota in Colorectal Cancer. Nutrients 2020, 12, 2512. [Google Scholar] [CrossRef]
- Flemer, B.; Herlihy, M.; O’Riordain, M.; Shanahan, F.; O’Toole, P.W. Tumour-Associated and Non-Tumour-Associated Microbiota: Addendum. Gut Microbes 2018, 9, 369–373. [Google Scholar] [CrossRef]
- Keeler, B.D.; Simpson, J.A.; Ng, O.; Padmanabhan, H.; Brookes, M.J.; Acheson, A.G.; IVICA Trial Grou. Randomized Clinical Trial of Preoperative Oral Versus Intravenous Iron in Anaemic Patients with Colorectal Cancer. Br. J. Surg. 2017, 104, 214–221. [Google Scholar] [CrossRef]
- Richards, T.; Baikady, R.R.; Clevenger, B.; Butcher, A.; Abeysiri, S.; Chau, M.; Macdougall, I.C.; Murphy, G.; Swinson, R.; Collier, T.; et al. Preoperative Intravenous Iron to Treat Anaemia before Major Abdominal Surgery (PREVENTT): A Randomised, Double-Blind, Controlled Trial. Lancet 2020, 396, 1353–1361. [Google Scholar] [CrossRef]
- Mahalhal, A.; Williams, J.M.; Johnson, S.; Ellaby, N.; Duckworth, C.A.; Burkitt, M.D.; Liu, X.; Hold, G.L.; Campbell, B.J.; Pritchard, D.M.; et al. Oral Iron Exacerbates Colitis and Influences the Intestinal Microbiome. PLoS ONE 2018, 13, e0202460. [Google Scholar]
- Liu, B.; Pan, X.; Liu, Z.; Han, M.; Xu, G.; Dai, X.; Wang, W.; Zhang, H.; Xie, L. Fecal Microbiota as a Noninvasive Biomarker to Predict the Tissue Iron Accumulation in Intestine Epithelial Cells and Liver. FASEB J. 2020, 34, 3006–3020. [Google Scholar] [CrossRef] [PubMed]
- Earth Microbiome Project. Illumina 16s PCR Protocols. 2020. Available online: https://www.earthmicrobiome.org/protocols-and-standards/16s/ (accessed on 29 June 2020).
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; Volume 2020. [Google Scholar]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic Biomarker Discovery and Explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Langille, M.G.I. Exploring Linkages between Taxonomic and Functional Profiles of the Human Microbiome. mSystems 2018, 3, e00163-17. [Google Scholar] [CrossRef]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical Analysis of Taxonomic and Functional Profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef]
- Storey, J.D. The Positive False Discovery Rate: A Bayesian Interpretation and the Q-Value. Ann. Statist. 2003, 31, 2013–2035. [Google Scholar] [CrossRef]
- Wilson, M.J.; Dekker, J.W.T.; Harlaar, J.J.; Jeekel, J.; Schipperus, M.; Zwaginga, J.J. The Role of Preoperative Iron Deficiency in Colorectal Cancer Patients: Prevalence and Treatment. Int. J. Colorectal Dis. 2017, 32, 1617–1624. [Google Scholar] [CrossRef]
- Gereklioglu, C.; Asma, S.; Korur, A.; Erdogan, F.; Kut, A. Medication Adherence to Oral Iron Therapy in Patients with Iron Deficiency Anemia. Pak. J. Med. Sci. 2016, 32, 604–607. [Google Scholar] [CrossRef]
- Brookes, M.J.; Boult, J.; Roberts, K.; Cooper, B.T.; Hotchin, N.A.; Matthews, G.; Iqbal, T.; Tselepis, C. A Role for Iron in Wnt Signalling. Oncogene 2008, 27, 966–975. [Google Scholar] [CrossRef] [PubMed]
- Radulescu, S.; Brookes, M.J.; Salgueiro, P.; Ridgway, R.A.; McGhee, E.; Anderson, K.; Ford, S.J.; Stones, D.H.; Iqbal, T.H.; Tselepis, C.; et al. Luminal Iron Levels Govern Intestinal Tumorigenesis After Apc Loss in Vivo. Cell Rep. 2012, 2, 270–282. [Google Scholar] [CrossRef]
- Chua, A.C.G.; Klopcic, B.R.S.; Ho, D.S.; Fu, S.K.; Forrest, C.H.; Croft, K.D.; Olynyk, J.K.; Lawrance, I.C.; Trinder, D. Dietary Iron Enhances Colonic Inflammation and IL-6/IL-11-Stat3 Signaling Promoting Colonic Tumor Development in Mice. PLoS ONE 2013, 8, e78850. [Google Scholar]
- Lee, T.; Clavel, T.; Smirnov, K.; Schmidt, A.; Lagkouvardos, I.; Walker, A.; Lucio, M.; Michalke, B.; Schmitt-Kopplin, P.; Fedorak, R.; et al. Oral Versus Intravenous Iron Replacement Therapy Distinctly Alters the Gut Microbiota and Metabolome in Patients with IBD. Gut 2017, 66, 863–871. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.M.; Jesus, E.C.; Lopes, A.; Aguiar, S., Jr.; Begnami, M.D.; Rocha, R.M.; Carpinetti, P.A.; Camargo, A.A.; Hoffmann, C.; Freitas, H.C.; et al. Tissue-Associated Bacterial Alterations in Rectal Carcinoma Patients Revealed by 16S rRNA Community Profiling. Front. Cell. Infect. Microbiol. 2016, 6, 179. [Google Scholar] [CrossRef]
- Flemer, B.; Lynch, D.B.; Brown, J.M.R.; Jeffery, I.B.; Ryan, F.J.; Claesson, M.J.; O’Riordain, M.; Shanahan, F.; O’Toole, P.W. Tumour-Associated and Non-Tumour-Associated Microbiota in Colorectal Cancer. Gut 2017, 66, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.J.; Yang, J.; Seo, H.; Lee, W.H.; Ho Lee, D.; Kym, S.; Park, Y.S.; Kim, J.G.; Jang, I.; Kim, Y.; et al. Colorectal Cancer Diagnostic Model Utilizing Metagenomic and Metabolomic Data of Stool Microbial Extracellular Vesicles. Sci Rep. 2020, 10, 2860. [Google Scholar] [CrossRef]
- Drago, L. Probiotics and Colon Cancer. Microorganisms 2019, 7, 66. [Google Scholar] [CrossRef]
- Ma, Q.; Li, Y.; Wang, J.; Li, P.; Duan, Y.; Dai, H.; An, Y.; Cheng, L.; Wang, T.; Wang, C.; et al. Investigation of Gut Microbiome Changes in Type 1 Diabetic Mellitus Rats Based on High-Throughput Sequencing. Biomed. Pharmacother. 2020, 124, 109873. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, F.; Lu, J.; Shi, J.; Guan, J.; Yan, F.; Li, B.; Huo, G. Probiotic Mixture of Lactobacillus Plantarum Strains Improves Lipid Metabolism and Gut Microbiota Structure in High Fat Diet-Fed Mice. Front. Microbiol. 2020, 11, 512. [Google Scholar] [CrossRef] [PubMed]
- Miethke, M.; Marahiel, M.A. Siderophore-Based Iron Acquisition and Pathogen Control. Microbiol. Mol. Biol. Rev. 2007, 71, 413–451. [Google Scholar] [CrossRef] [PubMed]
- Rivera, M. Bacterioferritin: Structure, Dynamics, and Protein–Protein Interactions at Play in Iron Storage and Mobilization. Acc. Chem. Res. 2017, 50, 331–340. [Google Scholar] [CrossRef]
- Torti, S.V.; Torti, F.M. Iron and Cancer: More Ore to be Mined. Nat. Rev. Cancer 2013, 13, 342–355. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Flint, H.J. Formation of Propionate and Butyrate by the Human Colonic Microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Di Costanzo, L.; Gomez, G.A.; Christianson, D.W. Crystal Structure of Lactaldehyde Dehydrogenase from Escherichia Coli and Inferences regarding Substrate and Cofactor Specificity. J. Mol. Biol. 2007, 366, 481–493. [Google Scholar] [CrossRef][Green Version]
- Iraporda, C.; Romanin, D.E.; Bengoa, A.A.; Errea, A.J.; Cayet, D.; Foligné, B.; Sirard, J.; Garrote, G.L.; Abraham, A.G.; Rumbo, M. Local Treatment with Lactate Prevents Intestinal Inflammation in the TNBS-Induced Colitis Model. Front. Immunol. 2016, 7, 651. [Google Scholar] [CrossRef]
- Algieri, F.; Rodriguez-Nogales, A.; Garrido-Mesa, J.; Camuesco, D.; Vezza, T.; Garrido-Mesa, N.; Utrilla, P.; Rodriguez-Cabezas, M.E.; Pischel, I.; Galvez, J. Intestinal Anti-Inflammatory Activity of Calcium Pyruvate in the TNBS Model of Rat Colitis: Comparison with Ethyl Pyruvate. Biochem. Pharmacol. 2016, 103, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Murata, K.; Moriyama, M. Isoleucine, an Essential Amino Acid, Prevents Liver Metastases of Colon Cancer by Antiangiogenesis. Cancer Res. 2007, 67, 3263–3268. [Google Scholar] [CrossRef] [PubMed]
- Konno, Y.; Ashida, T.; Inaba, Y.; Ito, T.; Tanabe, H.; Maemoto, A.; Ayabe, T.; Mizukami, Y.; Fujiya, M.; Kohgo, Y. Isoleucine, an Essential Amino Acid, Induces the Expression of Human Β Defensin 2 through the Activation of the G-Protein Coupled Receptor-ERK Pathway in the Intestinal Epithelia. Food Nutr. Sci. 2012, 3, 548–555. [Google Scholar] [CrossRef][Green Version]
- Kim, Y.; Hwang, S.W.; Kim, S.; Lee, Y.; Kim, T.; Lee, S.; Kim, S.J.; Yoo, H.J.; Kim, E.N.; Kweon, M. Dietary Cellulose Prevents Gut Inflammation by Modulating Lipid Metabolism and Gut Microbiota. Gut Microbes 2020, 11, 944–961. [Google Scholar] [CrossRef]
- Chen, H.; Lin, Y.; Wang, Y. Comparative Effects of Cellulose and Soluble Fibers (Pectin, Konjac Glucomannan, Inulin) on Fecal Water Toxicity Toward Caco-2 Cells, Fecal Bacteria Enzymes, Bile Acid, and Short-Chain Fatty Acids. J. Agric. Food Chem. 2010, 58, 10277–10281. [Google Scholar] [CrossRef] [PubMed]
- Augimeri, R.V.; Varley, A.J.; Strap, J.L. Establishing a Role for Bacterial Cellulose in Environmental Interactions: Lessons Learned from Diverse Biofilm-Producing Proteobacteria. Front. Microbiol. 2015, 6, 1282. [Google Scholar] [CrossRef] [PubMed]
- Premalatha, N.; Gopal, N.O.; Jose, P.A.; Anandham, R.; Kwon, S. Optimization of Cellulase Production by Enhydrobacter Sp. ACCA2 and its Application in Biomass Saccharification. Front. Microbiol. 2015, 6, 1046. [Google Scholar] [CrossRef] [PubMed]
- Kantor, E.D.; Lampe, J.W.; Navarro, S.L.; Song, X.; Milne, G.L.; White, E. Associations between Glucosamine and Chondroitin Supplement use and Biomarkers of Systemic Inflammation. J. Altern. Complement. Med. 2014, 20, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Kantor, E.D.; Newton, C.C.; Giovannucci, E.L.; McCullough, M.L.; Campbell, P.T.; Jacobs, E.J. Glucosamine use and Risk of Colorectal Cancer: Results from the Cancer Prevention Study II Nutrition Cohort. Cancer Causes Control 2018, 29, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Koliarakis, I.; Athanasakis, E.; Sgantzos, M.; Mariolis-Sapsakos, T.; Xynos, E.; Chrysos, E.; Souglakos, J.; Tsiaoussis, J. Intestinal Microbiota in Colorectal Cancer Surgery. Cancers 2020, 12, 3011. [Google Scholar] [CrossRef]

| Patient Characteristics | Oral Iron (n = 16) | Intravenous Iron (n = 24) |
|---|---|---|
| Age | 74.9 (7.4) | 74.9 (9.5) |
| Male | 8 (50%) | 16 (67%) |
| Female | 8 (50%) | 8 (33%) |
| Height, m | 1.66 (0.1) | 1.70 (0.09) |
| Weight, kg | 72.7 (17.4) | 79.3 (17.3) |
| Inclusion Hb, g/L | 100.3 (10.6) | 98.8 (13.1) |
| Recruitment ferritin, μg/L * | 24.5 (11.1–37.3) | 23 (10–48.3) |
| Recruitment transferrin saturation, % * | 2.9 (2.5–3.3) | 2.8 (2.4–3.3) |
| Duration of iron treatment, days * | 26.5 (15–43) | 23.5 (15–40.5) |
| Tumour Features | ||
| Tumour size, mm | 48.5 (25.3) | 42.4 (23.2) |
| Tumour Stage | ||
| T≤2 | 1 (6.25%) | 3 (12.5%) |
| T3 | 10 (62.5%) | 18 (75%) |
| T4 | 5 (31.25%) | 3 (12.5%) |
| Tumour Location | ||
| Right colon | 14 (87.5%) | 17 (71%) |
| Left colon | 2 (12.5%) | 7 (29%) |
| Preoperative Risk Assessment | ||
| ASA fitness status classification | ||
| I–II | 12 (75%) | 10 (42%) |
| III–IV | 4 (25%) | 14 (58%) |
| CR-POSSUM mortality score, % * | 3.6 (2.8–9.3) | 3.5 (2.6–8.6) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phipps, O.; Al-Hassi, H.O.; Quraishi, M.N.; Dickson, E.A.; Segal, J.; Steed, H.; Kumar, A.; Acheson, A.G.; Beggs, A.D.; Brookes, M.J. Oral and Intravenous Iron Therapy Differentially Alter the On- and Off-Tumor Microbiota in Anemic Colorectal Cancer Patients. Cancers 2021, 13, 1341. https://doi.org/10.3390/cancers13061341
Phipps O, Al-Hassi HO, Quraishi MN, Dickson EA, Segal J, Steed H, Kumar A, Acheson AG, Beggs AD, Brookes MJ. Oral and Intravenous Iron Therapy Differentially Alter the On- and Off-Tumor Microbiota in Anemic Colorectal Cancer Patients. Cancers. 2021; 13(6):1341. https://doi.org/10.3390/cancers13061341
Chicago/Turabian StylePhipps, Oliver, Hafid O. Al-Hassi, Mohammed N. Quraishi, Edward A. Dickson, Jonathan Segal, Helen Steed, Aditi Kumar, Austin G. Acheson, Andrew D. Beggs, and Matthew J. Brookes. 2021. "Oral and Intravenous Iron Therapy Differentially Alter the On- and Off-Tumor Microbiota in Anemic Colorectal Cancer Patients" Cancers 13, no. 6: 1341. https://doi.org/10.3390/cancers13061341
APA StylePhipps, O., Al-Hassi, H. O., Quraishi, M. N., Dickson, E. A., Segal, J., Steed, H., Kumar, A., Acheson, A. G., Beggs, A. D., & Brookes, M. J. (2021). Oral and Intravenous Iron Therapy Differentially Alter the On- and Off-Tumor Microbiota in Anemic Colorectal Cancer Patients. Cancers, 13(6), 1341. https://doi.org/10.3390/cancers13061341







