Modification of Homologous Recombination Deficiency Score Threshold and Association with Long-Term Survival in Epithelial Ovarian Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
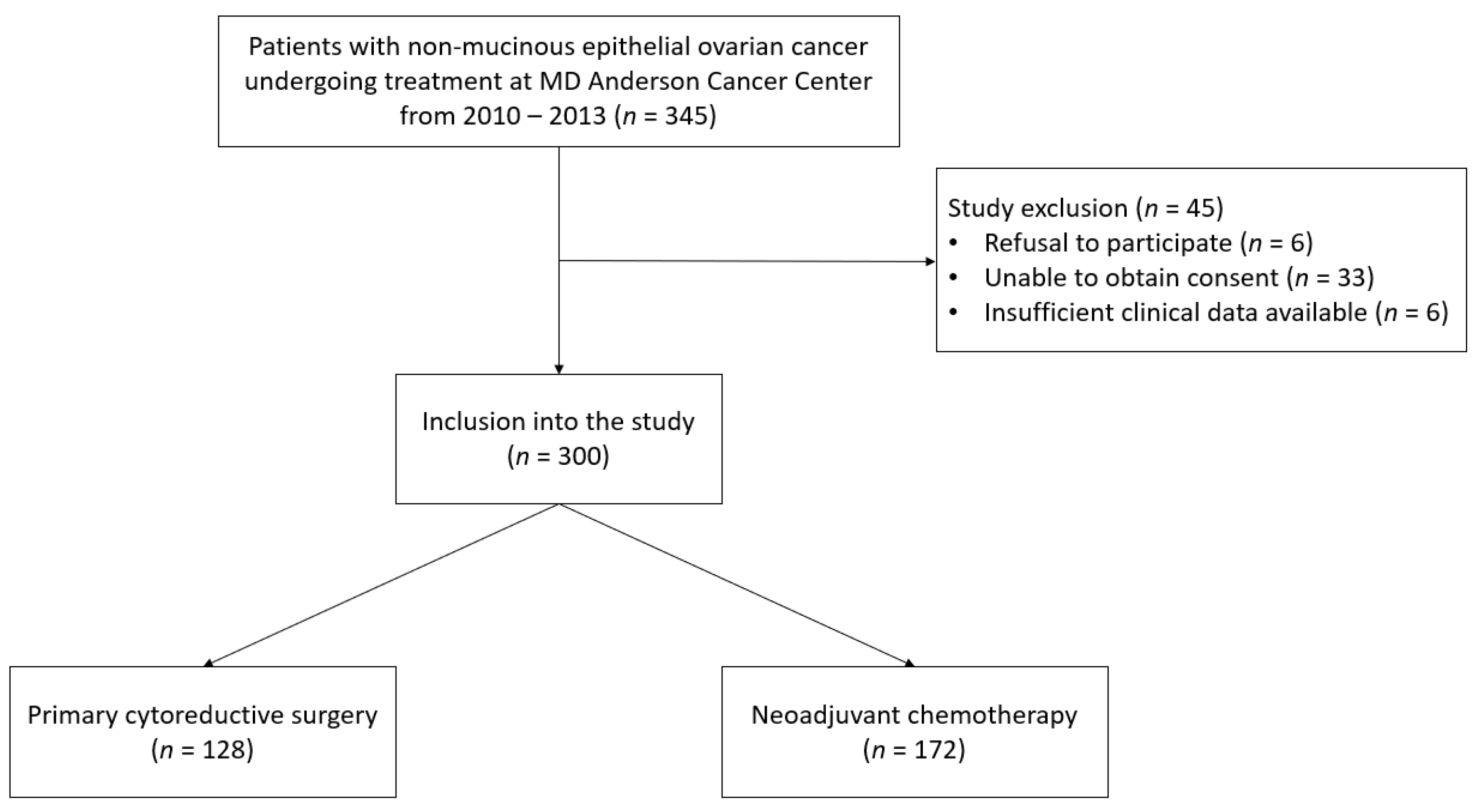
2.1. Patient Population
2.2. Blood Sample Collection and Molecular Testing
2.3. Tumor Testing
2.4. Clinical Data Collection
2.5. Statistical Analysis
3. Results
3.1. Patient Population
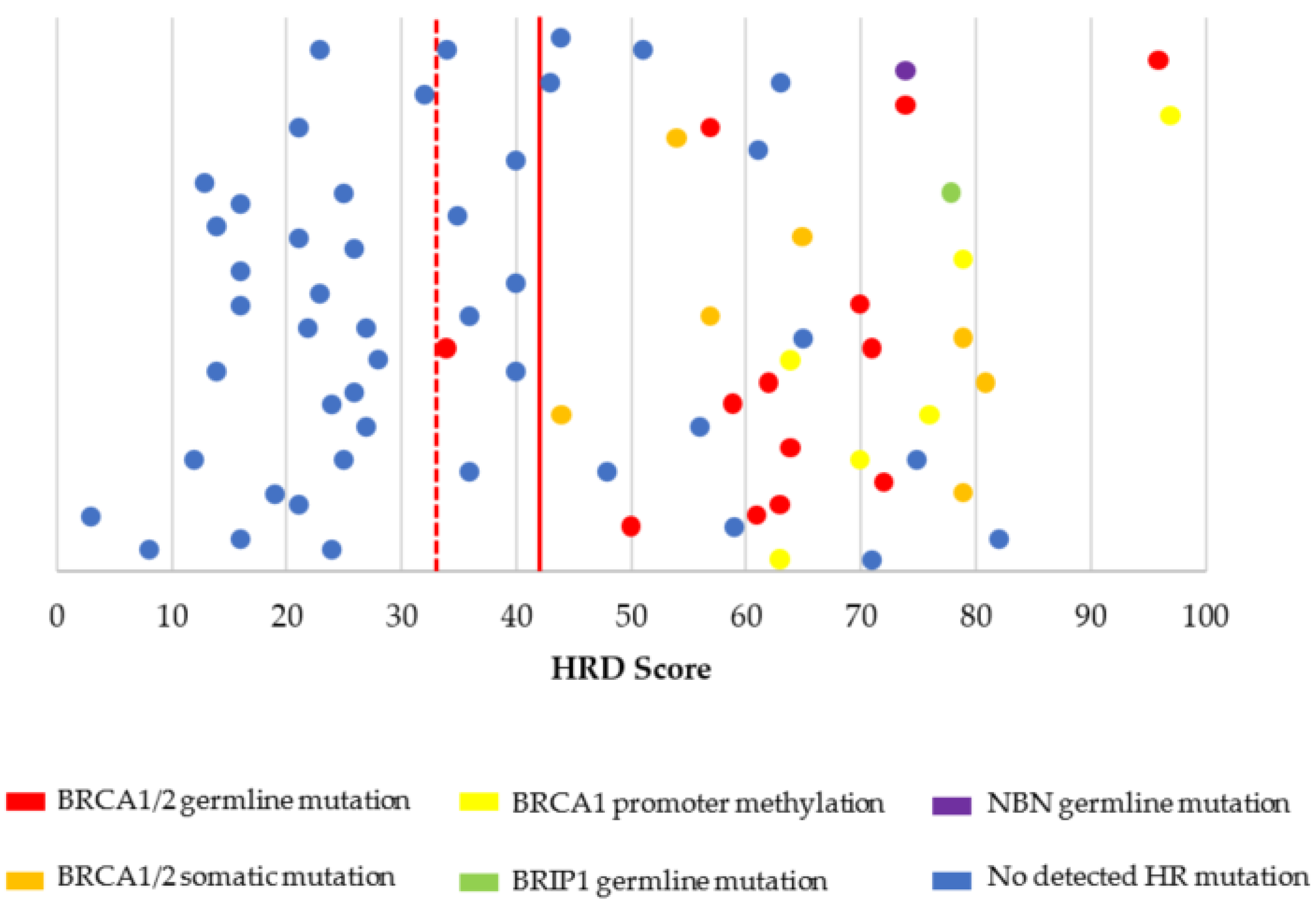
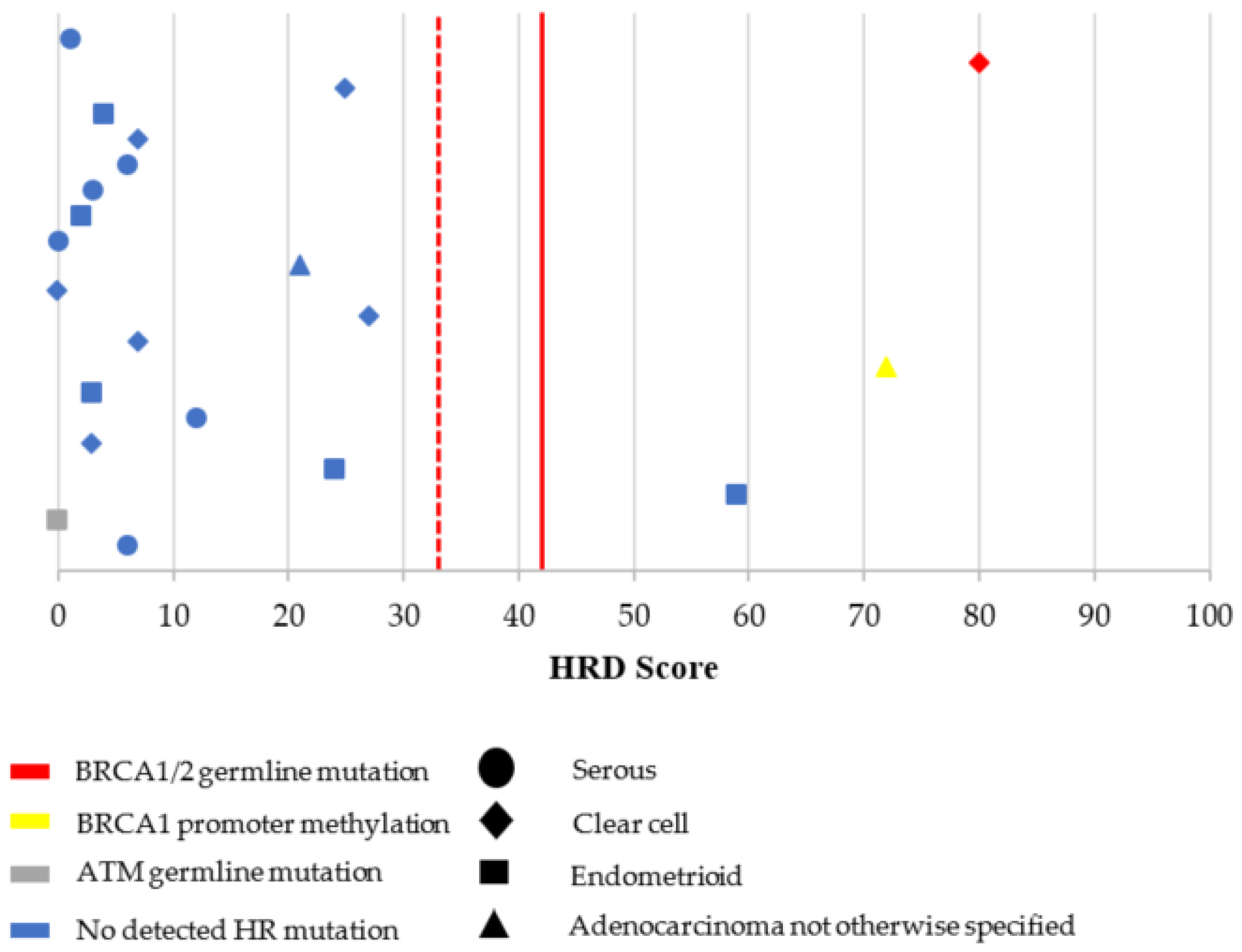
3.2. Characterization of HRD, MSI, and TMB
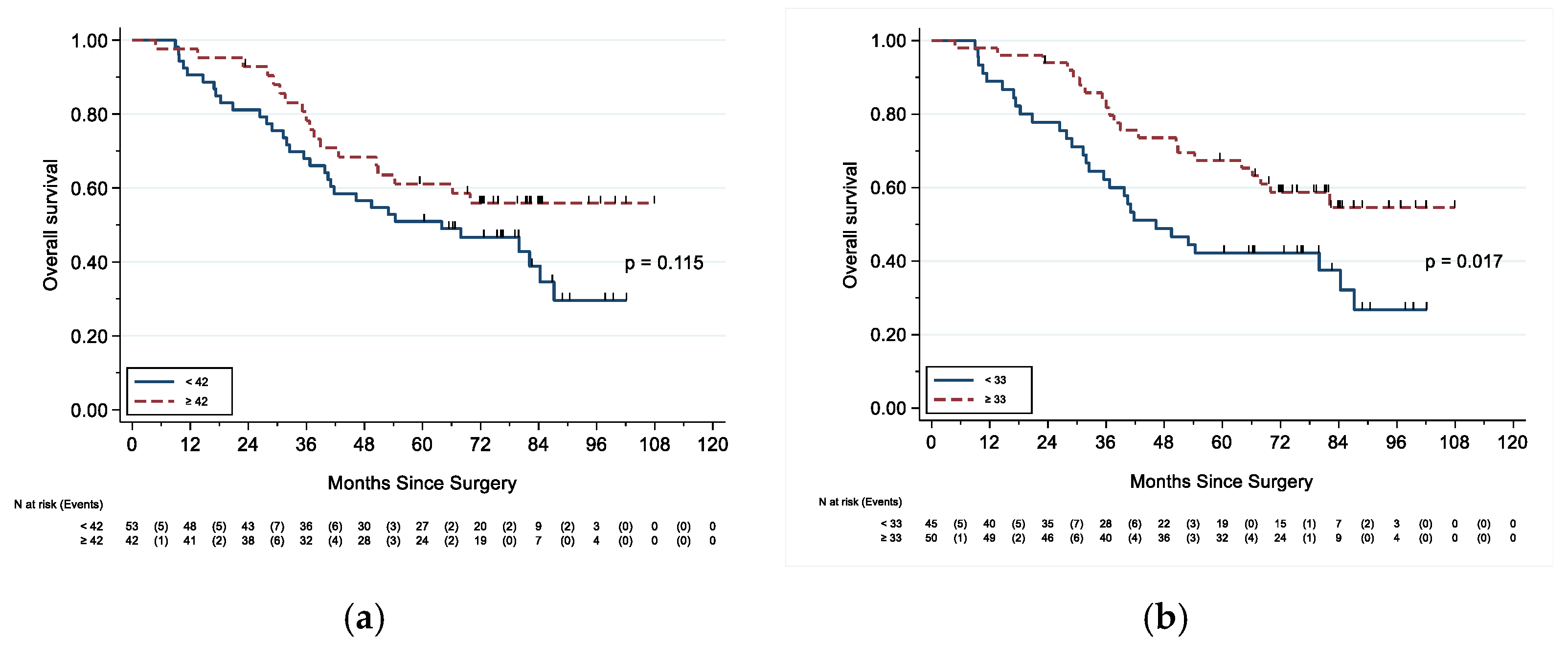
3.3. Survival
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Tumor Testing | n (%) |
|---|---|
| MSI status (n = 110) | |
| MSS | 106 (96.4%) |
| MSI | 1 (0.9%) |
| Failed testing | 3 (2.7%) |
| TMB (n = 94) | |
| Median (range) | 1.82 (0–30.4) |
| Low | 93 (98.9%) |
| High | 1 (1.1%) |
| Characteristic | N | Hazard Ratio (95% CI) | p | N | Hazard Ratio (95% CI) | p |
|---|---|---|---|---|---|---|
| Age | ||||||
| <65 ≥65 | 76 52 | Ref 2.57 (1.54–4.27) | <0.001 | 50 42 | Ref 2.88 (1.64–5.07) | <0.001 |
| Stage | ||||||
| I/II | 25 | Ref | 14 | Ref | ||
| III/IV | 95 | 9.58 (2.33–39.37) | 0.002 | 73 | 6.44 (1.56–26.58) | 0.010 |
| Residual disease after PCS | ||||||
| Optimal | 103 | Ref | 74 | Ref | ||
| Suboptimal (>1 cm) | 9 | 4.16 (1.93–8.95) | <0.001 | 7 | 3.57 (1.47–8.65) | 0.005 |
| Any BRCA1 mutation | ||||||
| No | 69 | Ref | 44 | Ref | ||
| Yes | 21 | 0.56 (0.25–1.26) | 0.160 | 19 | 0.35 (0.15–0.85) | 0.020 |
| Any BRCA2 mutation | ||||||
| No | 108 | Ref | 74 | Ref | ||
| Yes | 10 | 0.15 (0.02–1.08) | 0.059 | 10 | 0.11 (0.01–0.79) | 0.029 |
| Any BRCA1/2 mutation | ||||||
| No | 62 | Ref | 37 | Ref | ||
| Yes | 31 | 0.36 (0.17–0.77) | 0.008 | 29 | 0.19 (0.08–0.44) | <0.001 |
| gNon-BRCA HR mutation | ||||||
| Negative | 80 | Ref | 53 | Ref | ||
| Positive | 5 | 1.97 (0.70–5.59) | 0.200 | 4 | 1.95 (0.68–5.58) | 0.214 |
| BRCA1 promoter methylation | ||||||
| <15 | 86 | Ref | 60 | Ref | ||
| ≥15 | 7 | 1.14 (0.41–3.17) | 0.804 | 6 | 0.68 (0.21–2.20) | 0.516 |
| Overall (n = 128) | HGS (n = 92) | |||||
|---|---|---|---|---|---|---|
| Characteristic | N | Hazard Ratio (95% CI) | p | N | Hazard Ratio (95% CI) | p |
| Age | ||||||
| <65 ≥65 | 75 52 | Ref 1.87 (1.22–2.86) | 0.004 | 49 42 | Ref 1.82 (1.14–2.91) | 0.012 |
| Stage | ||||||
| I/II | 25 | Ref | 14 | Ref | ||
| III/IV | 94 | 2.98 (1.53–5.81) | 0.001 | 72 | 2.17 (1.04–4.29) | 0.038 |
| Residual disease after PCS | ||||||
| Optimal | 102 | Ref | 73 | Ref | ||
| Suboptimal (>1 cm) | 9 | 4.15 (1.97–8.72) | <0.001 | 7 | 3.10 (1.35–7.08) | 0.007 |
| Any BRCA1 mutation | ||||||
| No | 69 | Ref | 44 | Ref | ||
| Yes | 21 | 0.70 (0.38–1.29) | 0.254 | 19 | 0.47 (0.24–0.90) | 0.023 |
| Any BRCA2 mutation | ||||||
| No | 108 | Ref | 74 | Ref | ||
| Yes | 10 | 0.51 (0.20–1.26) | 0.143 | 10 | 0.35 (0.14–0.87) | 0.025 |
| Any BRCA1/2 mutation | ||||||
| No | 62 | Ref | 37 | Ref | ||
| Yes | 31 | 0.56 (0.33–0.98) | 0.041 | 29 | 0.30 (0.16–0.54) | <0.001 |
| gNon-BRCA HR mutation | ||||||
| Negative | 80 | Ref | 53 | Ref | ||
| Positive | 5 | 1.85 (0.67–5.11) | 0.237 | 4 | 2.99 (1.06–8.47) | 0.039 |
| BRCA1 promoter methylation | ||||||
| <15 | 85 | Ref | 59 | Ref | ||
| ≥15 | 7 | 0.78 (0.28–2.15) | 0.633 | 6 | 0.49 (0.15–1.57) | 0.229 |
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Matulonis, U.A. Management of newly diagnosed or recurrent ovarian cancer. Clin. Adv. Hematol. Oncol. 2018, 16, 426–437. [Google Scholar]
- McGuire, W.P.; Hoskins, W.J.; Brady, M.F.; Kucera, P.R.; Partridge, E.E.; Look, K.Y.; Clarke-Pearson, D.L.; Davidson, M. Cyclophosphamide and cisplatin compared with paclitaxel and cisplatin in patients with stage III and stage IV ovarian cancer. N. Engl. J. Med. 1996, 334, 1–6. [Google Scholar] [CrossRef]
- Ozols, R.F.; Bundy, B.N.; Greer, B.E.; Fowler, J.M.; Clarke-Pearson, D.; Burger, R.A.; Mannel, R.S.; DeGeest, K.; Hartenbach, E.M.; Baergen, R. Phase III trial of carboplatin and paclitaxel compared with cisplatin and paclitaxel in patients with optimally resected stage III ovarian cancer: A Gynecologic Oncology Group study. J. Clin. Oncol. 2003, 21, 3194–3200. [Google Scholar] [CrossRef]
- Ledermann, J.A.; Drew, Y.; Kristeleit, R.S. Homologous recombination deficiency and ovarian cancer. Eur. J. Cancer 2016, 60, 49–58. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef]
- Hoeijmakers, J.H. Genome maintenance mechanisms for preventing cancer. Nature 2001, 411, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Trenner, A.; Sartori, A.A. Harnessing DNA Double-Strand Break Repair for Cancer Treatment. Front. Oncol. 2019, 9, 1388. [Google Scholar] [CrossRef]
- Wang, Y.; Cortez, D.; Yazdi, P.; Neff, N.; Elledge, S.J.; Qin, J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000, 14, 927–939. [Google Scholar]
- Yuan, S.S.; Lee, S.Y.; Chen, G.; Song, M.; Tomlinson, G.E.; Lee, E.Y. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 1999, 59, 3547–3551. [Google Scholar] [PubMed]
- Kelland, L. The resurgence of platinum-based cancer chemotherapy. Nat. Rev. Cancer 2007, 7, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Konstantinopoulos, P.A.; Ceccaldi, R.; Shapiro, G.I.; D’Andrea, A.D. Homologous Recombination Deficiency: Exploiting the Fundamental Vulnerability of Ovarian Cancer. Cancer Discov. 2015, 5, 1137–1154. [Google Scholar] [CrossRef] [PubMed]
- Farmer, H.; McCabe, N.; Lord, C.J.; Tutt, A.N.; Johnson, D.A.; Richardson, T.B.; Santarosa, M.; Dillon, K.J.; Hickson, I.; Knights, C.; et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005, 434, 917–921. [Google Scholar] [CrossRef]
- Helleday, T. The underlying mechanism for the PARP and BRCA synthetic lethality: Clearing up the misunderstandings. Mol. Oncol. 2011, 5, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Hoppe, M.M.; Sundar, R.; Tan, D.S.P.; Jeyasekharan, A.D. Biomarkers for Homologous Recombination Deficiency in Cancer. J. Natl. Cancer Inst. 2018, 110, 704–713. [Google Scholar] [CrossRef] [PubMed]
- Coleman, R.L.; Oza, A.M.; Lorusso, D.; Aghajanian, C.; Oaknin, A.; Dean, A.; Colombo, N.; Weberpals, J.I.; Clamp, A.; Scambia, G.; et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017, 390, 1949–1961. [Google Scholar] [CrossRef]
- Pujade-Lauraine, E.; Ledermann, J.A.; Selle, F.; Gebski, V.; Penson, R.T.; Oza, A.M.; Korach, J.; Huzarski, T.; Poveda, A.; Pignata, S.; et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): A double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2017, 18, 1274–1284. [Google Scholar] [CrossRef]
- Ledermann, J.; Harter, P.; Gourley, C.; Friedlander, M.; Vergote, I.; Rustin, G.; Scott, C.L.; Meier, W.; Shapira-Frommer, R.; Safra, T.; et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: A preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 2014, 15, 852–861. [Google Scholar] [CrossRef]
- Mirza, M.R.; Monk, B.J.; Herrstedt, J.; Oza, A.M.; Mahner, S.; Redondo, A.; Fabbro, M.; Ledermann, J.A.; Lorusso, D.; Vergote, I.; et al. Niraparib Maintenance Therapy in Platinum-Sensitive, Recurrent Ovarian Cancer. N. Engl. J. Med. 2016, 375, 2154–2164. [Google Scholar] [CrossRef]
- Norquist, B.M.; Brady, M.F.; Harrell, M.I.; Walsh, T.; Lee, M.K.; Gulsuner, S.; Bernards, S.S.; Casadei, S.; Burger, R.A.; Tewari, K.S.; et al. Mutations in Homologous Recombination Genes and Outcomes in Ovarian Carcinoma Patients in GOG 218: An NRG Oncology/Gynecologic Oncology Group Study. Clin. Cancer Res. 2018, 24, 777–783. [Google Scholar] [CrossRef]
- Abkevich, V.; Timms, K.M.; Hennessy, B.T.; Potter, J.; Carey, M.S.; Meyer, L.A.; Smith-McCune, K.; Broaddus, R.; Lu, K.H.; Chen, J.; et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br. J. Cancer 2012, 107, 1776–1782. [Google Scholar] [CrossRef] [PubMed]
- Birkbak, N.J.; Wang, Z.C.; Kim, J.Y.; Eklund, A.C.; Li, Q.; Tian, R.; Bowman-Colin, C.; Li, Y.; Greene-Colozzi, A.; Iglehart, J.D.; et al. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov. 2012, 2, 366–375. [Google Scholar] [CrossRef] [PubMed]
- Popova, T.; Manie, E.; Rieunier, G.; Caux-Moncoutier, V.; Tirapo, C.; Dubois, T.; Delattre, O.; Sigal-Zafrani, B.; Bollet, M.; Longy, M.; et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res. 2012, 72, 5454–5462. [Google Scholar] [CrossRef]
- Timms, K.M.; Abkevich, V.; Hughes, E.; Neff, C.; Reid, J.; Morris, B.; Kalva, S.; Potter, J.; Tran, T.V.; Chen, J.; et al. Association of BRCA1/2 defects with genomic scores predictive of DNA damage repair deficiency among breast cancer subtypes. Breast Cancer Res. 2014, 16, 475. [Google Scholar] [CrossRef]
- Telli, M.L.; Timms, K.M.; Reid, J.; Hennessy, B.; Mills, G.B.; Jensen, K.C.; Szallasi, Z.; Barry, W.T.; Winer, E.P.; Tung, N.M.; et al. Homologous Recombination Deficiency (HRD) Score Predicts Response to Platinum-Containing Neoadjuvant Chemotherapy in Patients with Triple-Negative Breast Cancer. Clin. Cancer Res. 2016, 22, 3764–3773. [Google Scholar] [CrossRef] [PubMed]
- Ray-Coquard, I.; Pautier, P.; Pignata, S.; Pérol, D.; González-Martín, A.; Berger, R.; Fujiwara, K.; Vergote, I.; Colombo, N.; Mäenpää, J.; et al. Olaparib plus Bevacizumab as First-Line Maintenance in Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2416–2428. [Google Scholar] [CrossRef]
- Moore, K.N.; Secord, A.A.; Geller, M.A.; Miller, D.S.; Cloven, N.; Fleming, G.F.; Wahner Hendrickson, A.E.; Azodi, M.; DiSilvestro, P.; Oza, A.M.; et al. Niraparib monotherapy for late-line treatment of ovarian cancer (QUADRA): A multicentre, open-label, single-arm, phase 2 trial. Lancet Oncol. 2019, 20, 636–648. [Google Scholar] [CrossRef]
- Le, D.T.; Durham, J.N.; Smith, K.N.; Wang, H.; Bartlett, B.R.; Aulakh, L.K.; Lu, S.; Kemberling, H.; Wilt, C.; Luber, B.S.; et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017, 357, 409–413. [Google Scholar] [CrossRef]
- Pećina-Šlaus, N.; Kafka, A.; Salamon, I.; Bukovac, A. Mismatch Repair Pathway, Genome Stability and Cancer. Front. Mol. Biosci. 2020, 7, 122. [Google Scholar] [CrossRef]
- Marabelle, A.; Le, D.T.; Ascierto, P.A.; Di Giacomo, A.M.; De Jesus-Acosta, A.; Delord, J.-P.; Geva, R.; Gottfried, M.; Penel, N.; Hansen, A.R.; et al. Efficacy of Pembrolizumab in Patients With Noncolorectal High Microsatellite Instability/Mismatch Repair–Deficient Cancer: Results From the Phase II KEYNOTE-158 Study. J. Clin. Oncol. 2019, 38, 1–10. [Google Scholar] [CrossRef]
- Matulonis, U.A.; Shapira-Frommer, R.; Santin, A.D.; Lisyanskaya, A.S.; Pignata, S.; Vergote, I.; Raspagliesi, F.; Sonke, G.S.; Birrer, M.; Provencher, D.M.; et al. Antitumor activity and safety of pembrolizumab in patients with advanced recurrent ovarian cancer: Results from the phase II KEYNOTE-100 study. Ann. Oncol. 2019, 30, 1080–1087. [Google Scholar] [CrossRef]
- Goodman, A.M.; Kato, S.; Bazhenova, L.; Patel, S.P.; Frampton, G.M.; Miller, V.; Stephens, P.J.; Daniels, G.A.; Kurzrock, R. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol. Cancer Ther. 2017, 16, 2598–2608. [Google Scholar] [CrossRef]
- Judkins, T.; Leclair, B.; Bowles, K.; Gutin, N.; Trost, J.; McCulloch, J.; Bhatnagar, S.; Murray, A.; Craft, J.; Wardell, B.; et al. Development and analytical validation of a 25-gene next generation sequencing panel that includes the BRCA1 and BRCA2 genes to assess hereditary cancer risk. BMC Cancer 2015, 15, 215. [Google Scholar] [CrossRef]
- Hennessy, B.T.; Timms, K.M.; Carey, M.S.; Gutin, A.; Meyer, L.A.; Flake, D.D., 2nd; Abkevich, V.; Potter, J.; Pruss, D.; Glenn, P.; et al. Somatic mutations in BRCA1 and BRCA2 could expand the number of patients that benefit from poly (ADP ribose) polymerase inhibitors in ovarian cancer. J. Clin. Oncol. 2010, 28, 3570–3576. [Google Scholar] [CrossRef] [PubMed]
- Hodgson, D.R.; Dougherty, B.A.; Lai, Z.; Fielding, A.; Grinsted, L.; Spencer, S.; O’Connor, M.J.; Ho, T.W.; Robertson, J.D.; Lanchbury, J.S.; et al. Candidate biomarkers of PARP inhibitor sensitivity in ovarian cancer beyond the BRCA genes. Br. J. Cancer 2018, 119, 1401–1409. [Google Scholar] [CrossRef] [PubMed]
- Coleman, R.L.; Fleming, G.F.; Brady, M.F.; Swisher, E.M.; Steffensen, K.D.; Friedlander, M.; Okamoto, A.; Moore, K.N.; Efrat Ben-Baruch, N.; Werner, T.L.; et al. Veliparib with First-Line Chemotherapy and as Maintenance Therapy in Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2403–2415. [Google Scholar] [CrossRef]
- Telli, M.L.; Metzger, O.; Timms, K.; Evans, B.; Vogel, D.; Wei, H.; Jones, J.T.; Wenstrup, R.J.; McKee, M.D.; Sullivan, D.M.; et al. Evaluation of homologous recombination deficiency (HRD) status with pathological response to carboplatin +/- veliparib in BrighTNess, a randomized phase 3 study in early stage TNBC. J. Clin. Oncol. 2018, 36, 519. [Google Scholar] [CrossRef]
- Stronach, E.A.; Paul, J.; Timms, K.M.; Hughes, E.; Brown, K.; Neff, C.; Perry, M.; Gutin, A.; El-Bahrawy, M.; Steel, J.H.; et al. Biomarker Assessment of HR Deficiency, Tumor BRCA1/2 Mutations, and CCNE1 Copy Number in Ovarian Cancer: Associations with Clinical Outcome Following Platinum Monotherapy. Mol. Cancer Res. 2018, 16, 1103–1111. [Google Scholar] [CrossRef]
- Timms, K.; Zharkikh, A.; Perry, M.; Birkbak, N.; Szallasi, Z.; Gutin, A.; Richardson, A.; Lanchbury, J. Comparison between whole exome sequencing (WES) and single nucleotide polymorphism (SNP)-based tumor mutation burden analysis. J. Clin. Oncol. 2019, 37, 2634. [Google Scholar] [CrossRef]
- Siedel, J.H.; Ring, K.L.; Hu, W.; Dood, R.L.; Wang, Y.; Baggerly, K.; Gallagher, S.; Tshiaba, P.; Neff, C.; Timms, K.M.; et al. Clinical Significance of Homologous Recombination Deficiency Score Testing in Endometrial Cancer. Gynecol. Oncol. 2021. (manuscript in-press). [Google Scholar] [CrossRef] [PubMed]
- Harris, P.A.; Taylor, R.; Thielke, R.; Payne, J.; Gonzalez, N.; Conde, J.G. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform. 2009, 42, 377–381. [Google Scholar] [CrossRef]
- Balmus, G.; Pilger, D.; Coates, J.; Demir, M.; Sczaniecka-Clift, M.; Barros, A.C.; Woods, M.; Fu, B.; Yang, F.; Chen, E.; et al. ATM orchestrates the DNA-damage response to counter toxic non-homologous end-joining at broken replication forks. Nat. Commun. 2019, 10, 87. [Google Scholar] [CrossRef]
- Bakr, A.; Oing, C.; Köcher, S.; Borgmann, K.; Dornreiter, I.; Petersen, C.; Dikomey, E.; Mansour, W.Y. Involvement of ATM in homologous recombination after end resection and RAD51 nucleofilament formation. Nucleic Acids Res. 2015, 43, 3154–3166. [Google Scholar] [CrossRef] [PubMed]
- Bonneville, R.; Krook, M.A.; Kautto, E.A.; Miya, J.; Wing, M.R.; Chen, H.Z.; Reeser, J.W.; Yu, L.; Roychowdhury, S. Landscape of Microsatellite Instability Across 39 Cancer Types. JCO Precis. Oncol. 2017, 1, 1–15. [Google Scholar] [CrossRef]
- Deshpande, M.; Romanski, P.A.; Rosenwaks, Z.; Gerhardt, J. Gynecological Cancers Caused by Deficient Mismatch Repair and Microsatellite Instability. Cancers 2020, 12. [Google Scholar] [CrossRef]
- Pal, T.; Permuth-Wey, J.; Kumar, A.; Sellers, T.A. Systematic review and meta-analysis of ovarian cancers: Estimation of microsatellite-high frequency and characterization of mismatch repair deficient tumor histology. Clin. Cancer Res. 2008, 14, 6847–6854. [Google Scholar] [CrossRef]
- Morse, C.B.; Elvin, J.A.; Gay, L.M.; Liao, J.B. Elevated tumor mutational burden and prolonged clinical response to anti-PD-L1 antibody in platinum-resistant recurrent ovarian cancer. Gynecol. Oncol. Rep. 2017, 21, 78–80. [Google Scholar] [CrossRef] [PubMed]
- Strickland, K.C.; Howitt, B.E.; Shukla, S.A.; Rodig, S.; Ritterhouse, L.L.; Liu, J.F.; Garber, J.E.; Chowdhury, D.; Wu, C.J.; D′Andrea, A.D.; et al. Association and prognostic significance of BRCA1/2-mutation status with neoantigen load, number of tumor-infiltrating lymphocytes and expression of PD-1/PD-L1 in high grade serous ovarian cancer. Oncotarget 2016, 7, 13587–13598. [Google Scholar] [CrossRef]
- Segev, Y.; Zhang, S.; Akbari, M.R.; Sun, P.; Sellers, T.A.; McLaughlin, J.; Risch, H.A.; Rosen, B.; Shaw, P.; Schildkraut, J.; et al. Survival in women with ovarian cancer with and without microsatellite instability. Eur. J. Gynaecol. Oncol. 2015, 36, 681–684. [Google Scholar] [PubMed]
- Bi, F.; Chen, Y.; Yang, Q. Significance of tumor mutation burden combined with immune infiltrates in the progression and prognosis of ovarian cancer. Cancer Cell Int. 2020, 20, 373. [Google Scholar] [CrossRef]
- Chan, J.K.; Loizzi, V.; Lin, Y.G.; Osann, K.; Brewster, W.R.; DiSaia, P.J. Stages III and IV invasive epithelial ovarian carcinoma in younger versus older women: What prognostic factors are important? Obstet. Gynecol. 2003, 102, 156–161. [Google Scholar] [CrossRef]
- Bristow, R.E.; Tomacruz, R.S.; Armstrong, D.K.; Trimble, E.L.; Montz, F.J. Survival effect of maximal cytoreductive surgery for advanced ovarian carcinoma during the platinum era: A meta-analysis. J. Clin. Oncol. 2002, 20, 1248–1259. [Google Scholar] [CrossRef]
- Cass, I.; Baldwin, R.L.; Varkey, T.; Moslehi, R.; Narod, S.A.; Karlan, B.Y. Improved survival in women with BRCA-associated ovarian carcinoma. Cancer 2003, 97, 2187–2195. [Google Scholar] [CrossRef]
- Long, K.C.; Kauff, N.D. Hereditary ovarian cancer: Recent molecular insights and their impact on screening strategies. Curr. Opin. Oncol. 2011, 23, 526–530. [Google Scholar] [CrossRef]
- Moschetta, M.; George, A.; Kaye, S.B.; Banerjee, S. BRCA somatic mutations and epigenetic BRCA modifications in serous ovarian cancer. Ann. Oncol. 2016, 27, 1449–1455. [Google Scholar] [CrossRef]
- González-Martín, A.; Pothuri, B.; Vergote, I.; DePont Christensen, R.; Graybill, W.; Mirza, M.R.; McCormick, C.; Lorusso, D.; Hoskins, P.; Freyer, G.; et al. Niraparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2391–2402. [Google Scholar] [CrossRef]
- Norquist, B.M.; Harrell, M.I.; Brady, M.F.; Walsh, T.; Lee, M.K.; Gulsuner, S.; Bernards, S.S.; Casadei, S.; Yi, Q.; Burger, R.A.; et al. Inherited Mutations in Women With Ovarian Carcinoma. JAMA Oncol. 2016, 2, 482–490. [Google Scholar] [CrossRef]
- Yates, M.S.; Timms, K.; Daniels, M.S.; Oakley, H.D.; Munsell, M.F.; Lanchbury, J.S.; Lu, K.H. Evaluation of BRCA1/2 and homologous recombination defects in ovarian cancer and impact on clinical outcomes. J. Clin. Oncol. 2017, 35, 5511. [Google Scholar] [CrossRef]
| Clinicopathologic Characteristic | PCS (n = 128) | NACT (n = 172) | p | ||
|---|---|---|---|---|---|
| N | % | N | % | ||
| Age | 0.625 | ||||
| Median (range) | 61 (24–83) | 62 (25–95) | |||
| Race/ethnicity | 0.500 | ||||
| White | 97 | 75.8 | 123 | 72.4 | |
| African American/Black | 5 | 3.9 | 13 | 7.6 | |
| Asian | 9 | 7.0 | 15 | 8.8 | |
| Hispanic | 17 | 13.3 | 19 | 11.2 | |
| Unknown | 0 | 0 | 2 | N/A | |
| Disease site | 0.022 | ||||
| Fallopian tube | 10 | 7.9 | 6 | 3.5 | |
| Ovary | 95 | 75.4 | 117 | 68 | |
| Peritoneum | 21 | 16.7 | 49 | 28.5 | |
| Unknown | 2 | N/A | 0 | 0 | |
| Stage | <0.001 | ||||
| In Situ | 1 | 0.8 | 0 | 0.0 | |
| I | 11 | 9.2 | 0 | 0.0 | |
| II | 13 | 10.8 | 1 | 0.7 | |
| III | 82 | 68.3 | 72 | 53.3 | |
| IV | 13 | 10.8 | 60 | 44.4 | |
| Advanced | 0 | 0.0 | 2 | 1.5 | |
| Unknown/unstaged | 8 | N/A | 37 | N/A | |
| Histology | <0.001 | ||||
| Serous | |||||
| High grade | 92 | 71.9 | 155 | 90.1 | |
| Low grade | 16 | 12.5 | 12 | 6.9 | |
| Endometrioid | |||||
| High grade | 8 | 6.2 | 1 | 0.6 | |
| Low grade | 1 | 0.8 | 0 | 0.0 | |
| Clear cell | 9 | 7.0 | 2 | 1.2 | |
| Adenocarcinoma NOS | 2 | 1.6 | 2 | 1.2 | |
| Cytoreductive surgery | 0.356 | ||||
| Optimal | 103 | 92.0 | 139 | 88.5 | |
| R0 | 70 | 68.0 | 90 | 57.3 | |
| ≤1 cm | 18 | 16.1 | 29 | 18.5 | |
| Not specified | 15 | 13.4 | 20 | 12.7 | |
| Suboptimal (>1 cm) | 9 | 8.0 | 18 | 11.5 | |
| Unknown | 16 | N/A | 15 | N/A | |
| Follow-up (months) | 0.870 | ||||
| Alive at time of analysis | 68 | 53.1% | 45 | 26.2% | |
| Median (range) | 79.1 (18.6–114.9) | 79.2 (9.3–105.4) | |||
| Germline Testing | n (%) |
|---|---|
| gBRCA1/2 status (n = 122) | |
| gBRCA1/2 negative | 105 (86.1%) |
| gBRCA1 mutation 1 | 10 (8.2%) |
| gBRCA2 mutation 1 | 7 (5.7%) |
| Other HR gene mutations (n = 85) | |
| BRIP1 | 2 (2.4%) |
| ATM | 2 (2.4%) |
| NBN | 1 (1.2%) |
| MMR gene mutations 2 (n = 85) | 0 (0%) |
| Other germline mutations (n = 85) | |
| MUTYH | 2 (2.4%) |
| Tumor testing | n (%) |
| sBRCA1/2 status (n = 68) | |
| sBRCA1/2 negative | 59 (86.7%) |
| sBRCA1 mutation | 5 (7.4%) |
| sBRCA2 mutation | 3 (4.4%) |
| BRCA1 promoter methylation (n = 93) | |
| ≥15% | 7 (7.5%) |
| <15% | 86 (92.5%) |
| HRD score (n = 95) | |
| ≥42 | 42 (44.2%) |
| ≥33 | 50 (52.6%) |
| Overall (n = 128) | HGS (n = 92) | |||||
|---|---|---|---|---|---|---|
| Characteristic | N | Hazard Ratio (95% CI) | p | N | Hazard Ratio (95% CI) | p |
| HRD score | ||||||
| <42 | 53 | Ref | 35 | Ref | ||
| ≥42 | 42 | 0.63 (0.35–1.12) | 0.118 | 39 | 0.48 (0.26–0.90) | 0.021 |
| HRD score | ||||||
| <33 | 45 | Ref | 27 | Ref | ||
| ≥33 | 50 | 0.51 (0.29–0.90) | 0.019 | 47 | 0.31 (0.17–0.58) | <0.001 |
| Any HRD (HRD score ≥42) 1 | ||||||
| No | 42 | Ref | 26 | Ref | ||
| Yes | 49 | 0.59 (0.33–1.05) | 0.074 | 45 | 0.45 (0.24–0.85) | 0.014 |
| Any HRD (HRD score ≥33) 2 | ||||||
| No | 36 | Ref | 20 | Ref | ||
| Yes | 56 | 0.49 (0.27–0.87) | 0.014 | 52 | 0.29 (0.15–0.55) | <0.001 |
| Overall (n = 128) | HGS (n = 92) | |||||
|---|---|---|---|---|---|---|
| Characteristic | N | Hazard Ratio (95% CI) | p | N | Hazard Ratio (95% CI) | p |
| HRD score | ||||||
| <42 | 53 | Ref | 35 | Ref | ||
| ≥42 | 41 | 0.75 (0.47–1.21) | 0.246 | 38 | 0.57 (0.34–0.95) | 0.033 |
| HRD score | ||||||
| <33 | 45 | Ref | 27 | Ref | ||
| ≥33 | 49 | 0.72 (0.45–1.16) | 0.177 | 46 | 0.46 (0.27–0.77) | 0.003 |
| Any HRD (HRD score ≥42) 1 | ||||||
| No | 42 | Ref | 26 | Ref | ||
| Yes | 48 | 0.71 (0.44–1.16) | 0.170 | 54 | 0.57 (0.33–0.98) | 0.044 |
| Any HRD (HRD score ≥33) 2 | ||||||
| No | 36 | Ref | 20 | Ref | ||
| Yes | 55 | 0.66 (0.41–1.07) | 0.092 | 51 | 0.43 (0.24–0.75) | 0.003 |
| Overall Survival | Progression-Free Survival | |||||
|---|---|---|---|---|---|---|
| Characteristic | N | Hazard Ratio (95% CI) | p | N | Hazard Ratio (95% CI) | p |
| Any BRCA1 mutation | 67 | 0.76 (0.31–1.89) | 0.557 | 67 | 0.63 (0.29–1.35) | 0.233 |
| Any BRCA2 mutation | 84 | 0.19 (0.03–1.41) | 0.105 | 84 | 0.51 (0.18–1.42) | 0.194 |
| Any BRCA1/2 mutation | 68 | 0.44 (0.19–1.02) | 0.054 | 68 | 0.47 (0.24–0.92) | 0.028 |
| gNon-BRCA HR mutation | 62 | 0.93 (0.26–3.34) | 0.908 | 62 | 1.15 (0.33–4.09) | 0.825 |
| HRD score ≥42 | 78 | 0.69 (0.36–1.30) | 0.250 | 77 | 0.72 (0.42–1.24) | 0.233 |
| HRD score ≥33 | 78 | 0.43 (0.23–0.81) | 0.009 | 77 | 0.62 (0.36–1.06) | 0.078 |
| BRCA1p methylation ≥15 | 70 | 1.41 (0.49–4.00) | 0.522 | 69 | 1.11 (0.38–3.23) | 0.841 |
| Any HRD (HRD score ≥42) 1 | 73 | 0.68 (0.34–1.33) | 0.260 | 71 | 0.70 (0.38–1.27) | 0.240 |
| Any HRD (HRD score ≥33) 2 | 72 | 0.45 (0.23–0.89) | 0.022 | 72 | 0.60 (0.33–1.08) | 0.087 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
How, J.A.; Jazaeri, A.A.; Fellman, B.; Daniels, M.S.; Penn, S.; Solimeno, C.; Yuan, Y.; Schmeler, K.; Lanchbury, J.S.; Timms, K.; et al. Modification of Homologous Recombination Deficiency Score Threshold and Association with Long-Term Survival in Epithelial Ovarian Cancer. Cancers 2021, 13, 946. https://doi.org/10.3390/cancers13050946
How JA, Jazaeri AA, Fellman B, Daniels MS, Penn S, Solimeno C, Yuan Y, Schmeler K, Lanchbury JS, Timms K, et al. Modification of Homologous Recombination Deficiency Score Threshold and Association with Long-Term Survival in Epithelial Ovarian Cancer. Cancers. 2021; 13(5):946. https://doi.org/10.3390/cancers13050946
Chicago/Turabian StyleHow, Jeffrey A., Amir A. Jazaeri, Bryan Fellman, Molly S. Daniels, Suzanna Penn, Cara Solimeno, Ying Yuan, Kathleen Schmeler, Jerry S. Lanchbury, Kirsten Timms, and et al. 2021. "Modification of Homologous Recombination Deficiency Score Threshold and Association with Long-Term Survival in Epithelial Ovarian Cancer" Cancers 13, no. 5: 946. https://doi.org/10.3390/cancers13050946
APA StyleHow, J. A., Jazaeri, A. A., Fellman, B., Daniels, M. S., Penn, S., Solimeno, C., Yuan, Y., Schmeler, K., Lanchbury, J. S., Timms, K., Lu, K. H., & Yates, M. S. (2021). Modification of Homologous Recombination Deficiency Score Threshold and Association with Long-Term Survival in Epithelial Ovarian Cancer. Cancers, 13(5), 946. https://doi.org/10.3390/cancers13050946






