Identification and Validation of ERK5 as a DNA Damage Modulating Drug Target in Glioblastoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
2.1. Identification and Validation of ERK5 as a Novel Temozolomide Resistance Factor in GBM
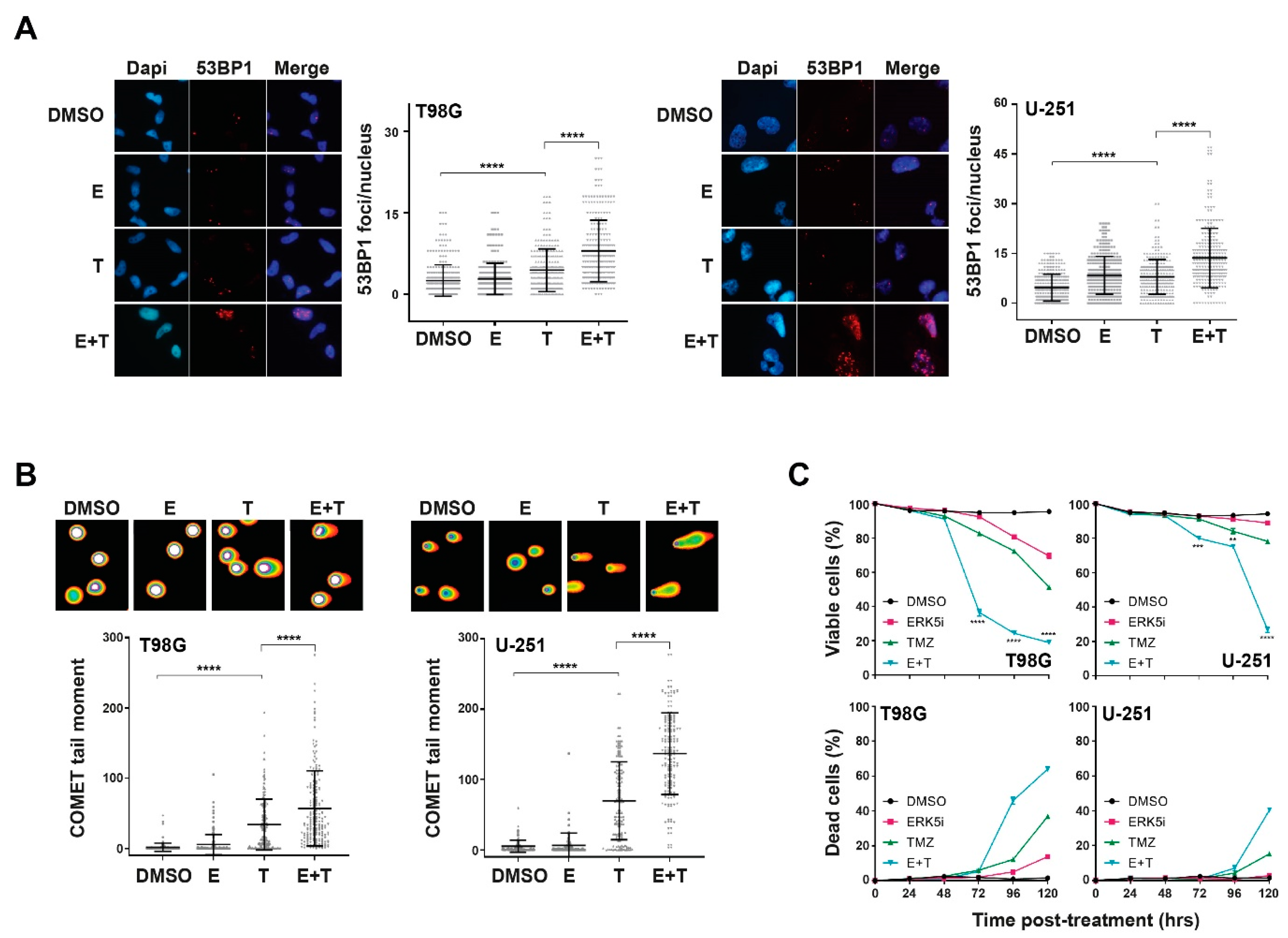
2.2. ERK5 Inhibition in Combination with TMZ Increases Cellular Levels of DNA Damage
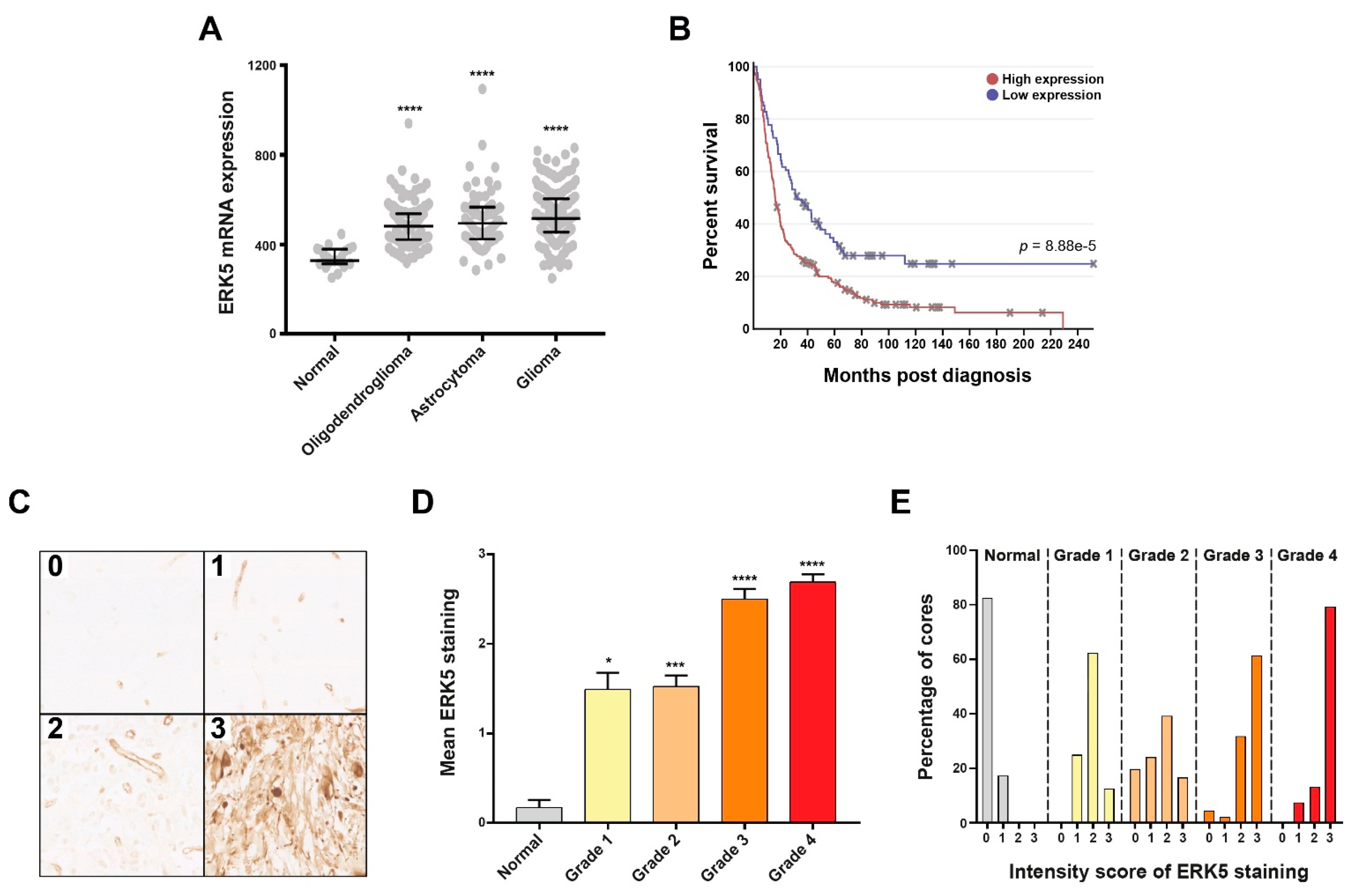
2.3. Target Validation of ERK5 in Glioma Patient Cohorts
3. Discussion
4. Materials and Methods
4.1. Cells and Cell Culture
4.2. RNAi Screening
4.3. Western Blotting
4.4. Immunofluorescence and COMET Assays
4.5. Immunohistochemistry and Scoring
4.6. FACS Analyses
4.7. Cell Survival Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brain, G.B.D. CNSCC Global, regional, and national burden of brain and other CNS cancer, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019, 18, 376–393. [Google Scholar] [CrossRef]
- Messaoudi, K.; Clavreul, A.; Lagarce, F. Toward an effective strategy in glioblastoma treatment. Part I: Resistance mechanisms and strategies to overcome resistance of glioblastoma to temozolomide. Drug Discov. Today 2015, 20, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Hegi, M.E.; Mason, W.P.; van den Bent, M.J.; Taphoorn, M.J.; Janzer, R.C.; Ludwin, S.K.; Allgeier, A.; Fisher, B.; Belanger, K.; et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009, 10, 459–466. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Cote, D.J.; Ascha, M.; Kruchko, C.; Barnholtz-Sloan, J.S. Adult Glioma Incidence and Survival by Race or Ethnicity in the United States From 2000 to 2014. JAMA Oncol. 2018, 4, 1254–1262. [Google Scholar] [CrossRef]
- Aldape, K.; Brindle, K.M.; Chesler, L.; Chopra, R.; Gajjar, A.; Gilbert, M.R.; Gottardo, N.; Gutmann, D.H.; Hargrave, D.; Holland, E.C.; et al. Challenges to curing primary brain tumours. Nat. Rev. Clin. Oncol. 2019, 16, 509–520. [Google Scholar] [CrossRef]
- Jiang, W.; Jin, G.; Cai, F.; Chen, X.; Cao, N.; Zhang, X.; Liu, J.; Chen, F.; Wang, F.; Dong, W.; et al. Extracellular signal-regulated kinase 5 increases radioresistance of lung cancer cells by enhancing the DNA damage response. Exp. Mol. Med. 2019, 51, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Pereira, D.M.; Gomes, S.E.; Borralho, P.M.; Rodrigues, C.M.P. MEK5/ERK5 activation regulates colon cancer stem-like cell properties. Cell Death Discov. 2019, 5, 68. [Google Scholar] [CrossRef] [PubMed]
- Simoes, A.E.; Rodrigues, C.M.; Borralho, P.M. The MEK5/ERK5 signalling pathway in cancer: A promising novel therapeutic target. Drug Discov. Today 2016, 21, 1654–1663. [Google Scholar] [CrossRef] [PubMed]
- Pereira, D.M.; Rodrigues, C.M.P. Targeted Avenues for Cancer Treatment: The MEK5-ERK5 Signaling Pathway. Trends Mol. Med. 2020, 26, 394–407. [Google Scholar] [CrossRef]
- Stecca, B.; Rovida, E. Impact of ERK5 on the Hallmarks of Cancer. Int. J. Mol. Sci. 2019, 20, 1426. [Google Scholar] [CrossRef]
- Finegan, K.G.; Perez-Madrigal, D.; Hitchin, J.R.; Davies, C.C.; Jordan, A.M.; Tournier, C. ERK5 is a critical mediator of inflammation-driven cancer. Cancer Res. 2015, 75, 742–753. [Google Scholar] [CrossRef]
- Kedika, S.R.; Shukla, S.P.; Udugamasooriya, D.G. Design of a dual ERK5 kinase activation and autophosphorylation inhibitor to block cancer stem cell activity. Bioorg. Med. Chem. Lett. 2020, 30, 127552. [Google Scholar] [CrossRef]
- Lochhead, P.A.; Tucker, J.A.; Tatum, N.J.; Wang, J.; Oxley, D.; Kidger, A.M.; Johnson, V.P.; Cassidy, M.A.; Gray, N.S.; Noble, M.E.M.; et al. Paradoxical activation of the protein kinase-transcription factor ERK5 by ERK5 kinase inhibitors. Nat. Commun. 2020, 11, 1383. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhao, Y.; Liu, Y.; Sun, D.; Zhen, Y.; Liu, J.; Fu, L.; Zhang, L.; Ouyang, L. Discovery of a Novel Dual-Target Inhibitor of ERK1 and ERK5 That Induces Regulated Cell Death to Overcome Compensatory Mechanism in Specific Tumor Types. J. Med. Chem. 2020, 63, 3976–3995. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.; Lemos, C.; Wortmann, L.; Eis, K.; Holton, S.J.; Boemer, U.; Moosmayer, D.; Eberspaecher, U.; Weiske, J.; Lechner, C.; et al. Discovery and Characterization of the Potent and Highly Selective (Piperidin-4-yl)pyrido[3,2- d]pyrimidine Based in Vitro Probe BAY-885 for the Kinase ERK5. J. Med. Chem. 2019, 62, 928–940. [Google Scholar] [CrossRef] [PubMed]
- Fleuren, E.D.; Zhang, L.; Wu, J.; Daly, R.J. The kinome ‘at large’ in cancer. Nat. Rev. Cancer 2016, 16, 83–98. [Google Scholar] [CrossRef]
- Ferguson, F.M.; Gray, N.S. Kinase inhibitors: The road ahead. Nat. Rev. Drug Discov. 2018, 17, 353–377. [Google Scholar] [CrossRef]
- Broustas, C.G.; Duval, A.J.; Chaudhary, K.R.; Friedman, R.A.; Virk, R.K.; Lieberman, H.B. Targeting MEK5 impairs nonhomologous end-joining repair and sensitizes prostate cancer to DNA damaging agents. Oncogene 2020, 39, 2467–2477. [Google Scholar] [CrossRef] [PubMed]
- Green, D.; Eyre, H.; Singh, A.; Taylor, J.T.; Chu, J.; Jeys, L.; Sumathi, V.; Coonar, A.; Rassl, D.; Babur, M.; et al. Targeting the MAPK7/MMP9 axis for metastasis in primary bone cancer. Oncogene 2020, 39, 5553–5569. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Cui, H.; Zhu, Z.; Wang, L. MicroRNA-200b-3p suppresses epithelial-mesenchymal transition and inhibits tumor growth of glioma through down-regulation of ERK5. Biochem. Biophys. Res. Commun. 2016, 478, 1158–1164. [Google Scholar] [CrossRef]
- Cook, S.J.; Tucker, J.A.; Lochhead, P.A. Small molecule ERK5 kinase inhibitors paradoxically activate ERK5 signalling: Be careful what you wish for. Biochem. Soc. Trans. 2020, 48, 1859–1875. [Google Scholar] [CrossRef] [PubMed]
- Hannus, M.; Beitzinger, M.; Engelmann, J.C.; Weickert, M.T.; Spang, R.; Hannus, S.; Meister, G. siPools: Highly complex but accurately defined siRNA pools eliminate off-target effects. Nucleic Acids Res. 2014, 42, 8049–8061. [Google Scholar] [CrossRef]
- Deng, X.; Elkins, J.M.; Zhang, J.; Yang, Q.; Erazo, T.; Gomez, N.; Choi, H.G.; Wang, J.; Dzamko, N.; Lee, J.D.; et al. Structural determinants for ERK5 (MAPK7) and leucine rich repeat kinase 2 activities of benzo[e]pyrimido-[5,4-b]diazepine-6(11H)-ones. Eur. J. Med. Chem. 2013, 70, 758–767. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.C.; Amantea, C.M.; Nomanbhoy, T.K.; Weissig, H.; Ishiyama, J.; Hu, Y.; Sidique, S.; Li, B.; Kozarich, J.W.; Rosenblum, J.S. ERK5 kinase activity is dispensable for cellular immune response and proliferation. Proc. Natl. Acad. Sci. USA 2016, 113, 11865–11870. [Google Scholar] [CrossRef]
- Bekker-Jensen, S.; Lukas, C.; Kitagawa, R.; Melander, F.; Kastan, M.B.; Bartek, J.; Lukas, J. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol. 2006, 173, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Shibata, A.; Jeggo, P.A. Roles for 53BP1 in the repair of radiation-induced DNA double strand breaks. DNA Repair 2020, 93, 102915. [Google Scholar] [CrossRef]
- Petermann, E.; Orta, M.L.; Issaeva, N.; Schultz, N.; Helleday, T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell 2010, 37, 492–502. [Google Scholar] [CrossRef]
- Yajima, H.; Lee, K.J.; Chen, B.P. ATR-dependent phosphorylation of DNA-dependent protein kinase catalytic subunit in response to UV-induced replication stress. Mol. Cell. Biol. 2006, 26, 7520–7528. [Google Scholar] [CrossRef]
- Gil Del Alcazar, C.R.; Todorova, P.K.; Habib, A.A.; Mukherjee, B.; Burma, S. Augmented HR Repair Mediates Acquired Temozolomide Resistance in Glioblastoma. Mol. Cancer Res. 2016, 14, 928–940. [Google Scholar] [CrossRef] [PubMed]
- Cicenas, J.; Kalyan, K.; Sorokinas, A.; Stankunas, E.; Levy, J.; Meskinyte, I.; Stankevicius, V.; Kaupinis, A.; Valius, M. Roscovitine in cancer and other diseases. Ann. Transl. Med. 2015, 3, 135. [Google Scholar] [CrossRef]
- Collis, S.J.; Barber, L.J.; Clark, A.J.; Martin, J.S.; Ward, J.D.; Boulton, S.J. HCLK2 is essential for the mammalian S-phase checkpoint and impacts on Chk1 stability. Nat. Cell Biol. 2007, 9, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Myers, K.N.; Barone, G.; Ganesh, A.; Staples, C.J.; Howard, A.E.; Beveridge, R.D.; Maslen, S.; Skehel, J.M.; Collis, S.J. The bornavirus-derived human protein EBLN1 promotes efficient cell cycle transit, microtubule organisation and genome stability. Sci. Rep. 2016, 6, 35548. [Google Scholar] [CrossRef]
- Zieve, G.W.; Turnbull, D.; Mullins, J.M.; McIntosh, J.R. Production of large numbers of mitotic mammalian cells by use of the reversible microtubule inhibitor nocodazole. Nocodazole accumulated mitotic cells. Exp. Cell Res. 1980, 126, 397–405. [Google Scholar] [CrossRef]
- Aparicio, T.; Baer, R.; Gautier, J. DNA double-strand break repair pathway choice and cancer. DNA Repair 2014, 19, 169–175. [Google Scholar] [CrossRef]
- Bonetti, D.; Colombo, C.V.; Clerici, M.; Longhese, M.P. Processing of DNA Ends in the Maintenance of Genome Stability. Front. Genet. 2018, 9, 390. [Google Scholar] [CrossRef] [PubMed]
- Scully, R.; Panday, A.; Elango, R.; Willis, N.A. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 2019, 20, 698–714. [Google Scholar] [CrossRef]
- Gusev, Y.; Bhuvaneshwar, K.; Song, L.; Zenklusen, J.C.; Fine, H.; Madhavan, S. The REMBRANDT study, a large collection of genomic data from brain cancer patients. Sci. Data 2018, 5, 180158. [Google Scholar] [CrossRef]
- Patil, A.A.; Sayal, P.; Depondt, M.L.; Beveridge, R.D.; Roylance, A.; Kriplani, D.H.; Myers, K.N.; Cox, A.; Jellinek, D.; Fernando, M.; et al. FANCD2 re-expression is associated with glioma grade and chemical inhibition of the Fanconi Anaemia pathway sensitises gliomas to chemotherapeutic agents. Oncotarget 2014, 5, 6414–6424. [Google Scholar] [CrossRef] [PubMed]
- Koncar, R.F.; Dey, B.R.; Stanton, A.J.; Agrawal, N.; Wassell, M.L.; McCarl, L.H.; Locke, A.L.; Sanders, L.; Morozova-Vaske, O.; Myers, M.I.; et al. Identification of Novel RAS Signaling Therapeutic Vulnerabilities in Diffuse Intrinsic Pontine Gliomas. Cancer Res. 2019, 79, 4026–4041. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. Targeting oncogenic Raf protein-serine/threonine kinases in human cancers. Pharmacol. Res. 2018, 135, 239–258. [Google Scholar] [CrossRef]
- Svilar, D.; Dyavaiah, M.; Brown, A.R.; Tang, J.B.; Li, J.; McDonald, P.R.; Shun, T.Y.; Braganza, A.; Wang, X.H.; Maniar, S.; et al. Alkylation sensitivity screens reveal a conserved cross-species functionome. Mol. Cancer Res. 2012, 10, 1580–1596. [Google Scholar] [CrossRef] [PubMed]
- Thaker, N.G.; Zhang, F.; McDonald, P.R.; Shun, T.Y.; Lewen, M.D.; Pollack, I.F.; Lazo, J.S. Identification of survival genes in human glioblastoma cells by small interfering RNA screening. Mol. Pharmacol. 2009, 76, 1246–1255. [Google Scholar] [CrossRef]
- Caragher, S.; Chalmers, A.J.; Gomez-Roman, N. Glioblastoma’s Next Top Model: Novel Culture Systems for Brain Cancer Radiotherapy Research. Cancers 2019, 11, 44. [Google Scholar] [CrossRef] [PubMed]
- Rominiyi, O.; Al-Tamimi, Y.; Collis, S.J. The ‘Ins and Outs’ of Early Preclinical Models for Brain Tumor Research: Are They Valuable and Have We Been Doing It Wrong? Cancers 2019, 11, 426. [Google Scholar] [CrossRef] [PubMed]
- Staples, C.J.; Barone, G.; Myers, K.N.; Ganesh, A.; Gibbs-Seymour, I.; Patil, A.A.; Beveridge, R.D.; Daye, C.; Beniston, R.; Maslen, S.; et al. MRNIP/C5orf45 Interacts with the MRN Complex and Contributes to the DNA Damage Response. Cell Rep. 2016, 16, 2565–2575. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carmell, N.; Rominiyi, O.; Myers, K.N.; McGarrity-Cottrell, C.; Vanderlinden, A.; Lad, N.; Perroux-David, E.; El-Khamisy, S.F.; Fernando, M.; Finegan, K.G.; et al. Identification and Validation of ERK5 as a DNA Damage Modulating Drug Target in Glioblastoma. Cancers 2021, 13, 944. https://doi.org/10.3390/cancers13050944
Carmell N, Rominiyi O, Myers KN, McGarrity-Cottrell C, Vanderlinden A, Lad N, Perroux-David E, El-Khamisy SF, Fernando M, Finegan KG, et al. Identification and Validation of ERK5 as a DNA Damage Modulating Drug Target in Glioblastoma. Cancers. 2021; 13(5):944. https://doi.org/10.3390/cancers13050944
Chicago/Turabian StyleCarmell, Natasha, Ola Rominiyi, Katie N. Myers, Connor McGarrity-Cottrell, Aurelie Vanderlinden, Nikita Lad, Eva Perroux-David, Sherif F. El-Khamisy, Malee Fernando, Katherine G. Finegan, and et al. 2021. "Identification and Validation of ERK5 as a DNA Damage Modulating Drug Target in Glioblastoma" Cancers 13, no. 5: 944. https://doi.org/10.3390/cancers13050944
APA StyleCarmell, N., Rominiyi, O., Myers, K. N., McGarrity-Cottrell, C., Vanderlinden, A., Lad, N., Perroux-David, E., El-Khamisy, S. F., Fernando, M., Finegan, K. G., Brown, S., & Collis, S. J. (2021). Identification and Validation of ERK5 as a DNA Damage Modulating Drug Target in Glioblastoma. Cancers, 13(5), 944. https://doi.org/10.3390/cancers13050944







